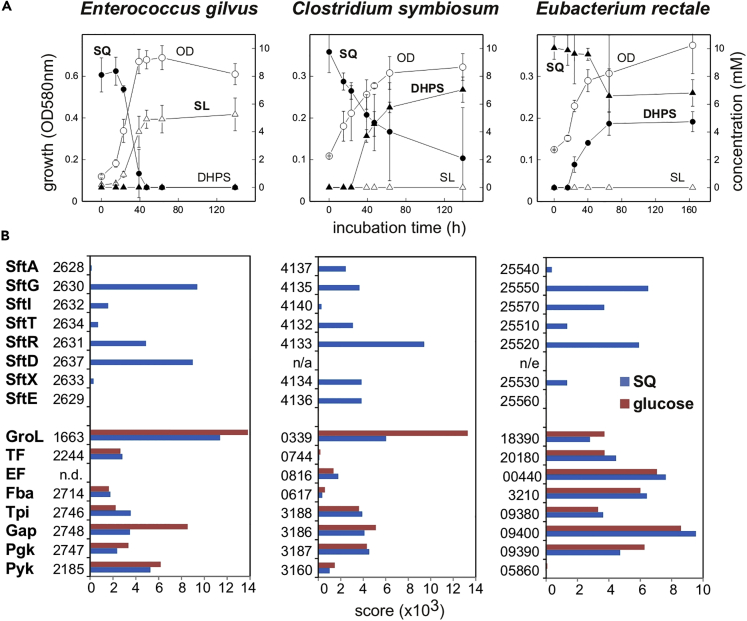
Figure 6.
Fermentation of SQ by Human Intestinal Bacteria via the SF-transaldolase Pathway
(A and B) Fermentation of SQ to SL or DHPS by human intestinal bacterial strains Enterococcus gilvus DSM15689, Clostridium symbiosum LT0011, and Eubacterium rectale DSM17629 (A) and proteomic confirmation of a strong, inducible expression of the Sft-proteins during growth with SQ (B). (A) E. gilvus produced SL during SQ fermentation and C. symbiosum and E. rectale produced DHPS but not SL. The used mineral salts medium had to be supplemented with yeast extract (0.1% w/v) for observing growth, and no complete turnover of the SQ provided (10 mM) was obtained, presumably due to depletion of supplements, except for E. gilvus. Triplicates (n = 3) are shown; the error bars indicate standard deviations. From replicate cultures, cellular biomass was collected at the end of the growth phase and submitted to total proteomic analyses in comparison to glucose-fermenting cells. (B) Differential proteomics confirmed a strongly inducible expression of the Sft-pathway genes during fermentation of SQ but not when the strains were grown with glucose. Each result was replicated at least once when starting from independent growth experiments. Abbreviations used: SftA, predicted SQ importer; SftG, SQG hydrolase (alpha-glucosidase); SftI, SQ isomerase; SftT, SF transaldolase; SftR, SLA reductase; SftD, SLA dehydrogenase; SftX, DUF4867; SftE, predicted SL/DHPS exporter; GroL, chaperon; TF, translation factor; EF, elongation factor; Fba, fructose bisphosphate aldolase; Tpi, triosephosphate isomerase; Gap, GAP dehydrogenase; Pyk, pyruvate kinase; n.d., not detected; n/e, not encoded in the respective sft-gene cluster (see Figure 5).