Summary
The human immune system is comprised of a diverse and interactive network of specialized cells localized in diverse tissues throughout the body, where they mediate protection against pathogens and environmental insults while maintaining tissue homeostasis. Although much of our understanding of human immunology has derived from studies of peripheral blood, recent work utilizing human tissue resources and innovative computational methods have employed a whole-body, systems-based approach, revealing tremendous complexity and heterogeneity of the immune system within individuals and across the population. In this review, we discuss how tissue localization, developmental and age-associated changes, and conditions of health and disease shape the immune response, as well as how improved understanding of interindividual and tissue-specific immunity can be leveraged for developing targeted therapeutics.
Subject Areas: Immunology, Complex System Biology
Graphical Abstract
Immunology; Complex System Biology
Introduction
The human immune system protects the body from infection by pathogens, exposure to environmental toxins and allergens, and cellular damage that may lead to cancer. The immune system also plays a critical role in maintaining homeostasis by mediating tissue repair and controlling inflammation. Unlike other organ systems that are contained within a specific tissue or structure (for example, the heart and blood vessels in the cardiovascular system), the immune system is comprised of a diverse population of specialized immune cells that provide surveillance to all tissues, forming a complex network across the human body. The two arms of the immune response comprise innate immunity, which provides a crucial immediate response triggered by signals from pathogens or dying cells, and adaptive immunity, which generates memory responses to specific antigens for long-term protective immunity. Cells of the innate immune system include macrophages and dendritic cells that largely reside in tissues, as well as neutrophils and other granulocytes that predominate in circulation. T and B lymphocytes form the cellular components of adaptive immunity and populate blood, lymphoid tissues, and non-lymphoid sites as circulating and tissue-resident subsets. Thus, understanding the complexity of the immune response requires examination of multiple cell subsets within diverse compartments across the body.
This vast and intricate system of cell-cell interactions across multiple anatomic sites has proven particularly challenging to study in humans. Although most of our understanding of human immunology is based on studies of peripheral blood, such analyses provide a limited view as the majority of innate and adaptive immune cells localize in tissues. Identifying differences in immune cells between blood and individual tissue sites has been accomplished with samples from biopsies and surgical resections, with the caveat that tissues are often associated with disease states (Alcántara-Hernández et al., 2017; Giesecke et al., 2014; McGovern et al., 2014; Medina et al., 2002; See et al., 2017; Sen et al., 2014; Simoni et al., 2017). By contrast, obtaining samples from deceased organ donors enables acquisition of multiple nondiseased tissue sites from an individual, such that tissue- and subset-specific influences on immune cells can be assessed (Boor et al., 2019; James et al., 2020; Schoettler et al., 2019; Senda et al., 2019; Snyder et al., 2019; Stewart et al., 2019). Analyzing multiple sites within an individual and among individuals can also provide insight into how the immune system functions as a network across diverse organ tissues within the human body across decades of life (Carpenter et al., 2018; Dogra et al., 2020; Granot et al., 2017; Meng et al., 2017; Sathaliyawala et al., 2013; Szabo et al., 2019a; Thome et al., 2014, Thome et al., 2016a; Yudanin et al., 2019). Advancements in cell profiling technology and computational techniques applied to these samples have proven indispensable for building a whole-body, system-based perspective of the human immune system, providing a new baseline from which to understand the immune response in disease. This review will examine recent progress in the field of human immunology and systems biology that have revealed new insights into the multiple levels of cellular and tissue interactions that underlie immune protection, regulation, and homeostasis and how they are altered due to age and disease.
Levels of Heterogeneity in the Human Immune System
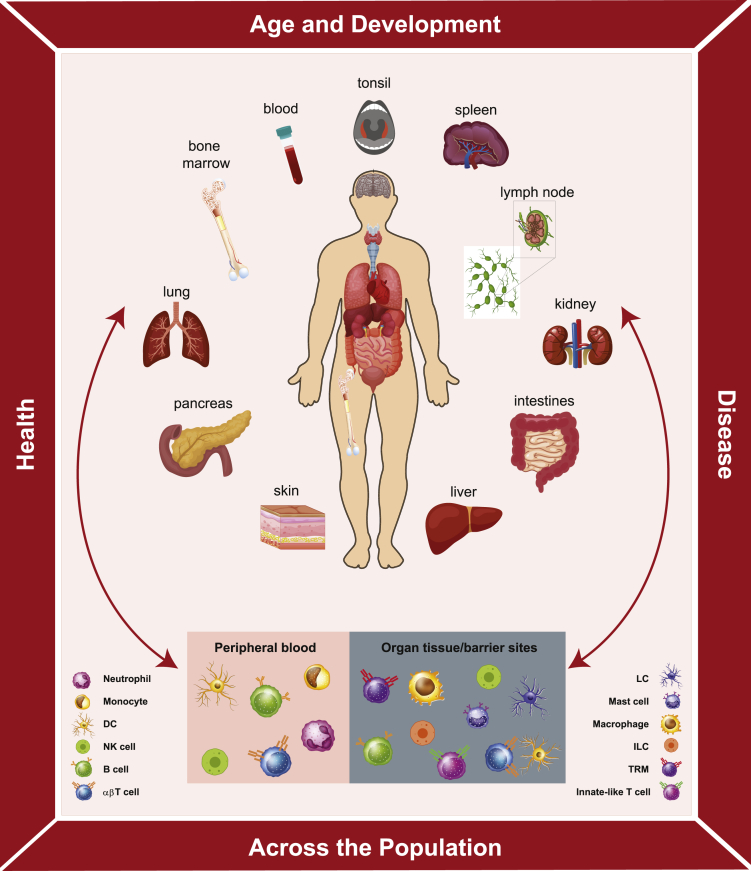
The immune system controls how we respond to pathogens, environmental antigens, cancer, and other insults; therefore, understanding the pathways, mechanism, cells, and molecules is central to human health. There is much to learn about the factors that impact the immune response, including variations among individuals over the broad range of ages of human life and as a feature of health and disease (Figure 1). Because the immune response is localized in multiple sites across the body and also in the blood, it is essential to sample key tissues that contain diverse immune cell populations including primary and secondary lymphoid organs (bone marrow, lymph nodes, spleen, tonsil), mucosal and barrier sites (skin, intestines, lungs), and metabolic organs, such as the liver and pancreas (Figure 1). Within each of these tissues are multiple innate and adaptive immune cell types and subsets; notably, similar immune cell lineages exhibit distinct features depending on whether they circulate in blood or localize in tissues. Adding to the complexity are tissue-specific adaptations that immune cells adopt within certain sites. Described below and in Figure 1 and Table 1 are the key cellular components of innate and adaptive immunity, their segregation in blood and different tissue sites, and their roles in the immune response.
Figure 1.
Immune System Heterogeneity of the Human Body
The human immune system is characterized by multilevel heterogeneity. Within an individual, immune cells are localized in multiple sites, including blood, primary and secondary lymphoid organs, organs of the respiratory and GI tract (lungs, intestines, pancreas liver), and barrier sites (skin, mucosal surfaces). Within tissues are diverse types of innate and adaptive immune cells—some that are present in circulation and migrate through blood and different tissues and others that establish and maintain residence within specific tissue sites. The composition, phenotype, function, and tissue-specific adaptations of diverse populations of immune cells differ between sites. Between individuals, immune responses vary depending on factors, such as age and conditions of health or disease. Understanding the whole body as a system requires investigation of the human immune system at all levels and integration of data from single cells to tissues to individuals.
Table 1.
Circulating and Tissue-Localized Immune Cell Types and Their Role in Immunity
| Immune Cell Type | Role in Immunitya | ||||
|---|---|---|---|---|---|
| Blood | Innate | Neutrophils |
|
||
| Eosinophils & Basophils |
|
||||
| Monocytes |
|
||||
| DCs | Plasmacytoid DC |
|
|||
| Natural killer (NK) cells |
|
||||
| Adaptive | Circulating T lymphocytes |
Naive T cell |
|
||
| TCM |
|
||||
| TEM |
|
||||
| TEMRA |
|
||||
| Treg |
|
||||
| Circulating B lymphocytes |
Naive B cell |
|
|||
| Plasmablast |
|
||||
| Memory B cell |
|
||||
| Tissue | Innate | Mast cells |
|
||
| Tissue macrophages |
|
||||
| DCs | Conventional DC |
|
|||
| Innate lymphoid cells (ILCs) | NK cell |
|
|||
| Helper-type ILC |
|
||||
| Adaptive | Tissue-resident T lymphocytes |
αβ T cell | TRM |
|
|
| Treg |
|
||||
| Tfh |
|
||||
| Innate-like T cell | γδ T cell |
|
|||
| iNKT cell |
|
||||
| MAIT cell |
|
||||
| Tissue-resident B lymphocytes |
Plasma cell |
|
|||
| Memory B cell |
|
||||
| Follicular B cell |
|
||||
| Marginal zone B cell |
|
||||
For references, see text.
In peripheral blood, innate immune cells comprise the majority of the immune cell compartment, with neutrophils being particularly abundant and T and B lymphocytes found in smaller proportions (Figure 1). Within organs and barrier sites, the innate and adaptive immune cells coordinate responses to provide tissue-specific immunity (Figure 1). The key tissue innate cells are macrophages, professional phagocytes that ingest cellular debris, pathogens, and other foreign substances. Long-lived tissue macrophages are established prenatally from embryonic precursors, self-renew in situ, and are highly specialized for their tissue of residence (Ginhoux and Guilliams, 2016; Gomez Perdiguero et al., 2015; Hashimoto et al., 2013; Yona et al., 2013). These include brain microglia, lung alveolar macrophages, spleen red pulp macrophages, and liver Kupffer cells (Ginhoux et al., 2010; Ginhoux and Jung, 2014; Hoeffel et al., 2012; Lavin et al., 2014; Merad et al., 2002; Nagelkerke et al., 2018) (Table 1).
Dendritic cells (DCs) are the innate immune cells that link innate and adaptive immunity through their role as antigen-presenting cells (APCs) crucial for T cell activation. Because of their unique ability to carry antigens from mucosal sites to T cell zones of tissue-draining lymph nodes, conventional DCs (cDCs), including cDC1 and cDC2 subsets perform the crucial task of presenting antigens to induce T cell responses (Table 1) (Audiger et al., 2017; Austyn, 2016; Bigley et al., 2015; Granot et al., 2017; Kashem et al., 2017; Klechevsky et al., 2008; Randolph et al., 2008). Circulating plasmacytoid DCs (pDCs) represent a small subset of DCs that infiltrate tissues to secrete large amounts of type I and type III interferons (IFNs) in response to viral infection and are particularly abundant in human tonsil at steady state (Bigley et al., 2015; Merad et al., 2013; Reizis et al., 2011b, Reizis et al., 2011a; Segura et al., 2013). Other innate cell types are innate lymphoid cells, which include natural killer (NK) cells abundant in human blood and tissues, as well as helper-type innate lymphoid cells (ILCs) ILC1, ILC2, and ILC3 that are mostly tissue-resident in different sites (Figure 1) (Brüggen et al., 2016; Dogra et al., 2020; Fuchs et al., 2013; Peng and Tian, 2017; Simoni et al., 2017; Yudanin et al., 2019). NK cells are important for anti-tumor and anti-viral immunity, whereas ILCs may act to promote tissue repair (Ishizuka et al., 2016; Sun and Lanier, 2011). Although innate immune cells have classically been described as having a rapid, short-lived response, studies in the past decade have shown that the function of myeloid and innate lymphoid cells can be shaped and enhanced by prior antigenic exposure through epigenetic and metabolic changes or modulation of surface receptor expression (Netea et al., 2020; O'Sullivan et al., 2015; Rodriguez et al., 2019; Sun et al., 2009; Wang et al., 2020).
The adaptive immune system is characterized by its diverse repertoire of antigen-specific receptors expressed by T and B lymphocytes and ability to retain immunological memory over decades. V(D)J recombination (recombination of receptor gene segments) and somatic hypermutation (mutation affecting the variable regions of immunoglobulin genes) allow a limited number of genes to generate a diverse repertoire of antigen receptors. T lymphocytes recognize specific antigens through interactions between their unique surface T cell receptor (TCR) and peptide-bound MHC molecules on APCs (Rossjohn et al., 2015). Conventional T cells are categorized into CD4+ helper and CD8+ cytotoxic T cells. Upon activation, naive CD4+ T cells differentiate into T helper cell lineages defined by their function and cytokine production, including Th1, Th2, Th9, Th17, Th22, T follicular helper cells (Tfh), and regulatory T cells (Treg) (for a review, see (Geginat et al., 2013)) (Table 1). CD8+ cytotoxic T cells comprise a less diverse group and, when activated, can secrete proinflammatory cytokines and cytotoxic mediators for killing of infected cells and tumor cells (Zhang and Bevan, 2011). Following the effector phase, during which expanded clones of antigen-specific T cell subsets coordinate antigen clearance, most T cells undergo apoptosis. However, a small number of T cells survive and differentiate into heterogeneous subsets of memory T cells, which provide long-term immunosurveillance and enhanced recall responses.
Diverse memory T cell subsets populate blood and virtually every tissue site; circulating subsets include central memory T cells (TCM) expressing the lymph node-homing receptor CCR7, effector memory T cells (TEM) that migrate through diverse tissues, and terminally differentiated effector T cells (TEMRA) (Sallusto et al., 2004; Thome et al., 2014), whereas tissue-resident memory T cells (TRM), characterized by their expression of CD69 (and CD103 in barrier sites), are noncirculating, localize within peripheral tissues, and provide optimal protective immunity to pathogens in situ (Clark et al., 2006; Heath and Carbone, 2013; Kumar et al., 2017; Masopust and Soerens, 2019; Mueller and Mackay, 2016; Sathaliyawala et al., 2013; Szabo et al., 2019b; Watanabe et al., 2015; Zhu et al., 2013). Other types of T cells with more restricted recognition properties and invariant TCR, such as γδ and mucosal-associated invariant T cells (MAIT) cells, also exhibit tissue residence (Table 1), and their role and interaction with conventional αβ T cells in mediating tissue immune responses is an active area of study (Bendelac et al., 2007; Chien et al., 2014; Xiao and Cai, 2017).
B cells can also be found in circulation and in tissues, but they are largely confined to lymphoid sites (lymph nodes, spleen, bone marrow) and comprise a relatively rare immune population in human nonlymphoid organs, often far outnumbered by T cells (Carpenter et al., 2018; Egbuniwe et al., 2015; Nihal et al., 2000; Sathaliyawala et al., 2013). B cells express a unique membrane-bound B cell antigen receptor (BCR) immunoglobulin that, when activated, is secreted as antibodies in plasma that can directly bind antigens. In secondary lymphoid organs (SLOs) and in isolated lymphoid follicles (ILFs) found along the length of the gastrointestinal (GI) tract and in Peyer patches (PPs), follicular B cells form clusters surrounded by a T cell zone, whereas marginal zone B cells populate the interface between nonlymphoid red pulp and lymphoid white pulp in the spleen and in other lymphoid tissues (Pillai and Cariappa, 2009). During an active immune response, B cell follicles form germinal centers that facilitate T-B cell interactions, immunoglobulin class switching, and B cell differentiation to antibody secreting plasma cells and memory B cells. Humoral immunity is maintained through maintenance of memory B cells in lymphoid sites, plasmablasts in circulation, and long-lived plasma cells in bone marrow (Nutt et al., 2015; Slifka et al., 1998).
Methods and Computational Approaches in Systems Immunology
Defining the heterogeneity of different immune cell lineages and their variability between and within sites, over age, and in disease requires high-dimensional experimental and computational methods. Assays of increasing resolution have led to new insights into immune cell populations on the protein, transcriptomic, and genomic level, including single-cell technologies that newly define heterogeneity of immune cells across tissues, among individuals, and over development (Figure 2). Cytometry is one of the most tried and true methods for analyzing immune populations at the population and single-cell level (Figure 2). Flow cytometry utilizes fluorophore-conjugated antibodies that bind to surface and intracellular proteins expressed by immune cells, which can then be analyzed based on the emission spectrum upon illumination with specific wavelengths. Previously, the number of parameters was limited by the spectral overlap of fluorescent markers, but with advances in instrumentation and fluorochromes, current technologies have the capability of analyzing up to 50 different parameters at high resolution (Dogra et al., 2020; Mair and Prlic, 2018; Saeys et al., 2016). Most recently, spectral flow cytometry, which captures the entire spectrum of fluorescence, has enabled the use of fluorochromes with closely overlapping emission spectra and allows for an ever increasing number of parameters (Nolan and Condello, 2013). Flow cytometry also enables isolation of populations defined based on these multiple parameters using different types of fluorescent-activated cells sorters.
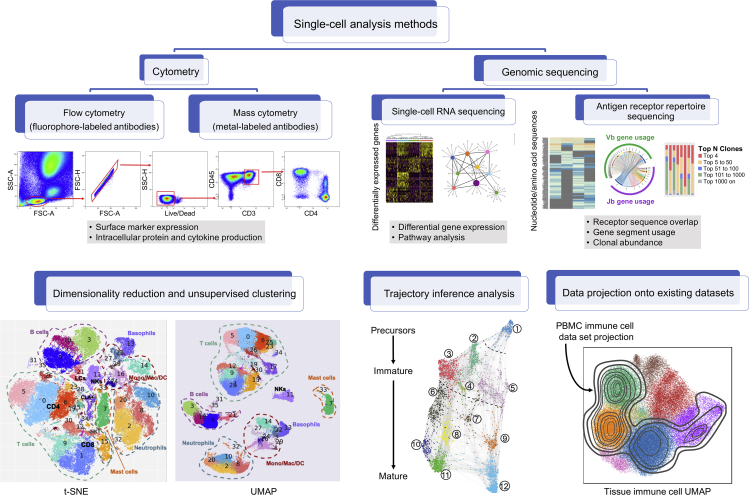
Figure 2.
Single-Cell Methods for Systems Immunology
Diagram shows the different single-cell technologies, such as cytometry and genomic sequencing techniques, and computational approaches that have been instrumental to systems-wide study of the human immune system. Flow cytometry and mass cytometry enable characterization of immune cell phenotype and function through quantification of cell surface proteins, intracellular proteins, and cytokine production. Single-cell RNA sequencing measures differentially expressed genes among single cells to elucidate heterogeneity within an immune population and identify distinct functional modules and regulatory networks. Antigen receptor sequencing characterizes lymphocyte repertoires, which inform the connectivity of adaptive immunity across diverse tissues within an individual through assessment of clonal overlap between sites, gene segment usage, and clonal abundance. Computational approaches for data visualization and analysis (bottom row) include methods for dimensionality reduction and unsupervised clustering (e.g. t-SNE, UMAP), trajectory inference analysis, and data projection onto existing datasets to directly compare immune parameters between tissues, individuals, under different conditions, and in health and disease.
An alternative to flow cytometry is mass cytometry, or cytometry by time-of-fight (CyTOF), which replaces fluorophore labels with heavy metal tags and analyzes cells using a time-of-flight mass spectrometer (Figure 2). Eliminating the issue of spectral overlap encountered in flow cytometry, CyTOF allows for analysis of up to 100 parameters although is often limited to 40–50 parameters (Amir et al., 2013; Miron et al., 2018; Nyman et al., 2017; Stewart et al., 2020). Studies using CyTOF technology have provided new insights in human CD8+ T cell activation, monocyte heterogeneity, DC development and interindividual variation, memory T cell maintenance in lymph nodes, remodeling of T cell populations following viral infection, and compartmentalization of immune cells in fetal tissues (Alcántara-Hernández et al., 2017; Hamers et al., 2019; Li et al., 2019; Miron et al., 2018; Newell et al., 2012; See et al., 2017; Sen et al., 2014). Limitations of CyTOF are that a smaller, less dynamic range in expression can be discerned compared with flow cytometry, and the cells, once analyzed, cannot be sorted for isolation.
Advances in detection of gene expression on the population and single-cell level has increased the depth and breadth at which we can analyze the human immune system. Whole transcriptome profiling by RNA sequencing (RNA-seq) generates read counts of all the transcripts within a population and differential gene expression analysis can reveal signatures that define specific subsets (Stark et al., 2019). RNA-seq has been used to define TRM as a distinct subset in mice and humans (Kumar et al., 2017; Mackay and Kallies, 2017; Mackay et al., 2016) and distinct features of human B cell subsets in blood and tissues (Weisel et al., 2020). Applying whole transcriptome profiling to single cells (scRNA-seq) enables identification and stratification of cell subsets based on differential gene expression and can lead to de novo discovery of new cell types and cell states, as well as tissue-specific adaptations of cell types within and across tissues (Figure 2). Since the introduction of scRNA-seq in 2009 (Tang et al., 2009), new technological advances in scRNA-seq methods including plate-based approaches and the 10X Genomics approach employing microdroplet-based systems enable rapid and efficient capture of high transcript numbers per cell (Bush et al., 2017; Hwang et al., 2018; Jaitin et al., 2014). Recent studies using scRNA-seq approaches have led to identification of cell types and progenitors, developmental processes, activation trajectories, and functional signatures for diverse immune cell lineages (Papalexi and Satija, 2018; Paul et al., 2015; Popescu et al., 2019; Schultze and Aschenbrenner, 2019; See et al., 2017; Stewart et al., 2019, Stewart et al., 2020; Szabo et al., 2019a; Villani et al., 2017; Yu et al., 2016).
T and B cells exhibit an additional level of genomic complexity in their expression of uniquely rearranged antigen receptor genes. Each T and B cell that develops expresses a distinct TCR or BCR that can be identified by high-throughput sequencing of the variable portion (containing the complementarity region 3 (CDR3) of the TCRβ chain for T cells and the IGH gene for B cells) (Bradley and Thomas, 2019; Chaudhary and Wesemann, 2018). Sequencing methods capturing beyond the CDR3 region also exist (Rosati et al., 2017). Common methods for high-throughput antigen receptor sequencing include PCR amplification of genomic DNA and sequencing or reverse transcription of mRNA transcripts to cDNA (Heather et al., 2018; Nielsen and Boyd, 2018). Other approaches include multiomic single-cell technologies, such as 10X Genomics single-cell immune profiling with V(D)J sequencing (Park et al., 2020), and innovative analysis methods that allow researchers to extract receptor sequences from RNA sequencing data (Bolotin et al., 2015, 2017). These approaches have enabled parallel analysis of TCR/BCR repertoire and transcription profile. Although the 10X Genomics approach involves a primer-based amplification of the antigen receptor locus, extraction of receptor sequences from RNA sequencing data ensures that there is no primer bias (Mose et al., 2016). However, in both methods, the data likely represent only a fraction of the total clonal diversity due to limited sampling. Sequencing TCR and BCR can identify how unique B and T cell clones of the adaptive immune response are expanded and maintained across the human body in health, as well as their role in inflammatory and autoimmune immune diseases and in cancer (Figure 2) (de Jong et al., 2018; James et al., 2020; Meng et al., 2017; Schoettler et al., 2019; Thome et al., 2014).
Along with the innovations in instrumentation for single-cell profiling, advancements in computational techniques have been crucial to analyze rapidly expanding datasets and extracting novel insights. Dimensionality reduction techniques have allowed for unbiased and more intuitive visualization of high-dimensional data. Common dimensionality reduction techniques include principle component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE), and Uniform Approximation and Projection (UMAP) method (Figure 2) (Becht et al., 2018; Luecken and Theis, 2019; van der Maaten and Hinton, 2008). Cluster analysis methods group cells based on similarity of gene expression profiles, providing structure to heterogeneous immune populations. Unsupervised clustering methods are particularly useful, because they provide an unbiased, data-driven approach to organizing unlabeled data that has led to discoveries of distinct immune subsets (Li et al., 2019; See et al., 2017; Sweatt et al., 2019). Longitudinal analyses of human immune responses are less readily accomplished compared with animal models; however, trajectory inference analysis, such as “pseudotime” or “pseudospace,” of single-cell data is a technique that orders individual cells along a trajectory based on expression profiles to infer a continuum of cell states from a static time point (Chen et al., 2018; Kunz et al., 2018; Saelens et al., 2019). This technique has been used to understand dynamic immune responses, capture transition states, define gene regulatory networks, and follow immune cell development and differentiation (Figure 2) (Bendall et al., 2014; Lonnberg et al., 2017; Paul et al., 2015; Setty et al., 2019; Stubbington et al., 2017; Velten et al., 2017). Finally, emerging computational strategies allow projection of data onto existing datasets, enabling direct comparison of immune responses under different conditions and in health and disease (Figure 2) (Szabo et al., 2019a).
Emerging single-cell technologies now enable simultaneous analysis of multiple data sources, including genomic DNA and mRNA transcription (Dey et al., 2015), gene and protein expression using DNA-labeled antibodies as in cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) (Peterson et al., 2017; Stoeckius et al., 2017), gene expression and TCR/BCR repertoire (Ferguson and Chen, 2020; James et al., 2020), and gene expression along with DNA methylation (Cheow et al., 2016; Macaulay et al., 2017). In addition, new technologies pairing single-cell transcriptomics and spatial profiling—spatial transcriptomics—enable deeper understanding of the role of tissue topography and environment in the function and organization of the human immune system (Eng et al., 2019; Moncada et al., 2020; Rodriques et al., 2019). As single-cell profiling technologies become more advanced and datasets become larger and more complex, the integration of systems biology is essential for identifying key features of the human immune system in health and disease.
Immunity in Space and Time
At the whole-body level, the integration of the multiple approaches described earlier has revealed new insights into the development, function, and maintenance of the immune response. We and others have used in-depth profiling that integrates flow or mass cytometry-based approaches with transcriptomics as applied to multiple sites obtained from individuals to assess how innate and adaptive immune cells are distributed and maintained in blood and tissue sites over decades of life (Carpenter et al., 2018; Dogra et al., 2020; Granot et al., 2017; Kumar et al., 2018, Kumar et al., 2017; Miron et al., 2018; Sathaliyawala et al., 2013; Senda et al., 2019; Thome et al., 2014, Thome et al., 2016a; Weisberg et al., 2019; Yudanin et al., 2019). These studies have revealed that the immune cell composition is largely a feature of the tissue site and that the distribution of subsets for each lineage type is also influenced by the developmental origin. Single-cell technologies have elucidated developmental and functional heterogeneity within lineages and how immune cells in blood relate to those in tissue sites. Finally, high-level analyses integrating multiple readouts are showing which aspects of the immune response change with age and the key drivers of individual difference between immune responses.
Innate Immune Cell Distribution, Maintenance, and Development
For innate cells, studies combining high-dimensional and basic flow cytometry methods, whole transcriptome profiling, and other approaches have shown that monocytes, granulocytes, DCs, and innate lymphoid cells, including NK cells and ILCs, exhibit tissue-intrinsic distribution. Monocytes and granulocytes are largely confined to blood and blood-rich tissues sites, and their distribution and frequencies are features of the specific site and do not show significant age-associated or interindividual variations (Carpenter et al., 2018; Granot et al., 2017). For DCs, studies in multiple tissues, including skin, lung, and intestine, and in SLOs, such as spleen, lymph nodes, and tonsil, have shown that the phenotype, function, localization, and composition of DC subsets are specialized to the tissue site (Bigley et al., 2015; Chu et al., 2012; Granot et al., 2017; Haniffa et al., 2012; Kashem et al., 2017; Klechevsky et al., 2008; Mittag et al., 2011; Sada-Ovalle et al., 2012; Segura et al., 2013; Steiniger, 2015). These tissue-intrinsic distribution patterns that are largely independent of age also apply to cDC subsets (in all sites and at highest frequencies in SLOs) and pDCs, which are rare subsets present in blood and lymphoid sites (Carpenter et al., 2018; Granot et al., 2017).
Studies of site-specific features of DCs have provided insights into their development and function. Mature cDC subsets, which upregulate MHC class II for antigen presentation and exhibit markers of tissue migration, are predominantly cDC2 and enriched in lymph nodes draining mucosal tissue sites, such as the lung and intestines (Granot et al., 2017), indicating active sites of DC-mediated immune surveillance during homeostasis. They identified bone-marrow-derived circulating cDC precursors present in both cord blood and adult peripheral blood and found that cDC1 and cDC2 populations are derived from distinct lineage-primed circulating precursors, which can be distinguished based on CD172a (Breton et al., 2016). Tissue-specific features of DCs have been identified, including Langerhans cells (LCs) in the skin, characterized by their expression of Langerin (CD207), and follicular DCs in B cell follicles of SLOs, essential for proper germinal center formation and memory B cell development (Heesters et al., 2014; Merad et al., 2008). Although these features of cDC tissue distribution are largely conserved over age and between individuals, interindividual heterogeneity among blood and skin cDC2s have also been found (Alcántara-Hernández et al., 2017), suggesting differences in DC-mediated surveillance between individuals. The developmental origin of DCs has been analyzed by scRNA-seq of blood DCs demonstrating a differentiation trajectory of human DCs, as well as precursors for the DC lineage and specific cDC subsets (Breton et al., 2016; See et al., 2017). Whether certain of these newly defined DC precursors are seeded or maintained in tissues remains to be established.
Innate lymphoid cells include NK cells that are abundant in many sites and subsets of helper-type ILC1, ILC2, and ILC3 that are rarer and mostly tissue-resident. A recent comprehensive analysis of NK cell subsets of different maturation and functional states demonstrated that characteristics of NK cell subsets are primarily driven by tissue site; mature, cytolytic, and terminally differentiated NK cells are enriched in circulation and in blood-rich tissues, such as the lung, bone marrow, and spleen, whereas less functionally mature NK cells are present in lymphoid sites and intestines and exhibit transcriptional and phenotypic features of tissue residency (Dogra et al., 2020). Analysis of multiple markers of NK cell development revealed that NK cells in lymph nodes and intestines are immature and also contain NK-like precursors with novel phenotypes (Dogra et al., 2020), suggesting that lymph nodes and intestines may be reservoirs for maintenance of immature NK cells for seeding and replenishing the mature NK cell pool. Complementary studies examining maintenance of NK cells and helper-type ILCs in diverse tissue sites demonstrated that innate lymphoid cell subsets display considerable phenotypic and transcriptional heterogeneity driven by tissue site and conserved across donors and age (Simoni et al., 2017; Yudanin et al., 2019). Together, these studies on cells of the innate immune system reveal tissue distribution based on site and developmental stage that is largely maintained over the lifetime of an individual.
Adaptive Immunity: T Cell Development, Differentiation, and Memory Maintenance
For adaptive immune cells, the majority of studies have focused on T cells, dissecting how the different subsets are distributed in tissue sites and the impact of tissue site and age in their functional regulation. T cells are unique among immune cells in that they have a specialized organ for their development and output—the thymus—which is highly active at birth, declines during childhood, and is nascently active in adulthood with evidence pointing to thymic shutdown in middle age (40–50 years) (Haynes et al., 2000; Thapa and Farber, 2019; Thome et al., 2016b). In the thymus, bone-marrow-derived T cell precursors undergo further development to mature T cells, involving rearrangement of T cell antigen receptor genes and selection for T cells that lack overt self-reactivity for export to the periphery. Applying scRNA-seq and systems analysis to T cell development in the human thymus has recently been reported across fetal development, childhood, and adult life (Park et al., 2020). Paired analysis of scRNA-seq data with TCR data revealed a developmental trajectory for human T cells starting from CD4−CD8− double-negative (DN) cells to mature CD4+ and CD8+ single positive (SP) cells and elucidated the process of TCRβ selection, providing evidence for progressive recombination of the TCRα allele starting with proximal V-J pairs followed by distal pairs (Park et al., 2020). In addition, Park, et al. described a diverging lineage of γδ T cells between the DN and DP stages and Tregs branching from αβ T cells (Park et al., 2020). This in-depth analysis revealed a developmental trajectory for human T cells that uniquely occurs within thymic tissue and prior to export of mature naive CD4+ and CD8+ T cells to peripheral sites.
Upon export from the thymus, mature T cells populate blood and virtually every tissue site in the body. Human T cell subsets also follow a tissue-dependent pattern of distribution and maintenance, but unlike innate cells, tissue intrinsic subset composition exhibits age-associated changes, as reviewed elsewhere (Kumar et al., 2018). Naive T cells are the predominant population in lymphoid tissue sites in early life and childhood, whereas memory T cells populate mucosal, exocrine, and barrier sites with the accumulation of antigen experience during childhood (Kumar et al., 2018; Thome et al., 2014, Thome et al., 2016a). As a result, for most of adult life, there is a stable segregation of naive T cells being only found in blood and lymphoid tissue (lymph node, spleen), whereas memory T cells predominate in virtually every nonlymphoid tissue examined, including mucosal sites (lungs, intestines), skin, exocrine tissues, liver, and brain (Kumar et al., 2017; Pallett et al., 2017; Smolders et al., 2018; Watanabe et al., 2015; Weisberg et al., 2019). The majority of these tissue memory cells comprise noncirculating TRM cells that exhibit distinct phenotypic and transcriptional profiles that enable their retention in tissue sites (Hombrink et al., 2016; Kumar et al., 2017); tissue-specific proportions of TRM in each site are stably maintained with age (Senda et al., 2019; Thome et al., 2014). Given that TRM are formed early in life and can be detected in mucosal sites in infants and children and during an active infection (Connors et al., 2018; Thome et al., 2016a), it is possible that TRM maintain long-term immunity for maintenance of tissue homeostasis.
TRM exhibit heterogeneity and tissue-specific properties that are starting to be defined using transcriptomics and single-cell approaches. In lymphoid sites, such as lymph nodes, tissue memory T cells exhibit increased expression of transcription factors and functional markers associated with stemness and denoting maintenance in a more quiescent state compared with memory T cells in spleen and bone marrow, which exhibit a more differentiated state with increased turnover (Miron et al., 2018). Conversely, TRM localized across the GI tract exhibit distinct phenotypes and metabolic signatures whether they localize to the small intestine, associated lymphoid tissue, or the pancreas (Weisberg et al., 2019), and unique localization of TRM in liver sinusoids requires specific adaptations (Holz et al., 2018; McNamara et al., 2017). Further elucidation of tissue memory T cell heterogeneity and tissue adaptations will be facilitated by applying scRNA-seq technologies.
Functional Regulation
Single-cell technologies are providing new insights into the diverse functions and subtypes of T cells and how these vary as a function of tissue site. A recent study using scRNA-seq profiling of resting and activated T cells isolated from blood, bone marrow, lung, and lung lymph node (LLN) from human donors identified functional modules conserved across all sites and specific signatures that distinguish blood and tissue T cells (Szabo et al., 2019a). Unsupervised clustering of scRNA-seq data from bone marrow, lung, and LLN from two organ tissue donors revealed three unique CD4+ T cell activation states distinguished by differential expression of IL2, TNF, and IL4R and two major functional states for CD8+ T cells characterized by differential expression of genes associated with pro-inflammatory cytokines and chemokines and cytotoxic mediators (Szabo et al., 2019a). Applying single-cell Hierarchical Poisson Factorization (scHPF) (Levitin et al., 2019) revealed activation and functional “modules” that were highly conserved across tissues and donors, including a proliferation module, an IFN response module, and two unique cytotoxicity modules. Moreover, projection of blood T cells profiles onto UMAP embeddings of T cells from tissue donors revealed that tissue T cells upregulated genes associated with cell structure, extracellular matrix, adhesion, and tissue residency, suggesting that T cells adopt structural changes that facilitate interactions with tissue matrix (Szabo et al., 2019a). Together, these analyses demonstrate that T cells adopt a spectrum of functional states when activated and are able to adapt when localized in peripheral tissues.
Functional heterogeneity of T cell subsets, such as Tregs and antigen-specific T cells, is also being elucidated by scRNA-seq profiling. A recent study of scRNA-seq profiling identified the activation trajectory of Tregs between draining lymph nodes and colon (James et al., 2020). They showed that there is a continuous range of functional states from resting Tregs in mesenteric lymph nodes to colonic Tregs, which express high levels of functional regulatory molecules (James et al., 2020). Variations in T cell functional states are hallmarks of diverse diseases, and scRNA-seq analysis can uncover new insights important for understanding disease pathogenesis and new therapeutic targets. A recent study identified specific functional states for Tregs and allergen-specific CD4+ T cells isolated from the blood of individuals with atopy and asthma, revealing type 2 cytokines, along with an interferon (IFN) response state (Seumois et al., 2020), similar to the IFN signature identified by scRNA-seq analysis of activated CD4+ T cells from blood and tissues (Szabo et al., 2019a). These findings highlight how scRNA-seq can reveal new cell states and identify whether they are associated with disease or are part of the healthy spectrum of functional states.
Clinical Applications
Advancements in sequencing technologies and computational techniques not only provide essential tools in understanding the human immune system in health, but also are crucial for the development of improved therapeutic strategies for human disease. In particular, vaccinology and cancer immunotherapies are two areas that have greatly benefited from our increased knowledge of human immunity as a system that spans the entire body.
Systems Vaccinology
A systems-biology-based approach to human immunology offers unique insights into vaccine development. In the clinical setting, neutralizing antibody titers measured in peripheral blood serve as the primary correlate of protection. However, immune responses within peripheral blood are not indicative of what is occurring or maintained at tissue sites. In the context of pathogen-specific responses, human-influenza-specific lung TRM have been shown to be abundant, polyfunctional, and diverse in their receptor sequences (Pizzolla et al., 2018; Purwar et al., 2011), whereas mouse studies of acute and persistent viral infections have revealed that a large portion of T cells reside in tissue as TRM and provide optimal protection long after viral clearance (see (Masopust and Soerens, 2019; Szabo et al., 2019b) for reviews). Thus, it is crucial to design strategies that will ensure the development and maintenance of immunity within the appropriate tissue sites.
There are two major challenges for generating vaccines that target tissue responses. First, optimal strategies for targeting durable and long-lasting tissue-resident responses need to be defined; and second, methods are needed for monitoring the efficacy of tissue-directed vaccines in peripheral blood. Although the former challenge requires more studies using animal models, progress in high-resolution analysis of blood has revealed that T cells with tissue signatures are detectable in circulation. For example, circulating forms of Tfh cells (resident in lymphoid follicles) are present in increased frequencies after vaccinations and can be used for assessing vaccine efficacy (Huber et al., 2020; Koutsakos et al., 2018). During steady state, T cells with a skin-homing phenotype have been detected in peripheral blood (Klicznik et al., 2019), and a small fraction of circulating T cells exhibit a transcriptional signature for tissue T cells (Szabo et al., 2019a). These findings suggest that perturbations in tissue responses may be possible to monitor in blood using sensitive single-cell technologies.
Innovative systems-based computational approaches to vaccinology have also enabled a more comprehensive understanding of the mechanisms that drive protective immunity. For instance, systems vaccinology approaches enable high-level analysis of innate and adaptive immune responses to vaccination (Hagan et al., 2015; Zak et al., 2014). Vaccine responses and efficacy can be further evaluated at the population level, addressing the challenge of designing vaccines that are effective among a diverse population (Koff et al., 2013; Li et al., 2017; Tsang et al., 2014). Combined data from serum analyte profiling, multiparameter cytometry, multiomics technologies, and adaptive immune receptor repertoire profiling identify shared molecular signatures and biomarkers to predict vaccine responsiveness and immunogenicity (Kazmin et al., 2017; Nakaya et al., 2015; Natrajan et al., 2019; Ramos, 2020; Sullivan et al., 2019; Team and Consortium, 2017). By incorporating molecular signatures of tissue responses, systems vaccinology approaches can inform development and monitoring of vaccines that ensure effective establishment of tissue immunity.
Cancer and Personalized Therapeutics
Understanding the human immune system beyond peripheral blood is essential for developing effective cancer immunotherapies, particularly therapeutic strategies that successfully target solid tumors. Progress has been made in defining the tumor-associated T cell populations in terms of their tissue residence and correlation to prognosis and responses to therapy. Recent studies of tumor infiltrating lymphocytes (TILs) have suggested that tissue localization is crucial for effective tumor killing. In fact, studies have shown that the expression of certain chemokines are positively correlated with the abundance of TILs and with postoperative survival in melanoma and colorectal cancer (Kistner et al., 2017; Martinet et al., 2012). In addition, TILs expressing TRM marker CD103 have been shown to be enriched in tumor tissue, particularly in cancers of epithelial origin, and often correlate with improved patient survival (Boddupalli et al., 2016; Djenidi et al., 2015; Edwards et al., 2018; Komdeur et al., 2017; Smazynski and Webb, 2018; Wang et al., 2016; Webb et al., 2015).
In recent years, adoptive T cell therapy has emerged as a promising option for treating malignancies. Among the various forms of adoptive T cell therapy, TCR-engineered T (TCR-T) cell therapy and chimeric antigen receptor T (CAR-T) cell therapy are two precision medicine strategies that use genetically modified T cells to treat cancers based on tumor antigen specificity (June, 2007; Kershaw et al., 2013). CAR-T cells, genetically engineered T cells that express an antigen-recognition domain and costimulatory signaling molecules, have proven remarkably successful in the treatment of hematological malignancies, especially when paired with checkpoint blockade (June and Sadelain, 2018; Kalos et al., 2011; Porter et al., 2011). However, their success in targeting solid tumors has been limited (Martinez and Moon, 2019; Newick et al., 2017). One major barrier to treatment efficacy is understanding how to target CAR-T cells to the affected tissue and maintain them there for elimination of tumor cells. This process requires identification of the homing receptors and residency markers that control tissue-specific trafficking and retention. Thus, understanding the role of unique tissue environments in determining human immune response is essential for designing cancer immunotherapies that can effectively reach target tissues and treat solid tumors.
An alternative to CAR-T cell therapy is TCR-T cell therapy, which has been shown to display greater sensitivity than CAR-T cell therapy, enabling more rapid destruction of tumor cells, as well as improved solid tumor penetration (Garber, 2018). TCR sequencing techniques have been crucial for understanding the immune landscape of tumors, as well as identifying TCR candidates for personalized precision immunotherapy. In particular, high-throughput paired sequencing of bulk DNA (pairSEQ) has enabled more rapid identification of TCR α and β chain sequences of T cells that recognize antigens unique to an individual tumor, which can then be used to engineer TCR-T cells (Howie et al., 2015; Pasetto et al., 2016; Ping et al., 2018).
Unfortunately, for both CAR-T cell and TCR-T cell therapy, a devastating consequence of unsuccessful clinical trials has been off-target effects in triggering inflammation at remote tissue sites such as the skin, gut, and pancreas (Kunert et al., 2013; Lamers et al., 2006; Linette et al., 2013; Morgan et al., 2013; Morgan et al., 2010; Parkhurst et al., 2011; van den Berg et al., 2015). Such inflammation may in part be due to the combination of adoptive T cell therapy and immune checkpoint inhibitor therapy. Because TRM express PD-1 during homeostasis and maintenance in tissues, anti-PD-1 therapy can potentially disrupt endogenous tissue T cell homeostasis (Weisberg et al., 2019). Given the crucial requirements for ensuring engineered T cells traffic not only to tissues but also to specific tumor tissue, approaching human immunology from a whole-body, system-based perspective is essential to facilitating more targeted strategies to enhance the efficacy and tissue specificity of engineered T cells for optimal anti-tumor responses.
Conclusions and Future Directions
The human immune system is a unique cellular network that spans the entire human body. Diverse and dynamic populations of specialized immune cells migrate within and across tissues to provide immune protection against infection and malignancy while ensuring tissue homeostasis. Studies of human tissues discussed in this review have highlighted the importance of tissue localization in determining the characteristics of immune cell interactions and functions. Integrating of systems biology approaches has shed light on the vast complexities and intricacies of the human immune system, providing new insights on immune heterogeneity, development, and function previously unexplored. As the field of systems immunology grows and progresses, one of the major challenges is consistency: with new technologies and computational methods emerging daily, it is crucial to develop principles of best practice when it comes to (1) how to analyze datasets such that results are reproducible when using different analysis pipelines, (2) how to define immune cell types and subtypes, and (3) how to integrate new insights into a growing framework of human immunology. In addition, as we amass large datasets, such information can be used to determine how age, sex, and race/ethnicity may influence immune responses, which is important for the development of personalized therapeutics and vaccine strategies. Although tremendous efforts have already been made in building a comprehensive systems-based perspective of the human immune system, advancements in single-cell profiling technologies, adaptive immune cell repertoire analysis, and computational strategies continue to pave new paths of discovery in the field of human immunology.
Acknowledgments
This work was supported by NIH grants AI AI106697 and AI128949 awarded to D.L.F. M.M.L.P. was supported by NIH T32GM007367.
Author Contributions
Conceptualization, Writing—Original Draft, Review and Editing, and Visualization were performed by M.M.L.P. and D.L.F.
References
- Alcántara-Hernández M., Leylek R., Wagar L.E., Engleman E.G., Keler T., Marinkovich M.P., Davis M.M., Nolan G.P., Idoyaga J. High-dimensional phenotypic mapping of human dendritic cells reveals interindividual variation and tissue specialization. Immunity. 2017;47:1037–1050.e6. doi: 10.1016/j.immuni.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir E.-A., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., Pe'er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audiger C., Rahman M.J., Yun T.J., Tarbell K.V., Lesage S. The importance of dendritic cells in maintaining immune tolerance. J. Immunol. 2017;198:2223–2231. doi: 10.4049/jimmunol.1601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austyn J.M. Dendritic cells in the immune system-history, lineages, tissues, tolerance, and immunity. Microbiol. Spectr. 2016;4:1–50. doi: 10.1128/microbiolspec.MCHD-0046-2016. [DOI] [PubMed] [Google Scholar]
- Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Bendall S.C., Davis K.L., Amir e.-A., Tadmor M.D., Simonds E.F., Chen T.J., Shenfeld D.K., Nolan G.P., Pe'er D. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A., Savage P.B., Teyton L. The biology of NKT cells. Annu. Rev. Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Bigley V., McGovern N., Milne P., Dickinson R., Pagan S., Cookson S., Haniffa M., Collin M. Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J. Leukoc. Biol. 2015;97:627–634. doi: 10.1189/jlb.1HI0714-351R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddupalli C.S., Bar N., Kadaveru K., Krauthammer M., Pornputtapong N., Mai Z., Ariyan S., Narayan D., Kluger H., Deng Y. Interlesional diversity of T cell receptors in melanoma with immune checkpoints enriched in tissue-resident memory T cells. JCI Insight. 2016;1:e88955. doi: 10.1172/jci.insight.88955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Davydov A.N., Frenkel F.E., Fanchi L., Zolotareva O.I., Hemmers S., Putintseva E.V., Obraztsova A.S., Shugay M. Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol. 2017;35:908–911. doi: 10.1038/nbt.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- Boor P.P.C., Bosma B.M., Tran K.T.C., van der Laan L.J.W., Hagenaars H., IJzermans J.N.M., Metselaar H.J., Kwekkeboom J. Characterization of antigen-presenting cell subsets in human liver-draining lymph nodes. Front. Immunol. 2019;10:441. doi: 10.3389/fimmu.2019.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P., Thomas P.G. Using T cell receptor repertoires to understand the principles of adaptive immune recognition. Annu. Rev. Immunol. 2019;37:547–570. doi: 10.1146/annurev-immunol-042718-041757. [DOI] [PubMed] [Google Scholar]
- Breton G., Zheng S., Valieris R., Tojal da Silva I., Satija R., Nussenzweig M.C. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J. Exp. Med. 2016;213:2861–2870. doi: 10.1084/jem.20161135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggen M.C., Bauer W.M., Reininger B., Clim E., Captarencu C., Steiner G.E., Brunner P.M., Meier B., French L.E., Stingl G. In situ mapping of innate lymphoid cells in human skin: evidence for remarkable differences between normal and inflamed skin. J. Invest. Dermatol. 2016;136:2396–2405. doi: 10.1016/j.jid.2016.07.017. [DOI] [PubMed] [Google Scholar]
- Bush E.C., Ray F., Alvarez M.J., Realubit R., Li H., Karan C., Califano A., Sims P.A. PLATE-Seq for genome-wide regulatory network analysis of high-throughput screens. Nat. Commun. 2017;8:105. doi: 10.1038/s41467-017-00136-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter D.J., Granot T., Matsuoka N., Senda T., Kumar B.V., Thome J.J.C., Gordon C.L., Miron M., Weiner J., Connors T. Human immunology studies using organ donors: impact of clinical variations on immune parameters in tissues and circulation. Am. J. Transplant. 2018;18:74–88. doi: 10.1111/ajt.14434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary N., Wesemann D.R. Analyzing immunoglobulin repertoires. Front. Immunol. 2018;9:462. doi: 10.3389/fimmu.2018.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Rénia L., Ginhoux F. Constructing cell lineages from single-cell transcriptomes. Mol. Aspects Med. 2018;59:95–113. doi: 10.1016/j.mam.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Cheow L.F., Courtois E.T., Tan Y., Viswanathan R., Xing Q., Tan R.Z., Tan D.S., Robson P., Loh Y.H., Quake S.R. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat. Methods. 2016;13:833–836. doi: 10.1038/nmeth.3961. [DOI] [PubMed] [Google Scholar]
- Chien Y.H., Meyer C., Bonneville M. γδ T cells: first line of defense and beyond. Annu. Rev. Immunol. 2014;32:121–155. doi: 10.1146/annurev-immunol-032713-120216. [DOI] [PubMed] [Google Scholar]
- Chu C.C., Ali N., Karagiannis P., Di Meglio P., Skowera A., Napolitano L., Barinaga G., Grys K., Sharif-Paghaleh E., Karagiannis S.N. Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL-10 and induce regulatory T cells that suppress skin inflammation. J. Exp. Med. 2012;209:935–945. doi: 10.1084/jem.20112583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.A., Chong B., Mirchandani N., Brinster N.K., Yamanaka K., Dowgiert R.K., Kupper T.S. The vast majority of CLA+ T cells are resident in normal skin. J. Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- Connors T.J., Baird J.S., Yopes M.C., Zens K.D., Pethe K., Ravindranath T.M., Ho S.H., Farber D.L. Developmental regulation of effector and resident memory T cell generation during pediatric viral respiratory tract infection. J. Immunol. 2018;201:432–439. doi: 10.4049/jimmunol.1800396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A., Jabbari A., Dai Z., Xing L., Lee D., Li M.M., Duvic M., Hordinsky M., Norris D.A., Price V. High-throughput T cell receptor sequencing identifies clonally expanded CD8+ T cell populations in alopecia areata. JCI Insight. 2018;3:e121949. doi: 10.1172/jci.insight.121949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S.S., Kester L., Spanjaard B., Bienko M., van Oudenaarden A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 2015;33:285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djenidi F., Adam J., Goubar A., Durgeau A., Meurice G., de Montpréville V., Validire P., Besse B., Mami-Chouaib F. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J. Immunol. 2015;194:3475–3486. doi: 10.4049/jimmunol.1402711. [DOI] [PubMed] [Google Scholar]
- Dogra P., Rancan C., Ma W., Toth M., Senda T., Carpenter D.J., Kubota M., Matsumoto R., Thapa P., Szabo P.A. Tissue determinants of human NK cell development, function, and residence. Cell. 2020;180:749–763.e13. doi: 10.1016/j.cell.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J., Wilmott J.S., Madore J., Gide T.N., Quek C., Tasker A., Ferguson A., Chen J., Hewavisenti R., Hersey P. CD103(+) tumor-resident CD8(+) T cells are associated with improved survival in immunotherapy-naive melanoma patients and expand significantly during anti-PD-1 treatment. Clin. Cancer Res. 2018;24:3036–3045. doi: 10.1158/1078-0432.CCR-17-2257. [DOI] [PubMed] [Google Scholar]
- Egbuniwe I.U., Karagiannis S.N., Nestle F.O., Lacy K.E. Revisiting the role of B cells in skin immune surveillance. Trends Immunol. 2015;36:102–111. doi: 10.1016/j.it.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Eng C.L., Lawson M., Zhu Q., Dries R., Koulena N., Takei Y., Yun J., Cronin C., Karp C., Yuan G.C. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568:235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson A., Chen K. Analysis of transcriptional profiling of immune cells at the single-cell level. Methods Mol. Biol. 2020;2111:47–57. doi: 10.1007/978-1-0716-0266-9_4. [DOI] [PubMed] [Google Scholar]
- Fuchs A., Vermi W., Lee J.S., Lonardi S., Gilfillan S., Newberry R.D., Cella M., Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K. Driving T-cell immunotherapy to solid tumors. Nat. Biotechnol. 2018;36:215–219. doi: 10.1038/nbt.4090. [DOI] [PubMed] [Google Scholar]
- Geginat J., Paroni M., Facciotti F., Gruarin P., Kastirr I., Caprioli F., Pagani M., Abrignani S. The CD4-centered universe of human T cell subsets. Semin. Immunol. 2013;25:252–262. doi: 10.1016/j.smim.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Giesecke C., Frölich D., Reiter K., Mei H.E., Wirries I., Kuhly R., Killig M., Glatzer T., Stölzel K., Perka C. Tissue distribution and dependence of responsiveness of human antigen-specific memory B cells. J. Immunol. 2014;192:3091–3100. doi: 10.4049/jimmunol.1302783. [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M.F., Geissmann F. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot T., Senda T., Carpenter D.J., Matsuoka N., Weiner J., Gordon C.L., Miron M., Kumar B.V., Griesemer A., Ho S.H. Dendritic cells display subset and tissue-specific maturation dynamics over human life. Immunity. 2017;46:504–515. doi: 10.1016/j.immuni.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan T., Nakaya H.I., Subramaniam S., Pulendran B. Systems vaccinology: enabling rational vaccine design with systems biological approaches. Vaccine. 2015;33:5294–5301. doi: 10.1016/j.vaccine.2015.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers A.A.J., Dinh H.Q., Thomas G.D., Marcovecchio P., Blatchley A., Nakao C.S., Kim C., McSkimming C., Taylor A.M., Nguyen A.T. Human monocyte heterogeneity as revealed by high-dimensional mass cytometry. Arterioscler. Thromb. Vasc. Biol. 2019;39:25–36. doi: 10.1161/ATVBAHA.118.311022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Shin A., Bigley V., McGovern N., Teo P., See P., Wasan P.S., Wang X.N., Malinarich F., Malleret B. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37:60–73. doi: 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Markert M.L., Sempowski G.D., Patel D.D., Hale L.P. The role of the thymus in immune reconstitution in aging, bone marrow transplantation, and HIV-1 infection. Annu. Rev. Immunol. 2000;18:529–560. doi: 10.1146/annurev.immunol.18.1.529. [DOI] [PubMed] [Google Scholar]
- Heath W.R., Carbone F.R. The skin-resident and migratory immune system in steady state and memory: innate lymphocytes, dendritic cells and T cells. Nat. Immunol. 2013;14:978–985. doi: 10.1038/ni.2680. [DOI] [PubMed] [Google Scholar]
- Heather J.M., Ismail M., Oakes T., Chain B. High-throughput sequencing of the T-cell receptor repertoire: pitfalls and opportunities. Brief. Bioinform. 2018;19:554–565. doi: 10.1093/bib/bbw138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesters B.A., Myers R.C., Carroll M.C. Follicular dendritic cells: dynamic antigen libraries. Nat. Rev. Immunol. 2014;14:495–504. doi: 10.1038/nri3689. [DOI] [PubMed] [Google Scholar]
- Hoeffel G., Wang Y., Greter M., See P., Teo P., Malleret B., Leboeuf M., Low D., Oller G., Almeida F. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med. 2012;209:1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz L.E., Prier J.E., Freestone D., Steiner T.M., English K., Johnson D.N., Mollard V., Cozijnsen A., Davey G.M., Godfrey D.I. CD8(+) T cell activation leads to constitutive formation of liver tissue-resident memory T cells that seed a large and flexible niche in the liver. Cell Rep. 2018;25:68–79.e4. doi: 10.1016/j.celrep.2018.08.094. [DOI] [PubMed] [Google Scholar]
- Hombrink P., Helbig C., Backer R.A., Piet B., Oja A.E., Stark R., Brasser G., Jongejan A., Jonkers R.E., Nota B. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 2016;17:1467–1478. doi: 10.1038/ni.3589. [DOI] [PubMed] [Google Scholar]
- Howie B., Sherwood A.M., Berkebile A.D., Berka J., Emerson R.O., Williamson D.W., Kirsch I., Vignali M., Rieder M.J., Carlson C.S. High-throughput pairing of T cell receptor α and β sequences. Sci. Transl. Med. 2015;7:301ra131. doi: 10.1126/scitranslmed.aac5624. [DOI] [PubMed] [Google Scholar]
- Huber J.E., Ahlfeld J., Scheck M.K., Zaucha M., Witter K., Lehmann L., Karimzadeh H., Pritsch M., Hoelscher M., von Sonnenburg F. Dynamic changes in circulating T follicular helper cell composition predict neutralising antibody responses after yellow fever vaccination. Clin. Transl. Immunol. 2020;9:e1129. doi: 10.1002/cti2.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018;50:96. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka I.E., Constantinides M.G., Gudjonson H., Bendelac A. The innate lymphoid cell precursor. Annu. Rev. Immunol. 2016;34:299–316. doi: 10.1146/annurev-immunol-041015-055549. [DOI] [PubMed] [Google Scholar]
- Jaitin D.A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James K.R., Gomes T., Elmentaite R., Kumar N., Gulliver E.L., King H.W., Stares M.D., Bareham B.R., Ferdinand J.R., Petrova V.N. Distinct microbial and immune niches of the human colon. Nat. Immunol. 2020;21:343–353. doi: 10.1038/s41590-020-0602-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June C.H. Adoptive T cell therapy for cancer in the clinic. J. Clin. Invest. 2007;117:1466–1476. doi: 10.1172/JCI32446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June C.H., Sadelain M. Chimeric antigen receptor therapy. N. Engl. J. Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalos M., Levine B.L., Porter D.L., Katz S., Grupp S.A., Bagg A., June C.H. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashem S.W., Haniffa M., Kaplan D.H. Antigen-presenting cells in the skin. Annu. Rev. Immunol. 2017;35:469–499. doi: 10.1146/annurev-immunol-051116-052215. [DOI] [PubMed] [Google Scholar]
- Kazmin D., Nakaya H.I., Lee E.K., Johnson M.J., van der Most R., van den Berg R.A., Ballou W.R., Jongert E., Wille-Reece U., Ockenhouse C. Systems analysis of protective immune responses to RTS, S malaria vaccination in humans. Proc. Natl. Acad. Sci. U S A. 2017;114:2425–2430. doi: 10.1073/pnas.1621489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw M.H., Westwood J.A., Darcy P.K. Gene-engineered T cells for cancer therapy. Nat. Rev. Cancer. 2013;13:525–541. doi: 10.1038/nrc3565. [DOI] [PubMed] [Google Scholar]
- Kistner L., Doll D., Holtorf A., Nitsche U., Janssen K.P. Interferon-inducible CXC-chemokines are crucial immune modulators and survival predictors in colorectal cancer. Oncotarget. 2017;8:89998–90012. doi: 10.18632/oncotarget.21286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klechevsky E., Morita R., Liu M., Cao Y., Coquery S., Thompson-Snipes L., Briere F., Chaussabel D., Zurawski G., Palucka A.K. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 2008;29:497–510. doi: 10.1016/j.immuni.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klicznik M.M., Morawski P.A., Höllbacher B., Varkhande S.R., Motley S.J., Kuri-Cervantes L., Goodwin E., Rosenblum M.D., Long S.A., Brachtl G. Human CD4+ CD103+ cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci. Immunol. 2019;4:eaav8995. doi: 10.1126/sciimmunol.aav8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff W.C., Burton D.R., Johnson P.R., Walker B.D., King C.R., Nabel G.J., Ahmed R., Bhan M.K., Plotkin S.A. Accelerating next-generation vaccine development for global disease prevention. Science. 2013;340:1232910. doi: 10.1126/science.1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komdeur F.L., Prins T.M., van de Wall S., Plat A., Wisman G.B.A., Hollema H., Daemen T., Church D.N., de Bruyn M., Nijman H.W. CD103+ tumor-infiltrating lymphocytes are tumor-reactive intraepithelial CD8+ T cells associated with prognostic benefit and therapy response in cervical cancer. Oncoimmunology. 2017;6:e1338230. doi: 10.1080/2162402X.2017.1338230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsakos M., Wheatley A.K., Loh L., Clemens E.B., Sant S., Nüssing S., Fox A., Chung A.W., Laurie K.L., Hurt A.C. Circulating TFH cells, serological memory, and tissue compartmentalization shape human influenza-specific B cell immunity. Sci. Transl. Med. 2018;10:eaan8405. doi: 10.1126/scitranslmed.aan8405. [DOI] [PubMed] [Google Scholar]
- Kumar B.V., Connors T.J., Farber D.L. Human T cell development, localization, and function throughout life. Immunity. 2018;48:202–213. doi: 10.1016/j.immuni.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.H., Lerner H. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017;20:2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert A., Straetemans T., Govers C., Lamers C., Mathijssen R., Sleijfer S., Debets R. TCR-engineered T cells meet new challenges to treat solid tumors: choice of antigen, T cell fitness, and sensitization of tumor milieu. Front. Immunol. 2013;4:363. doi: 10.3389/fimmu.2013.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz D.J., Gomes T., James K.R. Immune cell dynamics unfolded by single-cell technologies. Front. Immunol. 2018;9:1435. doi: 10.3389/fimmu.2018.01435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers C.H., Sleijfer S., Vulto A.G., Kruit W.H., Kliffen M., Debets R., Gratama J.W., Stoter G., Oosterwijk E. Treatment of metastatic renal cell carcinoma with autologous T-lymphocytes genetically retargeted against carbonic anhydrase IX: first clinical experience. J. Clin. Oncol. 2006;24 doi: 10.1200/JCO.2006.05.9964. e20–22. [DOI] [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin H.M., Yuan J., Cheng Y.L., Ruiz F.J., Bush E.C., Bruce J.N., Canoll P., Iavarone A., Lasorella A., Blei D.M. Gene signature identification from single-cell RNA-seq with hierarchical Poisson factorization. Mol. Syst. Biol. 2019;15:e8557. doi: 10.15252/msb.20188557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., van Unen V., Guo N., Abdelaal T., Somarakis A., Eggermont J., Mahfouz A., Chuva de Sousa Lopes S.M., Lelieveldt B.P.F., Koning F. Early-life compartmentalization of immune cells in human fetal tissues revealed by high-dimensional mass cytometry. Front. Immunol. 2019;10:1932. doi: 10.3389/fimmu.2019.01932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Sullivan N.L., Rouphael N., Yu T., Banton S., Maddur M.S., McCausland M., Chiu C., Canniff J., Dubey S. Metabolic phenotypes of response to vaccination in humans. Cell. 2017;169:862–877.e17. doi: 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linette G.P., Stadtmauer E.A., Maus M.V., Rapoport A.P., Levine B.L., Emery L., Litzky L., Bagg A., Carreno B.M., Cimino P.J. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. 2013;122:863–871. doi: 10.1182/blood-2013-03-490565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonnberg T., Svensson V., James K.R., Fernandez-Ruiz D., Sebina I., Montandon R., Soon M.S., Fogg L.G., Nair A.S., Liligeto U. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci. Immunol. 2017;2:eaal2192. doi: 10.1126/sciimmunol.aal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay I.C., Ponting C.P., Voet T. Single-cell multiomics: multiple measurements from single cells. Trends Genet. 2017;33:155–168. doi: 10.1016/j.tig.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay L.K., Kallies A. Transcriptional regulation of tissue-resident lymphocytes. Trends Immunol. 2017;38:94–103. doi: 10.1016/j.it.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Minnich M., Kragten N.A., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352:459–463. doi: 10.1126/science.aad2035. [DOI] [PubMed] [Google Scholar]
- Mair F., Prlic M. OMIP-044: 28-color immunophenotyping of the human dendritic cell compartment. Cytometry A. 2018;93:402–405. doi: 10.1002/cyto.a.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet L., Le Guellec S., Filleron T., Lamant L., Meyer N., Rochaix P., Garrido I., Girard J.P. High endothelial venules (HEVs) in human melanoma lesions: major gateways for tumor-infiltrating lymphocytes. Oncoimmunology. 2012;1:829–839. doi: 10.4161/onci.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Moon E.K. CAR T cells for solid tumors: new strategies for finding, infiltrating, and surviving in the tumor microenvironment. Front. Immunol. 2019;10:128. doi: 10.3389/fimmu.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D., Soerens A.G. Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol. 2019;37:521–546. doi: 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern N., Schlitzer A., Gunawan M., Jardine L., Shin A., Poyner E., Green K., Dickinson R., Wang X.N., Low D. Human dermal CD14⁺ cells are a transient population of monocyte-derived macrophages. Immunity. 2014;41:465–477. doi: 10.1016/j.immuni.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara H.A., Cai Y., Wagle M.V., Sontani Y., Roots C.M., Miosge L.A., O'Connor J.H., Sutton H.J., Ganusov V.V., Heath W.R. Up-regulation of LFA-1 allows liver-resident memory T cells to patrol and remain in the hepatic sinusoids. Sci. Immunol. 2017;2:eaaj1996. doi: 10.1126/sciimmunol.aaj1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina F., Segundo C., Campos-Caro A., González-García I., Brieva J.A. The heterogeneity shown by human plasma cells from tonsil, blood, and bone marrow reveals graded stages of increasing maturity, but local profiles of adhesion molecule expression. Blood. 2002;99:2154–2161. doi: 10.1182/blood.v99.6.2154. [DOI] [PubMed] [Google Scholar]
- Meng W., Zhang B., Schwartz G.W., Rosenfeld A.M., Ren D., Thome J.J.C., Carpenter D.J., Matsuoka N., Lerner H., Friedman A.L. An atlas of B-cell clonal distribution in the human body. Nat. Biotechnol. 2017;35:879–884. doi: 10.1038/nbt.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Ginhoux F., Collin M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol. 2008;8:935–947. doi: 10.1038/nri2455. [DOI] [PubMed] [Google Scholar]
- Merad M., Manz M.G., Karsunky H., Wagers A., Peters W., Charo I., Weissman I.L., Cyster J.G., Engleman E.G. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat. Immunol. 2002;3:1135–1141. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M., Kumar B.V., Meng W., Granot T., Carpenter D.J., Senda T., Chen D., Rosenfeld A.M., Zhang B., Lerner H. Human lymph nodes maintain TCF-1hi memory T cells with high functional potential and clonal diversity throughout life. J. Immunol. 2018;201:2132–2140. doi: 10.4049/jimmunol.1800716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag D., Proietto A.I., Loudovaris T., Mannering S.I., Vremec D., Shortman K., Wu L., Harrison L.C. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J. Immunol. 2011;186:6207–6217. doi: 10.4049/jimmunol.1002632. [DOI] [PubMed] [Google Scholar]
- Moncada R., Barkley D., Wagner F., Chiodin M., Devlin J.C., Baron M., Hajdu C.H., Simeone D.M., Yanai I. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 2020;38:333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- Morgan R.A., Chinnasamy N., Abate-Daga D., Gros A., Robbins P.F., Zheng Z., Dudley M.E., Feldman S.A., Yang J.C., Sherry R.M. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 2013;36:133–151. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R.A., Yang J.C., Kitano M., Dudley M.E., Laurencot C.M., Rosenberg S.A. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther. 2010;18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mose L.E., Selitsky S.R., Bixby L.M., Marron D.L., Iglesia M.D., Serody J.S., Perou C.M., Vincent B.G., Parker J.S. Assembly-based inference of B-cell receptor repertoires from short read RNA sequencing data with V'DJer. Bioinformatics. 2016;32:3729–3734. doi: 10.1093/bioinformatics/btw526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.N., Mackay L.K. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 2016;16:79–89. doi: 10.1038/nri.2015.3. [DOI] [PubMed] [Google Scholar]
- Nagelkerke S.Q., Bruggeman C.W., den Haan J.M.M., Mul E.P.J., van den Berg T.K., van Bruggen R., Kuijpers T.W. Red pulp macrophages in the human spleen are a distinct cell population with a unique expression of Fc-γ receptors. Blood Adv. 2018;2:941–953. doi: 10.1182/bloodadvances.2017015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya H.I., Hagan T., Duraisingham S.S., Lee E.K., Kwissa M., Rouphael N., Frasca D., Gersten M., Mehta A.K., Gaujoux R. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity. 2015;43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan M.S., Rouphael N., Lai L., Kazmin D., Jensen T.L., Weiss D.S., Ibegbu C., Sztein M.B., Hooper W.F., Hill H. Systems vaccinology for a live attenuated tularemia vaccine reveals unique transcriptional signatures that predict humoral and cellular immune responses. Vaccines (Basel) 2019;8:4. doi: 10.3390/vaccines8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Domínguez-Andrés J., Barreiro L.B., Chavakis T., Divangahi M., Fuchs E., Joosten L.A.B., van der Meer J.W.M., Mhlanga M.M., Mulder W.J.M. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020;20:375–388. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell E.W., Sigal N., Bendall S.C., Nolan G.P., Davis M.M. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newick K., O'Brien S., Moon E., Albelda S.M. CAR T cell therapy for solid tumors. Annu. Rev. Med. 2017;68:139–152. doi: 10.1146/annurev-med-062315-120245. [DOI] [PubMed] [Google Scholar]
- Nielsen S.C.A., Boyd S.D. Human adaptive immune receptor repertoire analysis-Past, present, and future. Immunol. Rev. 2018;284:9–23. doi: 10.1111/imr.12667. [DOI] [PubMed] [Google Scholar]
- Nihal M., Mikkola D., Wood G.S. Detection of clonally restricted immunoglobulin heavy chain gene rearrangements in normal and lesional skin: analysis of the B cell component of the skin-associated lymphoid tissue and implications for the molecular diagnosis of cutaneous B cell lymphomas. J. Mol. Diagn. 2000;2:5–10. doi: 10.1016/S1525-1578(10)60609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan J.P., Condello D. Spectral flow cytometry. Curr. Protoc. Cytom. 2013 doi: 10.1002/0471142956.cy0127s63. Chapter 1, Unit1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- Nyman T.A., Lorey M.B., Cypryk W., Matikainen S. Mass spectrometry-based proteomic exploration of the human immune system: focus on the inflammasome, global protein secretion, and T cells. Expert Rev. Proteomics. 2017;14:395–407. doi: 10.1080/14789450.2017.1319768. [DOI] [PubMed] [Google Scholar]
- O'Sullivan T.E., Sun J.C., Lanier L.L. Natural killer cell memory. Immunity. 2015;43:634–645. doi: 10.1016/j.immuni.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallett L.J., Davies J., Colbeck E.J., Robertson F., Hansi N., Easom N.J.W., Burton A.R., Stegmann K.A., Schurich A., Swadling L. IL-2high tissue-resident T cells in the human liver: sentinels for hepatotropic infection. J. Exp. Med. 2017;214:1567–1580. doi: 10.1084/jem.20162115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018;18:35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- Park J.E., Botting R.A., Domínguez Conde C., Popescu D.M., Lavaert M., Kunz D.J., Goh I., Stephenson E., Ragazzini R., Tuck E. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367:eaay3224. doi: 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst M.R., Yang J.C., Langan R.C., Dudley M.E., Nathan D.A., Feldman S.A., Davis J.L., Morgan R.A., Merino M.J., Sherry R.M. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther. 2011;19:620–626. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasetto A., Gros A., Robbins P.F., Deniger D.C., Prickett T.D., Matus-Nicodemos R., Douek D.C., Howie B., Robins H., Parkhurst M.R. Tumor- and neoantigen-reactive T-cell receptors can Be identified based on their frequency in fresh tumor. Cancer Immunol. Res. 2016;4:734–743. doi: 10.1158/2326-6066.CIR-16-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F., Arkin Y., Giladi A., Jaitin D.A., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Peng H., Tian Z. Diversity of tissue-resident NK cells. Semin. Immunol. 2017;31:3–10. doi: 10.1016/j.smim.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Peterson V.M., Zhang K.X., Kumar N., Wong J., Li L., Wilson D.C., Moore R., McClanahan T.K., Sadekova S., Klappenbach J.A. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- Pillai S., Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat. Rev. Immunol. 2009;9:767–777. doi: 10.1038/nri2656. [DOI] [PubMed] [Google Scholar]
- Ping Y., Liu C., Zhang Y. T-cell receptor-engineered T cells for cancer treatment: current status and future directions. Protein Cell. 2018;9:254–266. doi: 10.1007/s13238-016-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzolla A., Nguyen T.H., Sant S., Jaffar J., Loudovaris T., Mannering S.I., Thomas P.G., Westall G.P., Kedzierska K., Wakim L.M. Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J. Clin. Invest. 2018;128:721–733. doi: 10.1172/JCI96957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu D.M., Botting R.A., Stephenson E., Green K., Webb S., Jardine L., Calderbank E.F., Polanski K., Goh I., Efremova M. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D.L., Levine B.L., Kalos M., Bagg A., June C.H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwar R., Campbell J., Murphy G., Richards W.G., Clark R.A., Kupper T.S. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One. 2011;6:e16245. doi: 10.1371/journal.pone.0016245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos I. mSphere of influence: predicting immune responses and susceptibility to influenza virus-may the data Be with you. mSphere. 2020;5:e00085-20. doi: 10.1128/mSphere.00085-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph G.J., Ochando J., Partida-Sánchez S. Migration of dendritic cell subsets and their precursors. Annu. Rev. Immunol. 2008;26:293–316. doi: 10.1146/annurev.immunol.26.021607.090254. [DOI] [PubMed] [Google Scholar]
- Reizis B., Bunin A., Ghosh H.S., Lewis K.L., Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu. Rev. Immunol. 2011;29:163–183. doi: 10.1146/annurev-immunol-031210-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B., Colonna M., Trinchieri G., Barrat F., Gilliet M. Plasmacytoid dendritic cells: one-trick ponies or workhorses of the immune system? Nat. Rev. Immunol. 2011;11:558–565. doi: 10.1038/nri3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R.M., Suarez-Alvarez B., Lopez-Larrea C. Therapeutic epigenetic reprogramming of trained immunity in myeloid cells. Trends Immunol. 2019;40:66–80. doi: 10.1016/j.it.2018.11.006. [DOI] [PubMed] [Google Scholar]
- Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F., Macosko E.Z. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati E., Dowds C.M., Liaskou E., Henriksen E.K.K., Karlsen T.H., Franke A. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol. 2017;17:61. doi: 10.1186/s12896-017-0379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossjohn J., Gras S., Miles J.J., Turner S.J., Godfrey D.I., McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- Sada-Ovalle I., Talayero A., Chavéz-Galán L., Barrera L., Castorena-Maldonado A., Soda-Merhy A., Torre-Bouscoulet L. Functionality of CD4+ and CD8+ T cells from tonsillar tissue. Clin. Exp. Immunol. 2012;168:200–206. doi: 10.1111/j.1365-2249.2012.04573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens W., Cannoodt R., Todorov H., Saeys Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019;37:547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- Saeys Y., Van Gassen S., Lambrecht B.N. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 2016;16:449–462. doi: 10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Geginat J., Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- Sathaliyawala T., Kubota M., Yudanin N., Turner D., Camp P., Thome J.J., Bickham K.L., Lerner H., Goldstein M., Sykes M. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38:187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoettler N., Hrusch C.L., Blaine K.M., Sperling A.I., Ober C. Transcriptional programming and T cell receptor repertoires distinguish human lung and lymph node memory T cells. Commun. Biol. 2019;2:411. doi: 10.1038/s42003-019-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze J.L., Aschenbrenner A.C. Systems immunology allows a new view on human dendritic cells. Semin. Cell Dev. Biol. 2019;86:15–23. doi: 10.1016/j.semcdb.2018.02.017. [DOI] [PubMed] [Google Scholar]
- See P., Dutertre C.A., Chen J., Günther P., McGovern N., Irac S.E., Gunawan M., Beyer M., Händler K., Duan K. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356:eaag3009. doi: 10.1126/science.aag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Durand M., Amigorena S. Similar antigen cross-presentation capacity and phagocytic functions in all freshly isolated human lymphoid organ-resident dendritic cells. J. Exp. Med. 2013;210:1035–1047. doi: 10.1084/jem.20121103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N., Mukherjee G., Sen A., Bendall S.C., Sung P., Nolan G.P., Arvin A.M. Single-cell mass cytometry analysis of human tonsil T cell remodeling by varicella zoster virus. Cell Rep. 2014;8:633–645. doi: 10.1016/j.celrep.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda T., Dogra P., Granot T., Furuhashi K., Snyder M.E., Carpenter D.J., Szabo P.A., Thapa P., Miron M., Farber D.L. Microanatomical dissection of human intestinal T-cell immunity reveals site-specific changes in gut-associated lymphoid tissues over life. Mucosal Immunol. 2019;12:378–389. doi: 10.1038/s41385-018-0110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M., Kiseliovas V., Levine J., Gayoso A., Mazutis L., Pe'er D. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 2019;37:451–460. doi: 10.1038/s41587-019-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seumois G., Ramirez-Suastegui C., Schmiedel B.J., Liang S., Peters B., Sette A., Vijayanand P. Single-cell transcriptomic analysis of allergen-specific T cells in allergy and asthma. Sci. Immunol. 2020;5:eaba6087. doi: 10.1126/sciimmunol.aba6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni Y., Fehlings M., Kløverpris H.N., McGovern N., Koo S.L., Loh C.Y., Lim S., Kurioka A., Fergusson J.R., Tang C.L. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. 2017;46:148–161. doi: 10.1016/j.immuni.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifka M.K., Antia R., Whitmire J.K., Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity. 1998;8:363–372. doi: 10.1016/s1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- Smazynski J., Webb J.R. Resident memory-like tumor-infiltrating lymphocytes (TILRM): latest players in the immuno-oncology repertoire. Front. Immunol. 2018;9:1741. doi: 10.3389/fimmu.2018.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolders J., Heutinck K.M., Fransen N.L., Remmerswaal E.B.M., Hombrink P., Ten Berge I.J.M., van Lier R.A.W., Huitinga I., Hamann J. Tissue-resident memory T cells populate the human brain. Nat. Commun. 2018;9:4593. doi: 10.1038/s41467-018-07053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M.E., Finlayson M.O., Connors T.J., Dogra P., Senda T., Bush E., Carpenter D., Marboe C., Benvenuto L., Shah L. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci. Immunol. 2019;4:eaav5581. doi: 10.1126/sciimmunol.aav5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R., Grzelak M., Hadfield J. RNA sequencing: the teenage years. Nat. Rev. Genet. 2019;20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- Steiniger B.S. Human spleen microanatomy: why mice do not suffice. Immunology. 2015;145:334–346. doi: 10.1111/imm.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart B.J., Ferdinand J.R., Clatworthy M.R. Using single-cell technologies to map the human immune system - implications for nephrology. Nat. Rev. Nephrol. 2020;16:112–128. doi: 10.1038/s41581-019-0227-3. [DOI] [PubMed] [Google Scholar]
- Stewart B.J., Ferdinand J.R., Young M.D., Mitchell T.J., Loudon K.W., Riding A.M., Richoz N., Frazer G.L., Staniforth J.U.L., Vieira Braga F.A. Spatiotemporal immune zonation of the human kidney. Science. 2019;365:1461–1466. doi: 10.1126/science.aat5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbington M.J.T., Rozenblatt-Rosen O., Regev A., Teichmann S.A. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358:58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N.L., Eberhardt C.S., Wieland A., Vora K.A., Pulendran B., Ahmed R. Understanding the immunology of the Zostavax shingles vaccine. Curr. Opin. Immunol. 2019;59:25–30. doi: 10.1016/j.coi.2019.02.005. [DOI] [PubMed] [Google Scholar]
- Sun J.C., Beilke J.N., Lanier L.L. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Lanier L.L. NK cell development, homeostasis and function: parallels with CD8⁺ T cells. Nat. Rev. Immunol. 2011;11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt A.J., Hedlin H.K., Balasubramanian V., Hsi A., Blum L.K., Robinson W.H., Haddad F., Hickey P.M., Condliffe R., Lawrie A. Discovery of distinct immune phenotypes using machine learning in pulmonary arterial hypertension. Circ. Res. 2019;124:904–919. doi: 10.1161/CIRCRESAHA.118.313911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Levitin H.M., Miron M., Snyder M.E., Senda T., Yuan J., Cheng Y.L., Bush E.C., Dogra P., Thapa P. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 2019;10:4706. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Miron M., Farber D.L. Location, location, location: tissue resident memory T cells in mice and humans. Sci. Immunol. 2019;4:eaas9673. doi: 10.1126/sciimmunol.aas9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Team H.-C.S.P., Consortium H.-I. Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci. Immunol. 2017;2:eaal4656. doi: 10.1126/sciimmunol.aal4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa P., Farber D.L. The role of the thymus in the immune response. Thorac. Surg. Clin. 2019;29:123–131. doi: 10.1016/j.thorsurg.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome J.J., Bickham K.L., Ohmura Y., Kubota M., Matsuoka N., Gordon C., Granot T., Griesemer A., Lerner H., Kato T. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat. Med. 2016;22:72–77. doi: 10.1038/nm.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome J.J., Grinshpun B., Kumar B.V., Kubota M., Ohmura Y., Lerner H., Sempowski G.D., Shen Y., Farber D.L. Longterm maintenance of human naive T cells through in situ homeostasis in lymphoid tissue sites. Sci. Immunol. 2016;1:aah6506. doi: 10.1126/sciimmunol.aah6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome J.J., Yudanin N., Ohmura Y., Kubota M., Grinshpun B., Sathaliyawala T., Kato T., Lerner H., Shen Y., Farber D.L. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J.S., Schwartzberg P.L., Kotliarov Y., Biancotto A., Xie Z., Germain R.N., Wang E., Olnes M.J., Narayanan M., Golding H. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg J.H., Gomez-Eerland R., van de Wiel B., Hulshoff L., van den Broek D., Bins A., Tan H.L., Harper J.V., Hassan N.J., Jakobsen B.K. Case report of a fatal serious adverse event upon administration of T cells transduced with a MART-1-specific T-cell receptor. Mol. Ther. 2015;23:1541–1550. doi: 10.1038/mt.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L., Hinton G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- Velten L., Haas S.F., Raffel S., Blaszkiewicz S., Islam S., Hennig B.P., Hirche C., Lutz C., Buss E.C., Nowak D. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 2017;19:271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tian Z., Peng H. Tissue-resident memory-like ILCs: innate counterparts of TRM cells. Protein Cell. 2020;11:85–96. doi: 10.1007/s13238-019-0647-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Q., Milne K., Derocher H., Webb J.R., Nelson B.H., Watson P.H. CD103 and intratumoral immune response in breast cancer. Clin. Cancer Res. 2016;22:6290–6297. doi: 10.1158/1078-0432.CCR-16-0732. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Gehad A., Yang C., Scott L.L., Teague J.E., Schlapbach C., Elco C.P., Huang V., Matos T.R., Kupper T.S. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl. Med. 2015;7:279ra239. doi: 10.1126/scitranslmed.3010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb J.R., Milne K., Nelson B.H. PD-1 and CD103 are widely coexpressed on prognostically favorable intraepithelial CD8 T cells in human ovarian cancer. Cancer Immunol. Res. 2015;3:926–935. doi: 10.1158/2326-6066.CIR-14-0239. [DOI] [PubMed] [Google Scholar]
- Weisberg S.P., Carpenter D.J., Chait M., Dogra P., Gartrell-Corrado R.D., Chen A.X., Campbell S., Liu W., Saraf P., Snyder M.E. Tissue-resident memory T cells mediate immune homeostasis in the human pancreas through the PD-1/PD-L1 pathway. Cell Rep. 2019;29:3916–3932.e5. doi: 10.1016/j.celrep.2019.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel N.M., Weisel F.J., Farber D.L., Borghesi L.A., Shen Y., Ma W., Luning Prak N.T., Shlomchik M.J. Comprehensive analysis of B cell compartments across the human body reveals novel subsets and a gut resident memory phenotype. Blood. 2020 doi: 10.1182/blood.2019002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Cai J. Mucosal-associated invariant T cells: new insights into antigen recognition and activation. Front. Immunol. 2017;8:1540. doi: 10.3389/fimmu.2017.01540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Tsang J.C., Wang C., Clare S., Wang J., Chen X., Brandt C., Kane L., Campos L.S., Lu L. Single-cell RNA-seq identifies a PD-1. Nature. 2016;539:102–106. doi: 10.1038/nature20105. [DOI] [PubMed] [Google Scholar]
- Yudanin N.A., Schmitz F., Flamar A.L., Thome J.J.C., Tait Wojno E., Moeller J.B., Schirmer M., Latorre I.J., Xavier R.J., Farber D.L. Spatial and temporal mapping of human innate lymphoid cells reveals elements of tissue specificity. Immunity. 2019;50:505–519.e4. doi: 10.1016/j.immuni.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak D.E., Tam V.C., Aderem A. Systems-level analysis of innate immunity. Annu. Rev. Immunol. 2014;32:547–577. doi: 10.1146/annurev-immunol-032713-120254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bevan M.J. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Peng T., Johnston C., Phasouk K., Kask A.S., Klock A., Jin L., Diem K., Koelle D.M., Wald A. Immune surveillance by CD8αα+ skin-resident T cells in human herpes virus infection. Nature. 2013;497:494–497. doi: 10.1038/nature12110. [DOI] [PMC free article] [PubMed] [Google Scholar]