Abstract
Using microbial culturomics, three Bacillus strains were isolated, identified and characterized following the taxonogenomics strategy. Bacillus dakarensis strain Marseille-P3515T (=CSURP3515), Bacillus sinesaloumensis strain Marseille-P3516T (=CSURP3516), and Bacillus massiliogabonensis strain Marseille-P2639T (=CSURP2639) were isolated from human stool samples. The phylogenetic analysis, phenotypic characteristics and genotypic data presented here prove that these three bacteria are different from previously known bacterial species with standing in nomenclature and represent new Bacillus species.
Keywords: Africa, Bacillus sp., culturomics, human stool, taxonogenomics
Introduction
Genus Bacillus was created in 1872 by Ferdinand Julius Cohn [1]. To date, there are 379 species and seven subspecies with validly published names. Most Bacillus species are environmental bacteria found in food, soil, freshwater and the sea. Other Bacillus species may be saprophytic [2] or endophytic on plants [3]. Two species are important in public health, namely Bacillus cereus (associated with food poisoning) and Bacillus anthracis (responsible for anthrax) [4,5].
Bacteria involved in normal physiological functions and with a predisposition to human diseases should be studied for better understanding [6]. The culturomics method that isolates bacteria under different culture conditions is complemented in our laboratory by the systematic sequencing of the 16S rRNA gene, which allows us to explore the microbial diversity of the human gut [[7], [8], [9]]. The new species, which we report here, were described using a combination of genotypic and phenotypic characteristics, following a previously described taxonogenomics strategy [10,11].
We present the details of the isolation and taxonogenomics characterization of strain Marseille-P3515T, strain Marseille-P3516T and strain Marseille-P2639T as type strains of Bacillus dakarensis sp. nov., Bacillus sinesaloumensis sp. nov. and Bacillus massiliogabonensis sp. nov., respectively.
Isolation and growth conditions
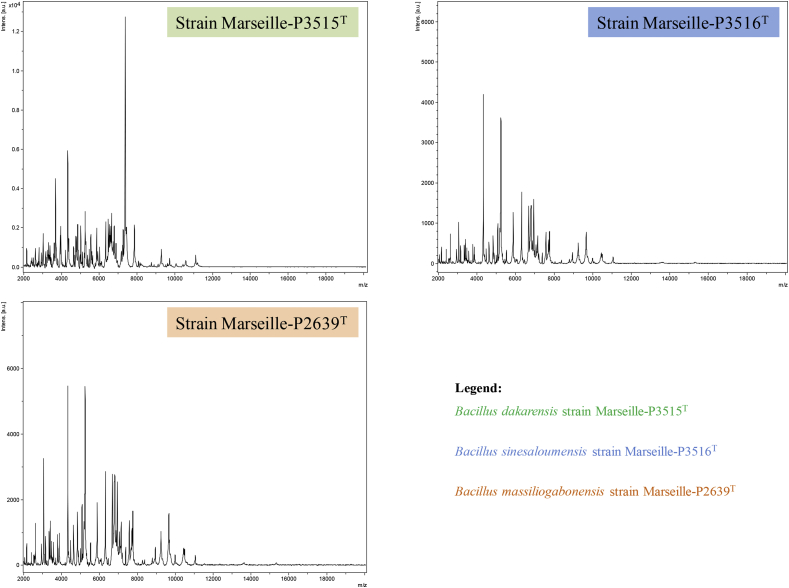
In 2016, three stool samples were collected with the aim of studying halophilic bacteria involved in human gut functioning [12]. Marseille-P3515T and Marseille-P3516T were isolated from the stool samples of a 17-year-old boy and a 10-year-old girl, respectively, both living in Ndiop, a rural area in Senegal. Strain Marseille-P2639T was isolated from the stool sample of a 16-year-old boy living in Gabon. After numerous attempts at identification by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry, no reliable recognition was obtained for the three strains. The screening was performed on a Microflex LT spectrometer (Bruker Daltonics, Bremen, Germany) as previously described [13]. The spectra obtained (Fig. 1) were imported into MALDI Biotyper 3.0 software and analysed against the Bruker database, which is permanently improved with the MEPHI database (https://www.mediterranee-infection.com/urms-data-base). The strains (Marseille-P3515T, Marseille-P3516T and Marseille-P2639T) were first isolated after 1–2 days of pre-incubation of stool samples in aerobic conditions in blood-culture bottles enriched with 5% rumen fluid sterilized by filtration at 0.2 μm and seeded on 5% sheep-blood Columbia agar (bioMérieux, Marcy l’Étoile, France) under aerobic conditions at 37°C.
Fig. 1.
MALDI-TOF MS reference spectrum of the three new species described above. The reference spectra were generated by comparison of spectra from 12 individual colonies for each species.
Strain identification
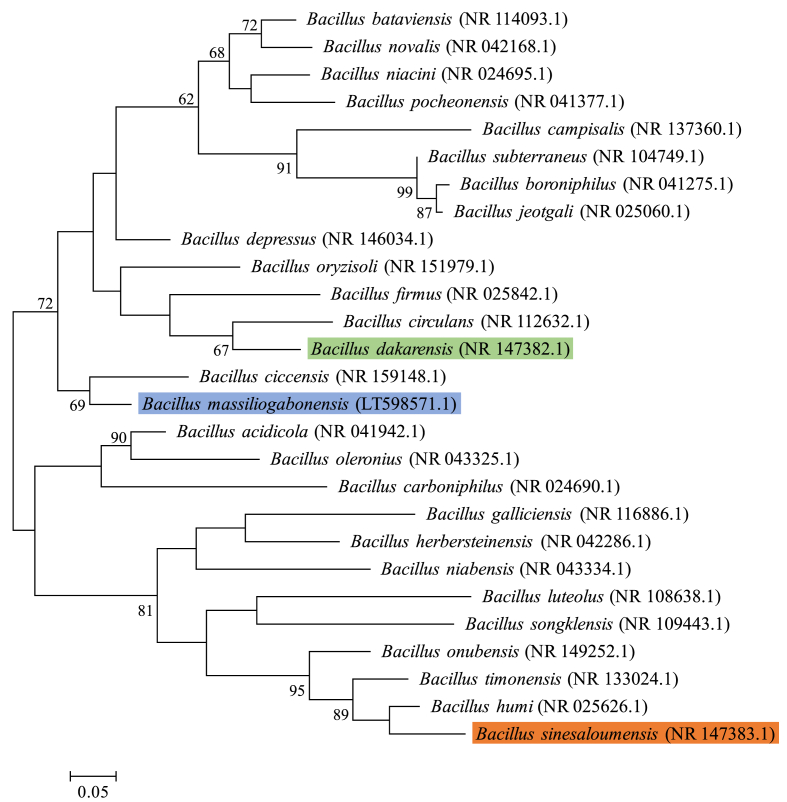
After missing identification using MALDI-TOF, the 16S rRNA genes for each strain were sequenced to classify these bacteria. These genes were amplified using universal primer pairs fD1 and rP2 (Eurogentec, Angers, France) and sequenced with the Big Dye® Terminator v1.1 Cycle Sequencing Kit and 3500xL Genetic Analyzer capillary sequencer (Thermofisher, Saint-Aubin, France), as previously described [14]. CodonCode Aligner software was used for assembly and to correct the 16S rRNA nucleotide sequences (http://www.codoncode.com). The sequences of each strain were submitted for BLAST in the NCBI database to determine the phylogenetically closest species with standing in nomenclature. It is in this context that we found that strain Marseille-P3515T exhibited a 97.96% sequence identity with Bacillus circulans strain NBRC 13626 (GenBank Accession no.: AY724690), strain Marseille-P3516T showed a 98.51% sequence identity with Bacillus humi strain LMG 22167 (AJ627210), and strain Marseille-P2639T displayed 98.41% sequence identity with Bacillus ciccensis strain 105-2 (KP965576). These values are below the threshold value recommended (<98.7% sequence similarity of the 16S rRNA gene) by authors to delineate new bacterial species within a genus without performing DNA–DNA hybridization [15,16]. Based on this observation, we declare that these strains are new members of the genus Bacillus belonging to the family Bacillaceae within the phylum Firmicutes (Fig. 2).
Fig. 2.
Phylogenetic tree highlighting the position of three new bacterial species relative to their most closely related and validly published type strains. GenBank accession numbers of 16S rRNA are indicated in parentheses. Sequences were aligned using MUSCLE with default parameters, phylogenetic inference were obtained using the maximum likelihood method and the MEGA 7 software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 1000 times to generate a majority consensus tree. The scale bar indicates a 5% nucleotide sequence divergence.
Phenotypic characteristics
The strains from Senegal were easily grown in an aerobic atmosphere. Apparent colonies were obtained after 24 h of incubation at 37°C on 5% sheep’s blood–Columbia agar medium (bioMérieux). Strains Marseille-P3515T and Marseille-P3516T were recovered from human stool sample from a Senegalese village named Ndiop. Their colonies appear rounded, beige and shiny with a mean diameter of 1.2 mm. Cells were Gram-positive bacteria, rod-shaped and catalase positive. In addition, the oxidase reaction test was positive for these two strains, which were mobile and spore forming. These strains were cultured on halophilic media with NaCl concentrations of 50, 75, 100 and 150 g/L of NaCl. In parallel, the growth of bacteria was tested on media at different pH (pH 6, 6.5, 7, 7.5 and 8). Tests have shown that the two strains grow better in 48 h at pH 7.5, at 75 g/L of NaCl and 37°C. These data indicate that strains Marseille-P3515T and Marseille-P3516T are halophilic bacteria.
Strain Marseille-P2639T was endospore forming and motile. It was a Gram-negative bacterium that exhibited catalase activity. Bacterial cells did not have an oxidase reaction. They measured 3.7 μm in length and 0.8 μm in diameter. Strain Marseille-P2639T was an aerobic bacterium that grew between 23°C and 45°C in <1 day of incubation. Colonies of strain Marseille-P2639T were white with a mean diameter of 3 mm on 5% sheep’s blood-enriched Columbia agar. Strain Marseille-P2639T is a bacterium that weakly tolerates salt concentrations >50 g/L of NaCl but is able to grow on media with a pH ranging from 6 to 10. The optimal growth temperature is 37°C under aerobic conditions.
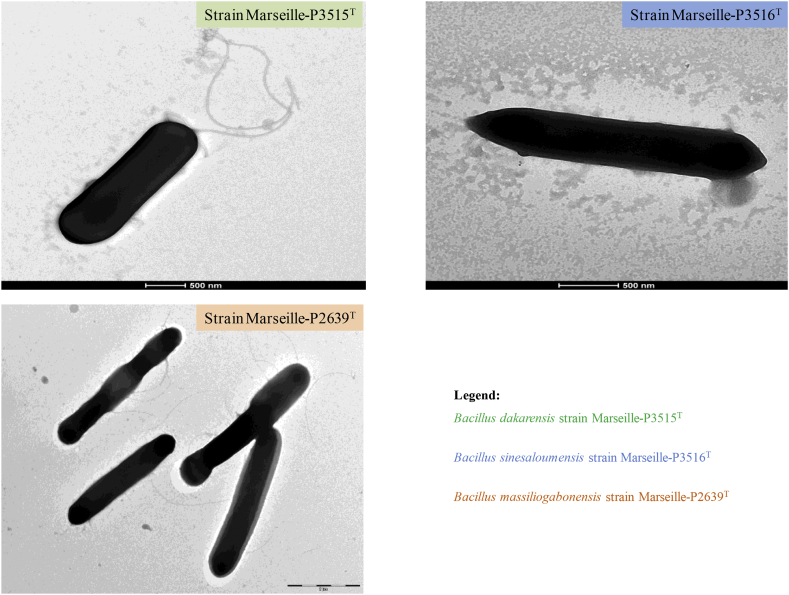
The shape of these bacteria was highlighted with the Tecnai G20 transmission electron microscope (FEI Company) (Fig. 3). The biochemical characteristics of these strains were tested using the API ZYM and API 50 CH strips (bioMérieux) and are presented in Table 1. A comparative study of the differential characteristics of these strains with other closely related species is displayed in Table 2.
Fig. 3.
Transmission electron microscopy of three new Bacillus species. Cells are observed on Tecnai G20 transmission electron microscope operated at 200 keV. Scales are displayed on the figures.
Table 1.
Phenotypic characterization of Bacillus dakarensis sp. nov., strain Marseille-P3515T, Bacillus sinesaloumensis sp. nov., strain Marseille-P3516T and Bacillus massiliogabonensis sp. nov., strain Marseille-P2639T sp. nov., based on analytical profile index (API) tests
| Tests | Number | Characteristics | Marseille-P3515T | Marseille-P3516T | Marseille-P2639T |
|---|---|---|---|---|---|
| API ZYM | 2 | Alkaline phosphatase | − | − | + |
| 3 | Esterase (C4) | + | + | + | |
| 4 | Esterase lipase (C8) | − | − | − | |
| 5 | Lipase (C14) | − | − | − | |
| 6 | Leucine arylamidase | − | − | − | |
| 7 | Valine arylamidase | − | − | − | |
| 8 | Cystine arylamidase | − | − | − | |
| 9 | Trypsin | − | − | − | |
| 10 | α-chymotrypsin | − | − | + | |
| 11 | Acid phosphatase | + | − | + | |
| 12 | Naphthol-AS-BI-phosphohydrolase | − | − | − | |
| 13 | α-galactosidase | − | + | − | |
| 14 | β-galactosidase | − | + | − | |
| 15 | β-glucuronidase | − | + | − | |
| 16 | α-glucosidase | − | + | − | |
| 17 | β-glucosidase | − | + | − | |
| 18 | N-acetyl-β-glucosaminidase | − | + | − | |
| 19 | α-mannosidase | − | − | − | |
| 20 | α-fucosidase | − | − | − | |
| API 50CH | 1 | Glycerol | − | − | + |
| 2 | Erythritol | − | − | − | |
| 3 | d-arabinose | + | − | − | |
| 4 | l-arabinose | + | − | − | |
| 5 | d-ribose | − | − | − | |
| 6 | d-xylose | + | − | − | |
| 7 | l-xylose | − | − | − | |
| 8 | d-Adonitol | + | − | − | |
| 9 | Methyl βd-xylopyranoside | − | − | − | |
| 10 | d-galactose | − | − | + | |
| 11 | d-glucose | − | − | − | |
| 12 | d-fructose | − | − | − | |
| 13 | d-mannose | − | − | − | |
| 14 | l-sorbose | − | − | − | |
| 15 | l-rhamnose | + | − | − | |
| 16 | Dulcitol | − | − | − | |
| 17 | Inositol | + | − | − | |
| 18 | d-mannitol | − | − | − | |
| 19 | d-sorbitol | − | − | − | |
| 20 | Methyl αd-mannopyranoside | − | − | − | |
| 21 | Methyl αd-glucopyranoside | − | − | + | |
| 22 | N-acetyl-glucosamine | − | − | + | |
| 23 | Amygdalin | − | − | + | |
| 24 | Arbutin | − | − | + | |
| 25 | Esculin ferric citrate | + | + | + | |
| 26 | Salicin | − | − | + | |
| 27 | d-cellobiose | − | − | − | |
| 28 | d-maltose | − | − | − | |
| 29 | d-lactose | − | − | − | |
| 30 | d-melibiose | + | − | − | |
| 31 | Sucrose | − | − | − | |
| 32 | d-trehalose | − | − | + | |
| 33 | Inulin | − | − | + | |
| 34 | d-melezitose | − | − | + | |
| 35 | d-raffinose | − | − | + | |
| 36 | Starch | − | − | + | |
| 37 | Glycogen | − | − | + | |
| 38 | Xylitol | + | − | − | |
| 39 | Gentiobiose | − | − | − | |
| 40 | d-turanose | − | − | − | |
| 41 | d-lyxose | + | − | − | |
| 42 | d-tagalose | − | − | − | |
| 43 | d-fucose | + | − | − | |
| 44 | l-fucose | − | − | − | |
| 45 | d-arabitol | − | − | − | |
| 46 | l-arabitol | − | − | + | |
| 47 | Potassium gluconate | − | − | − | |
| 48 | Potassium 2-ketogluconate | − | − | − | |
| 49 | Potassium 5-ketogluconate | − | − | − |
Table 2.
Differential characteristics of Bacillus dakarensis strain Marseille-P3515T, Bacillus sinesaloumensis strain Marseille-P3516T and Bacillus massiliogabonensis strain Marseille-P2639T compared with Bacillus ndiopicus strain FF3T and Bacillus dielmoensis strain FF4T
| Property | P3515T | P3516T | P2639T | FF3T | FF4T |
|---|---|---|---|---|---|
| Cell diameter (μm) | 0.5–1 | 1–1.9 | 0.7–1 | 0.8–1.6 | 0.5–0.8 |
| Oxygen requirement | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic |
| Gram stain | + | + | − | + | + |
| Motility | + | + | − | + | + |
| Endospore formation | + | + | + | + | − |
| Production of: | |||||
| Alkaline phosphatase | − | − | + | + | + |
| Acid phosphatase | + | − | + | − | + |
| Catalase | + | + | + | + | + |
| Oxidase | + | + | − | − | − |
| β-Galactosidase | − | + | − | − | + |
| α-Glucosidase | − | + | − | − | + |
| Esterase | + | + | + | + | + |
| Esterase lipase | − | − | − | + | + |
| Naphthol-AS-BI-phosphohydrolase | − | − | − | − | + |
| N-acetyl-β-glucosaminidase | − | + | − | − | − |
| Utilization of: | |||||
| Potassium 5-ketogluconate | − | − | − | − | − |
| d-Xylose | + | − | − | − | − |
| d-Fructose | − | − | − | − | − |
| d-Glucose | − | − | − | − | − |
| d-Mannose | − | − | − | − | − |
| Habitat | Human sample | Human sample | Stool sample | Human skin | Human skin |
Note: +, positive result; −, negative result; NA, data not available.
Genome sequencing
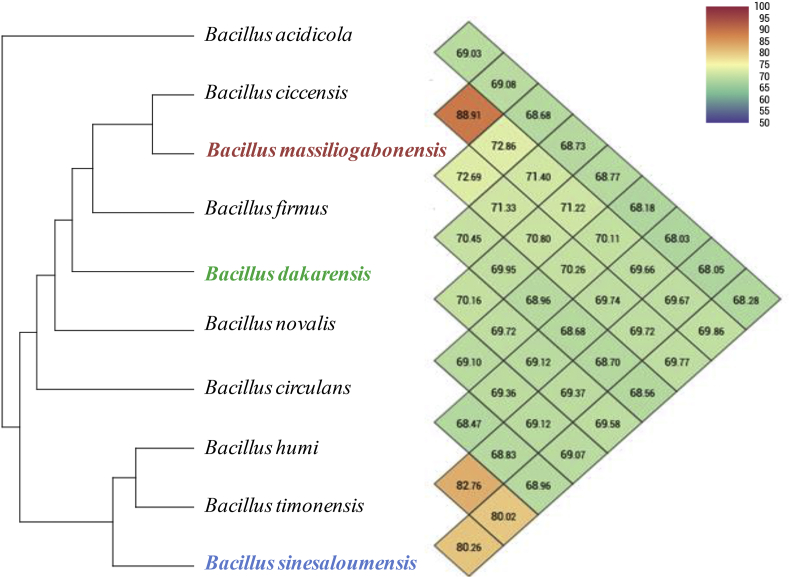
Genomic DNA was extracted using the EZ1 biorobot (Qiagen, Courtaboeuf, France) with the EZ1 DNA tissue Kit and then sequenced using MiSeq technology (Illumina, San Diego, CA, USA) with the Nextera Mate Pair sample prep kit and Nextera XT Paired end (Illumina), as previously described [17]. The assembly was performed with a pipeline incorporating different softwares (Velvet [18], Spades [19] and Soap Denovo [20]), and trimmed data (MiSeq and Trimmomatic [21] softwares) or untrimmed data (only MiSeq software). GapCloser was used to reduce assembly gaps. Scaffolds <800 bp and scaffolds with a depth value < 25% of the mean depth were removed. The best assembly was selected using different criteria (number of scaffolds, N50, number of N). The degree of genomic similarity of these three strains with closely related species was estimated using OrthoANI software [22]. OrthoANI values among Bacillus species (Fig. 4) ranged from 68.03% between Bacillus acidicola and B. humi to 88.91% between B. ciccensis and B. massiliogabonensis. These values were <95%, the threshold value suggested for delineating new bacterial species in an OrthoANI comparison [23].
Fig. 4.
Heatmap generated with OrthoANI values calculated using the OAT software for Bacillus dakarensis strain Marseille-P3515T, Bacillus sinesaloumensis strain Marseille-P3516T and Bacillus massiliogabonensis strain Marseille-P2639T among other related Bacillus species with standing in nomenclature.
Genome properties
The genome of strain Marseille-P3515T is 5 482 351 bp long with 38.6 mol% G + C content (Table 3). It is composed of 11 scaffolds (composed of 62 contigs). Of the 5282 predicted genes, 5040 are protein-coding genes and 242 are RNAs (15 genes are 5S rRNA, 7 genes are 16S rRNA, 9 genes are 23S rRNA, 211 genes are tRNA genes). A total of 3600 genes (71.43%) were assigned as putative function (by cogs or by NR blast); 206 genes were identified as ORFans (4.09%). The remaining genes were annotated as hypothetical proteins (1014 genes, ≥20.12%).
Table 3.
Nucleotide content and gene count levels of genomes of Bacillus dakarensis strain Marseille-P3515T, Bacillus sinesaloumensis strain Marseille-P3516T and Bacillus massiliogabonensis strain Marseille-P2639T
| Attribute |
B. dakarensis |
B. sinesaloumensis |
B. massiliogabonensis |
|||
|---|---|---|---|---|---|---|
| Value | % Of totala | Value | % Of totala | Value | % Of totala | |
| Size (bp) | 5 482 351 | 100 | 4 556 426 | 100 | 5 224 786 | 100 |
| G + C content (bp) | 2 057 335 | 38.6 | 1 711 281 | 37.9 | 1 944 649 | 37.9 |
| Total of genes | 5282 | 100 | 4466 | 100 | 5234 | 100 |
| RNA genes | 242 | 4.5 | 143 | 3.2 | 189 | 3.6 |
| Coding sequence size (bp) | 4 474 355 | 81.6 | 3 886 129 | 85.3 | 4 337 909 | 83.0 |
| Protein-coding genes | 504 | 100 | 4323 | 100 | 5045 | 100 |
| Protein assigned to COGs | 3246 | 64.4 | 2741 | 63.4 | 3063 | 60.7 |
| Genes with peptide signals | 595 | 11.8 | 478 | 11.0 | 561 | 11.1 |
| Genes with transmembrane helices | 1189 | 23.6 | 1183 | 27.3 | 1278 | 25.3 |
| Genes associated with mobilome | 2147 | 42.6 | 1714 | 39.6 | 2068 | 40.9 |
| Genes associated with virulence | 951 | 18.8 | 790 | 18.2 | 948 | 18.7 |
Total is based on total number of protein-coding genes in annotated genome.
Strain Marseille-P3516T has a genome size of 4 556 426 bp long with 37.9 mol% G + C content (Table 3). It is composed of three scaffolds with 26 contigs. Of the 4466 predicted genes, 4323 are protein-coding genes and 143 are RNAs. These RNA genes are divided into 12 5S rRNA genes, 13 16S rRNA genes, 13 23S rRNA genes and 105 tRNA genes. Strain Marseille-P3516T possesses 3095 genes (71.59%) as putative function (by cogs or by NR blast) and 130 genes as ORFans (3.01%); 868 genes (20.08%) were annotated as hypothetical proteins.
The genome of strain Marseille-P2639T is 5 224 786 bp in length with 37.9 mol% G + C content and is composed of nine scaffolds with 69 contigs (Table 3). Overall, in 5234 predicted genes, 5045 genes encode proteins and 189 genes are RNAs (12 genes are 5S rRNA, 18 genes are 16S rRNA, 16 genes are 23S rRNA, 143 genes are tRNA genes). A total of 3476 genes (68.9%) were assigned as putative function (by the cogs or by NR blast), 179 genes were recognized as ORFans (3.55%) and the remaining 1151 genes (22.81%) were annotated as hypothetical proteins.
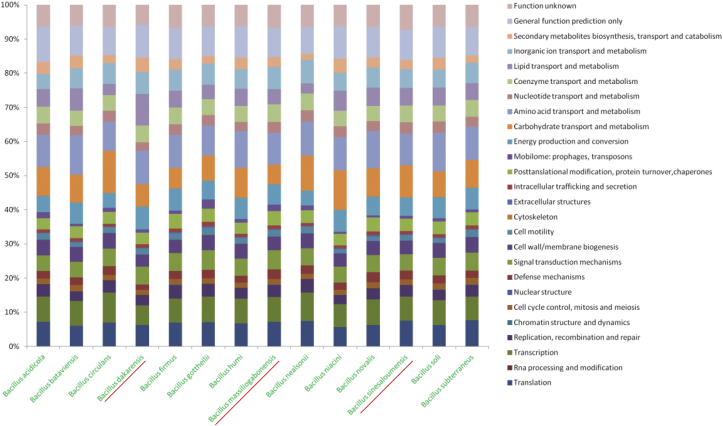
The digital DNA–DNA hybridization between the genomes of three new Bacillus species and other available genomes of the phylogenetically closest species, was calculated using the Genome-to-Genome Distance Calculator online calculator with formula 2 (Table 4). A 70% threshold is fixed by Meier-Kolthoff et al., to differentiate two distinct species [24]. DNA–DNA hybridization values ranged from 21.5% between B. ciccensis and Bacillus firmus to 34.9% between Bacillus sinesaloumensis and Bacillus acidicola, which supported the previous data indicating the classification of these as a new bacterial species (Table 4). Likewise, repartition of genes into the 25 general COG categories is illustrated in Table 5 and Fig. 5. The average percentage nucleotide identity calculated using the Average Genomic Identity of Orthologous Gene Sequences (AGIOS) in-house software [10] and number of orthologous genes of B. dakarensis, B. sinesaloumensis and B. massiliogabonensis shared with others species are displayed in Table 6.
Table 4.
Genome comparison between three new bacterial strains and closely related species using GGDC and formula 2 (dDDH estimates based on identities over HSP length), upper right. The inherent uncertainty in assigning dDDH values from intergenomic distances is presented in the form of confidence intervals
| Bdak | Bmas | Bsin | Baci | Bcic | Bcir | Bfir | Bnov | Btim | |
|---|---|---|---|---|---|---|---|---|---|
| Bdak | 100% | 24.3% ± 4.8 | 27.5% ± 4.9 | 28.4% ± 4.9 | 24.5% ± 4.8 | 28.5% ± 4.9 | 22.7% ± 4.7 | 24.3% ± 4.8 | 23.2% ± 4.8 |
| Bmas | 100% | 31.5% ± 4.9 | 32.1% ± 4.9 | 37.6% ± 5 | 30.3% ± 4.9 | 22.0% ± 4.7 | 26.3% ± 4.8 | 25.9% ± 4.8 | |
| Bsin | 100% | 34.9% ± 4.9 | 27.8% ± 4.8 | 27.1% ± 4.8 | 26.4% ± 4.9 | 32.1% ± 5 | 23.8% ± 4.7 | ||
| Baci | 100% | 32.7% ± 4.9 | 33.9% ± 4.9 | 27.3% ± 4.9 | 29.3% ± 4.9 | 29.3% ± 4.9 | |||
| Bcic | 100% | 28.9% ± 4.8 | 21.5% ± 4.7 | 25.6% ± 4.8 | 25.0% ± 4.8 | ||||
| Bcir | 100% | 23.4% ± 4.8 | 32.6% ± 4.9 | 25.0% ± 4.8 | |||||
| Bfir | 100% | 22.2% ± 4.7 | 22.6% ± 4.7 | ||||||
| Bnov | 100% | 26.2% ± 4.8 | |||||||
| Btim | 100% |
dDDH, digital DNA–DNA hybridization; HSP, high scoring pair; GGDC, genome-to-genome distance calculator.
Bacillus dakarensis strain Marseille-P3515T (Bdak), Bacillus massiliogabonensis strain Marseille-P2639T (Bmas), Bacillus sinesaloumensis strain Marseille-P3516T (Bsin), Bacillus acidicola strain DSM 14745T (Baci), Bacillus ciccensis strain KCTC 33663T (Bcic), Bacillus circulans strain NBRC 13626 (Bcir), Bacillus firmus strain NBRC 15306 (Bfir), Bacillus novalis strain NBRC 102450 (Bnov) and Bacillus timonensis strain Marseille-P162 (Btim).
Table 5.
Number of genes associated with 25 general COG functional categories
| Code |
Bacillus sinesaloumensis |
Bacillus dakarensis |
Bacillus massiliogabonensis |
Description | |||
|---|---|---|---|---|---|---|---|
| Value | % Of total | Value | % Of total | Value | % Of total | ||
| [J] | 236 | 5.45 | 237 | 4.70 | 250 | 4.95 | Translation |
| [A] | 0 | 0 | 0 | 0 | 0 | 0 | RNA processing and modification |
| [K] | 219 | 5.06 | 217 | 4.30 | 255 | 5.05 | Transcription |
| [L] | 104 | 2.40 | 111 | 2.20 | 126 | 2.49 | Replication, recombination and repair |
| [B] | 1 | 0.02 | 1 | 0.01 | 1 | 0.01 | Chromatin structure and dynamics |
| [D] | 52 | 1.20 | 56 | 1.11 | 58 | 1.14 | Cell cycle control, mitosis and meiosis |
| [Y] | 0 | 0 | 0 | 0 | 0 | 0 | Nuclear structure |
| [V] | 79 | 1.82 | 58 | 1.15 | 98 | 1.94 | Defence mechanisms |
| [T] | 151 | 3.49 | 197 | 3.90 | 198 | 3.92 | Signal transduction mechanisms |
| [M] | 122 | 2.82 | 132 | 2.61 | 139 | 2.75 | Cell wall/membrane biogenesis |
| [N] | 53 | 1.22 | 69 | 1.36 | 72 | 1.42 | Cell motility |
| [Z] | 0 | 0 | 0 | 0 | 1 | 0.01 | Cytoskeleton |
| [W] | 8 | 0.18 | 9 | 0.17 | 8 | 0.15 | Extracellular structures |
| [U] | 29 | 0.67 | 40 | 0.79 | 34 | 0.67 | Intracellular trafficking and secretion |
| [O] | 113 | 2.61 | 128 | 2.53 | 149 | 2.95 | Post-translational modification, protein turnover, chaperones |
| [X] | 26 | 0.60 | 32 | 0.63 | 66 | 1.30 | Mobilome: prophages, transposons |
| [C] | 167 | 3.86 | 254 | 5.03 | 208 | 4.12 | Energy production and conversion |
| [G] | 294 | 6.80 | 245 | 4.86 | 201 | 3.98 | Carbohydrate transport and metabolism |
| [E] | 291 | 6.73 | 368 | 7.30 | 329 | 6.52 | Amino acid transport and metabolism |
| [F] | 100 | 2.31 | 94 | 1.86 | 111 | 2.20 | Nucleotide transport and metabolism |
| [H] | 156 | 3.60 | 187 | 3.71 | 178 | 3.52 | Coenzyme transport and metabolism |
| [I] | 158 | 3.65 | 345 | 6.84 | 160 | 3.17 | Lipid transport and metabolism |
| [P] | 172 | 3.97 | 237 | 4.70 | 227 | 4.49 | Inorganic ion transport and metabolism |
| [Q] | 81 | 1.87 | 164 | 3.25 | 98 | 1.94 | Secondary metabolites biosynthesis, transport and catabolism |
| [R] | 278 | 6.43 | 363 | 7.20 | 300 | 5.94 | General function prediction only |
| [S] | 230 | 5.32 | 220 | 4.36 | 239 | 4.73 | Function unknown |
| _ | 1582 | 36.59 | 1794 | 35.59 | 1982 | 39.28 | Not in COGs |
COG, cluster of orthologous groups.
Fig. 5.
Distribution of functional classes of predicted genes according to the COG of proteins of Bacillus dakarensis strain Marseille-P3515T, Bacillus sinesaloumensis strain Marseille-P3516T and Bacillus massiliogabonensis strain Marseille-P2639T among other related Bacillus species.
Table 6.
Numbers of orthologous proteins shared between genomes (bold)a
| Genomes | Bacillus circulans | NCIMB8773 | Marseille-P3515 | 105-2 | WCC4585 | IAM12464 |
|---|---|---|---|---|---|---|
| Bacillus circulans | 4950 | 1829 | 1857 | 1842 | 2031 | 2037 |
| Bacillus lentus strain NCIMB8773 | 60.91 | 4088 | 1754 | 1698 | 1830 | 1808 |
| Bacillus dakarensis strain Marseille-P3515 | 61.84 | 67.91 | 5040 | 1816 | 2099 | 2244 |
| Bacillus acidicola strain 105-2 | 60.54 | 67.96 | 67.56 | 4876 | 1839 | 1976 |
| Bacillus gottheilii strain WCC4585 | 61.69 | 67.52 | 70.24 | 67.66 | 4450 | 2285 |
|
Bacillus firmus strain IAM12464 |
59.95 |
58.64 |
60.36 |
59.11 |
61.28 |
4922 |
|
PB1NCIMB |
LMG21833 |
AM31D |
LMG22167 |
Marseille-P3516 |
IFO15566 |
|
| Bacillus methanolicus strain PB1NCIMB | 3410 | 1704 | 1524 | 1627 | 1628 | 1715 |
| Bacillus bataviensis strain LMG21833 | 61.99 | 5207 | 1754 | 2168 | 2086 | 2449 |
| Bacillus krulwichiae strain AM31D | 53.99 | 55.98 | 4596 | 1774 | 1793 | 1880 |
| Bacillus humi strain LMG22167 | 55.03 | 58.02 | 66.45 | 4842 | 2316 | 2198 |
| Bacillus sinesaloumensis strain Marseille-P3516 | 55.00 | 57.56 | 66.84 | 81.22 | 4323 | 2161 |
|
Bacillus niacini strain IFO15566 |
55.76 |
60.61 |
66.02 |
68.48 |
68.34 |
5952 |
|
Marseille-P2639 |
LMG21831 |
LMG21837 |
LMG21838 |
A1-2 |
DSM13966 |
|
| Bacillus massiliogabonensis strain Marseille-P2639 | 5045 | 2208 | 2364 | 2338 | 2369 | 1904 |
| Bacillus drentensis strain LMG21831 | 70.62 | 5043 | 2707 | 2796 | 2107 | 1902 |
| Bacillus novalis strain LMG21837 | 70.47 | 77.50 | 5425 | 3014 | 2246 | 1958 |
| Bacillus soli strain LMG21838 | 70.39 | 79.26 | 80.82 | 5340 | 2244 | 1944 |
| Bacillus eiseniae strain A1-2 | 72.08 | 69.53 | 69.15 | 69.22 | 5468 | 1870 |
| Bacillus subterraneus strain DSM13966 | 60.44 | 60.27 | 60.17 | 60.27 | 60.07 | 3465 |
Average percentage similarity of nucleotides corresponding to orthologous protein shared between genomes and numbers of proteins per genome (bold).
Conclusion
Based on unique phenotypic characteristics, including API test strips, MALDI-TOF spectra, and phylogenetic and genomic analyses such as 16S rRNA sequence similarity <98.7% and OrthoANI values < 95% with the phylogenetically closest species with standing in nomenclature, we propose strains Marseille-P3515T, Marseille-P3516T and Marseille-P2639T, respectively, as being the type strains of Bacillus dakarensis sp. nov., Bacillus sinesaloumensis sp. nov. and Bacillus massiliogabonensis sp. nov., which are new species in the genus Bacillus.
Description of Bacillus dakarensis sp. nov.
Bacillus dakarensis (da.ka.ren’sis, N.L. masc. adj. dakarensis of Dakar, the name of the capital of Senegal where the stool sample was collected). The colonies of the strain appear beige and circular on blood agar with a mean diameter of 1.2 mm. The cells are mobile and spore-forming. They are Gram-positive bacilli and present positive oxidase and positive catalase activities. The draft genome size of strain Marseille-P3515T is about 5.33 Mb with a 38.6 mol% of G + C content. The 16S rRNA gene sequence and whole-genome shotgun sequence of B. dakarensis strain Marseille-P3515T were deposited in GenBank under accession numbers LT671589 and FTOZ00000000, respectively. The type strain Marseille-P3515T (=CSURP3515) was isolated from the stool sample of 17-year-old-boy living in Senegal.
Description of Bacillus sinesaloumensis sp. nov.
Bacillus sinesaloumensis (si.ne.sa.lou.men’sis, N.L. masc. adj. sinesaloumensis of Sine-Saloum, a former administrative region of Senegal where the village of Ndiop is located, from which this strain was sampled). Colonies grow on 5% sheep blood Colombia agar plate after 24 h of incubation under aerobic conditions. They are shiny, beige and 2 mm in diameter. The cells are Gram-positive bacteria, mobile and spore-forming. Oxidase and catalase activities were positive. The DNA G + C content of the type strain is 37.9 mol% in a genome sequence length of 4.52 Mb. Its type strain, Marseille-P3516T (= CSURP3516T), was isolated from a 10-year-old girl from Ndiop, a rural area in Senegal.
Description of Bacillus massiliogabonensis sp. nov.
Bacillus massiliogabonensis (mas.si.li.ga.bo.nen’sis: NL. masc. adj. a composed name designating Marseille and Gabon, the city and the country where the strain and the stool specimen was characterized and collected, respectively). The strain grows at temperatures ranging from 23°C to 45°C in aerobic conditions with an optimum temperature of 37°C. Colonies with a white aspect had a mean diameter of 3 mm on blood agar medium. The strain Marseille-P2639T is a Gram-negative bacterium and exhibits positive catalase and negative oxidase activities. The genome of the Marseille-P2639T strain was 5.13 Mb with 37.9 mol% of G + C content. The potential pathogenicity of the type strain Marseille-P2639T (=CSURP2639) is unknown. It was isolated from the stool sample of a healthy 16-year-old Gabonese boy.
Nucleotide sequence accession number
Table 7 shows the16S rRNA gene and genome sequence accession numbers deposited in GenBank for these three new bacterial species:
Table 7.
Accession numbers of Bacillus dakarensis strain Marseille-P3515T, Bacillus sinesaloumensis strain Marseille-P3516T and Bacillus massiliogabonensis strain Marseille-P2639T
| Species | Strain number | 16S rRNA number | Genome accession number |
|---|---|---|---|
| Bacillus dakarensis | Marseille-P3515 | LT671589 | FTOZ00000000 |
| Bacillus sinesaloumensis | Marseille-P3516 | LT671591 | FTOX00000000 |
| Bacillus massiliogabonensis | Marseille-P2639 | LT598571 | FZRJ00000000 |
Conflict of interest
None to declare.
Funding sources
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the programme Investissements d’avenir, reference ANR-10-IAHU-03, the Région Provence-Alpes-Côte d'Azur and European funding FEDER PRIMI.
Ethics and consent
The study and consent procedures were approved by the ethics committee of the Institut Hospitalo-Universitaire Méditerrannée Infection (No. 2011-11), the National ethics committee of Gabon (No. 0023/2013/SG/CNE) and IFR48 of Marseille France (No. 09-022). The volunteers gave a written consent.
Acknowledgements
The authors thank Catherine Robert for sequencing the genome, Aurelia Caputo for submitting the genomic sequence to GenBank and Carine Couderc for producing the MALDI-TOF reference spectrum.
References
- 1.Cohn F. Untersuchungen über Bakterien. Beitrage zur Biologie der Pflanzen Heft. 1872;1:127–224. [Google Scholar]
- 2.Lopez M.S., Hodde M.K., Chamakura K.R., Kuty Everett G.F. Complete genome of Bacillus megaterium podophage page. Genome Announc. 2014;2(2):e00332-14. doi: 10.1128/genomeA.00332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y.Z., Chen W.F., Li M., Sui X.H., Liu H.C., Zhang X.X. Bacillus endoradicis sp. nov., an endophytic bacterium isolated from soybean root. Int J Syst Evol Microbiol. 2012;62:359–363. doi: 10.1099/ijs.0.028936-0. [DOI] [PubMed] [Google Scholar]
- 4.Jernigan J.A., Stephens D.S., Ashford D.A., Omenaca C., Topiel M.S., Galbraith M. Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States. Emerg Infect Dis. 2001;7:933–944. doi: 10.3201/eid0706.010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottone E.J. Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev. 2010;23:382–398. doi: 10.1128/CMR.00073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. https://www.nature.com/articles/nature06244 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 8.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 11.Fournier P.E., Lagier J.C., Dubourg G., Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Seck E.H., Diop A., Armstrong N., Delerce J., Fournier P.E., Raoult D. Microbial culturomics to isolate halophilic bacteria from table salt: genome sequence and description of the moderately halophilic bacterium Bacillus salis sp. nov. New Microbe. New Infect. 2018;23:28–38. doi: 10.1016/j.nmni.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo C.I., Fall B., Sambe-Ba B., Diawara S., Gueye M.W., Mediannikov O., Sokhna C. MALDI-TOF mass spectrometry: a powerful tool for clinical microbiology at Hôpital Principal de Dakar, Senegal (West Africa) PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0145889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morel A.S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.P. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis. 2015;34:561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 15.Meier-Kolthoff J.P., Göker M., Spröer C., Klenk H.P. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- 16.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 17.Lo C.I., Sankar S.A., Fall B., Ba B.S., Diawara S., Gueye M.W. High-quality draft genome sequence and description of Haemophilus massiliensis sp. nov. Stand Genom Sci. 2016;11:31. doi: 10.1186/s40793-016-0150-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee I., Ouk Kim Y., Park S.C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 23.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]