Summary
Microtubules are cytoskeletal components involved in pivotal eukaryotic functions such as cell division, ciliogenesis, and intracellular trafficking. They assemble from αβ-tubulin heterodimers and disassemble in a process called dynamic instability, which is driven by GTP hydrolysis. Structures of the microtubule and of soluble tubulin have been determined by cryo-EM and by X-ray crystallography, respectively. Altogether, these data define the mechanism of tubulin assembly-disassembly at atomic or near-atomic level. We review here the structural changes that occur during assembly, tubulin switching from a curved conformation in solution to a straight one in the microtubule core. We also present more subtle changes associated with GTP binding, leading to tubulin activation for assembly. Finally, we show how cryo-EM and X-ray crystallography are complementary methods to characterize the interaction of tubulin with proteins involved either in intracellular transport or in microtubule dynamics regulation.
Subject Areas: Biochemistry Methods, Cell Biology, Structural Biology
Graphical Abstract
Biochemistry Methods, Cell Biology ,Structural Biology
Microtubules are filaments of the eukaryotic cytoskeleton that assemble and disassemble in a process called dynamic instability (Desai and Mitchison, 1997; Mitchison and Kirschner, 1984). The αβ-tubulin heterodimer (hereafter called tubulin), identified more than 50 years ago as the protein that binds colchicine (Borisy and Taylor, 1967; Wilson and Friedkin, 1967), was found as the main component of cytoplasmic microtubules (Weisenberg et al., 1968). The α and β subunits share more than 40% sequence identity (Little et al., 1981) and, as expected from this similarity (Chothia and Lesk, 1986), they eventually turned out to have very similar structures (Nogales et al., 1998). In addition, each tubulin subunit binds a nucleotide. A GTP molecule is bound to α-tubulin and is neither hydrolyzed nor exchanged, whereas either GTP or GDP can be found at the β-tubulin nucleotide binding site (Geahlen and Haley, 1977), defining GTP-tubulin and GDP-tubulin, respectively. Only GTP-tubulin assembles in microtubules on its own, and microtubule assembly is accompanied by GTP hydrolysis (Carlier, 1989).
Despite the high degree of conservation across species (Little et al., 1981), a variety of tubulin molecules can be found in a single organism. Indeed, nine genes code for the α chain and nine genes code for the β chain in humans (Janke and Magiera, 2020; Roll-Mecak, 2020). In addition to isotypic heterogeneity, which results directly from a genetic contribution, post-translational modifications (PTMs) of tubulin have been described. Most of them target the C-terminal tail of both subunits, which are the main disordered regions of tubulin. They include the addition of chains of glutamate (polyglutamylation) or glycine residues (polyglycylation) (reviewed in Janke, 2014). Another modification is detyrosination of α-tubulin, which removes the last amino acid of this subunit, an amino acid that may be added back to regenerate the original sequence (Valenzuela et al., 1981). However, PTMs do not only concern the C-terminal tails. For instance, acetylation takes place on Lys 40 of α-tubulin (L'Hernault and Rosenbaum, 1985) and the acetyl group may then be removed (Hubbert et al., 2002). The combination of the differential expression of α- and β-tubulin genes and of the PTMs collectively define a “tubulin code” conferring specialized functions to the microtubule cytoskeleton (Janke and Magiera, 2020; Roll-Mecak, 2020).
Here, we will review advances in the mechanism of tubulin based on structural data. First, we will mention the structural methods used to study tubulin and microtubules, highlighting their complementarity, with a focus on methods currently available to determine the structure of unassembled tubulin. Then we will show how the structures enlighten the mechanism of microtubule dynamics, from tubulin activation upon GTP binding to structural changes associated with assembly, and its regulation by various proteins that bind to tubulin or to microtubules. We will finally discuss areas where further progress in our understanding may soon become possible.
Complementary Methods for the Structural Study of Tubulin and Binding Partners
The microtubule is a cylindrical assembly of tubulin with a diameter of about 25 nm and a typical length of several micrometers. In vivo, it most often comprises 13 parallel protofilaments in which tubulin molecules are arranged head to tail, defining a (fast growing) plus end terminated by β-tubulin and a (slower growing) minus end exposing α-tubulin. As such, it cannot be crystallized but is well suited for cryo-EM studies (reviewed in Downing and Nogales, 1998; Manka and Moores, 2018a; Nogales and Kellogg, 2017). Actually, it has fully benefited from the advances of this method, the resolution having improved from 20 to about 3 Å in less than 20 years (Nogales et al., 1999; Zhang et al., 2018). Cryo-EM is also suitable for studying the interaction with microtubule-associated proteins (or MAPs, taken in this review in a broad sense, i.e., proteins that bind to soluble or microtubular tubulin), although the resolution of the MAP partner is usually lower than that of the microtubule. In contrast, the size of soluble tubulin, about 100 kDa, makes this protein a better candidate for X-crystallography structural studies, provided that the propensity of tubulin to self-assemble into heterogeneous species is managed (see next section). Cryo-EM and X-crystallography are highly complementary for studying microtubule dynamics structurally, as will be illustrated in this review with the tubulin conformational changes during assembly and with the interaction of MAPs with tubulin and/or microtubules. The study of microtubule-targeting agents is another example of this complementarity; this last aspect was reviewed recently (Steinmetz and Prota, 2018).
Microtubules and MAPs have also been studied by NMR spectroscopy as this method allows one to study flexible segments of a protein, even when embedded in an assembly as large as the microtubule. Tubulin itself has been produced as an NMR-labeled protein (Wall et al., 2016) to characterize the dynamicity of its C-terminal tails, which are disordered and hence usually not seen in cryo-EM or X-ray crystallography studies. The NMR data show that the C-terminal tails behave differently in comparison with isolated peptides of the same sequence, indicating an interaction with the ordered regions of tubulin (Wall et al., 2016). However, NMR spectroscopy has so far been mainly used to study the interaction of NMR-labeled MAPs with unlabeled tubulin or microtubules. Probably the best example of such studies concerns Tau, an intrinsically disordered neuronal protein that stabilizes axonal microtubules (Lippens et al., 2016; Melková et al., 2019). Because many Tau residues remain NMR visible even in the presence of tubulin complexes, a model for the enhancement of microtubule assembly by this protein could be proposed from the NMR characterization of such Tau-tubulin complexes (Gigant et al., 2014). Recent near-atomic cryo-EM data on Tau-decorated microtubules (Kellogg et al., 2018) allowed discussion of the validity of the NMR-based model (Lippens and Gigant, 2019).
Preventing the Self-Association of Tubulin Leads to Its Crystallization
Because of the physiological importance of tubulin, there have been many attempts to determine its structure. A milestone in this quest was the discovery that stathmin increases the frequency of microtubule catastrophes (the transition from growth to shrinkage) (Belmont and Mitchison, 1996), which led to the identification of a stable 2:1 tubulin:stathmin assembly (Curmi et al., 1997; Jourdain et al., 1997) and, eventually, to crystallization of T2R complexes made by tubulin with the stathmin-like domain (SLD) of the RB3 protein (Figure 1A) (Gigant et al., 2000). Higher-resolution diffracting crystals from T2R were then obtained, either after cleavage of the C-terminal tails of tubulin (Nawrotek et al., 2011) or by adding to T2R the tubulin tyrosine ligase (TTL) enzyme (Prota et al., 2013b). Eventually, these findings provided the basis for the design of SLDs binding one to four tubulin heterodimers and the structures of the corresponding complexes were determined (Mignot et al., 2012).
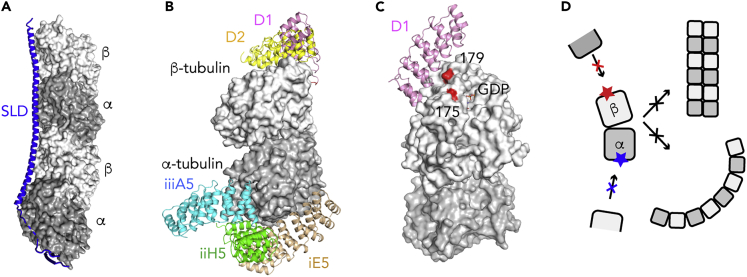
Figure 1.
Crystallization Helpers Prevent Tubulin Self-Assembly
(A) The T2R complex, composed of two tubulin heterodimers, with the α subunits in dark gray and the β subunits in light gray, and one SLD of the RB3 protein (blue).
(B) The binding site of tubulin crystallization chaperones selected from libraries of artificial proteins to target either the β subunit (e.g., D1 and D2 DARPins) or the α subunit (e.g., iE5, iiH5, and iiiA5 αReps).
(C) Assembly-blocked mutants of tubulin. In yeast tubulin, the T175R, V179R double mutation in β-tubulin impairs longitudinal contacts between tubulin molecules, favoring crystallization. The position of the mutated residues is shown on mammalian tubulin, because residue 175 was not traced in the yeast tubulin model. The D1 binding site is shown as a reference.
(D) The crystallization helpers target tubulin surfaces (red and blue stars) to prevent the tubulin self-association into straight (top, right) or curved (bottom, right) assemblies.
Although fruitful, the use of SLDs as tubulin-crystallization helpers prevents the structural study of complexes with proteins whose binding site on tubulin overlaps with that of SLDs; such is the case, for instance, of kinesins. It was therefore useful to elicit additional tubulin binders whose site does not overlap, for instance, with tubulin residues exposed on the outer surface of microtubules. Such is the case of some Designed Ankyrin Repeat Proteins (DARPins) (Plückthun, 2015) selected to bind to tubulin immobilized with its α subunit longitudinal surface (i.e., a surface engaged in tubulin-tubulin interactions along a protofilament) facing the surface of the wells of microtiter plates (Pecqueur et al., 2012). The structures of several such tubulin-DARPin complexes were determined and the DARPin-binding sites were found to be on the β-tubulin longitudinal surface (Figure 1B) (Campanacci et al., 2019b; Mignot et al., 2012; Pecqueur et al., 2012), i.e., the one at the opposite end of this protein with respect to the surface facing the microtiter plate when DARPins were elicited. In an experiment reciprocal to the previous one, αReps, artificial proteins based on a consensus sequence of a HEAT-like repeated motif (Urvoas et al., 2010), were selected to bind to tubulin interacting with a high-affinity, slowly dissociating DARPin (Ahmad et al., 2016) immobilized on a microtiter plate. Indeed, the binding sites of three such αReps were found on, or close to, the α-tubulin longitudinal surface (Figure 1B) (Campanacci et al., 2019a, Campanacci et al., 2019b).
The three families of proteins just described allow one to determine the structures of tubulin or of tubulin complexes with MAPs because they form monodisperse assemblies with tubulin that crystallize. Since it is now possible to design and produce mutants of yeast tubulin that disfavor self-assembly (Figure 1C) (Johnson et al., 2011), it is also possible to use such mutants in crystallization experiments. Such was the case when the structure of the complex with tubulin of a TOG domain of the XMAP215/Stu2p catalysts for microtubule elongation was determined (Ayaz et al., 2012). The defining factor of these crystallization helpers, either tubulin-binding proteins or mutations, is that they prevent the longitudinal self-association of tubulin in assemblies in which tubulin is curved (e.g., rings or helices) or straight (as in the microtubule) (Figure 1D). In the next section, we will describe more deeply these two general conformations, focusing on the structural changes associated with microtubule assembly.
The Tubulin Conformational Changes Associated with Microtubule Assembly
Cryo-EM analysis of the microtubule core defines a straight conformation of tubulin, in which the α and β subunits along a protofilament are aligned (or almost aligned; Figure 2A) (Alushin et al., 2014; Nogales et al., 1999). In contrast, in all the X-ray structures determined so far, non-microtubular tubulin is curved: in addition to a translation, as in straight protofilaments, a rotation is needed to superpose the α and β subunits of the tubulin heterodimer (Gigant et al., 2000). The rotation angle is variable, depending on the structure considered, with values ranging from about 9° to as much as 18° (Campanacci et al., 2019b) (Figure 2A). The curvature may be modulated by tubulin-binding proteins, as postulated in the case of a KIF2A kinesin construct, which forms a 2:1 tubulin:KIF2A complex (Trofimova et al., 2018). In this structure, the curvature angle of the tubulin heterodimer bound to the kinesin motor domain is about 12°, whereas that of the tubulin molecule interacting with the KIF2A N-terminal “neck” is greater than 15°. However, the angle value observed in one particular crystal context should be interpreted with caution. Indeed, tubulin in complex with crystallization helpers that bind to the distal α or β subunit longitudinal surface, hence that are unlikely to influence the tubulin hinge, shows variable curvatures, arguing for a crystal packing effect modulating curvature.
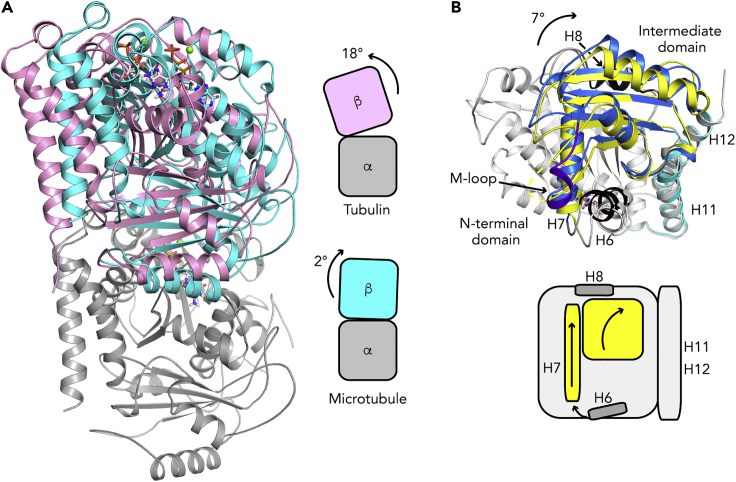
Figure 2.
Tubulin Conformational Changes Associated with Microtubule Assembly
(A) Relative orientation of tubulin subunits in soluble and in microtubular tubulins. (Left) The structure of tubulin bound to an αRep (pdb id 6GWC, β-tubulin in pink) is compared with that of tubulin in GMPCPP-microtubules (pdb id 6DPU, cyan). The N-terminal domains of the α subunits have been superposed on their secondary structural elements and only α-tubulin of 6GWC is shown (gray). (Right) Schematic illustration of the rotation angles needed to align the α and β subunits in these two structures.
(B) Structural changes in the β subunit. (Top) The β-tubulin N-terminal domains of soluble tubulin (pdb id 6S8K) and of the microtubule (pdb id 6DPU) have been superposed using their secondary structural elements; only that of soluble tubulin is shown. The C-terminal helices coincide, whereas the intermediate domains are misaligned by about 7°, which also leads to a translation of the H7 helix. Soluble tubulin is in gray, except for its intermediate domain, which is in yellow. Microtubular tubulin is colored blue (intermediate domain, with the M-loop in purple blue), black (H6 and H8 helices), and cyan (C-terminal helices). (Bottom) Schematic illustration of the movements.
The curved-to-straight conformational change is accompanied by rearrangements within the tubulin subunits (Figure 2B). Based on the structure of tubulin in Zn-induced sheets of antiparallel straight protofilaments (Nogales et al., 1998) and on that in T2R, we reported that the intermediate domain of the tubulin subunits moves with respect to a larger ensemble comprising the N-terminal domain and the C-terminal H11-H11′-H12 helices (Ravelli et al., 2004). The intermediate domain movement could be characterized as a rotation of its β sheet and a translation of the H7 helix. Essentially the same conclusion can be derived from the comparison of the higher resolution data that are now available, reaching 3.1 Å in the case of the microtubule (pdb id 6DPU; Zhang et al., 2018) and 1.52 Å in that of a tubulin-DARPin complex (pdb id 6S8K; La Sala et al., 2019). Interestingly, although the H7 helix is part of the intermediate domain, the adjacent H6 and H8 helices hardly belong to any of the two domains. When tubulin switches from curved to straight, H6 acts as a hinge between these two domains (Ravelli et al., 2004), whereas the H8 helix, which is a major contributor to inter-subunit contacts along a protofilament, moves less than the intermediate domain (Dorléans et al., 2009).
The curved-to-straight transition of tubulin and the associated domain movements within subunits give rise to microtubule-specific contacts between tubulin molecules. First, there is a marked increase of the surface engaged in longitudinal contacts between tubulin subunits in straight protofilaments (Nogales et al., 1999) compared with, e.g., that seen in the T2R complex (Gigant et al., 2000). The straightening of protofilaments also allows lateral interactions (between tubulins of adjacent protofilaments) to be established. One of the key elements for lateral contacts is the so-called M-loop (“M” for microtubule) (Nogales et al., 1999), embedded in the intermediate domain. This loop undergoes a disordered-to-helical conformational change during microtubule assembly (Figure 2B) (Prota et al., 2013a). Conversely, the straight-to-curved switch during disassembly results in the loss of lateral contacts and in the lowering of longitudinal ones, part of it being due to an H6-H7 loop conformation in curved tubulin that is not compatible with straight protofilaments. Taken together, these changes provide a rationale for rapid microtubule disassembly, which is characteristic of dynamic instability (Ravelli et al., 2004).
Although high-resolution views of straight and curved tubulin have been pictured, representing two extreme points of its structural cycle, tubulin oligomers are expected to adopt intermediate conformations upon assembly and disassembly. Cryo-EM analysis identified curved sheets at microtubule ends and led to a model in which tubulin molecules undergo a slow conformational change between the tip of the sheet and the beginning of the microtubule core (Chrétien et al., 1995; Guesdon et al., 2016). Intermediate conformations have also been visualized in GMPCPP-induced macrotubes of tubulin (Wang and Nogales, 2005). It should, however, be noted that, in these studies, the resolution remains too low to make it possible to discriminate tubulin subunits. Therefore, neither the degree of the curvature within heterodimers nor the respective orientation of domains in the subunits could be determined.
How GTP Binding Activates Tubulin for Microtubule Assembly
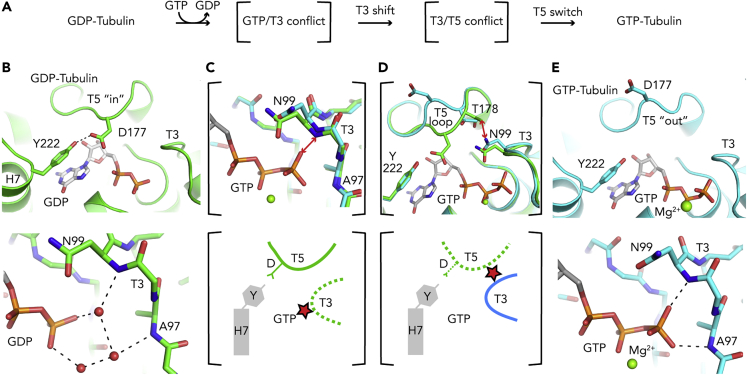
The activation of tubulin for microtubule assembly upon GTP binding is a key step in microtubule dynamics. Because of the curved-to-straight transition from soluble to microtubular tubulin, an obvious hypothesis is that GTP induces a straightening of tubulin upon binding, allowing microtubule-specific contacts to be made, and hence incorporation in the microtubule. However, this proposal has been challenged. Indeed, small molecule compounds that only bind to curved tubulin have similar affinities for GTP- and GDP-tubulin (Barbier et al., 2010; Rice et al., 2008). Relatedly, X-ray structural studies indicate that tubulin has the same overall curved conformation regardless of the bound nucleotide (Nawrotek et al., 2011; Pecqueur et al., 2012). The question that arises is therefore how does GTP activate tubulin. When the first high-resolution structures of soluble GDP- and GTP-tubulin became available, we proposed an activation mechanism based on a conformational switch of the β-tubulin T5 loop (Figure 3A) (Nawrotek et al., 2011). This loop is at the longitudinal end of the β subunit, nearby the exchangeable nucleotide binding site, and is involved in contacts within protofilaments. With bound GDP, structural data define a T5 loop “in” conformation, in which the T5 residue Asp177 (sequential numbering) interacts with the hydroxyl of Tyr222, the first residue of the H7 helix (Figure 3B). The binding of GTP leads to a cascade of structural changes (Figures 3C and 3D), including a movement of the T3 loop to optimize the interaction with the nucleotide (Figure 3E). The T3 movement triggers the destabilization of T5, characterized by the breakage of the Asp177-Tyr222 interaction and a flipping of T5 in an “out” conformation, making Asp177 point to the solvent. Such a conformation for a tubulin molecule at a protofilament plus end may favor its elongation, the acidic Asp177 side chain being able to interact with basic residues of the α-tubulin longitudinal surface of an incoming heterodimer.
Figure 3.
How GTP Binding Activates Tubulin for Microtubule Assembly
(A) A cascade of rearrangements occurs in the β-tubulin nucleotide binding site upon GTP binding.
(B) With bound GDP, the T5 loop adopting an “in” conformation is prone to interact with the H7 helix Tyr222 residue (Top), and the interaction of GDP with the T3 loop is mediated by water molecules (red spheres) (bottom).
(C) Following GTP binding, GTP would come too close to the NH amide of Asn99 from T3 (2.2 Å distance), illustrated by a double red arrow in the superposed structures of GDP- and GTP-tubulin (top; GDP is not shown) and by a red star in the schematic drawing (bottom).
(D) The conformation of T3 in GTP-tubulin is not compatible with the T5 “in” structure, as it would lead to a steric conflict between Asn99 and Thr178 (double red arrow and red star in the top and bottom panels, respectively).
(E) In GTP-tubulin, T5 has switched to an “out” conformation (top) and T3 interacts directly with the GTP γ-phosphate (bottom). GDP-tubulin is in green and GTP-tubulin in cyan. Selected hydrogen bonds are shown as dashed black lines in (B) and (E).
This model emphasizes the contribution of Tyr222 in the GTP-induced activation of tubulin. Tyr222 is conserved in animals; relatedly, it has been shown that a Y222F mutation leads to a congenital developmental disorder named circumferential skin creases Kunze type (CSC-KT) and that this mutation compromises microtubule dynamics (Isrie et al., 2015). Interestingly, a Phe residue is found at this position in β-tubulin of plants and of unicellular eukaryotes. Accordingly, tubulin purified from Chlamydomonas reinhardtii cilia is less dynamic than mammalian brain tubulin (Orbach and Howard, 2019). Additional characterization is, however, needed to understand the effect of the Y222F substitution on microtubule dynamics.
Insights into the Mechanism of MAPs Specific of Soluble Tubulin
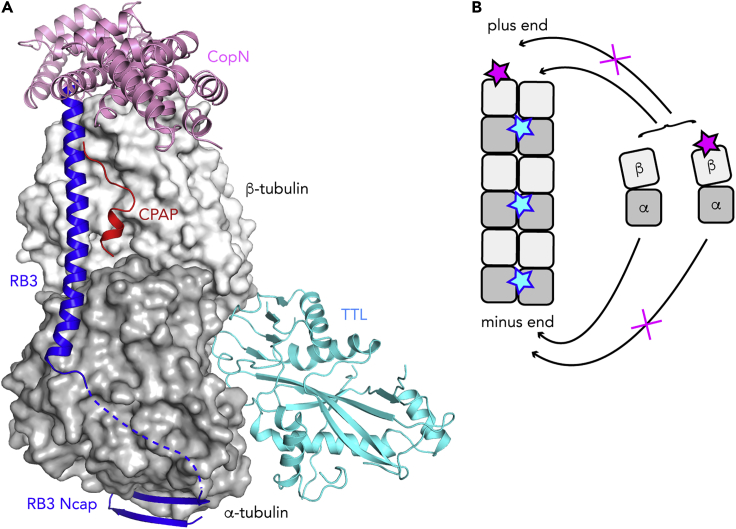
The possibility to crystallize tubulin, using either stabilizing proteins or assembly-blocked mutants, also opens up the opportunity to crystallize tubulin-MAP complexes and to determine their structure. The T2R complex was the first reported case of such a study, the SLD being both the crystallization helper and the tubulin-binding factor (Figure 1A) (Gigant et al., 2000). Since then, several X-ray structures of tubulin in complex with a MAP have been determined. These studies are complementary to cryo-EM of MAP-decorated microtubules. The complementarity of the two methods is the most obvious for proteins that bind either to soluble or to microtubular tubulin but not to both. Hence only one type of assembly, with tubulin in the appropriate oligomeric state, can be formed. Structural data are expected to give a rationale for this discrimination. For instance, among the proteins that are specific of soluble tubulin, the Chlamydia pneumoniae CopN virulence factor targets the β subunit surface that is engaged in longitudinal contacts along a protofilament (Figure 4A) (Campanacci et al., 2019a). In doing so, it prevents these contacts from being made; hence, it inhibits microtubule assembly with a specific effect at the plus end (Figure 4B). These properties have been reproduced with DARPins selected to bind β-tubulin (Figures 1B and 1D) (Pecqueur et al., 2012) and are also shared by the PN2-3 domain of the centrosomal CPAP protein. In this last case, although the structure of CPAP constructs in complex with tubulin has been determined (Sharma et al., 2016; Zheng et al., 2016), only about 15 residues that bind to the lateral outer surface of tubulin could be traced in the electron density map and no binding to the longitudinal β-tubulin surface was seen (Figure 4A). There is an obvious reason for this: these structures have been determined from a ternary complex with a DARPin, which occupies this surface (Pecqueur et al., 2012). This apparent limit was advantageously exploited in a competition experiment for tubulin binding between CPAP constructs and the DARPin. Together with competition with other tubulin ligands of known binding site, either drugs or an SLD Ncap peptide, and with inhibition of tubulin nucleotide exchange experiments, these results establish that the PN2-3 domain caps the longitudinal β-tubulin surface (Cormier et al., 2009; Sharma et al., 2016; Zheng et al., 2016). Relatedly, CPAP tracks and caps the microtubule plus end and inhibits its elongation. In addition, it increases the frequency of rescues (the transition from shrinkage to growth) and decreases that of catastrophes, enhancing microtubule stability (Sharma et al., 2016).
Figure 4.
MAPs Specific of Soluble Tubulin Prevent Microtubule Assembly
(A) Composite figure of the structure of tubulin in complex with MAPs specific of soluble tubulin. The cellular proteins RB3 and CPAP and the bacterial protein CopN would overlap on tubulin, whereas TTL binds at a distinct site. Note that the SLD of RB3 binds two tubulin molecules (see Figure 1A), only one being shown here.
(B) Schematic drawing illustrating the specific effect of CopN (pink star) as an inhibitor of the microtubule plus end elongation. At that end, both free and CopN-associated tubulin molecules can elongate free protofilaments (right protofilament). Once a CopN molecule is incorporated (left protofilament), it blocks the binding of additional tubulins and impairs microtubule elongation through a capping mechanism. In contrast, at the minus end, the tubulin-CopN complex cannot be incorporated and elongation will occur if enough free tubulin is available. At that end, inhibition will be observed at high CopN concentrations through a tubulin sequestration mechanism. The steric conflict that prevents TTL from binding to the microtubule (cyan stars) is also illustrated. The microtubule is schematized by two protofilaments of three tubulins.
The SLDs, through their N-terminal β-hairpin (Ncap; Figure 4A), also inhibit tubulin longitudinal contacts, in this case by capping the longitudinal surface of the α subunit (Ravelli et al., 2004), as do α-tubulin-specific αReps (Figures 1B and 1D) (Campanacci et al., 2019b). In addition, the SLD C-terminal helix stabilizes the two tubulin molecules of the complex in a curved conformation, not compatible with their incorporation in the microtubule core. Accordingly, an SLD construct limited to this C-terminal helix still efficiently inhibits microtubule assembly (Redeker et al., 2000; Steinmetz et al., 2000) but, different from a full-length SLD, remains compatible with the formation of curved assemblies (Steinmetz et al., 2000). In the case of tubulin tyrosine ligase (TTL), which adds a C-terminal tyrosine residue back to α-tubulin, modeling indicates that binding to microtubules would in particular lead to steric hindrance with a neighboring protofilament (Figure 4B) (Prota et al., 2013b), in agreement with the observation that TTL inhibits microtubule assembly in vitro (Szyk et al., 2011). More importantly, these results explain the selective modification of unassembled tubulin. Because the tubulin tyrosine carboxypeptidases are more active on microtubules (Aillaud et al., 2017; Nieuwenhuis et al., 2017), newly assembled microtubules are enriched in tyrosinated tubulin, whereas detyrosinated tubulin is a marker of long-persistent microtubules.
The Mechanism of MAPs that Bind to Microtubules, as Deduced from Their Complexes with Tubulin
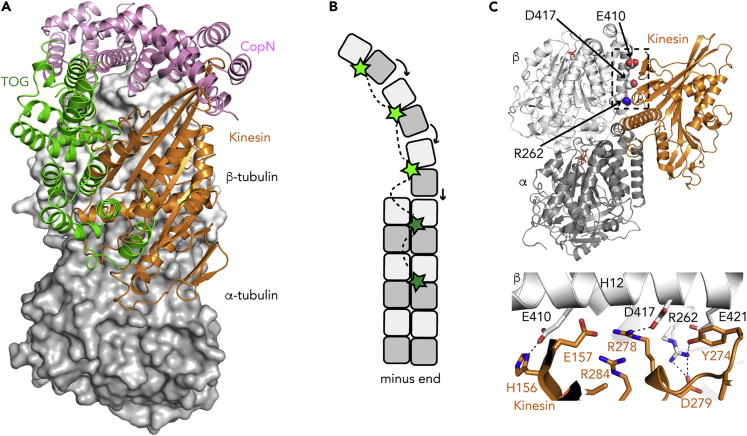
The MAPs described in the previous section bind to tubulin in a way that prevents microtubule assembly. The case of TOG domain-containing microtubule polymerases is more complex, as these proteins autonomously track the microtubule plus end (Brouhard et al., 2008), a feature potentiated by EB proteins (Li et al., 2012; Matsuo et al., 2016). TOG proteins regulate microtubule dynamics, e.g., enhancing assembly or promoting rescue. They often comprise arrays of TOG domains. For instance, XMAP215 family members are composed of five TOG domains, whereas the yeast homolog Stu2p is a homodimer containing two TOG domains per monomer. Structures of tubulin in complex with yeast TOG proteins have been determined. The constructs used contained either one TOG domain, forming a 1:1 complex with tubulin (Ayaz et al., 2012, Ayaz et al., 2014), or two TOG domains, leading to a 2:1 tubulin:construct assembly (Nithianantham et al., 2018). In all these structures, tubulin is curved, with the TOG domain interacting with both tubulin subunits (Figure 5A). Interestingly, a protein construct with a single TOG domain has been shown to inhibit microtubule assembly (Ayaz et al., 2012). The presence of several TOG domains, each one binding a tubulin heterodimer to form an assembly in which tubulin longitudinal contacts are favored (Nithianantham et al., 2018), is required for the polymerase activity. These results agree with a model in which TOG1 and TOG2 (and possibly TOG3) of XMAP215 capture soluble (curved) tubulin molecules, whereas TOG4 and TOG5, together with the C-terminal region, anchor the TOG protein to the microtubule plus end and might favor contacts between protofilaments, hence enhancing microtubule assembly (Figure 5B) (Byrnes and Slep, 2017).
Figure 5.
The Mechanism of MAPs that Bind to the Microtubule, as Deduced from Their Complexes with Tubulin
(A) Composite figure of the structure of tubulin bound to a TOG domain and to a kinesin motor domain. CopN is shown as a reference.
(B) A model for the microtubule polymerase activity of the TOG domain-containing XMAP215 protein. TOGs 1–3 (bright green) capture soluble tubulin and favor longitudinal contacts (arrows), whereas TOG4 and 5 (dark green) and the C-terminal domain (not shown) anchor XMAP215 at the microtubule plus end, possibly enhancing inter-protofilament interactions.
(C) Structural basis for the effect of disease-related tubulin mutations on the interaction with kinesins. (Top) Overview of the structure of tubulin bound to kinesin-1. β-Tubulin residues whose mutation is found in CFEOM3 are highlighted. (Bottom) Close-up of the part framed in the top panel. Selected interactions, involving tubulin residues mutated in CFEOM3, are shown as dashed lines.
At first glance, the usefulness of X-ray crystallography to study MAP-tubulin complexes may seem less obvious in the case of kinesins. The typical function of these motor proteins is to translocate along microtubules to carry loads. The microtubule is thus their natural binding partner and, accordingly, cryo-EM is a method of choice to study kinesin-decorated microtubules structurally. There is, however, a class of kinesins for which the structural study of complexes with soluble tubulin is fully justified, that of kinesin-13s. Indeed, in addition to their function in intracellular trafficking, some kinesins regulate microtubule dynamics (Drummond, 2011). Among these kinesins, kinesin-13s are particular because they are not motile but target microtubule ends to induce disassembly. Therefore, at some stage of their cycle, they are associated with detached tubulin (Friel and Howard, 2011). X-ray structures indicate that kinesin-13s stabilize curved conformations of tubulin and of tubulin assemblies (Trofimova et al., 2018; Wang et al., 2017) and that this feature is an important contribution to the mechanism of microtubule disassembly. Similar results have been obtained using a cryo-EM approach (Benoit et al., 2018).
Given that the resolution of the kinesin partner in cryo-EM maps is usually lower than that of the microtubule (Kellogg et al., 2017; Locke et al., 2017), X-ray crystallography of tubulin-kinesin complexes may also bring valuable information in the case of motile kinesins. This approach is further warranted because biochemical data indicate that motile kinesins can also bind to soluble tubulin (Alonso et al., 2007). So far, two X-ray structures of the motor domain of a motile kinesin have been determined in complex with tubulin, those of the human kinesin-1 KIF5B in its two high-affinity states for microtubules, either without nucleotide or in an ATP-like form (Cao et al., 2014; Gigant et al., 2013). The kinesin-binding site on tubulin overlaps with that of TOG proteins (Figure 5A). These structural data support the ATP hydrolysis mechanism that has been proposed from the high-resolution structure of the isolated kinesin-5 Eg5 captured in an ATP-like state (Parke et al., 2010). Concomitantly to a cryo-EM study (Shang et al., 2014), they have also enabled the identification of three subdomains within the kinesin motor domain. These subdomains reorganize along the kinesin cycle, linking the nucleotide state to movement. This model has become a standard way of interpreting the structural mechanism of kinesins, beyond those of class 1 (Atherton et al., 2017b; Locke et al., 2017).
The structural study of kinesin bound to soluble tubulin is also useful to understand the effect of mutations at the molecular level. For instance, mutations in the β-tubulin TUBB3 isotype cause a disease termed congenital fibrosis of the extraocular muscles type 3 (CFEOM3). Among these, the R262C/H, E410K, and D417H/N substitutions were found to impair the interaction of kinesins with microtubules (Tischfield et al., 2010). These in vivo data have been confirmed in vitro using purified tubulin mutants (Minoura et al., 2016; Uchimura et al., 2006). These β-tubulin residues, mutated in the disease, do indeed interact with the motor domain of kinesins, as deduced from the crystal structure of kinesin-1 bound to tubulin (Cao et al., 2014; Gigant et al., 2013). β-Tubulin residues Arg262 and Asp417 make salt bridges with the kinesin residues Asp279 and Arg278, respectively, whereas β-Glu410 interacts with His156 (Figure 5C). Therefore, structural data give a rationale for the effect of these TUBB3 mutations on kinesin motility.
MAPs Whose Structure Has Been Determined as a Complex with Microtubules but Not with Tubulin
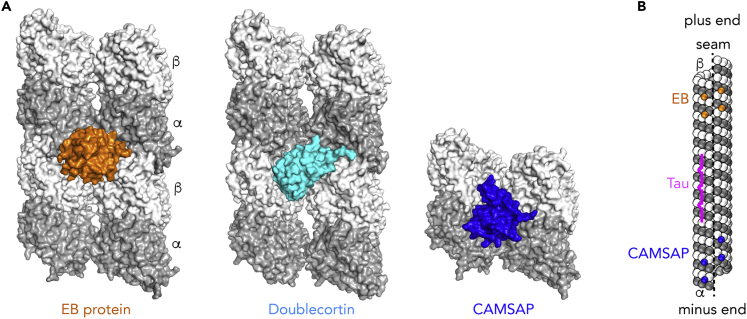
Finally, several families of MAPs have been shown to be better candidates for cryo-EM than for X-ray crystallography studies (reviewed in Manka and Moores, 2018a; Nogales and Zhang, 2016). This is in particular the case of MAPs that bind at the junction between protofilaments (Figure 6A), a site that is not fully formed in soluble tubulin complexes whose structure has been determined. The binding of these MAPs is usually not uniform along the length of the microtubule. For instance, EB proteins bind to the ends of growing microtubules (Duellberg et al., 2013; Mimori-Kiyosue et al., 2000), whereas CAMSAP proteins specifically target the minus end (Goodwin and Vale, 2010; Jiang et al., 2014). In the case of doublecortin, binding to growing microtubule ends (Bechstedt and Brouhard, 2012) and to the microtubule lattice has been reported (Ettinger et al., 2016). It should, however, be noted that the binding preference is not absolute, allowing a uniform decoration of the entire microtubule length at higher MAP concentrations, a prerequisite for determining the cryo-EM structure of MAP-decorated microtubules.
Figure 6.
Examples of MAPs Whose Structure Has Been Determined as a Complex with Microtubules but Not with Tubulin
(A) Proteins that bind at the inter-protofilament interface. EB proteins (Maurer et al., 2012; Zhang et al., 2015) and doublecortin (Fourniol et al., 2010) bind at the corner of four tubulin heterodimers, whereas the CKK domain of CAMSAP proteins (Atherton et al., 2017a) targets two adjacent tubulin molecules.
(B) Schematic drawing of a 13-protofilament microtubule. EBs bind preferentially to the growing ends of microtubules (shown here at the plus end), whereas CAMSAP targets and stabilizes the minus end. The binding mode of these proteins implies that they do not target the microtubule seam (dashed line), where interactions between adjacent protofilaments are heterotypic (α-tubulin interacting with β-tubulin and vice versa). Because they bind at the corner of four tubulin subunits, these proteins can modulate both longitudinal and lateral interactions. In contrast, the microtubule-binding repeats of Tau (magenta) bind at the crest of one protofilament, with a major effect on longitudinal contact stabilization.
These proteins may stabilize microtubules, as in the case of doublecortin, a protein that also favors nucleation of 13-protofilament microtubules but does not promote elongation (Moores et al., 2006). CAMSAP proteins also stabilize microtubules, either on their own (Jiang et al., 2014) or by preventing disassembly catalyzed by kinesin-13 kinesins (Goodwin and Vale, 2010). The binding at the junction between protofilaments, hence cross-linking them, seems an ideal place for stabilizing microtubules. Another obvious option to fulfill this function is to strengthen longitudinal contacts, as does the Tau protein (Figure 6B) (Kellogg et al., 2018). Interestingly, similar to doublecortin, EB proteins bind at the corner of four tubulin heterodimers, but they interfere with microtubule dynamics in a different manner. Although a mild increase of microtubule growth has been measured in in vitro experiments, the most obvious effect of EBs is to enhance catastrophe frequency, making microtubules more dynamic (Duellberg et al., 2013). Structural and biochemical data suggest that EBs favor GTP hydrolysis and stabilize a transient GTP-hydrolysis intermediate state of assembled tubulin (Zhang et al., 2015). In the context of the cell, the effect of EBs is even more complex as they recruit to the ends of growing microtubules a range of proteins that themselves regulate microtubule dynamics (Akhmanova and Steinmetz, 2015). Clearly, the structural study of proteins interacting mostly with microtubules is at its beginning and, as more cases will be elucidated, the corresponding mechanisms will be clarified.
Perspectives
X-ray crystallography and cryo-EM, by, respectively, defining structures of soluble tubulin and of the microtubule, provide the proper framework to study microtubule dynamics and its regulation by MAPs and by small molecule compounds. Despite continuous advances, additional data are still needed. Some of them might soon become available. For instance, the recent identification of crystallization chaperones targeting α-tubulin represents an opportunity to study tubulin in complex with MAPs that bind to the β subunit. This approach has already allowed the characterization of the structural mechanism of the C. pneumoniae CopN protein (Campanacci et al., 2019a). In addition, methods to overexpress tubulin in insect cells have been recently developed (Minoura et al., 2013; Vemu et al., 2016). These methods make the study of the effects of mutations possible (Minoura et al., 2016; Roostalu et al., 2020; Ti et al., 2016) and pave the way for the validation of structural models through a mutational approach. Clearly, the study of microtubule dynamics and of its regulation continues to benefit from the development of such tools.
An example where additional structural investigation is needed concerns the consequences of GTP hydrolysis that accompanies microtubule assembly. GTP hydrolysis converts the microtubule to a metastable structure and is therefore at the basis of its dynamic instability (Desai and Mitchison, 1997). Because of this hydrolysis, analogs of GTP and of GTP hydrolysis transition states are commonly used in the structural study of microtubules as surrogates of GTP and of its hydrolysis products. But it is not always clear which state they emulate and to what extent they do so. Another level of complexity comes from the fact that most structures of the microtubule have been determined with a marker (e.g., a kinesin) used to distinguish between the α and β subunits during image analysis, a protein which itself may influence microtubule structure (Peet et al., 2018). As a consequence, the effect of GTP hydrolysis remains a matter of debate (Alushin et al., 2014; Estévez-Gallego et al., 2020; Manka and Moores, 2018b). The recent characterization of a tubulin mutant unable to hydrolyze GTP (Roostalu et al., 2020) and the determination of the structure of undecorated microtubules (Zhang et al., 2018) open the possibility to establish the structure of bona fide GTP-microtubules, although these would be constituted of a mutated tubulin.
At least two additional outstanding challenges remain for the understanding of the structural mechanism of microtubule function. The first one concerns tubulin's PTMs (Janke and Magiera, 2020; Roll-Mecak, 2020). Many tubulin-modifying enzymes have been identified, and their structure, either isolated or bound to inhibitors or to tubulin peptides, has been determined. But the structure of a PTM “writer” or “eraser” in complex with tubulin has been determined only in the case of TTL (Prota et al., 2013b). Such data are, however, required to establish how these enzymes modify tubulin and, if applicable, their specificity for soluble or microtubular tubulin. The structural basis for the recognition of modified tubulin by PTM “readers” is also poorly understood. One difficulty is that many PTMs concern the C-terminal tail of both tubulin subunits and, in particular, their polyglutamylation and polyglycylation. These tubulin tails are flexible and probably remain so even in complex with a PTM writer, eraser, or reader. Although NMR spectroscopy may bring some information (Wall et al., 2016), classical structural biology at high resolution may not be the best method for this objective.
A second pending challenge is the characterization of transient or sparsely populated tubulin structures. These include “defects” in the microtubule lattice, which may be involved in rescue events (Aumeier et al., 2016; de Forges et al., 2016), and also tubulin in a curved-to-straight intermediate conformation, be it before microtubule incorporation or at microtubule ends. These last structures are the binding site of various MAPs targeting one or the other end of the microtubule (Akhmanova and Steinmetz, 2019). Although the binding mode of some of them has been determined from decorated microtubules, as in the case of CAMSAP (Atherton et al., 2017a) and EBs (Maurer et al., 2012), the end-binding preference remains poorly understood in most cases. For instance, it is not clear how TOG domains 4 and 5 of XMAP215 target the lattice at the microtubule plus end rather than free tubulin. More fundamentally, the structural pathway of tubulin during assembly, from curved tubulin in solution to its incorporation at microtubule ends and then in the microtubule core, has not been established. Such data would be helpful to further characterize GTP hydrolysis that accompanies microtubule assembly. They are also needed for a better understanding of the mechanism of microtubule nucleation, either templated or from a pure tubulin solution. One possibility to meet this challenge is based on cryo-electron tomography, which does not require the averaging needed in cryo-EM but remains for the time being a low-resolution method (Guesdon et al., 2016; McIntosh et al., 2018). Another possibility would be to design scaffolds for tubulin complexes that mimic transient assemblies. Artificial tubulin-binding proteins might be useful in this case. For instance, tandem repeats of a DARPin (Pecqueur et al., 2012) or of an αRep (Campanacci et al., 2019b), in which two copies of the protein are covalently linked, can bind two tubulin heterodimers and inhibit microtubule assembly more efficiently than the monomeric parent constructs. It remains to be determined whether these assemblies will incorporate tubulin in an intermediate conformation or whether they just represent starting points for the design of larger, more faithful intermediate-like structures. Whatever the case, such assemblies would be well suited to X-ray crystallography or cryo-EM studies that would further enhance our understanding of microtubule assembly.
Acknowledgments
This work has been supported by the Fondation ARC pour la Recherche sur le Cancer and by CNRS.
Author Contributions
M.K. and B.G. wrote the manuscript. All authors reviewed the manuscript.
References
- Ahmad S., Pecqueur L., Dreier B., Hamdane D., Aumont-Nicaise M., Plückthun A., Knossow M., Gigant B. Destabilizing an interacting motif strengthens the association of a designed ankyrin repeat protein with tubulin. Sci. Rep. 2016;6:28922. doi: 10.1038/srep28922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aillaud C., Bosc C., Peris L., Bosson A., Heemeryck P., Van Dijk J., Le Friec J., Boulan B., Vossier F., Sanman L.E. Vasohibins/SVBP are tubulin carboxypeptidases (TCPs) that regulate neuron differentiation. Science. 2017;358:1448–1453. doi: 10.1126/science.aao4165. [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Steinmetz M.O. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015;16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Steinmetz M.O. Microtubule minus-end regulation at a glance. J. Cell Sci. 2019;132:jcs227850. doi: 10.1242/jcs.227850. [DOI] [PubMed] [Google Scholar]
- Alonso M.C., Drummond D.R., Kain S., Hoeng J., Amos L., Cross R.A. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin G.M., Lander G.C., Kellogg E.H., Zhang R., Baker D., Nogales E. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton J., Jiang K., Stangier M.M., Luo Y., Hua S., Houben K., van Hooff J.J.E., Joseph A.P., Scarabelli G., Grant B.J. A structural model for microtubule minus-end recognition and protection by CAMSAP proteins. Nat. Struct. Mol. Biol. 2017;24:931–943. doi: 10.1038/nsmb.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton J., Yu I.M., Cook A., Muretta J.M., Joseph A., Major J., Sourigues Y., Clause J., Topf M., Rosenfeld S.S. The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry. Elife. 2017;6:e27793. doi: 10.7554/eLife.27793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumeier C., Schaedel L., Gaillard J., John K., Blanchoin L., Thery M. Self-repair promotes microtubule rescue. Nat. Cell Biol. 2016;18:1054–1064. doi: 10.1038/ncb3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P., Munyoki S., Geyer E.A., Piedra F.A., Vu E.S., Bromberg R., Otwinowski Z., Grishin N.V., Brautigam C.A., Rice L.M. A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. Elife. 2014;3:e03069. doi: 10.7554/eLife.03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P., Ye X., Huddleston P., Brautigam C.A., Rice L.M. A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier P., Dorléans A., Devred F., Sanz L., Allegro D., Alfonso C., Knossow M., Peyrot V., Andreu J.M. Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers. J. Biol. Chem. 2010;285:31672–31681. doi: 10.1074/jbc.M110.141929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechstedt S., Brouhard G.J. Doublecortin recognizes the 13-protofilament microtubule cooperatively and tracks microtubule ends. Dev. Cell. 2012;23:181–192. doi: 10.1016/j.devcel.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L.D., Mitchison T.J. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Benoit M., Asenjo A.B., Sosa H. Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s. Nat. Commun. 2018;9:1662. doi: 10.1038/s41467-018-04044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisy G.G., Taylor E.W. The mechanism of action of colchicine. Binding of colchincine-3H to cellular protein. J. Cell Biol. 1967;34:525–533. doi: 10.1083/jcb.34.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., Hyman A.A. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes A.E., Slep K.C. TOG-tubulin binding specificity promotes microtubule dynamics and mitotic spindle formation. J. Cell Biol. 2017;216:1641–1657. doi: 10.1083/jcb.201610090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanacci V., Urvoas A., Cantos-Fernandes S., Aumont-Nicaise M., Arteni A.A., Velours C., Valerio-Lepiniec M., Dreier B., Plückthun A., Pilon A. Insight into microtubule nucleation from tubulin-capping proteins. Proc. Natl. Acad. Sci. U S A. 2019;116:9859–9864. doi: 10.1073/pnas.1813559116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanacci V., Urvoas A., Consolati T., Cantos-Fernandes S., Aumont-Nicaise M., Valerio-Lepiniec M., Surrey T., Minard P., Gigant B. Selection and characterization of artificial proteins targeting the tubulin α subunit. Structure. 2019;27:497–506 e494. doi: 10.1016/j.str.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Wang W., Jiang Q., Wang C., Knossow M., Gigant B. The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement. Nat. Commun. 2014;5:5364. doi: 10.1038/ncomms6364. [DOI] [PubMed] [Google Scholar]
- Carlier M.F. Role of nucleotide hydrolysis in the dynamics of actin filaments and microtubules. Int. Rev. Cytol. 1989;115:139–170. doi: 10.1016/s0074-7696(08)60629-4. [DOI] [PubMed] [Google Scholar]
- Chothia C., Lesk A.M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D., Fuller S.D., Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J. Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier A., Clément M.J., Knossow M., Lachkar S., Savarin P., Toma F., Sobel A., Gigant B., Curmi P.A. The PN2-3 domain of centrosomal P4.1-associated protein implements a novel mechanism for tubulin sequestration. J. Biol. Chem. 2009;284:6909–6917. doi: 10.1074/jbc.M808249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curmi P.A., Andersen S.S., Lachkar S., Gavet O., Karsenti E., Knossow M., Sobel A. The stathmin/tubulin interaction in vitro. J. Biol. Chem. 1997;272:25029–25036. doi: 10.1074/jbc.272.40.25029. [DOI] [PubMed] [Google Scholar]
- de Forges H., Pilon A., Cantaloube I., Pallandre A., Haghiri-Gosnet A.M., Perez F., Poüs C. Localized mechanical stress promotes microtubule rescue. Curr. Biol. 2016;26:3399–3406. doi: 10.1016/j.cub.2016.10.048. [DOI] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Dorléans A., Gigant B., Ravelli R.B., Mailliet P., Mikol V., Knossow M. Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. Proc. Natl. Acad. Sci. U S A. 2009;106:13775–13779. doi: 10.1073/pnas.0904223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing K.H., Nogales E. Tubulin and microtubule structure. Curr. Opin. Cell Biol. 1998;10:16–22. doi: 10.1016/s0955-0674(98)80082-3. [DOI] [PubMed] [Google Scholar]
- Drummond D.R. Regulation of microtubule dynamics by kinesins. Semin. Cell Dev. Biol. 2011;22:927–934. doi: 10.1016/j.semcdb.2011.09.021. [DOI] [PubMed] [Google Scholar]
- Duellberg C., Fourniol F.J., Maurer S.P., Roostalu J., Surrey T. End-binding proteins and Ase1/PRC1 define local functionality of structurally distinct parts of the microtubule cytoskeleton. Trends Cell Biol. 2013;23:54–63. doi: 10.1016/j.tcb.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Estévez-Gallego J., Josa-Prado F., Ku S., Buey R.M., Balaguer F.A., Prota A.E., Lucena-Agell D., Kamma-Lorger C., Yagi T., Iwamoto H. Structural model for differential cap maturation at growing microtubule ends. Elife. 2020;9:e50155. doi: 10.7554/eLife.50155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger A., van Haren J., Ribeiro S.A., Wittmann T. Doublecortin is excluded from growing microtubule ends and recognizes the GDP-microtubule lattice. Curr. Biol. 2016;26:1549–1555. doi: 10.1016/j.cub.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourniol F.J., Sindelar C.V., Amigues B., Clare D.K., Thomas G., Perderiset M., Francis F., Houdusse A., Moores C.A. Template-free 13-protofilament microtubule-MAP assembly visualized at 8 Å resolution. J. Cell Biol. 2010;191:463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friel C.T., Howard J. The kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. EMBO J. 2011;30:3928–3939. doi: 10.1038/emboj.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geahlen R.L., Haley B.E. Interactions of a photoaffinity analog of GTP with the proteins of microtubules. Proc. Natl. Acad. Sci. U S A. 1977;74:4375–4377. doi: 10.1073/pnas.74.10.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B., Curmi P.A., Martin-Barbey C., Charbaut E., Lachkar S., Lebeau L., Siavoshian S., Sobel A., Knossow M. The 4 Å X-ray structure of a tubulin:stathmin-like domain complex. Cell. 2000;102:809–816. doi: 10.1016/s0092-8674(00)00069-6. [DOI] [PubMed] [Google Scholar]
- Gigant B., Landrieu I., Fauquant C., Barbier P., Huvent I., Wieruszeski J.M., Knossow M., Lippens G. Mechanism of Tau-promoted microtubule assembly as probed by NMR spectroscopy. J. Am. Chem. Soc. 2014;136:12615–12623. doi: 10.1021/ja504864m. [DOI] [PubMed] [Google Scholar]
- Gigant B., Wang W., Dreier B., Jiang Q., Pecqueur L., Plückthun A., Wang C., Knossow M. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat. Struct. Mol. Biol. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- Goodwin S.S., Vale R.D. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143:263–274. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guesdon A., Bazile F., Buey R.M., Mohan R., Monier S., Garcia R.R., Angevin M., Heichette C., Wieneke R., Tampe R. EB1 interacts with outwardly curved and straight regions of the microtubule lattice. Nat. Cell Biol. 2016;18:1102–1108. doi: 10.1038/ncb3412. [DOI] [PubMed] [Google Scholar]
- Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.F., Yao T.P. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Isrie M., Breuss M., Tian G., Hansen A.H., Cristofoli F., Morandell J., Kupchinsky Z.A., Sifrim A., Rodriguez-Rodriguez C.M., Dapena E.P. Mutations in either TUBB or MAPRE2 cause circumferential skin creases kunze type. Am. J. Hum. Genet. 2015;97:790–800. doi: 10.1016/j.ajhg.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C. The tubulin code: molecular components, readout mechanisms, and functions. J. Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 2020;21:307–326. doi: 10.1038/s41580-020-0214-3. [DOI] [PubMed] [Google Scholar]
- Jiang K., Hua S., Mohan R., Grigoriev I., Yau K.W., Liu Q., Katrukha E.A., Altelaar A.F., Heck A.J., Hoogenraad C.C. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell. 2014;28:295–309. doi: 10.1016/j.devcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Johnson V., Ayaz P., Huddleston P., Rice L.M. Design, overexpression, and purification of polymerization-blocked yeast αβ-tubulin mutants. Biochemistry. 2011;50:8636–8644. doi: 10.1021/bi2005174. [DOI] [PubMed] [Google Scholar]
- Jourdain L., Curmi P., Sobel A., Pantaloni D., Carlier M.F. Stathmin: a tubulin-sequestering protein which forms a ternary T2S complex with two tubulin molecules. Biochemistry. 1997;36:10817–10821. doi: 10.1021/bi971491b. [DOI] [PubMed] [Google Scholar]
- Kellogg E.H., Hejab N.M.A., Howes S., Northcote P., Miller J.H., Diaz J.F., Downing K.H., Nogales E. Insights into the distinct mechanisms of action of taxane and non-taxane microtubule stabilizers from cryo-EM structures. J. Mol. Biol. 2017;429:633–646. doi: 10.1016/j.jmb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E.H., Hejab N.M.A., Poepsel S., Downing K.H., DiMaio F., Nogales E. Near-atomic model of microtubule-tau interactions. Science. 2018;360:1242–1246. doi: 10.1126/science.aat1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault S.W., Rosenbaum J.L. Chlamydomonas α-tubulin is posttranslationally modified by acetylation on the ε-amino group of a lysine. Biochemistry. 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- La Sala G., Olieric N., Sharma A., Viti F., de Asis Balaguer Perez F., Huang L., Tonra J.R., Lloyd G.K., Decherchi S., Díaz J.F. Structure, thermodynamics, and kinetics of plinabulin binding to two tubulin isotypes. ChemBioChem. 2019;5:2969–2986. [Google Scholar]
- Li W., Moriwaki T., Tani T., Watanabe T., Kaibuchi K., Goshima G. Reconstitution of dynamic microtubules with Drosophila XMAP215, EB1, and Sentin. J. Cell Biol. 2012;199:849–862. doi: 10.1083/jcb.201206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens G., Gigant B. Elucidating Tau function and dysfunction in the era of cryo-EM. J. Biol. Chem. 2019;294:9316–9325. doi: 10.1074/jbc.REV119.008031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens G., Landrieu I., Smet C., Huvent I., Gandhi N.S., Gigant B., Despres C., Qi H., Lopez J. NMR meets Tau: insights into its function and pathology. Biomolecules. 2016;6:28. doi: 10.3390/biom6020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M., Krauhs E., Ponstingl H. Tubulin sequence conservation. Biosystems. 1981;14:239–246. doi: 10.1016/0303-2647(81)90031-9. [DOI] [PubMed] [Google Scholar]
- Locke J., Joseph A.P., Peña A., Möckel M.M., Mayer T.U., Topf M., Moores C.A. Structural basis of human kinesin-8 function and inhibition. Proc. Natl. Acad. Sci. U S A. 2017;114:E9539–E9548. doi: 10.1073/pnas.1712169114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manka S.W., Moores C.A. Microtubule structure by cryo-EM: snapshots of dynamic instability. Essays Biochem. 2018;62:737–751. doi: 10.1042/EBC20180031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manka S.W., Moores C.A. The role of tubulin-tubulin lattice contacts in the mechanism of microtubule dynamic instability. Nat. Struct. Mol. Biol. 2018;25:607–615. doi: 10.1038/s41594-018-0087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y., Maurer S.P., Yukawa M., Zakian S., Singleton M.R., Surrey T., Toda T. An unconventional interaction between Dis1/TOG and Mal3/EB1 in fission yeast promotes the fidelity of chromosome segregation. J. Cell Sci. 2016;129:4592–4606. doi: 10.1242/jcs.197533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer S.P., Fourniol F.J., Bohner G., Moores C.A., Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., O'Toole E., Morgan G., Austin J., Ulyanov E., Ataullakhanov F., Gudimchuk N. Microtubules grow by the addition of bent guanosine triphosphate tubulin to the tips of curved protofilaments. J. Cell Biol. 2018;217:2691–2708. doi: 10.1083/jcb.201802138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melková K., Zapletal V., Narasimhan S., Jansen S., Hritz J., Škrabana R., Zweckstetter M., Ringkjøbing Jensen M., Blackledge M., Žídek L. Structure and functions of microtubule associated proteins Tau and MAP2c: similarities and differences. Biomolecules. 2019;9:105. doi: 10.3390/biom9030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot I., Pecqueur L., Dorléans A., Karuppasamy M., Ravelli R.B., Dreier B., Plückthun A., Knossow M., Gigant B. Design and characterization of modular scaffolds for tubulin assembly. J. Biol. Chem. 2012;287:31085–31094. doi: 10.1074/jbc.M112.383869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Shiina N., Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Minoura I., Hachikubo Y., Yamakita Y., Takazaki H., Ayukawa R., Uchimura S., Muto E. Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett. 2013;587:3450–3455. doi: 10.1016/j.febslet.2013.08.032. [DOI] [PubMed] [Google Scholar]
- Minoura I., Takazaki H., Ayukawa R., Saruta C., Hachikubo Y., Uchimura S., Hida T., Kamiguchi H., Shimogori T., Muto E. Reversal of axonal growth defects in an extraocular fibrosis model by engineering the kinesin-microtubule interface. Nat. Commun. 2016;7:10058. doi: 10.1038/ncomms10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Moores C.A., Perderiset M., Kappeler C., Kain S., Drummond D., Perkins S.J., Chelly J., Cross R., Houdusse A., Francis F. Distinct roles of doublecortin modulating the microtubule cytoskeleton. EMBO J. 2006;25:4448–4457. doi: 10.1038/sj.emboj.7601335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrotek A., Knossow M., Gigant B. The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J. Mol. Biol. 2011;412:35–42. doi: 10.1016/j.jmb.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis J., Adamopoulos A., Bleijerveld O.B., Mazouzi A., Stickel E., Celie P., Altelaar M., Knipscheer P., Perrakis A., Blomen V.A. Vasohibins encode tubulin detyrosinating activity. Science. 2017;358:1453–1456. doi: 10.1126/science.aao5676. [DOI] [PubMed] [Google Scholar]
- Nithianantham S., Cook B.D., Beans M., Guo F., Chang F., Al-Bassam J. Structural basis of tubulin recruitment and assembly by microtubule polymerases with tumor overexpressed gene (TOG) domain arrays. Elife. 2018;7:e38922. doi: 10.7554/eLife.38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Kellogg E.H. Challenges and opportunities in the high-resolution cryo-EM visualization of microtubules and their binding partners. Curr. Opin. Struct. Biol. 2017;46:65–70. doi: 10.1016/j.sbi.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Whittaker M., Milligan R.A., Downing K.H. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Nogales E., Wolf S.G., Downing K.H. Structure of the αβ tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Nogales E., Zhang R. Visualizing microtubule structural transitions and interactions with associated proteins. Curr. Opin. Struct. Biol. 2016;37:90–96. doi: 10.1016/j.sbi.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach R., Howard J. The dynamic and structural properties of axonemal tubulins support the high length stability of cilia. Nat. Commun. 2019;10:1838. doi: 10.1038/s41467-019-09779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke C.L., Wojcik E.J., Kim S., Worthylake D.K. ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism. J. Biol. Chem. 2010;285:5859–5867. doi: 10.1074/jbc.M109.071233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecqueur L., Duellberg C., Dreier B., Jiang Q., Wang C., Plückthun A., Surrey T., Gigant B., Knossow M. A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc. Natl. Acad. Sci. U S A. 2012;109:12011–12016. doi: 10.1073/pnas.1204129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet D.R., Burroughs N.J., Cross R.A. Kinesin expands and stabilizes the GDP-microtubule lattice. Nat. Nanotechnol. 2018;13:386–391. doi: 10.1038/s41565-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plückthun A. Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 2015;55:489–511. doi: 10.1146/annurev-pharmtox-010611-134654. [DOI] [PubMed] [Google Scholar]
- Prota A.E., Bargsten K., Zurwerra D., Field J.J., Diaz J.F., Altmann K.H., Steinmetz M.O. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science. 2013;339:587–590. doi: 10.1126/science.1230582. [DOI] [PubMed] [Google Scholar]
- Prota A.E., Magiera M.M., Kuijpers M., Bargsten K., Frey D., Wieser M., Jaussi R., Hoogenraad C.C., Kammerer R.A., Janke C. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J. Cell Biol. 2013;200:259–270. doi: 10.1083/jcb.201211017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli R.B., Gigant B., Curmi P.A., Jourdain I., Lachkar S., Sobel A., Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Redeker V., Lachkar S., Siavoshian S., Charbaut E., Rossier J., Sobel A., Curmi P.A. Probing the native structure of stathmin and its interaction domains with tubulin. Combined use of limited proteolysis, size exclusion chromatography, and mass spectrometry. J. Biol. Chem. 2000;275:6841–6849. doi: 10.1074/jbc.275.10.6841. [DOI] [PubMed] [Google Scholar]
- Rice L.M., Montabana E.A., Agard D.A. The lattice as allosteric effector: structural studies of αβ- and γ-tubulin clarify the role of GTP in microtubule assembly. Proc. Natl. Acad. Sci. U S A. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A. The tubulin code in microtubule dynamics and information encoding. Dev. Cell. 2020;54:7–20. doi: 10.1016/j.devcel.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J., Thomas C., Cade N.I., Kunzelmann S., Taylor I.A., Surrey T. The speed of GTP hydrolysis determines GTP cap size and controls microtubule stability. Elife. 2020;9:e51992. doi: 10.7554/eLife.51992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Z., Zhou K., Xu C., Csencsits R., Cochran J.C., Sindelar C.V. High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation. Elife. 2014;3:e04686. doi: 10.7554/eLife.04686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Aher A., Dynes N.J., Frey D., Katrukha E.A., Jaussi R., Grigoriev I., Croisier M., Kammerer R.A., Akhmanova A. Centriolar CPAP/SAS-4 imparts slow processive microtubule growth. Dev. Cell. 2016;37:362–376. doi: 10.1016/j.devcel.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M.O., Kammerer R.A., Jahnke W., Goldie K.N., Lustig A., van Oostrum J. Op18/stathmin caps a kinked protofilament-like tubulin tetramer. EMBO J. 2000;19:572–580. doi: 10.1093/emboj/19.4.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M.O., Prota A.E. Microtubule-targeting agents: strategies to hijack the cytoskeleton. Trends Cell Biol. 2018;28:776–792. doi: 10.1016/j.tcb.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Szyk A., Deaconescu A.M., Piszczek G., Roll-Mecak A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 2011;18:1250–1258. doi: 10.1038/nsmb.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti S.C., Pamula M.C., Howes S.C., Duellberg C., Cade N.I., Kleiner R.E., Forth S., Surrey T., Nogales E., Kapoor T.M. Mutations in human tubulin proximal to the kinesin-binding site alter dynamic instability at microtubule plus- and minus-ends. Dev. Cell. 2016;37:72–84. doi: 10.1016/j.devcel.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield M.A., Baris H.N., Wu C., Rudolph G., Van Maldergem L., He W., Chan W.M., Andrews C., Demer J.L., Robertson R.L. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofimova D., Paydar M., Zara A., Talje L., Kwok B.H., Allingham J.S. Ternary complex of Kif2A-bound tandem tubulin heterodimers represents a kinesin-13-mediated microtubule depolymerization reaction intermediate. Nat. Commun. 2018;9:2628. doi: 10.1038/s41467-018-05025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura S., Oguchi Y., Katsuki M., Usui T., Osada H., Nikawa J., Ishiwata S., Muto E. Identification of a strong binding site for kinesin on the microtubule using mutant analysis of tubulin. EMBO J. 2006;25:5932–5941. doi: 10.1038/sj.emboj.7601442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urvoas A., Guellouz A., Valerio-Lepiniec M., Graille M., Durand D., Desravines D.C., van Tilbeurgh H., Desmadril M., Minard P. Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins (αRep) based on thermostable HEAT-like repeats. J. Mol. Biol. 2010;404:307–327. doi: 10.1016/j.jmb.2010.09.048. [DOI] [PubMed] [Google Scholar]
- Valenzuela P., Quiroga M., Zaldivar J., Rutter W.J., Kirschner M.W., Cleveland D.W. Nucleotide and corresponding amino acid sequences encoded by α and β tubulin mRNAs. Nature. 1981;289:650–655. doi: 10.1038/289650a0. [DOI] [PubMed] [Google Scholar]
- Vemu A., Atherton J., Spector J.O., Szyk A., Moores C.A., Roll-Mecak A. Structure and dynamics of single-isoform recombinant neuronal human tubulin. J. Biol. Chem. 2016;291:12907–12915. doi: 10.1074/jbc.C116.731133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall K.P., Pagratis M., Armstrong G., Balsbaugh J.L., Verbeke E., Pearson C.G., Hough L.E. Molecular determinants of tubulin's C-terminal tail conformational ensemble. ACS Chem. Biol. 2016;11:2981–2990. doi: 10.1021/acschembio.6b00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.W., Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Cantos-Fernandes S., Lv Y., Kuerban H., Ahmad S., Wang C., Gigant B. Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin. Nat. Commun. 2017;8:70. doi: 10.1038/s41467-017-00091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberg R.C., Borisy G.G., Taylor E.W. The colchicine-binding protein of mammalian brain and its relation to microtubules. Biochemistry. 1968;7:4466–4479. doi: 10.1021/bi00852a043. [DOI] [PubMed] [Google Scholar]
- Wilson L., Friedkin M. The biochemical events of mitosis. II. The in vivo and in vitro binding of colchicine in grasshopper embryos and its possible relation to inhibition of mitosis. Biochemistry. 1967;6:3126–3135. doi: 10.1021/bi00862a021. [DOI] [PubMed] [Google Scholar]
- Zhang R., Alushin G.M., Brown A., Nogales E. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., LaFrance B., Nogales E. Separating the effects of nucleotide and EB binding on microtubule structure. Proc. Natl. Acad. Sci. U S A. 2018;115:E6191–E6200. doi: 10.1073/pnas.1802637115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Ramani A., Soni K., Gottardo M., Zheng S., Ming Gooi L., Li W., Feng S., Mariappan A., Wason A. Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length. Nat. Commun. 2016;7:11874. doi: 10.1038/ncomms11874. [DOI] [PMC free article] [PubMed] [Google Scholar]