Abstract
MicroRNAs (miRNAs) exert critical roles in the majority of biological and pathological processes. Recent studies have associated miR-150 with a number of different cancer types. However, little is known about miR-150 targets in cervical cancer. In the present study, the HeLa human cervical cancer cell line was transfected with hsa-miR-150-5p mimics, hsa-miR-150-5p inhibitors or miRNA controls. miR-150 was predicted to bind the 3′untranslated region (3′UTR) of the CDKN1B gene, which encodes the cyclin-dependent kinase inhibitor 1B (p27Kip1). The direct binding between miR-150 and the 3′UTR of CDKN1B was confirmed using dual-luciferase reporter assays. The effects of miR-150 on CDKN1B mRNA expression, p27Kip1 protein expression, cell cycle and cell proliferation were determined using reverse-transcription quantitative PCR, western blot analysis, flow cytometry and WST-8 assays, respectively. miR-150 was demonstrated to directly target the 3′UTR of CDKN1B in transfected HeLa cells. The expression of CDKN1B mRNA and p27Kip1 protein was reduced by miR-150 mimics, and increased by miR-150 inhibitors. Moreover, the overexpression of miR-150 promoted cell cycle progression from the G0/G1 to the S phase and led to a significant increase in HeLa cell proliferation. The results of the present study indicated that miR-150 promotes HeLa cell cycle progression and proliferation via the suppression of p27Kip1 expression.
Keywords: microRNA-150, cervical cancer, HeLa cell, CDKN1B, p27Kip1, cell cycle
Introduction
Cervical cancer is a frequent diagnosis in women worldwide; however, its incidence is decreasing in developed countries thanks to early diagnosis (1,2). Although comprehensive cervical cancer genomic profiles have been reported (3), the underlying genomic mechanisms remain poorly understood. MicroRNAs (miRNAs) represent a family of abundant endogenous, noncoding RNA molecules, ~22 nucleotides in length (4,5), which post-transcriptionally repress the expression of target genes by typically binding the 3′untranslated regions (3′UTRs) of messenger RNA (mRNA) (6,7). It is becoming increasingly evident that now miRNAs play a role in virtually every pathophysiological process (8,9). One particular miRNA, namely miR-150, reportedly influences the progression of many cancer types (10–17). Notably, the expression of miR-150 in cervical carcinoma is significantly higher than that in normal cervical tissue from healthy donors, and its expression in cervical tissue correlates with cancer stage progression (18). Moreover, the overexpression of miR-150 promotes proliferation in cervical cancer by targeting FOXO4, P2RX7, and SRCIN1 (18–20). FOXO4 regulates many cellular pathways, including oxidative stress signaling, longevity, insulin signaling, cell cycle progression, and apoptosis (21). In addition, FOXO4 regulates the expression of cyclin D1, p27, BIM, and FASL (21). The activation of the P2X7 receptor mediates apoptosis via the caspase-9 mitochondrial pathway (22). SRCIN1 is another tumor suppressor that plays a vital role in tumor cells (23). Furthermore, microRNA-150 promotes cell proliferation, migration, and invasion of cervical cancer by targeting PDCD4 (24). However, the exact identities of the molecular targets of miR-150 in cervical cancer remain uncertain.
We predicted that miR-150 binds the 3′UTR of CDKN1B using the TargetScan database (http://www.targetscan.org/). The CDKN1B gene encodes the cyclin-dependent kinase inhibitor 1B (p27Kip1), which integrates extracellular signals that negatively regulate the cell cycle progression at the G1 stage (25,26). In this study, we aimed to investigate the interaction between miR-150 and CDKN1B, via dual-luciferase reporter assay in the HeLa human cervical cancer cell line. We also carried out oligonucleotide transfection and western blot analysis to examine the effect of miR-150 on HeLa cell proliferation.
Materials and methods
Cell culture
The HeLa human cervical cancer cell line was obtained from the RIKEN Cell Bank (Tsukuba, Japan) and cultured in complete Dulbecco's Modified Eagle's medium (Sigma-Aldrich; Merck KGaA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) and 1% antibiotic/antimycotic solution (Thermo Fisher Scientific, Inc.) at 37°C in a humidified 5% CO2 incubator.
Dual-luciferase reporter assay
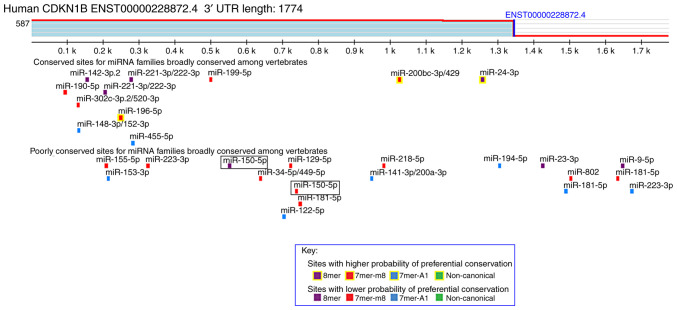
To explore the mechanism through which miR-150 promotes tumor progression, we used TargetScan as a miRNA target prediction algorithm. Based on the frequencies of miR-150 sites in the 3′UTRs of mRNAs, more than 100 mRNAs were predicted to be regulated by miR-150. In this study, we focused on CDKN1, which encodes a cell cycle regulator. As shown in Fig. 1, this analysis revealed putative 8- and 7-mer binding sites for miR-150 in the 3′UTR of the CDKN1B transcript. The restriction sites in the psiCHECK-2 vector multiple cloning region are SgfI, XhoI, PmeI, and NotI. Therefore, two pairs of wild-type and mutant 3′UTR sequences of CDKN1B with XhoI and NotI restriction enzyme digestion sites were chemically synthesized (Table I). The oligo DNAs of the paired sequences were annealed and digested by XhoI and NotI restriction enzymes. These synthesized sequences were integrated into the psiCHECK-2 vector (Promega Corporation), and the recombinant plasmids were transformed into One Shot chemically competent E. coli (Thermo Fisher Scientific, Inc.) and purified according to the manufacturer's instructions. The recombinant plasmids were named WT1, MUT1, WT2, and MUT2 (Table I). HeLa cells were co-transfected with the recombinant plasmids and 100 nM miR-150 mimics or miRNA control for 48 h using Lipofectamine 3000 Transfection reagent (Thermo Fisher Scientific, Inc.). After transfection, both firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega Corporation). Luciferase activities were normalized to Renilla luciferase. This analysis was performed in triplicate.
Figure 1.
Predicted binding site between miR-150 and 3′UTR of CDKN1B based on TargetScan 7.2 database. miR, microRNA; UTR, untranslated region.
Table I.
Chemically synthesized sequences for dual-luciferase reporter assay
| Sequence | XhoI site | 3′UTR of CDKN1B | NotI site | ||
|---|---|---|---|---|---|
| WT-1 (5′-3′) | GG | CTCGAG | AAGUUUAUUCUCAUUUGGGAGA | GCGGCCGC | GG |
| WT-1 (3′-5′) | CC | GAGCTC | UUCAAAUAAGAGUAAACCCUCU | CGCCGGCG | CC |
| MUT-1 (5′-3′) | GG | CTCGAG | AAGUUUAUUCUCAUAACCCUCU | GCGGCCGC | GG |
| MUT-1 (3′-5′) | CC | GAGCTC | UUCAAAUAAGAGUAUUGGGAGA | CGCCGGCG | CC |
| WT-2 (5′-3′) | GG | CTCGAG | AAAAUCCGAGGUGCUUGGGAGU | GCGGCCGC | GG |
| WT-2 (3′-5′) | CC | GAGCTC | UUUUAGGCUCCACGAACCCUCA | CGCCGGCG | CC |
| MUT-2 (5′-3′) | GG | CTCGAG | AAAAUCCGAGGUGCAACCCUCU | GCGGCCGC | GG |
| MUT-2 (3′-5′) | CC | GAGCTC | UUUUAGGCUCCACGUUGGGAGA | CGCCGGCG | CC |
WT, wildtype; MUT, mutant; UTR, untranslated region.
Oligonucleotide transfection
HeLa cells were transfected with 100 nM hsa-miR-150-5p mimics (miR-150 mimics), 100 nM hsa-miR-150-5p inhibitors (miR-150 inhibitors), or 100 nM miRNA control (Bioneer) for 72 h using Lipofectamine RNAiMAX Transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions.
Reverse-transcription quantitative (RT-q) PCR
The expression level of miR-150 and CDKN1B mRNA was quantified by real-time PCR. Total RNA from transfected HeLa cells was extracted using the miRNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. The quantity and quality of total genomic RNA were evaluated using the NanoDrop Lite spectrophotometer (Thermo Fisher Scientific, Inc.). For miRNA analysis, TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.) was used according to the manufacturer's protocol. The program was the following: 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. Quantitative PCR (qPCR) on miRNA was performed using TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific, Inc.) and TaqMan miRNA assays (Thermo Fisher Scientific, Inc.) for hsa-miR-150-5p (Assay ID, 000473) and RNU6B (Assay ID, 001093) in accordance with the manufacturer's instructions. RNU6B was used as a reference gene. The cycling conditions were as follows; 95°C for 20 sec, followed by 40 cycles at 95°C for 1 sec and 60°C for 20 sec. For mRNA analysis, the extracted total RNA was reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Real-time PCR was performed using the PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions, starting with 100 ng of total RNA. The cycling conditions were as follows; 95°C for 20 sec, followed by 40 cycles at 95°C for 3 sec and 60°C for 30 sec. Primer sequences are shown in Table II. GAPDH was quantified as a reference gene. All real-time PCR experiments were conducted with a StepOnePlus instrument (Thermo Fisher Scientific, Inc.). The average cycle threshold value for the housekeeping gene was used to normalize the raw cycle threshold data and calculate ΔCq. The 2−ΔΔCq method was used to determine the relative expression levels of miR-150 and CDKN1B. All experiment was performed in triplicate.
Table II.
Primer sequences for reverse-transcription quantitative PCR.
| Gene name | Orientation | Primer sequences |
|---|---|---|
| CDKN1B | Forward | 5′-GGCCTCAGAAGACGTCAAAC-3′ |
| Reverse | 5′-CAGGATGTCCATTCCATGAAG-3′ | |
| GAPDH | Forward | 5′-GCACCGTCAAGGCTGAGAAC-3′ |
| Reverse | 5′-TGGTGAAGACGCCAGTGGA-3′ |
Western blotting
Transfected HeLa cells were homogenized in cold EzRIPA Lysis buffer (ATTO). A 100-µg protein extract was separated by SDS-PAGE using a 10% polyacrylamide gel, and the proteins were transferred onto polyvinylidene difluoride membranes (Merck Millipore). After blocking with 5% non-fat dry milk at room temperature for 1 h, the membranes were incubated with primary antibodies against p27Kip1 (1:1,000; cat. no. 3686; Cell Signaling Technology, Inc.) and β-actin (1:1,000; cat. no. 8457; Cell Signaling Technology, Inc.) overnight at 4°C. Subsequently, the membranes were washed and exposed to goat anti-rabbit-IgG HRP (1:1,000; cat. no. 7074, Cell Signaling Technology, Inc.) for 1 h at room temperature, which was followed by imaging with a chemiluminescent substrate. This assay was performed in triplicate.
Cell cycle analysis
To analyze the cell cycle, transfected cells were collected, washed with phosphate-buffered saline (PBS), and fixed in cold 70% ethanol at 4°C overnight. Fixed cells were washed with PBS and incubated with 250 µg/ml RNase A and 50 µg/ml propidium iodide for 30 min. The DNA content was determined using a Cell Lab Quanta SC flow cytometer (Beckman Coulter) and the cell cycle profile was determined with ModFit LT, which is the most comprehensive flow cytometric DNA cell cycle analysis software available (Verity Software House). This assay was performed in triplicate.
Cell proliferation assay
Cell proliferation of transfected HeLa cells was determined using the 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium (WST-8) assay with the Cell Counting Kit-8 (CCK-8; Dojindo Laboratories) every 24 h according to the manufacturer's instructions. CCK-8 solutions were added to the cell culture, and cells were incubated at 37°C for 2 h. Later, the absorbance was measured at 450 nm. This assay was performed in triplicate.
Statistical analysis
Results are expressed as the mean ± standard deviation. Statistical significance for the experiments was determined using Dunn's test, a Mann-Whitney's U test, or a one-way factorial ANOVA test. P<0.05 was considered to indicate a statistically significant difference. StatMate V software was used to perform statistical analyses.
Results
miR-150 directly targets the 3′UTR of CDKN1B in transfected HeLa cells
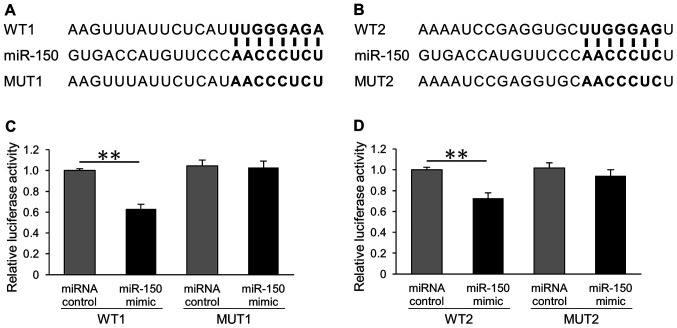
miRNAs repress the expression of target genes by binding the 3′UTR of target genes. The target sites of miRNA-150 were investigated using the TargetScan miRNA target prediction database. CDKN1B is a predicted target gene of miR-150. Two predicted miR-150-binding sites in the 3′UTR of CDKN1B were identified. To verify these two binding sites, we designed recombinant plasmids, including wild-type or mutant 3′UTR sequences of CDKN1B (Fig. 2A and B). Then, dual-luciferase reporter assays were performed to investigate whether miR-150 could influence the expression of luciferase via binding to the predicted sequences. Our results showed that miR-150 directly downregulated CDKN1B in the HeLa cells (Fig. 2C and D). Conversely, there was no effect on the reporters with mutant sequences.
Figure 2.
miR-150 directly targets the 3′-UTR of CDKN1B. (A) Sequences of the miR-150 binding sites in the 3′-UTR of wild-type CDKN1B (WT1) and a CDKN1B mutant containing eight mutated nucleotides in the 3′-UTR of CDKN1B (MUT1). The predicted miR-150-binding sites are presented in bold. (B) Sequences of the miR-150 binding sites in the 3′UTR of wild-type CDKN1B (WT2) and a CDKN1B mutant containing 7 mutated nucleotides in the 3′UTR of CDKN1B (MUT2). The predicted miR-150-binding sites are presented in bold. (C) Luciferase activity assays of the WT1 or MUT1 reporter in HeLa cells transfected with miR-150 mimics or miRNA controls. (D) Luciferase activity assays of the WT2 or MUT2 reporter in HeLa cells transfected with miR-150 mimics or miRNA controls. Firefly and Renilla luciferase activities were measured. Luciferase activities were normalized to Renilla luciferase. Significant differences were assessed using the Mann-Whitney U test. **P<0.01. Bars represent the mean ± standard deviation of three experiments. miR, microRNA; UTR, untranslated region; WT, wild-type; MUT, mutant; miR, microRNA.
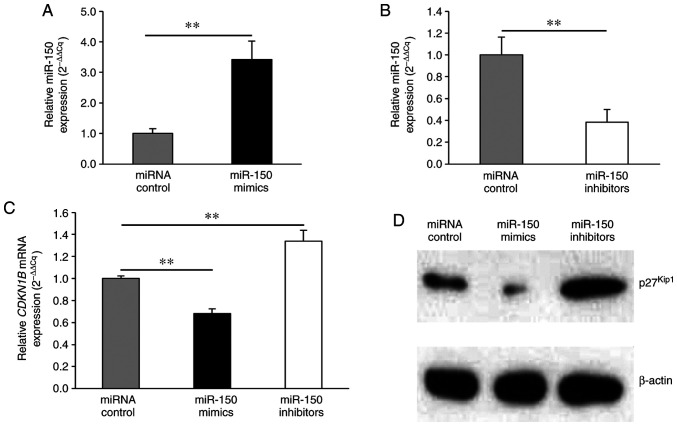
miR-150 reduces CDKN1B mRNA expression and p27Kip1 protein expression in transfected HeLa cells
To investigate the molecular details of miR-150 effects, HeLa cells were transfected with miR-150 mimics, miR-150 inhibitors, or miRNA control. Firstly, qPCR for miRNA showed that miR-150 expression levels were significantly higher and lower in the miR-150 mimic-transfected and miR-150 inhibitor-transfected cells, respectively, relative to the controls (Fig. 3A, B). Since miR-150 directly targeted the 3′UTR of the CDKN1B gene, the expression levels of CDKN1B mRNA and p27Kip1 protein were evaluated after the transfection of miR-150 mimics or inhibitors using qPCR and western blot analysis, respectively. miR-150 mimics significantly decreased CDKN1B mRNA expression levels (Fig. 3C). In contrast, miR-150 inhibitors significantly upregulated CDKN1B mRNA expression levels (Fig. 3C). Consistent with the RT-PCR results, the protein levels of p27Kip1 were reduced by miR-150 mimics and increased by miR-150 inhibitors (Fig. 3D).
Figure 3.
miR-150 downregulates CDKN1B mRNA and p27Kip1 expression. Total RNA and protein were extracted from transfected HeLa cells. (A) HeLa cells were transfected with miRNA controls or miR-150 mimics. (B) HeLa cells were transfected with miRNA controls or miR-150 inhibitors. Average cycle threshold value for the endogenous control RNU6B was used to normalize the raw cycle threshold data and calculate ΔCq. The 2−ΔΔCq method was used to determine the relative expression levels of miR-150. Significant differences were assessed using the Mann-Whitney U test. (C) The average cycle threshold value for the housekeeping gene GAPDH was used to normalize the raw cycle threshold data and calculate ΔCq. The 2−ΔΔCq method was used to determine the relative expression levels of CDKN1B. Significant differences were assessed using Dunn's test. **P<0.01. Bars represent the mean ± standard deviation of three experiments. (D) Representative western blot analysis shows p27Kip1 protein expression in transfected HeLa cells. β-actin was used as a loading control for western blot analysis. miR, microRNA.
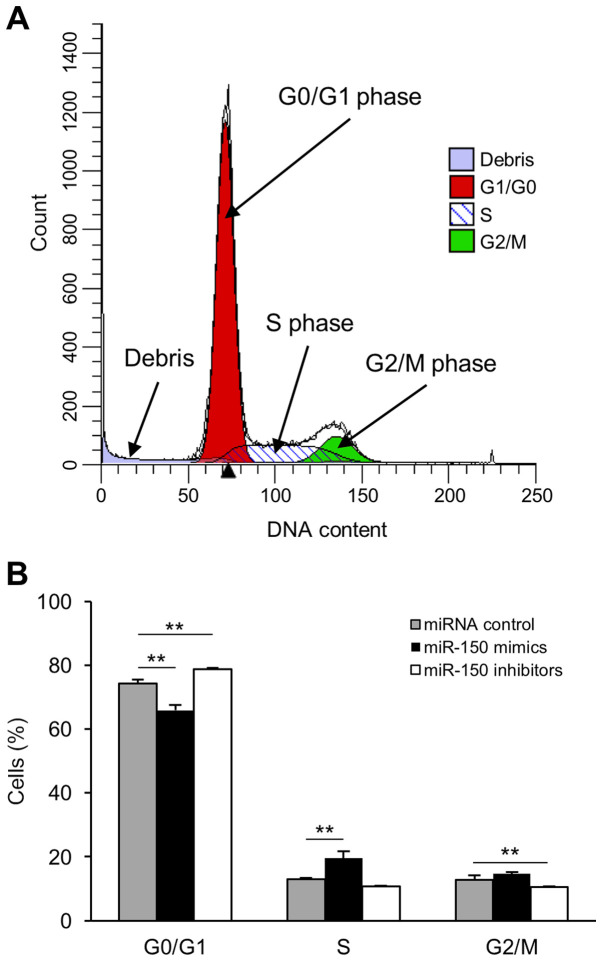
miR-150 facilitates cell cycle progression in transfected HeLa cells
To investigate the role of miR-150 in cervical cancer cells, we analyzed cell cycle progression in transfected HeLa cells using a flow cytometer (Fig. 4A). There was an increased number of cells in the S phase in miR-150 mimic-transfected cells compared to that in miRNA control-transfected cells (Fig. 4B). In contrast, transfection of miR-150 inhibitors induced G0/G1 phase arrest (Fig. 4B).
Figure 4.
miR-150 promotes cell cycle progression. HeLa cells were transfected with miR-150 mimics, miR-150 inhibitors or miRNA controls for 72 h, and the number of cells in each phase of the cell cycle was measured using a flow cytometer and determined with ModFit LT software. (A) Representative DNA content graphic profiles of the cell cycle are presented. ModFit LT program allows to scan the histogram for peaks and to determine the number of cell cycles based on the sizes and positions. Debris is excluded from cell cycle determination by ModFit LT algorithm. The X-axis shows the DNA content. The red region, diagonal line, and green area indicate the G0/G1 phase, S phase, G2/M phase, respectively. (B) The percentages of cells in different cell cycle phases were analyzed and presented as a histogram. Significant differences were assessed using Dunn's test. **P<0.01. Bars represent the mean ± standard deviation of three experiments. miR, microRNA.
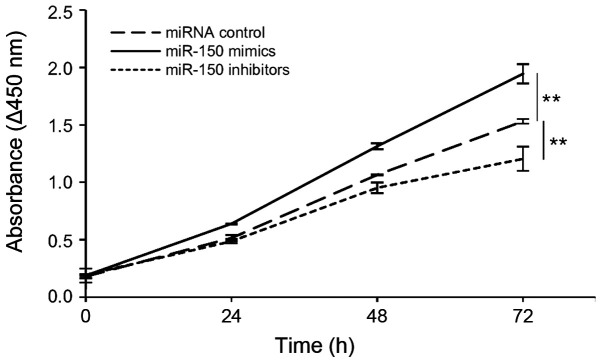
miR-150 promotes cell proliferation in transfected HeLa cells
We hypothesized that miR-150 levels are involved in cell proliferation in cervical cancer. The proliferation of transfected HeLa cells was determined by measuring the absorbance at 450 nm based on a colorimetric assay. Our results showed that the overexpression of miR-150 led to a significant increase in cell proliferation, whereas expressing miR-150 inhibitors led to a significant decrease in HeLa cell proliferation (Fig. 5).
Figure 5.
miR-150 promotes cell proliferation. HeLa cells were transfected with miR-150 mimics, miR-150 inhibitors or miRNA controls for 72 h, and then, cell proliferation was determined using CCK-8 every 24 h. Significant differences were assessed using a one-way factorial ANOVA test. **P<0.01. Bars represent the mean ± standard deviation of three experiments. miR, microRNA.
Discussion
miRNAs play critical roles in many diverse biological and pathological processes, such as cell proliferation, migration, apoptosis, and the pathogenesis of cancer (8,9). Among them, recent studies have reported the influence of miR-150 on many types of cancers (10–17). Previous studies have reported decreased levels of miR-150 in colorectal cancer (27,28), liver cancer (29), pancreatic cancer (12), esophageal squamous cell carcinoma (30), and ovarian cancer (14). In contrast, the upregulated expression of miR-150 has been found in breast cancer (13), non-small cell lung cancer (31–33), gastric cancer (34), and prostate cancer (15). A previous report indicated that miR-150 expression levels are significantly higher in cervical carcinoma cells than in para-carcinoma tissues (18). Moreover, the expression of miR-150 in samples from patients with cervical carcinoma significantly exceeded that of normal cervical tissue from healthy donors. Moreover, miR-50 expression correlated with stage progression (18). However, results depict a controversial picture regarding the targets of miR-150 in cervical cancer. Moreover, the consensus regarding a hypothetical influence of miR-150 on cervical cancer is lacking.
In the present study, we analyzed the effect of miR-150 on cervical cancer cells in vitro. We first investigated the target sites of miRNA-150 using the TargetScan miRNA target prediction database and predicted two binding sites on the 3′UTR of CDKN1B. Further, the results of dual-luciferase reporter assays showed that miR-150 directly bound and downregulated CDKN1B in HeLa cells. Notably, miR-150 significantly decreased CDKN1B mRNA expression levels, and western blot analysis showed that the transfection of miR-150 mimics reduced p27Kip1 protein levels. The p27Kip1 protein transduces extracellular signals of cell cycle regulators that negatively regulate G1 cell cycle progression (25,26).
To investigate the role of miR-150 in HeLa cells, we analyzed cell cycle progression by flow cytometry. In miR-150 mimic-transfected cells, a higher proportion of cells was in the S phase. Moreover, the transfection of miR-150 inhibitors induced cell cycle arrest at the G1/G0 phase. These results suggested that miR-150 promotes cell cycle progression from the G0/G1 to S phase in cervical cancer cells by suppressing the p27Kip1 function.
We then measured transfected HeLa cell numbers using CCK-8, via a sensitive colorimetric assay for the determination of cell proliferation. The cells transfected with miR-150 mimics proliferated faster than control cells, whereas the cells transfected with miR-150 inhibitors grew slower than control cells. These findings suggest that miR-150 promotes cell cycle progression and cell proliferation by suppressing p27Kip1 expression. Our results validated previously reported direct targets of miR-150 in cervical cancer, namely FOXO4, P2RX7, and SRCIN1 (18–20). However, our results also suggest that miR-150 promotes proliferation by directly targeting CDKN1B. Considering the well-known pleiotropic effects of miRNA, a single miRNA could bind several hundred target mRNAs (35). Although the present study provides a clearer understanding of the function and molecular mechanism of miR-150 in HeLa cells, more comprehensive in vitro and in vivo studies are needed to decipher the role of miR-150 in cervical cancer.
In conclusion, our results indicate that miR-150 promotes cell cycle progression and cell proliferation of HeLa human cervical cancer cells by directly suppressing p27Kip1 expression. We identified CDKN1B as a novel target of miR-150 in cervical cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
WO conceived and designed all the experiments. WO and KH performed the experiments. HT, KI, YY, AK, TN and NY analyzed the experimental data. TN and NY contributed to writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Kagawa Prefectural University of Health Sciences, Japan.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Hagemann T, Bozanovic T, Hooper S, Ljubic A, Slettenaar VI, Wilson JL, Singh N, Gayther SA, Shepherd JH, Van Trappen PO. Molecular profiling of cervical cancer progression. Br J Cancer. 2007;96:321–328. doi: 10.1038/sj.bjc.6603543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Lai EC. miRNAs: Whys and wherefores of miRNA-mediated regulation. Curr Biol. 2005;15:R458–R460. doi: 10.1016/j.cub.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Ren X, Zhang X. Role of microRNA-150 in solid tumors. Oncol Lett. 2015;10:11–16. doi: 10.3892/ol.2015.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng J, Yang Y, Zhang P, Wang F, Ma Y, Qin H, Wang Y. miR-150 functions as a tumour suppressor in human colorectal cancer by targeting c-Myb. J Cell Mol Med. 2014;18:2125–2134. doi: 10.1111/jcmm.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srivastava SK, Bhardwaj A, Singh S, Arora S, Wang B, Grizzle WE, Singh AP. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011;32:1832–1839. doi: 10.1093/carcin/bgr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Chen Y, Wu W, Ouyang N, Chen J, Li H, Liu X, Su F, Lin L, Yao Y. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PLoS One. 2013;8:e80707. doi: 10.1371/journal.pone.0080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin M, Yang Z, Ye W, Xu H, Hua X. MicroRNA-150 predicts a favorable prognosis in patients with epithelial ovarian cancer, and inhibits cell invasion and metastasis by suppressing transcriptional repressor ZEB1. PLoS One. 2014;9:e103965. doi: 10.1371/journal.pone.0103965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dezhong L, Xiaoyi Z, Xianlian L, Hongyan Z, Guohua Z, Bo S, Shenglei Z, Lian Z. miR-150 is a factor of survival in prostate cancer patients. J BUON. 2015;20:173–179. [PubMed] [Google Scholar]
- 16.Zhang J, Luo N, Luo Y, Peng Z, Zhang T, Li S. MicroRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb. Int J Oncol. 2012;40:747–756. doi: 10.3892/ijo.2011.1242. [DOI] [PubMed] [Google Scholar]
- 17.Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, Ogawa R, Harata K, Fujii Y. MicroRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537–542. [PubMed] [Google Scholar]
- 18.Li J, Hu L, Tian C, Lu F, Wu J, Liu L. MicroRNA-150 promotes cervical cancer cell growth and survival by targeting FOXO4. BMC Mol Biol. 2015;16:24. doi: 10.1186/s12867-015-0052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou L, Qi X, Potashkin JA, Abdul-Karim FW, Gorodeski GI. MicroRNAs miR-186 and miR-150 down-regulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3′-untranslated region of the gene that decrease steady-state levels of the transcript. J Biol Chem. 2008;283:28274–2886. doi: 10.1074/jbc.M802663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Han S. miR-150-5p promotes the proliferation and epithelial-mesenchymal transition of cervical carcinoma cells via targeting SRCIN1. Pathol Res Pract. 2019;215:738–747. doi: 10.1016/j.prp.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Boura E, Silhan J, Herman P, Vecer J, Sulc M, Teisinger J, Obsilova V, Obsil T. Both the N-terminal loop and wing W2 of the forkhead domain of transcription factor Foxo4 are important for DNA binding. J Biol Chem. 2007;282:8265–8275. doi: 10.1074/jbc.M605682200. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Li X, Wang L, Feng YH, Zeng R, Gorodeski G. Antiapoptotic effects of estrogen in normal and cancer human cervical epithelial cells. Endocrinology. 2004;145:5568–5579. doi: 10.1210/en.2004-0807. [DOI] [PubMed] [Google Scholar]
- 23.Cabodi S, del Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Integrin signalling adaptors: Not only figurants in the cancer story. Nat Rev Cancer. 2010;10:858–870. doi: 10.1038/nrc2967. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Wang J, Li J, Wang X, Song W. MicroRNA-150 promotes cell proliferation, migration, and invasion of cervical cancer through targeting PDCD4. Biomed Pharmacother. 2018;97:511–517. doi: 10.1016/j.biopha.2017.09.143. [DOI] [PubMed] [Google Scholar]
- 25.Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massagué J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-X. [DOI] [PubMed] [Google Scholar]
- 26.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 27.Pizzini S, Bisognin A, Mandruzzato S, Biasiolo M, Facciolli A, Perilli L, Rossi E, Esposito G, Rugge M, Pilati P, et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics. 2013;14:589. doi: 10.1186/1471-2164-14-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y, Zhang P, Wang F, Zhang H, Yang J, Peng J, Liu W, Qin H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61:1447–1453. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 29.Sun W, Zhang Z, Wang J, Shang R, Zhou L, Wang X, Duan J, Ruan B, Gao Y, Dai B, et al. MicroRNA-150 suppresses cell proliferation and metastasis in hepatocellular carcinoma by inhibiting the GAB1-ERK axis. Oncotarget. 2016;7:11595–11608. doi: 10.18632/oncotarget.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokobori T, Suzuki S, Tanaka N, Inose T, Sohda M, Sano A, Sakai M, Nakajima M, Miyazaki T, Kato H, Kuwano H. miR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104:48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Ouyang R, Wang Z, Zhou W, Chen H, Jiang Y, Zhang Y, Li H, Liao M, Wang W, et al. miR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci Rep. 2016;6:39001. doi: 10.1038/srep39001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin QW, Sun XF, Yang GT, Li XB, Wu MS, Zhao J. Increased expression of microRNA-150 is associated with poor prognosis in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:842–846. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang N, Wei X, Xu L. miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett. 2013;587:2346–2351. doi: 10.1016/j.febslet.2013.05.059. [DOI] [PubMed] [Google Scholar]
- 34.Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K, Ren G, Su T, Pan Y, Feng B, et al. miR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene EGR2. Biochem Biophys Res Commun. 2010;392:340–345. doi: 10.1016/j.bbrc.2009.12.182. [DOI] [PubMed] [Google Scholar]
- 35.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.