Summary
Light-inducible gene expression systems represent powerful methods for studying the functional roles of dynamic gene expression. Here, we developed an optimized light-inducible Gal4/UAS gene expression system for mammalian cells. We designed photoactivatable (PA)-Gal4 transcriptional activators based on the concept of split transcription factors, in which light-dependent interactions between Cry2-CIB1 PA-protein interaction modules can reconstitute a split Gal4 DNA-binding domain and p65 transcription activation domain. We developed a set of PA-Gal4 transcriptional activators (PA-Gal4cc), which differ in terms of induced gene expression levels following pulsed or prolonged light exposure, and which have different activation/deactivation kinetics. These systems offer optogenetic tools for the precise manipulation of gene expression at fine spatiotemporal resolution in mammalian cells.
Subject Areas: Optical Imaging, Molecular Genetics, Cell Biology
Graphical Abstract
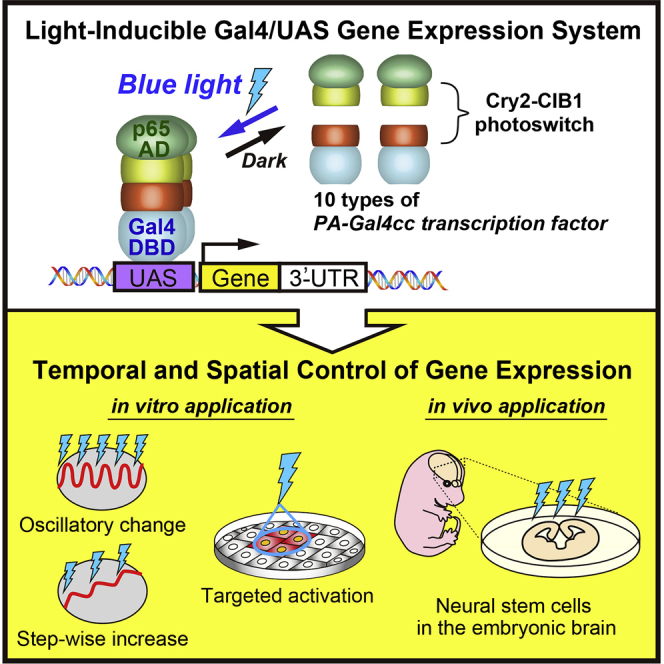
Highlights
-
•
Photoactivatable (PA)-Gal4cc transcription factors are developed in mammalian cells
-
•
The PA-Gal4cc activities are controlled by blue light
-
•
The PA-Gal4cc allows precise temporal and spatial control of gene expression
-
•
The PA-Gal4cc can be applied to various types of cells in vitro and in vivo
Optical Imaging; Molecular Genetics; Cell Biology
Introduction
Over the course of development, homeostatic maintenance, and environmental responses of multicellular organisms, gene expression patterns in cells are dynamically altered. To precisely analyze the functional roles of dynamic gene expression changes, tools that allow spatiotemporal control at fine resolution are needed. Light-control systems are powerful methods to artificially regulate cellular functions at fine spatiotemporal resolution, including gene expression control (Crefcoeur et al., 2013; Hallett et al., 2016; Horner et al., 2017; Imayoshi et al., 2013; Konermann et al., 2013; Motta-Mena et al., 2014; Pathak et al., 2017; Polstein and Gersbach, 2012; Shimizu-Sato et al., 2002; Wang et al., 2012; Yazawa et al., 2009; Quejada et al., 2017; Yamada et al., 2018; Liu et al., 2012; Muller et al., 2013). By applying diverse types of photoactivatable (PA) molecules originally cloned from plants, fungi, and bacteria, such as light-switchable enzymes or protein interaction modules, the application of these optogenetic tools has expanded to studies of the regulation of many different cellular functions and biological activities.
The Gal4/UAS system is a binary gene expression system primarily used in Drosophila, although it has also been applied to zebrafish and mammalian model organisms (Fischer et al., 1988; Brand and Perrimon, 1993). Gal4, a transcription factor originally cloned from yeast, contains a DNA-binding domain (DBD) and a transcription activation domain (AD), and binds to a specific sequence, UAS (upstream activation sequence). It activates transcription from a basal promoter placed downstream of UAS. The Gal4/UAS system has two major advantages. First, the UAS results in the expression of downstream genes at much higher levels than endogenous tissue-specific promoters. Therefore, this amplification process allows for high levels of gene expression. Second, expression vectors or transgenic animals with Gal4 and UAS constructs are widely used in many different research models. By combining these substantial resources, it is possible to induce expression of the gene of interest in a desired cell type/tissue at a high level at the desired time.
For the precise manipulation of gene expression at fine spatiotemporal resolution in mammalian cells, we designed a blue light-inducible Gal4/UAS gene expression system based on the concept of split transcription factors. In this system, light-dependent interactions between Cry2-CIB1 PA-protein interaction modules can reconstitute a split Gal4 DBD and p65 transcription AD. We adapted the Arabidopsis thaliana-derived blue light-responsive heterodimer formation module consisting of the cryptochrome 2 (Cry2) photoreceptor and its specific binding protein cryptochrome-interacting basic-helix-loop-helix 1 (CIB1) (Wu and Yang, 2010; Kennedy et al., 2010; Taslimi et al., 2016). This was because the Cry2-CIB1 PA-protein interaction system is efficient and reversible and had therefore already been exploited in previously developed PA gene expression systems (Hallett et al., 2016; Taslimi et al., 2016; Quejada et al., 2017; Yamada et al., 2018).
Here, we optimized light-inducible Gal4/UAS gene expression systems via comprehensive functional screening of candidate constructs of the PA-Gal4 transcriptional activator (PA-Gal4cc) in mammalian cells. Each PA-Gal4cc has a different light-induced transcription efficacy and activation/deactivation kinetics. The conventional Gal4/UAS system is widely used in different research models, such as expression vectors or transgenic animals with UAS regulatory sequences; therefore our light-controlled PA-Gal4cc transcriptional activators will allow the optogenetic manipulation of genes of interest in broad fields of biology.
Results
Functional Screening of Optimized PA-Gal4cc Transcription Factors
Previously, several light-inducible Gal4/UAS systems were developed using yeast cells. However, some such systems optimized in yeast cells (Hallett et al., 2016) do not function efficiently in mammalian cells (Figure S1 and our unpublished data). Therefore, we carried out functional screening of candidate PA-Gal4 transcriptional activator constructs in the immortalized human embryonic kidney cell line HEK293T (Figures 1 and S2–S13 and Table S1). To avoid a possibility that saturated expression of the stable reporter product might mask the differences of light-induced gene expression levels, we applied the destabilized luciferase reporter Ub-NLS-luc2 (Figures 1B and S14) and placed the Ascl1 3′ UTR sequence just downstream of Ub-luc. This is known to result in a shorter mRNA half-life and prevent accumulation of the reporter activity in the measured cells (Imayoshi et al., 2013; Luker et al., 2003; Voon et al., 2005; Masamizu et al., 2006). We used two types of the Gal4 DBD, because existence of internal dimerization domain reportedly inhibits nuclear localization of the transcription factor in combination with the light-induced dimerization system (Pathak et al., 2017). In the short version, for constructs of the Gal4 DBD, we used the sequences containing Gal4 residues 1–65. The long version constructs of Gal4 DBD contain its original dimerization domain in addition to the DBD (residues 1–147). For functional screening of these candidate PA-Gal4 transcriptional activator constructs, we used the short or long Gal4 constructs as the split DBD, together with the transcription AD of p65 (p65 AD). We confirmed the strong activity of p65 AD with a comparison to VP16 and VP64 AD (Figure S15) (Wang et al., 2012). In addition to the Cry2-CIB1 system, we also screened constructs of PA-Gal4 activators using other optical dimer formation systems, such as Magnet (Kawano et al., 2015) (Figure S10), tunable light-controlled interacting protein tags (TULIPs) (Strickland et al., 2012) (Figure S11), and original light-inducible dimer/improved light-inducible dimer (oLID/iLID) (Guntas et al., 2015; Hallett et al., 2016) (Figures S12 and S13). However, most constructs did not yield efficient light-inducible transcriptional activity in our functional screening studies. Therefore, we focused on PA-Gal4 constructs using the Cry2-CIB1 system (Figures 1 and S2–S9 and Tables S1–S4).
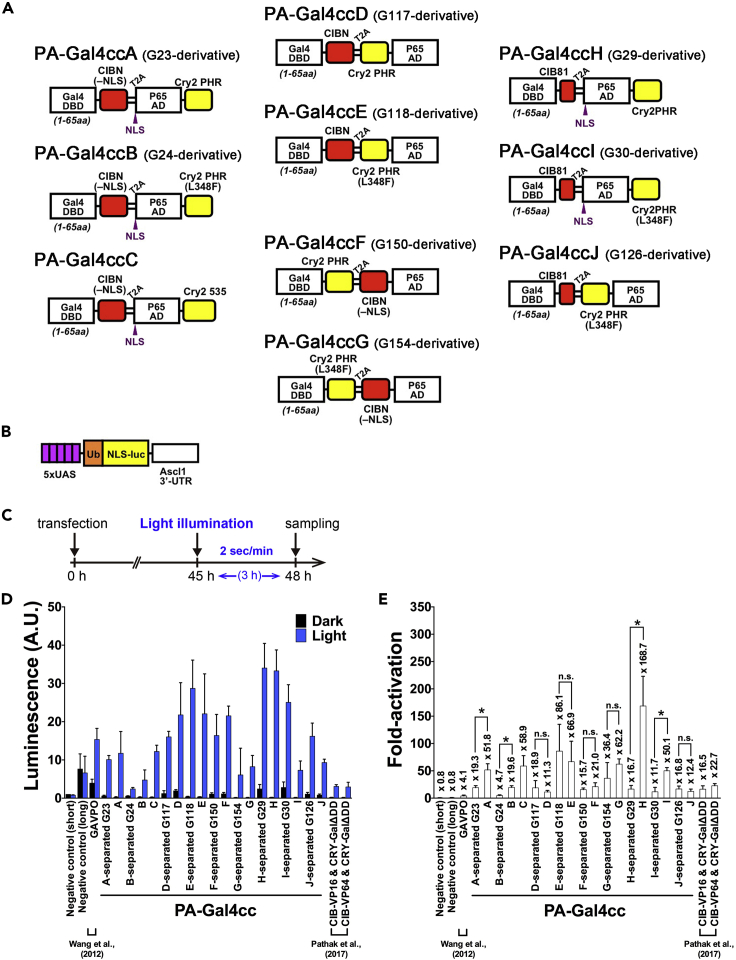
Figure 1.
Generation of the Photoactivatable (PA)-Gal4cc Transcriptional Activators
(A) Schematic illustration of the PA-Gal4cc constructs. Yellow boxes indicate Cry2 variants, and red boxes indicate CIB1 variants adapted in this study. Codon optimization for efficient expression in mammalian cells was performed for all Cry2 and CIB1 derivatives.
(B) The reporter construct used in this experiment consisted of 5x UAS, Ub-NLS-luc2, and Ascl1 3′ UTR sequences.
(C) Experimental time course.
(D) Validation of light-dependent regulation of the PA-Gal4cc constructs in transiently transfected HEK293T cells. Ten selected candidate construct pairs that showed low basal background and significant induction (e.g., “PA-Gal4cc-A ~ J-separated” constructs) were modified as single expression plasmids, in which the PA-module-tethered Gal4 DBD and p65 AD were co-expressed together with a T2A self-cleaving peptide (i.e., PA-Gal4cc-A ~ J). The pEF-Gal4 DBD short and pEF-p65 AD and pEF-Gal4 DBD long and pEF-p65 AD without any PA dimer formation molecules were co-transfected as the negative control (short) and the negative control (long), respectively.
(E) Fold-increase of luciferase activity (light/dark). The previously developed PA-Gal4 transcription activators (Wang et al., 2012; Pathak et al., 2017) were included for comparison. PHR, photolyase homology region; NLS, nuclear localization signal. The data represent mean values ±standard deviation (SD) (n = 9) from three independent experiments; Each experiment consisted of three replicates. Luciferase assay data of the negative control (short) in the dark were used for the correction of data of each construct. The values in bar graphs and summary of the statistical comparisons were also displayed in Table S1. ∗p < 0.05; two-tailed Student's t test between the results of each separated and T2A construct pair.
Arabidopsis thaliana-derived Cry2 and CIB1 were originally regulatory components of development and growth in plants, acting via circadian clock control. Cry2 has two domains, the N-terminal photolyase homology region (PHR) and the cryptochrome C-terminal extension. PHR is a domain that noncovalently binds to the chromophore flavin adenine dinucleotide (FAD). Cry2 binds the basic-helix-loop-helix (bHLH) transcription factor CIB1 in a blue light-specific manner. Truncated versions of the Cry2 and CIB1 essential domains act as a blue light-dependent heterodimer formation module, and several point mutations of Cry2 result in faster or slower photocycles (Kennedy et al., 2010; Liu et al., 2012; Taslimi et al., 2016; Yamada et al., 2018; Hughes et al., 2012). Because these Cry2/CIB1 variants and their respective pairs have different binding affinities, kinetics, and background activity in the dark, we undertook detailed investigations of combinations of the different Gal4 DBD, p65 AD, and Cry2/CIB1 variants (Figures 1 and S2–S9).
Of the 180 tested constructs using the Cry2-CIB1 system, 64 showed light-dependent increases (>5-fold) of the luciferase transcription reporter (Figures 1 and S2–S9). This was more common for the construct sets incorporating the short version of Gal4 DBD. Of the 64 light-inducible Gal4 activator constructs, 16 yielding a >5-fold increase and 33 with a >10-fold increase contained the short version of Gal4 DBD. This might due in part to the nuclear clearing phenotype of Cry2-fused proteins (Pathak et al., 2017), which was reported to be dependent on the presence of a dimerization domain contained within Cry2-fused proteins. In contrast, the long version of the Gal4 DBD construct has an inherent dimerization domain.
We selected 10 construct pairs for subsequent validation (PA-Gal4cc-separated A ~ J in Figure 1 and Table S1) because of their low levels of background activity in the dark and their consistent light-induced gene expression. Importantly, the selected pairs exhibited lower background activity than GAVPO (Wang et al., 2012; Imayoshi et al., 2013) in the dark (Tables S1–S3). In the construct screening experiments, PA-module-tethered Gal4 DBD and p65 AD were expressed separately from the two independent expression plasmids. When the PA-Gal4 construct was expressed from a single expression plasmid in which the PA-module-tethered Gal4 DBD and p65 AD were co-expressed together with a T2A self-cleaving peptide (Kim et al., 2011), light-induced transcriptional activity was preserved or increased (Figures 1D and 1E and Table S1). This could be due to the improved simultaneous expression efficiency of the PA-module-tethered Gal4 DBD and p65 AD in each transfected cell using T2A-based bicistronic expression vectors. We finally selected these 10 constructs on the T2A vectors and designated them “PA-Gal4ccA ~ PA-Gal4ccJ” (Figure 1A).
In the candidate construct screening studies for PA-Gal4cc, the cells were exposed to pulsed blue light (e.g., 2-s pulse every minute) for only 3 h before cell sampling (Figures 2A–2C). When cells were exposed to similar blue light pulses (2-s pulse every minute) for longer time periods (e.g., 24 h), the induced transcription reporter activity for PA-Gal4cc was increased (Figures 2D–2F). Most of the constructs had essentially similar or superior activities to the light-insensitive constitutively active Gal4 transcriptional activator Gal4-VN8x6 (Salghetti et al., 2000) (Figures 2E and Tables S2 and S3). The rank order of the degree of induced gene expression between the PA-Gal4cc constructs was mostly same for 3- and 24-h illumination (Figure S16).
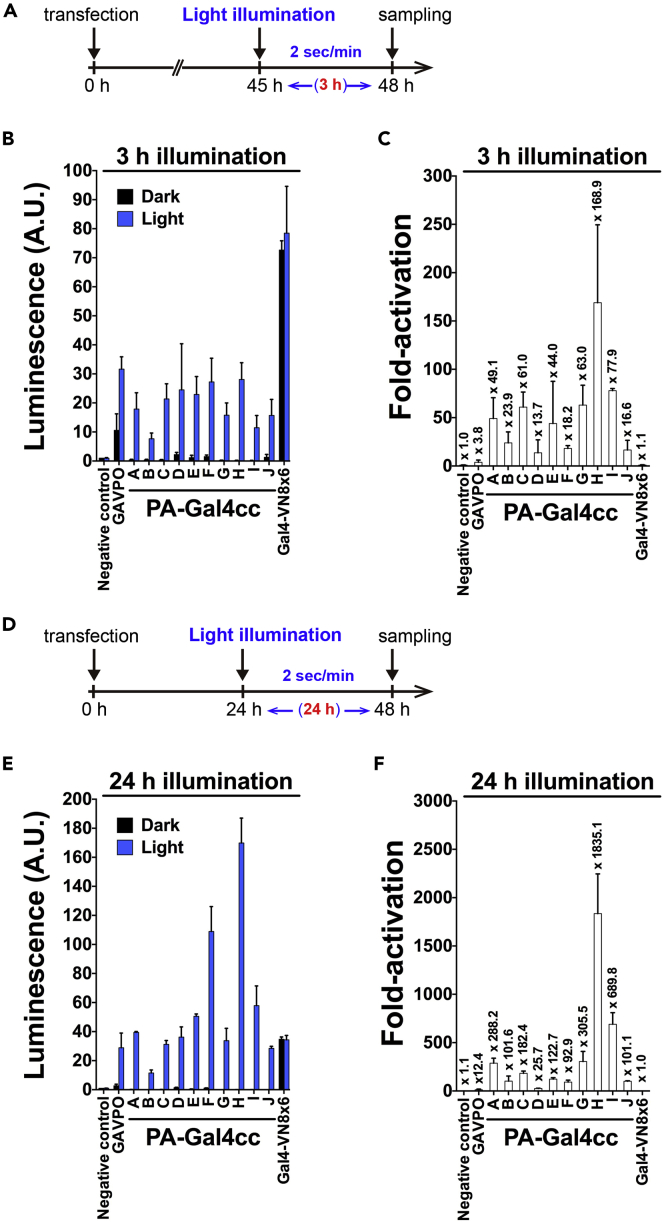
Figure 2.
Comparison of Two Different Light Exposure Protocols to Activate PA-Gal4cc-Mediated Transcription
(A) Illumination protocol used for the luciferase assay is indicated.
(B and C) Validation of light-induced luciferase reporter activities by PA-Gal4cc constructs in transiently transfected HEK293T cells. Measured luciferase activities (B) and fold-increase of luciferase activity (Light/Dark) (C). The pEF-Gal4-VN8x6 plasmid was used for expressing the light-insensitive constitutively active Gal4 transcriptional activator. The pEF-Gal4 DBD short and pEF-p65 AD without any PA dimer formation molecules were co-transfected as the negative control. The rank order of the degree of fold activation was as follows: PA-Gal4cc-H, I, G, C, A, E, B, F, J, D. The data represent mean values ± SD (n = 9) from three independent experiments; each experiment consisted of three replicates.
(D) Illumination protocol with prolonged exposure used for the luciferase assay is indicated. The light wavelength and radiant energy were the same as (A).
(E and F) Validation of light-induced luciferase reporter activities by PA-Gal4cc constructs in transiently transfected HEK293T cells. Measured luciferase activities (E) and fold-increase of luciferase activity (Light/Dark) (F). The rank order of the degree of fold activation was as follows: PA-Gal4cc-H, I, G, A, C, E, B, J, F, D. The data represent mean values ± SD (n = 6) from three independent experiments; each experiment consisted of duplicates. The values in bar graphs and summary of the statistical comparisons were also displayed in Tables S2 and S3. The rank orders of PA-Gal4cc in the two experiments were summarized in Figure S16.
Light Dose-Dependent Transcriptional Activity of PA-Gal4cc
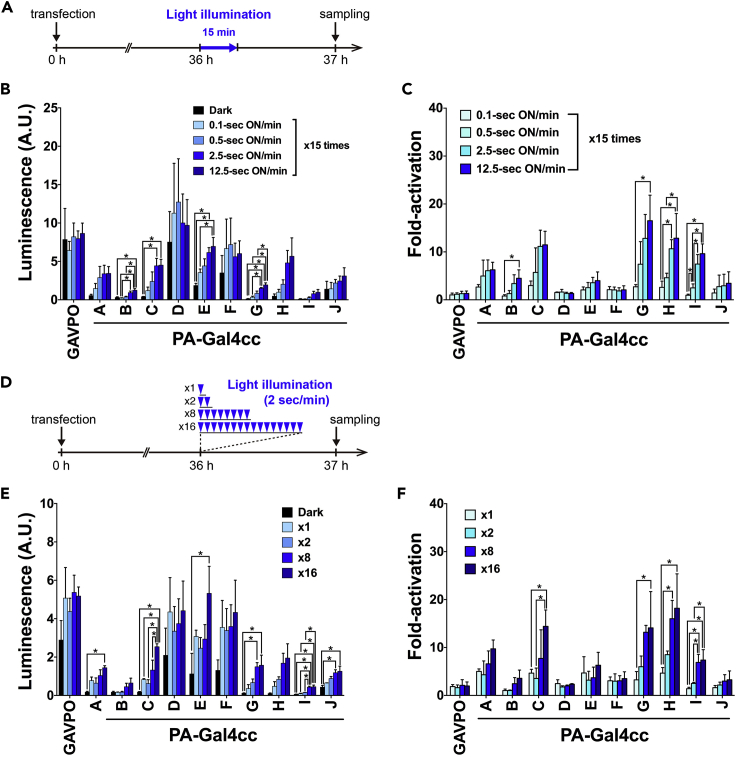
One important advantage of a light-inducible gene expression system is the ease of tuning gene expression levels by modifying illumination protocols. We investigated the effects of (1) the duration of the blue light on-phase in the on-off cycle (Figure 3A) and (2) the number of blue light pulses applied (Figure 3D).
Figure 3.
Light-Dose-Dependent Control of PA-Gal4cc Transcriptional Activity
(A) Schematic representation of experimental conditions for testing the effects of the duration of blue light on-phase in the on-off cycle.
(B and C) Blue light duration-dependent increase in measured luciferase activities (B) and fold-increase of luciferase activity (Light/Dark) (C) in PA-Gal4cc transiently transfected HEK293T cells. The data represent mean values ± SD (n = 8) from three independent experiments.
(D) Schematic representation of experimental conditions for analyzing the effects of the number of applied blue light pulses.
(E and F) Light pulse number-dependent increase in measured luciferase activities (E) and fold-increase of luciferase activity (Light/Dark) (F) in PA-Gal4cc transiently transfected HEK293T cells. The data represent mean values ± SD (n = 7) from three independent experiments. The data represent mean ± SD. ∗p < 0.05; one-way ANOVA followed by Tukey's posthoc test.
We observed an expected blue light duration-dependent increase of luciferase reporter activity in PA-Gal4cc-transduced HEK293T cells (Figures 3B and 3C), indicating that fine control of downstream gene expression was achieved by changing the duration of blue light illumination in the on-phase of the cycle. The duration-dependent significant changes were observed in PA-Gal4ccB, G, H, and I (Figure 3C). We also observed increased luciferase reporter activity dependent on the number of light pulses (Figures 3E and 3F). The light pulse number-dependent significant changes were observed in PA-Gal4ccC, G, H, and I (Figure 3F). However, sensitivity to limited duration or numbers of blue light pulses and linear responses to multiple exposures varied between the PA-Gal4cc constructs. For instance, PA-Gal4ccA, D, E, F, and J were sensitive to short pulses or a single pulse of blue light but did not show further significant increases on multiple pulses. In contrast, PA-Gal4ccB, C, G, H, and I exhibited increased reporter gene expression depending on the duration or number of pulses of blue light. In both cases, fine control of gene expression levels with GAVPO was difficult to achieve in the transient transfection experiments using HEK293T cells (Figure 3). This is because the leaky activity of GAVPO in the dark was high and significant luciferase activity was already induced before exposure to blue light.
Temporal Characteristics of PA-Gal4cc
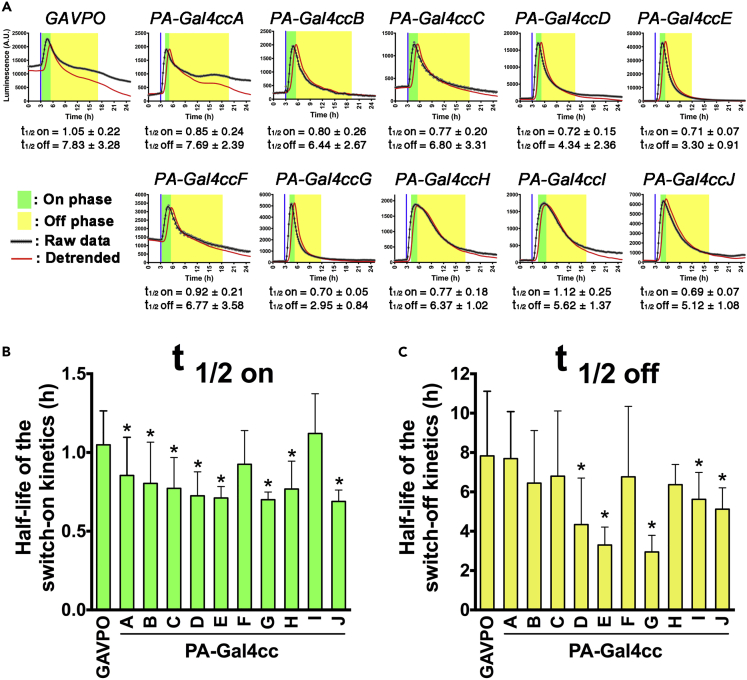
Because of the rapid activation and deactivation kinetics of the previously developed PA-Tet-OFF/ON system (Yamada et al., 2018), in which the same Cry2-CIB1 switch was applied, it might be expected that PA-Gal4cc could be used for dynamic control of downstream gene expression. The original Cry2 is rapidly activated by exposure to light, and then spontaneously dissociates from CIB1 with a half-life of ~5.5 min (Kennedy et al., 2010; Taslimi et al., 2016). We validated the temporal characteristics of each PA-Gal4cc construct by exposure to short pulses of light (2 min) and monitored luciferase reporter expression levels in real time (Figure 4A).
Figure 4.
Temporal Features of the PA-Gal4cc Transcriptional Activators
(A) HEK293T cell transfected with the PA-Gal4cc constructs and 5x UAS-Ub-NLS-luc2-Ascl1 3′ UTR reporter were exposed to a single blue light pulse. The timing of blue light exposure is indicated by vertical blue lines. The blue light was applied to cells 30 h after the transfection. The transcription on- and off-phases are highlighted in green and yellow, respectively.
(B and C) Using the single light pulse data set, kymograph analysis was used to determine the half-lives of the switch-on (B) and switch-off (C) kinetics of the PA-Gal4cc transcriptional activators. The data represent mean ± SD. ∗p < 0.05; one-way ANOVA followed by Dunnett's posthoc test (GAVPO versus each PA-Gal4cc). The rank order of the half-life of the switch-on/off kinetics between the PA-Gal4cc constructs was summarized in Figure S18.
Analyzing HEK293T cells transiently transfected with PA-Gal4cc and UAS-Ub-NLS-luc2 reporter we found that the temporal patterns of blue light pulse-induced luciferase activity was different for the different constructs (Figures 4 and S17). When we compared the on-kinetics of PA-Gal4cc constructs with GAVPO, most of the tested constructs, with the exception of PA-Gal4ccF and I, had significantly lower values than GAVPO (Figure 4B). Light-induced gene expression did not cease rapidly in cells transiently transfected with GAVPO and PA-Gal4cc. However, the off-kinetics of PA-Gal4ccD, E, G, I, and J were significantly shorter than GAVPO, indicating that the former are excellent candidates for rapid and dynamic gene expression control (Figure 4C).
We conducted similar experiments using normal, stable luciferase reporters (luc2 in Figure S17A). The activation and deactivation kinetics of light-induced gene expression were extended when the reporter is constructed with normal, stable luciferase (Yamada et al., 2018). Indeed, we observed extended activation and deactivation kinetics of light-induced gene expression with PA-Gal4cc (Figures S17B and S17C). The rank order of the on/off-kinetics of the different PA-Gal4cc constructs was also mostly preserved among these reported constructs with different half-lives (Figure S18).
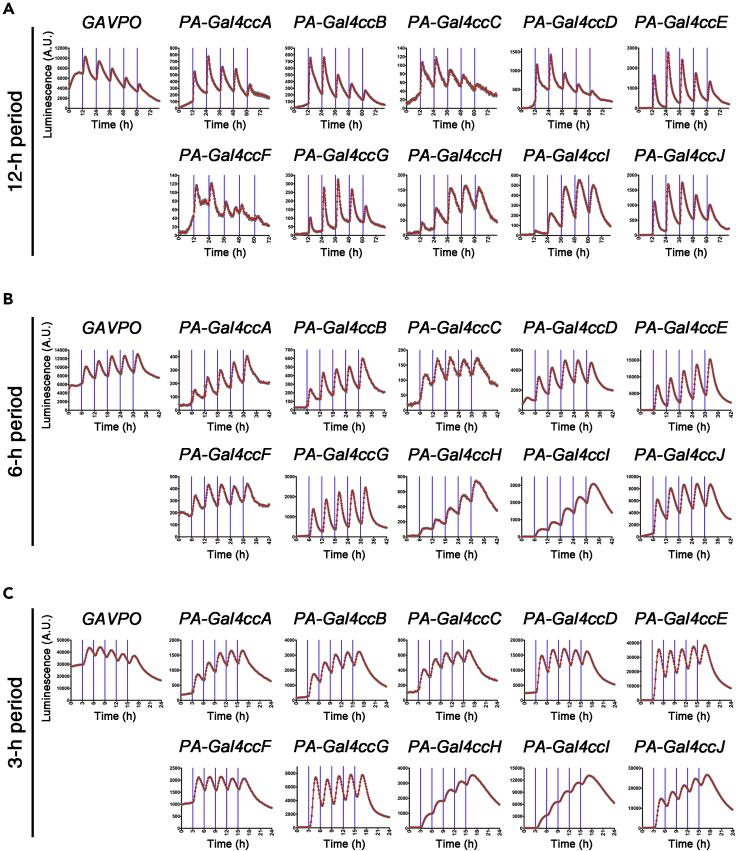
In the earlier construct validation studies (Figures 1, 2, and 3) we had analyzed the mass effects of multiple blue light pulses. When we periodically applied short-term blue light pulses with different periods at 3, 6, and 12 h and monitored the luciferase reporter expression level in real time, different types of dynamic gene expression patters were induced among the PA-Gal4cc and destabilized/normal luciferase reporter constructs (Figures 5 and S19). For example, in the case of destabilized UAS-Ub-NLS-luc2, experiments with 12-h periodic illumination, most of the PA-Gal4cc induced oscillatory gene expression with minimum accumulation of the reporter (Figure 5A). Under blue light irradiation with a 6-h period, a stepwise increase in luciferase reporter activity was observed in PA-Gal4ccH- or I-transfected cells (Figure 5B). In addition to PA-Gal4ccH and I, PA-Gal4cc J also induced stepwise increase of the reporter under blue light irradiation with a 3-h period (Figure 5C). In contrast, PA-Gal4ccE and G still induced oscillatory expression with 3-h periodic illumination (Figure 5C). These differences in the induced gene expression patterns might be attributable to the different activation/deactivation kinetics of each PA-Gal4cc. For instance, PA-Gal4ccE and G had a significantly shorter half-life of off-kinetics and would more easily induce oscillatory expression of downstream genes (Figures 4 and 5).
Figure 5.
Periodic Activation of the PA-Gal4cc Transcriptional Activators
(A–C) Transiently transfected HEK293T cells, in which PA-Gal4cc and 5x UAS-Ub-NLS-luc2-Ascl1 3′ UTR reporter had been introduced via lipofection, were repeatedly exposed to blue light pulses at 12- (A), 6- (B), or 3-h (C) intervals. The timing of blue light exposure is indicated by vertical blue lines. The first blue light illumination was initiated 24 h after the transfection. Experiments were repeated at least three times with similar results.
Reporter expression dynamics were also changed when we used a more stable reporter construct (i.e., UAS-normal luc2-reporter) (Figure S19A). Most of the PA-Gal4cc constructs showed a stepwise increase type of reporter expression under blue light illumination with a 3-h period (Figure S19D). Thus, by changing the reporter protein half-lives as well as the light exposure pattern, different gene expression patterns (e.g., oscillatory change or stepwise increase) can be induced with the same PA-Gal4cc. For example, under blue light irradiation with a 3-h period, PA-Gal4ccE induced an oscillatory pattern with the unstable UAS-Ub-NLS-luc2-reporter, but a stepwise increase pattern with the stable UAS-luc2-reporter.
Application of PA-Gal4cc together with Lentivirus Vectors
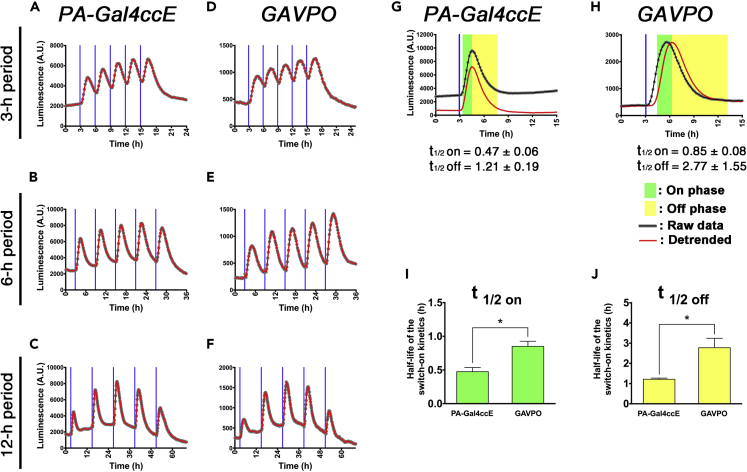
To reduce experimental variability due to different cellular transfection efficiencies, we used lentivirus vectors to stably express PA-Gal4ccE, one of the fastest cycling PA-Gal4ccs, in HEK293T cells and to integrate the reporter construct (Figures 6 and S20). Consistent with the co-transient transfection data of PA-Gal4ccE and destabilized luciferase reporter, the reporter activity was greatly enhanced in the stable PA-Gal4ccE cells exposed to blue light relative to cells left in the dark. When we applied blue light pulses with different periods, robust oscillatory expression was induced at 3, 6, and 12 h (Figures 6A–6C, S20E, S20G, and S20I). However, to develop stable cells manifesting reliable blue light responsiveness, multiple rounds of selections with drug/fluorescence-activated cell sorting were required to purify transduced cells that have higher copy numbers of lentivirus vectors and expression levels of PA-Gal4ccE (Figure S21). In contrast, when we generated GAVPO-expressing stable cells, one single round of drug selection was sufficient (Figure S21). Furthermore, these stable cells in which GAVPO and the UAS-Ub-NLS-luc2-reporter were integrated with lentivirus vectors exhibited efficient and reliable blue light-inducible gene expression and also showed rapid activation/deactivation kinetics (Figures 6D–6F, S20B, S20D, S20F, S20H, and S20J). Although stably transfected GAVPO is significantly slower than PA-Gal4ccE (Figures 6G–6J), the reliability and temporal kinetics were dramatically improved compared with the results of transient transfection experiments (Figure S20). These findings indicate that GAVPO is more suitable for experiments in which stable cells expressing this factor at not-too-high levels can be prospectively screened and identified. In contrast, due to the lower background activity of PA-Gal4cc, this is more suitable for transient transfection experiments where the rigorous control of PA transcription factor expression levels is more difficult and transfection efficiencies are more variable between cells.
Figure 6.
Light-Induced Gene Expression Control with PA-Gal4cc-Expressing Lentiviral Vectors
(A–F) The PA-Gal4ccE or GAVPO lentiviral vector-transduced HEK293T cells were repeatedly exposed to blue light pulses at 3- (A and D), 6- (B and E), or 12-h (C and F) intervals. The reporter construct consisted of 5x UAS, Ub-NLS-luc2, and Hes1 3′ UTR sequences. The timing of blue light exposure is indicated by vertical blue lines. Experiments were repeated at least three times with similar results.
(G and H) PA-Gal4ccE (G)- and GAVPO (H)-introduced HEK293T cells were exposed to a single blue light pulse.
(I and J) Using the single light pulse data set, kymograph analysis was used to determine the half-lives of the switch-on (I) and switch-off (J) kinetics of light-induced gene expression. The data represent mean ± SD. ∗p < 0.05; two-tailed Student's t test.
Targeted Activation of PA-Gal4cc in Spatially Restricted Cells
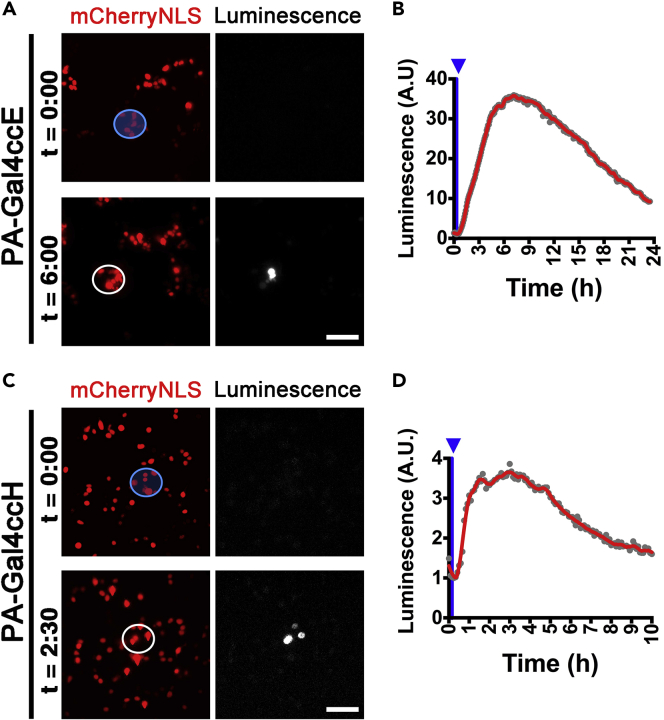
Next, we examined whether we could spatially control gene expression in targeted cells. To test this, we equipped a bioluminescence imaging microscope with a digital mirror device (DMD) to stimulate the targeted cells. We tested PA-Gal4ccE and H in such spatial control gene expression experiments. After exposure to a blue light pulse, bioluminescence imaging revealed that luciferase expression in PA-Gal4cc-transfected HEK293T cells with the UAS-Ub-NLS-luc2 reporter occurred in the areas determined by the DMD device (Figure 7). These results indicated that spatial control of gene expression is feasible using the PA-Gal4cc/UAS-system.
Figure 7.
Spatially Controlled Regulation of PA-Gal4cc Transcriptional Activators by Patterned Light Illumination
(A–D) Targeted cell populations were illuminated by patterned light generated by a digital mirror device (DMD). The patterned light, indicated by blue circles, was applied to HEK293T cells in which the PA-Gal4ccE (A and B) and PA-Gal4ccH (C and D) were transiently introduced by lipofection. (B and D) Light-induced reporter expression was quantified after patterned light illumination in the white circled regions. The timing of blue light exposure is indicated by blue arrowheads. Experiments were repeated at least three times with similar results. 86.0% ± 19.0% and 83.3% ± 28.9% showed light-induced reporter expressions in the PA-Gal4ccE- and PA-Gal4ccH-transfected cells, respectively. Scale bars, 100 μm.
Validation of PA-Gal4cc in Brain Slice Cultures
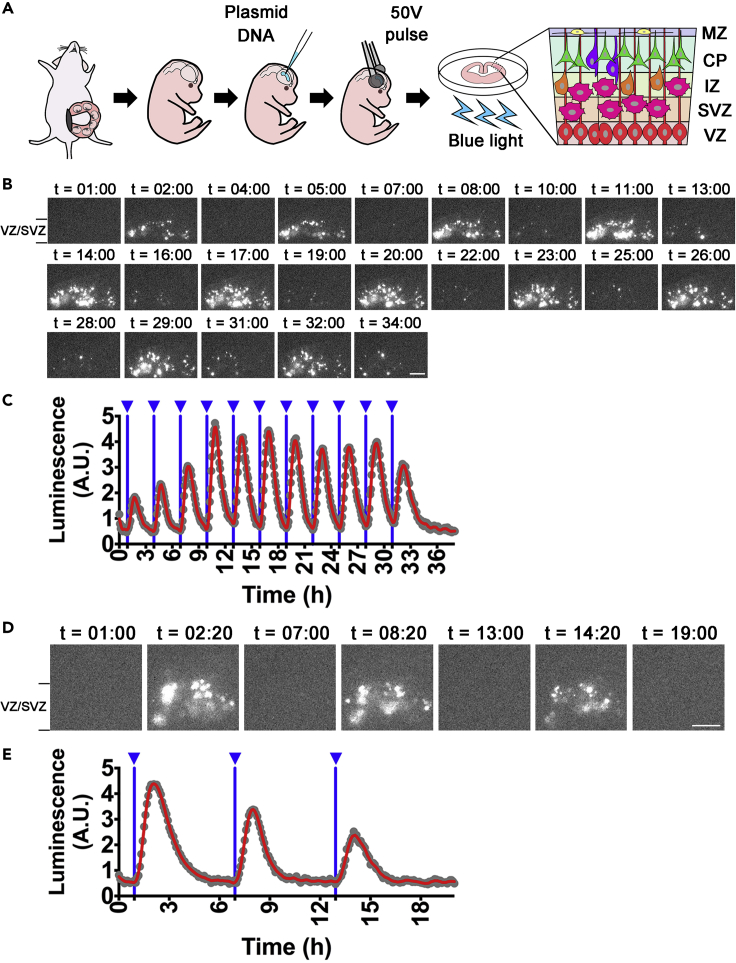
Finally, we tested the ability of the PA-Gal4cc/UAS system to induce light-triggered gene expression in tissues other than cultured cell lines. To this end, we examined PA-Gal4cc activity in neural stem/progenitor cells of the developing mouse forebrain (Figure 8). We transfected the PA-Gal4ccE expression plasmid together with the UAS-Ub-NLS-luc2 reporter into neural stem/progenitor cells using ex utero electroporation methods. When tissue slices derived from the electroporated brain were periodically illuminated by blue light at 3- (Figures 8B and 8C) and 6-h (Figures 8D and 8E) periods, oscillatory reporter expression was observed in the ventricular/subventricular zone where the neural stem/progenitor cells are preferentially located (Imayoshi and Kageyama, 2014a, 2014b). These findings suggest that PA-Gal4cc can be introduced into cells by various different methods, including electroporation, as well as lipofection and with lentiviral vectors. We also documented efficient blue light-induced gene expression in primary cultured cells, such as neural stem/progenitor cells of acutely prepared embryonic brain slices.
Figure 8.
Optogenetic Manipulation of Gene Expression in Brain Slices
(A) PA-Gal4ccE with 5x UAS-Ub-NLS-luc2-Ascl1 3′ UTR reporter was introduced into neural stem/progenitor cells of the developing brain by ex utero electroporation. The electroporated brain was immediately extracted from the embryo and sliced on tissue culture membrane.
(B–E) Blue light was periodically applied to the slice over a 3- (B and C) and a 6-h period (D and E) and reporter activity was monitored. Blue light-induced luciferase expression was observed in the neural stem/progenitor cells of the ventricular and subventricular zones (VZ/SVZ). Scale bars, 200 μm. MZ, marginal zone; CP, cortical plate; IZ, intermediate zone.
Discussion
Here we describe a set of improved PA-Gal4cc transcriptional activators for the spatiotemporal control of gene expression in mammalian cells. To develop PA-Gal4cc transcriptional activators, we carried out functional screening by investigating the following parameters of the candidate constructs: (1) Gal4 DBD elements, (2) Cry2/CIB1 truncation and point mutations, (3) Cry2/CIB1 configuration (i.e., N-terminal or C-terminal fusion), and (4) expression vector structures necessary for efficient expression in target cells. We finally selected 10 PA-Gal4cc transcriptional activators (PA-Gal4cc A ~ J) with different light-induced transcription efficacy and activation/deactivation kinetics. Importantly, all selected PA-Gal4cc had low background activity in the dark, achieving reliable dynamic gene expression control with minimal leaky transcription before light exposure. In our PA-Gal4cc, PA-module-fused Gal4 DBD and p65 AD are co-expressed together with a T2A self-cleaving peptide. The IRES sequence, another tool for co-expression of two polypeptides, is sometimes used for the reconstitution of synthetic transcription factors by light (Quejada et al., 2017). The size of the IRES sequence is much greater than the DNA sequence encoding the T2A peptide, and its integration in expression vectors reduces the level of gene expression. This may limit the application of PA transcription factors to viral vectors in which the size of the inserted sequence is limited and a shorter sequence is preferred for preparing high-titer virus purified products. One concern when using the T2A peptide is that the residue peptides of cleaved T2A may change the properties of the expressed functional molecules, in this case, the efficiency of the reconstitution of Cry2/CIB1-fused Gal4 DBD and p65AD. However, this concern was not relevant for our PA-Gal4cc because the light-induced transcriptional activity was preserved in the separately expressed vectors and T2A-based bicistronic expression vectors (Figure 1 and Tables S1–S3).
In our construct screening, we used p65 AD as a transcription AD. In previous reports that developed the light-activatable Gal4/UAS system in mammalian cells, VP16 AD or VP64 AD were applied (Pathak et al., 2017; Quejada et al., 2017). Although they identified constructs that showed strong light-induced gene expressions, the structures of the identified optimal constructs were different from the constructs developed in our study, in terms of the applied Cry2/CIB1 truncation and point mutations and Cry2/CIB1 configurations. These results indicate that, when the different kinds of molecular elements are applied in development of synthetic light-reconstitutable transcription factors, rigorous functional screenings must be required to identify the optimal constructs.
For reliable and fine cellular gene expression control by light, high sensitivity to light, large dynamic range of induced gene expression, and low background transcription activity in the dark are required. To reduce the effects of photo-toxicity resulting from exposure to intense light, high sensitivity is essential for achieving gene expression control using lower-power and/or short-duration light pulses. A requirement for prolonged light exposure also reduces the temporal resolution of gene expression control. To artificially control the magnitude of gene expression as well as the timing (e.g., initiation and termination), light-inducible gene expression systems having large dynamic ranges are needed. Here, we validated more than 200 candidate PA-Gal4 transcriptional activator constructs. The sensitivity to light and dynamic range of induced gene expression levels varied depending on the PA modules used, and their different temporal features, such as activation/deactivation kinetics. Indeed, the selected 10 PA-Gal4cc-A ~ J constructs had different light-induced transcription efficacies and temporal features (Figures 2, 3, 4, 5, S17, and S19 and Tables S1–S4). For example, PA-Gal4ccE or G had significantly faster on/off-kinetics than the other PA-Gal4ccs and is therefore more suitable for generating oscillatory gene expression patterns (Figures 5, S18, and S19 and Table S4). In contrast, PA-Gal4ccH and I and are more suitable for inducing accumulated-type gene expression, such as stepwise increases, and this might be partially attributed to relatively slower on/off-kinetics of PA-Gal4ccH and I than that of PA-Gal4ccE and G (Figures 5, S18, and S19 and Table S4). Regarding the maximum light-induced gene expression level, PA-Gal4ccH showed higher values than other the other PA-Gal4ccs (Figures 2 and S16 and Table S4). In terms of light sensitivity, PA-Gal4ccD and E are very sensitive and can be fully activated by very dim light or a small number of light pulses (Figure 3 and Table S4). All things considered, PA-Gal4ccE could be the first choice when the experiment type needs rapid activation/deactivation of gene expressions and/or the blue light illumination power or exposure time is limited. In contrast, when the experiment type prefers higher induced gene expression levels and prolonged repeated light exposures are permitted, PA-Gal4ccH is more suitable.
In the selected PA-Gal4cc constructs, different sets of Cry2 and CIB1variants were used. However, the light-induced transcription activation/deactivation kinetics of the PA-Gal4cc constructs did not closely correlate with the reported photocycle differences of the Cry2/CIB1 variant pairs. For example, PA-Gal4ccE, the fastest cycling PA-Gal4cc, has a Cry2 PHR module with the L348F slow photocycle mutation (~24-min half-life). The off-kinetics of PA-Gal4ccE was significantly shorter than that of PA-Gal4ccD, in which a wild-type Cry2 PHR module (~5.5-min half-life) was integrated and the remaining construct structure is identical to PA-Gal4ccE (Figures 4, 5, S16, S18, and S19). Similarly, PA-Gal4ccA and B, F and G, and H and I have related construct structures except for a Cry2 PHR L348 point mutation difference. However, we did not observe the expected effects of the Cry2 PHR slow photocycle mutation on the off-kinetics of light-induced gene expression within each pair of PA-Gal4cc constructs. In addition, we also observed significant differences in the induced gene expression levels (Figures 1, 2, and 3). Thus, this Cry2 L348 point mutation might also change other features of the reconstituted PA-Gal4, such as the overall 3D structure, binding affinity for the UAS sequence, and efficiency in recruiting the transcriptional machinery. In the PA-Gal4ccH and I constructs, the truncated short version of CIB1, CIB81, was integrated. PA-Gal4ccH and I showed longer on-kinetics and preferentially induced the stepwise increase pattern of light-induced gene expression (Figures 4, 5, S16, and S19). Although the detailed temporal characteristics of this CIB1 variant have not been analyzed (Taslimi et al., 2016), CIB81 may slowly generate heterodimer complexes with Cry2.
Because of these different characteristics of our multiple PA-Gal4cc transcriptional activators, we can induce different types of gene expression patterns just by changing the selection of PA-Gal4cc-series variants even under the same blue light illumination protocols (Figures 5 and S19). This would contribute to the analysis of the functional roles of different gene expression dynamics. Some types of transcription factors can change their functional roles in the context of self-renewal and fate determination of stem cells by altering their gene expression dynamics (e.g., oscillatory versus sustained). These phenomena were discovered by the application of light-induced gene expression systems (Imayoshi et al., 2013; Imayoshi and Kageyama, 2014a).
In conclusion, we optimized the light-controllable Gal4/UAS gene expression system in mammalian cells by developing sets of PA-Gal4cc transcriptional activators. These allow the induction of different types of gene expression dynamics at fine spatiotemporal resolution in several types of mammalian cells. This technology will contribute to the systematic analysis of dynamic changes in cellular gene expression.
Limitations of the Study
The PA-Gal4cc constructs can be introduced into cells by different methods, including lipofection, electroporation, and by use of lentiviral vectors. We also demonstrated efficient light-triggered gene expression in neural stem/progenitor cells in the developing brain. Because the Gal4/UAS system is commonly used in Drosophila, we attempted to develop transgenic flies specifically expressing Drosophila-codon-optimized PA-Gal4ccE and G in mushroom body neurons (Figure S22). However, blue light-inducible gene expression was not observed in such transgenic PA-Gal4ccE-expressing flies due to too high a background reporter gene expression before exposure to light. In addition, transgenic PA-Gal4ccG-expressing flies showed only limited light-induced transcriptional activity in adult mushroom body neurons. Furthermore, light-induced activity of PA-Gal4ccE and G in the Drosophila S2 cell line was weak (Figure S23). This failure of application of our PA-Gal4cc in Drosophila could be attributed to the original optimization of the PA-Gal4cc for the human cell line HEK293T. These results imply that efficacy of light-induced transcription may be different in different cellular contexts and rigorous optimization processes are needed in different cell types and model organisms of interest.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Itaru Imayoshi (imayoshi.itaru.2n@kyoto-u.ac.jp).
Materials Availability
All unique materials generated in this study are available from the lead Contact upon request.
Data and Code Availability
Requests for custom scripts and raw data can be directed to the Lead Contact, Itaru Imayoshi (imayoshi.itaru.2n@kyoto-u.ac.jp).
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank all members of the Imayoshi lab, Kageyama lab, and SK project for their support and Brian Kuhlman for generous gift of the PA-Gal4 constructs optimized in yeast cells. We are also grateful to Mami Matsumoto, Mai Takakura, Yoko Kimura, and Ikuko Iwata for technical help. This work was supported by Grant-in-Aid for Scientific Research on Young Scientists (A) (Japan Society for the Promotion of Science [JSPS] 15H05570) (I.I.), (B) (JSPS 15K18362) (M.Y.), (B) (JSPS 17K14950) (M.Y.), and (A) (JSPS 17H04984) (Y.H.); Grant-in-Aid for Scientific Research on Innovative Area (JSPS 15H01489) (I.I.), (JSPS 16H01424) (I.I.), (JSPS 16H06529) (I.I.), and (JSPS 16H01274) (Y.H.); Grant-in-Aid for Scientific Research on challenging Exploratory Research (JSPS 26640011) (I.I.); Grant-in-Aid for Scientific Research (B) (JSPS 18H02449) (I.I.) from the Ministry of Education, Culture, Sports, Science and the Technology of Japan (MEXT); by Japan Science and Technology Agency (JST) PRESTO program (JPMJPR14F3) (I.I.) and CREST program (JPMJCR1752) (I.I.), (JPMJCR1921) (I.I.), and the Program for Technological Innovation of Regenerative Medicine (JP18bm0704020, I.I.), Brain/MINDS (19dm0207090h0001, I.I.) from the Japanese Agency for Medical research and Development (AMED). I.I. also thanks the Leading Initiative for Excellent Young Researchers program of MEXT and the Waksman Foundation of Japan Inc, the Cell Science Research Foundation, and Tokyo Biochemical Research Foundation for support.
Author Contributions
M.Y. and I.I. conceived the project and designed the experiments. M.Y., S.C.N., and I.I. performed the experiments. Y.S. conducted data analysis. Y.H. produced and provided the transgenic flies. M.Y. and I.I. wrote the manuscript with inputs from all other authors.
Declaration of Interests
The authors have no competing interests.
Published: September 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101506.
Supplemental Information
References
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Crefcoeur R.P., Yin R., Ulm R., Halazonetis T.D. Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat. Commun. 2013;4:1779. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- Fischer J.A., Giniger E., Maniatis T., Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Guntas G., Hallett R.A., Zimmerman S.P., Williams T., Yumerefendi H., Bear J.E., Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U S A. 2015;112:112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett R.A., Zimmerman S.P., Yumerefendi H., Bear J.E., Kuhlman B. Correlating in vitro and in vivo activities of light-inducible dimers: a cellular optogenetics guide. ACS Synth. Biol. 2016;5:53–64. doi: 10.1021/acssynbio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner M., Muller K., Weber W. Light-responsive promoters. Methods Mol. Biol. 2017;1651:173–186. doi: 10.1007/978-1-4939-7223-4_13. [DOI] [PubMed] [Google Scholar]
- Hughes R.M., Vrana J.D., Song J., Tucker C.L. Light-dependent, dark-promoted interaction between Arabidopsis cryptochrome 1 and phytochrome B proteins. J. Biol. Chem. 2012;287:22165–22172. doi: 10.1074/jbc.M112.360545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I., Isomura A., Harima Y., Kawaguchi K., Kori H., Miyachi H., Fujiwara T., Ishidate F., Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Kageyama R. bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron. 2014;82:9–23. doi: 10.1016/j.neuron.2014.03.018. [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Kageyama R. Oscillatory control of bHLH factors in neural progenitors. Trends Neurosci. 2014;37:531–538. doi: 10.1016/j.tins.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Kawano F., Suzuki H., Furuya A., Sato M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 2015;6:6256. doi: 10.1038/ncomms7256. [DOI] [PubMed] [Google Scholar]
- Kennedy M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., Tucker C.L. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y., Kim M.K., Shin B.A., Choi S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M.D., Trevino A., Hsu P.D., Heidenreich M., Cong L., Platt R.J., Scott D.A., Church G.M., Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Gomez G., Lin S., Lin S., Lin C. Optogenetic control of transcription in zebrafish. PLoS One. 2012;7:e50738. doi: 10.1371/journal.pone.0050738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker G.D., Pica C.M., Song J., Luker K.E., Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat. Med. 2003;9:969–973. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- Masamizu Y., Ohtsuka T., Takashima Y., Nagahara H., Takenaka Y., Yoshikawa K., Okamura H., Kageyama R. Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc. Natl. Acad. Sci. U S A. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena L.B., Reade A., Mallory M.J., Glantz S., Weiner O.D., Lynch K.W., Gardner K.H. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K., Engesser R., Schulz S., Steinberg T., Tomakidi P., Weber C.C., Ulm R., Timmer J., Zurbriggen M.D., Weber W. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013;41:e124. doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak G.P., Spiltoir J.I., Hoglund C., Polstein L.R., Heine-Koskinen S., Gersbach C.A., Rossi J., Tucker C.L. Bidirectional approaches for optogenetic regulation of gene expression in mammalian cells using Arabidopsis cryptochrome 2. Nucleic Acids Res. 2017;45:e167. doi: 10.1093/nar/gkx260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein L.R., Gersbach C.A. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J. Am. Chem. Soc. 2012;134:16480–16483. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quejada J.R., Park S.E., Awari D.W., Shi F., Yamamoto H.E., Kawano F., Jung J.C., Yazawa M. Optimized light-inducible transcription in mammalian cells using Flavin Kelch-repeat F-box1/GIGANTEA and CRY2/CIB1. Nucleic Acids Res. 2017;45:e172. doi: 10.1093/nar/gkx804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti S.E., Muratani M., Wijnen H., Futcher B., Tansey W.P. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. U S A. 2000;97:3118–3123. doi: 10.1073/pnas.050007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J.M., Quail P.H. A light-switchable gene promoter system. Nat. Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- Strickland D., Lin Y., Wagner E., Hope C.M., Zayner J., Antoniou C., Sosnick T.R., Weiss E.L., Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taslimi A., Zoltowski B., Miranda J.G., Pathak G.P., Hughes R.M., Tucker C.L. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat. Chem. Biol. 2016;12:425–430. doi: 10.1038/nchembio.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon D.C., Subrata L.S., Baltic S., Leu M.P., Whiteway J.M., Wong A., Knight S.A., Christiansen F.T., Daly J.M. Use of mRNA- and protein-destabilizing elements to develop a highly responsive reporter system. Nucleic Acids Res. 2005;33:e27. doi: 10.1093/nar/gni030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen X., Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- Wu L., Yang H.Q. CRYPTOCHROME 1 is implicated in promoting R protein-mediated plant resistance to Pseudomonas syringae in Arabidopsis. Mol. Plant. 2010;3:539–548. doi: 10.1093/mp/ssp107. [DOI] [PubMed] [Google Scholar]
- Yamada M., Suzuki Y., Nagasaki S.C., Okuno H., Imayoshi I. Light control of the tet gene expression system in mammalian cells. Cell Rep. 2018;25:487–500.e6. doi: 10.1016/j.celrep.2018.09.026. [DOI] [PubMed] [Google Scholar]
- Yazawa M., Sadaghiani A.M., Hsueh B., Dolmetsch R.E. Induction of protein-protein interactions in live cells using light. Nat. Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for custom scripts and raw data can be directed to the Lead Contact, Itaru Imayoshi (imayoshi.itaru.2n@kyoto-u.ac.jp).