Abstract
A subset of TLRs is specialised in the detection of incoming pathogens by sampling endosomes for nucleic acid contents. Among them, TLR3 senses the abnormal presence of double-stranded RNA in the endosomes and initiates a potent innate immune response via activation of NF-κB and IRF3. Nevertheless, mechanisms governing TLR3 regulation remain poorly defined. To identify new molecular players involved in the TLR3 pathway, we performed a genome-wide screen using CRISPR/Cas9 technology. We generated TLR3+ reporter cells carrying a NF-κB-responsive promoter that controls GFP expression. Cells were next transduced with a single-guide RNA (sgRNA) library, subjected to sequential rounds of stimulation with poly(I:C) and sorting of the GFP-negative cells. Enrichments in sgRNA estimated by deep sequencing identified genes required for TLR3-induced activation of NF-κB. Among the hits, five genes known to be critically involved in the TLR3 pathway, including TLR3 itself and the chaperone UNC93B1, were identified by the screen, thus validating our strategy. We further studied the top 40 hits and focused on the transcription factor aryl hydrocarbon receptor (AhR). Depletion of AhR had a dual effect on the TLR3 response, abrogating IL-8 production and enhancing IP-10 release. Moreover, in primary human macrophages exposed to poly(I:C), AhR activation enhanced IL-8 and diminished IP-10 release. Overall, these results reveal AhR plays a role in the TLR3 cellular innate immune response.
Keywords: TLR, TLR3, AhR, innate immunity, genetic screen
Introduction
Deciphering the mechanisms underlying innate sensing is critical due to its central role in fighting pathogens and in the initiation of adaptive immune responses. Important attention to the field has also been fuelled by the need to manipulate the immune response and design appropriate immunotherapies to fight diseases, including cancers. Among the numerous receptors specialised in innate sensing, TLRs represent in humans a family of 10 members that are type I transmembrane proteins sharing a similar organisation in domains and in shape.1 A subset of the TLRs, called nucleic acid-sensing TLRs (NAS TLRs), has been selected by evolution to react to pathogens, such as viruses, by sensing the abnormal location of nucleic acids within the endocytic pathway. Thus, they are also often referred to as endosomal TLRs. Microbial nucleic acids are generally not directly accessible to cell surface receptors. They can be found in the endocytic pathway following phagocytosis of apoptotic bodies of virus-infected cells or upon degradation of microbes.2 Since NAS TLRs do not biochemically discriminate between host- and pathogen-derived ligands, it implies that self-detection by endosomal TLRs is limited by tight regulation of their activity.
The various NAS TLRs exhibit different specificities and expression patterns. TLR3 detects double-stranded RNA (dsRNA)3 and stands out among the NAS TLRs for several of its features. In humans, it is the only TLR for which a non-redundant function has been attributed: homozygous deleterious mutations on TLR3 are strongly associated with recurrent herpes simplex virus infections, leading to dramatic encephalitis.4 Unlike other NAS TLRs whose expression is restricted to a few immune cell types, TLR3 is expressed in various cell types, including myeloid cells (macrophages, microglia and dendritic cells), but also epithelial cells, vessel endothelial cells, fibroblasts and hepatocytes.5 Activation of the other TLRs leads to the formation of a supramolecular complex containing the adaptor MyD88, called the myddosome. This complex constitutes a signalling platform, leading to the production of type I IFN and inflammatory cytokines. In contrast, TLR3 activation is MyD88 independent and leads to the recruitment of TRIF, an adaptor specific of this pathway, with the exception of TLR4 which can use both.6,7 Whether TRIF also forms a supramolecular complex remains elusive. Ultimately, TLR3 activation cascade leads to the production of inflammatory cytokines and type I IFNs.8 Given the potent response that TLR3 engagement can elicit, ligands of TLR3 are currently tested in clinical trials to boost the immune response in immunotherapy protocols. As a reflection of this potency, TLR3 appears to be tightly regulated at different levels to prevent its activation against the self, as indicated by its contribution to the pathogenesis of autoimmune rheumatoid arthritis.9–11
Our current knowledge of the regulation of NAS TLRs and of their precise involvement in the immune response remains, however, rather superficial. While the intracellular trafficking route of the NAS TLR7 and 9 has been studied in detail,12–15 the trafficking of TLR3 remains to be better established. All the NAS TLRs associate upon neo-synthesis with the chaperone UNC93B1 in the endoplasmic reticulum (ER), which is central in their post-ER trafficking towards endosomes.12,13 In the absence of functional UNC93B1, none of the NAS TLR can function.16–19 TLR3, like the other NAS TLRs, has to reach the endosome where it becomes functional by cleavage of its ectodomain and can then encounter its dsRNA ligand.20–23 Regulation of TLR3 can thus occur by controlling its access to the endosomal compartment where its proteolytic maturation can take place. Moreover, spatial dissociation of sensing, that is, TLR3 binding to dsRNA, and signalling through TRIF in a different set of endosomes represent an attractive level of regulation that remains to be demonstrated.24,25 Overall, knowledge about the post-translational regulation of TLR3 to prevent activation by self-nucleic acids remains rather superficial.
Schematically, TLR3 signalling leads to the recruitment of TRIF and TRAFs adaptors and activation of two signalling cascades.8,26,27 First, TRIF and TRAF6 initiate a cascade leading to the activation of NF-κB and the production of inflammatory cytokines.27,28 In parallel, TRIF and TRAF3 recruitment leads to IRF3 phosphorylation inducing its dimerisation and nuclear translocation where it drives the expression of type I IFN.29,30 Despite these studies, we still have only partial knowledge of the molecular players involved in TLR3 signalling and of their regulation.
The development of CRIPSR/Cas9 technology combined with lentiviral delivery of single-guide RNA (sgRNA) is the method of choice to perform gene editing for systematic genetic screens in mammalian cells.31,32 Given the lack of knowledge regarding the regulation of the TLR3 pathway and the great potential for new molecules that could precisely control its signalling, we developed a genome-wide screen to identify new players controlling the TLR3 pathway.
Materials and methods
Cell culture
The near-haploid myeloid cell line KBM7, derived from human myeloid leukaemia (kindly provided by Prof. T. Brummelkamp) was cultured in Iscove’s Modified Dulbecco’s Medium plus 10% FCS and penicillin/streptomycin in 5% CO2 at 37°C. 293FT (cat. #R70007 RRID: CVCL_6911 Thermo Fisher Scientific, Waltham, MA) cells were cultured in DMEM plus 10% FCS and penicillin/streptomycin in 5% CO2 at 37°C. Macrophages were obtained as published in Decalf et al.33 Plasmapheresis residues were obtained from healthy adult donors (EFS Blood Bank, Paris, France). Peripheral PBMCs were separated using Ficoll-Paque (GE Healthcare, Chicago, IL), and monocytes were isolated by positive selection using CD14 magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) and differentiated into macrophages for 7 d in RPMI (Gibco, Life Technologies, Grand Island, NY) supplemented with 5% FCS (BioWest, Nuaillé, France), 5% human serum AB (Sigma–Aldrich, St Louis, MO), penicillin/streptomycin (Gibco) and 50 ng/ml M-CSF (ImmunoTools, Friesoythe, Germany).
Abs and reagents
Abs
The Abs used in this study were: rabbit polyclonal Ab anti-aryl hydrocarbon receptor (AhR; BML-SA210; WB 1/1000; Enzo Life Sciences, Inc., Farmingdale, NY); mouse mAb (clone C4) anti-actin (MAB1501; WB 1/5000; Merck, Darmstadt, Germany); goat Ab anti-rabbit IgG linked to HRP (7074; WB 1/3000; Cell Signaling Technology, Danvers, MA); horse Ab anti-mouse IgG linked to HRP (7076; WB 1/3000; Cell Signaling Technology); mouse Ab anti-TLR3 (40F9.6; WB 1/1000; kindly provided by Innate Pharma, Marseille, France); rat monoclonal Ab (Clone YL1/2) anti-tubulin alpha (MCA77G; WB 1/2500; Bio-Rad, Hercules, CA); and goat polyclonal Ab anti-rat IgG linked to HRP (112-036-062; Jackson ImmunoResearch, West Grove, PA).
Reagents
The reagents used in this study were: poly(I:C) low molecular mass (tlrl-picw; InvivoGen, San Diego, CA), recombinant human TNF-α (T6674; Sigma–Aldrich), 6-formylindolo[3,2-b]carbazole (FICZ; BML-GR206-0100; Enzo Life Sciences), StemRegenin 1 (SR-1; T1831; Bertin Bioreagent, Montigny le Bretonneux, France) and concanamycin B.34
Plasmids
pTRH1 NF-κB destabilized copepod GFP (dscGFP) was a gift from Prof. Hidde Ploegh.35 Its promoter contains four repeats of the NF-κB binding site GGGACTTTCC; pTRH1 NF-κB RFP was produced from pTRH1 NF-κB dscGFP by introducing TagRFP in place of dscGFP by overlapping PCR (forward 5′-gtcaaagcttaccatggtgtctaagggcgaaga-3′; reverse 5′-tgacgtcgactattaattaagtttgtgcccca -3′) and the restriction sites HindIII and SalI. pTRIP-SFFV TLR3-HA 2A mTagBFP2 salI was produced from hTLR3-4HA22 and pTRIP-SFFV-GFP by successive overlapping PCRs. pHRSIN pSFFV FLAG NLS Cas9 NLS pGK Hygro was a gift from Prof. Paul Lehner. To generate targeted sgRNA targeting lentivectors, we used the lentiCRISPR v2 (gifted from Prof. Feng Zhang; plasmid # 52961; Addgene, Watertown, MA36). As recommended, we digested the BsMB1 sites before annealing with the appropriate oligos. Supplemental Table S1 displays the list of sgRNA used to control the candidates from the screen (*sgRNA further used in the article).
CRISPR screen
The screen was performed as described.37 The Cas9 enzyme was stably transduced in KBM7 cells. The MOI of the GeCKO v2 sgRNA library A and B (generously deposited by Prof. Feng Zhang32; #1000000047; Addgene) was estimated on cells exposed to various volumes of viruses and cultured with or without puromycin before counting the remaining cells. At d 1, 100 million cells were transduced at a MOI of 0.09 and 0.25 for each screen. At d 2, puromycin was added (2 µg/ml) to remove untransduced cells from the suspension. At d 7, cells were stimulated twice by 35 µg/ml of poly(I:C) 4 h apart. At d 8, 16 h after the last stimulation, cells were enriched by FACS on the 5% GFP low cells. This enrichment was performed three times successively, each time following a stimulation of the cells, at d 8, 16 and 22. From each sort, genomic DNA was extracted (Gentra Puregene Cell Kit 158767; Qiagen, Valencia, CA) from both the sorted cells and a control-unselected pool of mutagenised cells. sgRNA amplification, sequencing and identification were performed as described.38
Library amplification
The GeCKO library was amplified according to the protocol from Feng Zhang’s laboratory.36
Production of lentivector
The lentivector used to transduce the GeCKO library, or the other constructs, was produced as described.39 Viral particles were produced by transfection of 293FT cells in six-well plates with 3 µg DNA and 8 µl TransIT-293 (Mirus Bio, Madison, WI) per well. Cells were transfected with 0.4 µg CMV-VSVG, 1 µg psPAX2 and 1.6 µg of the plasmids of interest. One d after transfection, medium was removed, and fresh DMEM was added. Viral supernatants were harvested 1 d later, filtered at 0.45 µM and used freshly or aliquoted and frozen at –80°C. For TLR3 transduction, ∼40 ml of viruses were ultra-centrifuged for 90 min at 120,000 g using a Beckman LE80K ultracentrifuge (Beckman Coulter, Brea, CA). The pellet was then re-suspended in 500 µl of DMEM. The KBM7 cells were spinoculated for 2 h at 1200 g before being mixed with the virus.
Flow cytometry
The GFP expression and the cytokine bead assays were analysed on a FACSVERSE (BD Biosciences, San Jose, CA; laser 488 nm, filter 527/32) for the GFP. The RFP expression was analysed on a LSRFORTESSA (BD Biosciences; laser 561 nm, filter 610/20). The sorts for the screen were performed on a FACSARIA III (BD Biosciences) or the S3 Cell Sorter (Bio-Rad).
RNA isolation and quantitative RT-PCR and oligonucleotides
The total RNA from the samples was extracted from cultured cells using Nucleospin RNA II from (740955.5; Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. The RNA samples were then reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (4368814; Thermo Fisher Scientific). Quantitative PCR was then performed using the LightCycler 480 SYBR Green I Master (04887352001; Roche, Basel, Switzerland) according to the manufacturer’s instructions. The oligonucleotides used for cytochrome P450 family 1 subfamily A member 1 (CYP1A1) mRNA were: forward, 5′-AGGCCCTGATTACCCAGAAT-3′; reverse, 5′-TCCCAGCTCAGCTCAGTACC-3′.
Immunoblotting
Cells were lysed in RIPA 2× (100 mM Tris, pH 7.4; 300 mM NaCl; Triton 2%; SDS 0.2%; deoxycholate 1%; 1× protease inhibitor cocktail; 11873580001; Sigma–Aldrich). After 30 min of incubation on ice, samples were centrifuged 10 min at 16,100 g at 4°C. Protein quantification was performed on the supernatant by micro BCA (23235; Thermo Fisher Scientific). The samples were normalised for their protein content. LDS sample buffer (NP0008; Thermo Fisher Scientific) containing 5% β-mercaptoethanol was added to the samples. After 10 min at 95°C, samples were loaded and resolved on Mini-PROTEAN TGX Precast Protein gradient polyacrylamide gels (4–15% or 4–20%; Bio-Rad). Samples were subsequently transferred to a polyvinylidene difluoride membrane (#1704156; Bio-Rad) and blocked with 5% (w/v) skim-milk powder in PBS containing 0.1% Tween-20 (PBS-Tween) for 1 h at room temperature (20°C). Membranes were then probed with an appropriate dilution of primary Ab overnight (14 h) at 4°C. Membranes were washed three times in PBS-Tween before incubation in diluted secondary Ab for 1 h at room temperature. Membranes were washed as before and developed with ECL (#1705061; Bio-Rad; or 34096; Thermo Fisher Scientific) using a Bio-Rad Chemi Doc system setting for signal accumulation. Pictures were analysed and prepared using the Image Lab (Bio-Rad) software.
Cytokines secretion assay
IP-10 and IL-8 concentration was measured after 16 h of stimulation using IP-10 of IL-8 cytometric bead assay (558280 and 558277; BD Biosciences) according to the manufacturer’s protocol.
Statistics and analysis
All statistics were performed as described using GraphPad Prism v6 (GraphPad Software, Inc., San Diego, CA). The analysis of the IP-10 and IL-8 cytometric assay was performed on FCAP Array (BD Biosciences). Graphs were generated with either GraphPad Prism v6 or RStudio (RStudio, Inc., Boston, MA).
Results
A genome-wide screen identifies genes required for TLR3-induced response
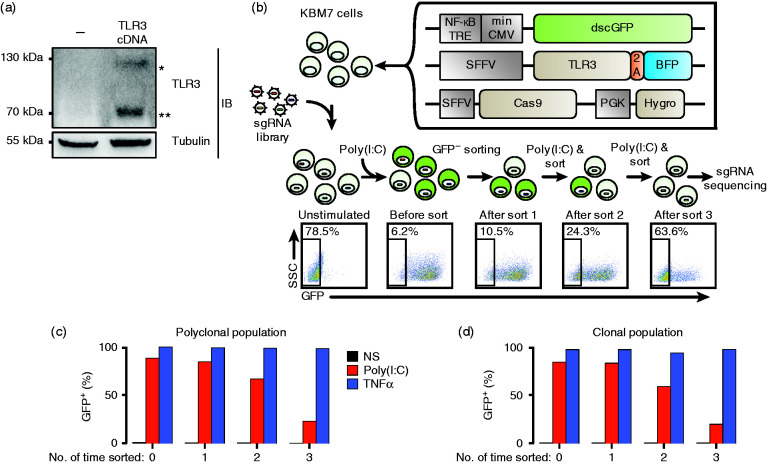
One of the early steps following TLR3 signalling is the activation of the NF-κB transcription factor that is translocated to the nucleus. To monitor TLR3 activation easily, we created a near-haploid KBM7 cell line transduced with an NF-κB reporter construct made of a minimal CMV promoter with four NF-κB binding sites driving the expression of the dscGFP (which has a shorter half-life than the widely used eGFP) or the RFP. The KBM7 reporter cells did not respond to poly(I:C) but did to TNF-α, another NF-κB activator that is independent of TLR/TRIF pathways (Supplemental Figure S1a). We thus transduced the KBM7 reporter cells with TLR3 cDNA and controlled by Western blot that TLR3 was correctly expressed and processed generating a 70 kDa fragment (Figure 1a).22 Next, to select for KBM7 cells capable of a high GFP response upon TLR3 activation but still able to return after a few days to a GFP-negative state, we performed two rounds of cell sorting. First, following poly(I:C) treatment, GFP-positive cells were sorted by flow cytometry, and second, after a week of culture, cells that had returned to a GFP-negative state were sorted. The resulting cells were transduced with a third lentivirus encoding the Cas9 enzyme (see Figure 1b) and selected with hygromycin. These cells, referred to as KBM7Rep cells, were maintained in bulk and, in parallel, subcloned.
Figure 1.
Outline of the genome-wide CRISPR/Cas9 forward genetic screen to identify genes required for TLR3 signalling. (a) Transduction of KBM7 cells with TLR3 cDNA leads to the expression of both full-length (*) and cleaved/mature (**) TLR3. Immunoblot of cell lysates of KBM7 cells complemented by the indicated cDNA was revealed by an anti-TLR3 mAb. As a loading control, tubulin was revealed with a specific mAb. (b) Design of the forward genetic screen in human near-haploid KBM7 cells. Creation of the NF-κB reporter KBM7 cell line by transduction of three independent expression cassettes encoding for dscGFP, TLR3 and Cas9. DscGFP expression was controlled by a NF-κB dependent promoter. Cells were transduced with the lentiviral GeCKO v2 sgRNA library (122,411 sgRNAs). Cells that were successfully transduced were selected with puromycin. After selection, the population was split into two; half was not sorted to represent the entire library, while the remaining population was sorted by flow cytometry after poly(I:C) stimulation to enrich for dscGFP negative cells. After three rounds of poly(I:C) stimulation/enrichment, the DNA from the enriched populations (rounds 1, 2 and 3) was harvested, and enriched sgRNAs were identified by sequencing and compared to an unsorted library. FACS plots obtained after each sort on dscGFP-negative cells show progressive enrichment rates of dscGFP-negative cells. (c) and (d) Proportion of cells responding to poly(I:C) as judged by dscGFP expression before and after sorting during the sequential enrichment process. Polyclonal or clonal cell populations were stimulated with poly(I:C; twice at 35 µg/ml, 4 h apart) or TNF-α (10 ng/ml) for 16 h before sorting at each round. In both cases, the proportion of dscGFP negative cells increased at each round of enrichment. sgRNA: single-guide RNA; NF-κB TRE: NF-κB transcription responsive element; min CMV: minimal CMV; dscGFP: destabilized copepod GFP; SFFV: spleen focus-forming virus promoter; 2A: peptide bond skipping sequence; BFP: blue fluorescent protein: PGK: phosphoglycerate kinase promoter; Hygro: hygromycin resistance gene.
Next, we verified that the KBM7Rep cells properly responded to poly(I:C) by monitoring the production of IP-10 and IL-8 (Supplemental Figure S1b) and the activation of the NF-κB reporter (Supplemental Figure S1c). The absence of response to poly(I:C) exposure of the original KBM7 cell line, unless transduced with TLR3 cDNA, indicated that the responses we monitored were not the result of other RNA sensors such as MDA5 or RIGI in our conditions.
TLR3 signalling is strictly dependent on the acidic pH present in the endosomal pathway.22,40 Inhibition of the proton pump responsible for the acidification of the endosomes can be achieved very rapidly and efficiently with the macrolide ConB.34 ConB exposure of KBM7Rep cells totally abrogated poly(I:C)-induced GFP expression, but did not when cells were stimulated by TNF-α (Supplemental Figure S1c). Moreover, IP-10 and IL-8 production by KBM7Rep cells in response to poly(I:C) required TLR3 expression and was totally abrogated by ConB exposure (Supplemental Figure S1b).
Next, we controlled that the Cas9 introduced in the reporter cells was active using an sgRNA targeting the β2 microglobulin by following cell surface expression of MHC I,41 which dramatically decreased in a large proportion of the population, revealing high rates of gene editing (not shown). We concluded that our reporter cells were fitted for the screen and decided to run two screens in parallel: one with the cells maintained as a bulk population, and the other with a clonal one.
We performed a genome-wide CRISPR/Cas9 forward genetic screen using the lentiviral GeCKO v2 sgRNA library to transduce our reporter cells. The library contained 122,411 sgRNAs targeting 19,050 genes. We aimed for a low MOI (< 0.3) to limit the possibility of two viruses infecting the same cell.32,38 We designed the screen to enrich for loss of function of the response to poly(I:C). Cells that were successfully transduced were selected with puromycin and enriched for GFP-negative cells after poly(I:C) stimulation (Figure 1b). For both polyclonal and clonal cell populations, a progressive enrichment in GFP-negative cells took place along the rounds of stimulation/sorting (Figure 1c and d).
Genome-wide screen identified expected and new genes required for TLR3 signalling
The sgRNA abundance of the enriched populations obtained after one, two or three rounds of stimulation/sorting was estimated twice by deep sequencing and compared to a control unsorted population for each of the two screens (Figure 2). We chose a cut-off, considering only genes identified with a –log(P-value) > 4 found in more than 2/12 total estimations of enrichment generated with the two screens (Figure 2a). Strikingly, the most significantly enriched genes identified were TLR3 itself and UNC93B1. The fact that both hits possessed the lowest P value indicated that our whole screening process was correct to probe for genes involved in TLR3 signalling (Figure 2b). Moreover, we also identified two genes involved in the NF-κB pathway: CHUK and RELA. Both genes are known members of the NF-κB pathway that have already been extensively studied in the context of TLR signalling. Finally, using a different cut-off to analyse the data (i.e. a –log(P value) > 3) found in at least 5/12 enrichment analyses performed, in addition to the already mentioned four hits, we found the TICAM1 gene coding for TRIF protein (Supplemental Figure S2).
Figure 2.
Genome-wide screen of the TLR3 pathway identifies expected and new genes. (a) –log P value of the results of sgRNA enrichments identified by sequencing, corresponding to 42 genes. After one, two or three rounds of enrichment, the DNA from the enriched cell population was harvested, and enriched sgRNAs were identified by sequencing and comparison to an unsorted library. This determination was carried out for the two independent screens performed in parallel, and sequencing was performed twice generating a total of 12 enriched cell populations. Genes are presented when –log(P value) > 4 for at least 2/12 comparisons. Each dot represents the gene enrichments calculated from one condition. The RSA algorithm was used to identify the significantly enriched genes targeted in the selected cells. Red: known key members of TLR3 pathway; blue: further validated genes. (b) Plot illustrating the hits from the genetic screen. Mean log(P values) determined with the clonal population as a function of those found with polyclonal population. Each dot is a gene. Red squares: known key members of the TLR3 pathway; light blue triangles: genes further validated; grey dots: genes further tested; density colours: remaining genes out of the 42 found in panel (a).
We concluded that the genome-wide screen performed was likely to identify new genes involved in the TLR3 pathway. Thus, we selected the 35 genes encoding proteins identified in Figure 2a for further studies.
DLX1, SOD1 and AhR are required for IL-8 production by poly(I:C) stimulated cells
For each of the 35 hits, we selected and cloned two sgRNAs in a lentivector encoding the Cas9 enzyme. (Selection of the sgRNAs was based on the sgRNAs found enriched in the screen.) The sgRNAs encoding lentiviral vectors were transduced in KBM7Rep cells, selected in puromycin-containing medium and maintained as bulk populations. Cells were exposed for 16 h to increasing doses of poly(I:C) or to a single dose of TNF-α and then analysed by FACS to monitor NF-κB activity by following GFP expression (Figure 3a). Confirming the results of the screen, the sgRNA targeting UNC93B1 and TLR3 were the most active at inhibiting TLR3 signalling as judged by the levels of GFP expression (Figure 3a and b). Interestingly, sgRNA targeting DLX1, SOD1 and AhR grouped together with the best hits (Figure 3a). We next examined IL-8 and IP-10 production. In response to poly(I:C) stimulation and compared to the Cas9 control, the DLX1, SOD1 and AhR sgRNA-transduced cell lines exhibited a residual IL-8 production, similar to UNC93B1 sgRNA-transduced cells (Figure 3c). While the IP-10 response was abrogated in cells transduced with the UNC93B1 sgRNA, the cells transduced with AhR or SOD1 sgRNA demonstrated an unexpected increase in IP-10 production (although to a lesser extent for SOD1 sgRNA), while the DLX1 sgRNA did not modify the IP-10 response (Figure 3d).
Figure 3.
DLX1, SOD1 and AhR are required for a proper TLR3 response. (a) Heat map of the GFP expression results obtained with KBM7Rep cells transduced with the various sgRNAs (n = 3 independent experiments). KBM7Rep cells transduced with the indicated sgRNAs were exposed to increasing doses of poly(I:C) or TNF-α (10 ng/ml) for 16 h before measurement of the GFP expression by FACS. The cluster of genes containing two sgRNAs targeting UNC93B1 and one targeting TLR3 is magnified. red: positive controls; blue: genes further studied; grey: negative controls. (b) NF-κB activity or (c) IL-8 and (d) IP-10 were measured by FACS or cytometric bead assay (CBA), respectively, in response to poly(I:C) exposure of the indicated cells (two-way ANOVA between one sgRNA condition and ctrl Cas9 ctrl). (e), (f) and (g) The indicated parameters were measured in the absence or presence of TNF-α (n = 3 independent experiments). *, **, *** or ****: significantly different from ctrl Cas9 (Friedman test, with Dunn’s multiple comparison test between sgRNA conditions and Cas9 ctrl). UNC: UNC93B1. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
The specificity of the effects observed on the TLR3 pathway was assayed in parallel by following the response to TNF-α. While UNC93B1 sgRNA-transduced cells retained their responsiveness in all cases, the DLX1, SOD1 and AhR sgRNA-transduced cell lines remained only slightly impaired in their NF-κB and IL-8 responses to TNF-α (Figure 3e and f), as the differences were statistically non-significant. In contrast, the IP-10 response to TNF-α was significantly higher in AhR sgRNA-transduced cells compared to control cells (Figure 3g), indicating that the effects of AhR sgRNA are not restricted to TLR3 signalling.
Next, we selected AhR for further study, since its absence generated the strongest phenotype. AhR is a ligand-induced transcription factor endowed with a large immunomodulatory capacity on different immune cell types.42 AhR agonists direct bind and free AhR from its cytosolic complex made of hepatitis B virus X-associated protein 2 and a dimer of heat shock protein 90,43,44 allowing for its translocation to the nucleus where it can exert its transcription factor activity on several genes.45 These include the up-regulation of CYP1A1.46 The availability of the AhR agonist FICZ and the AhR antagonist SR-1 enables functional interrogation of AhR in the TLR3 response, complementing the genetic approach.
AhR-Deficient cells produce less IL-8 and more IP-10 than wild type cells in response to TLR3 stimulation
We first verified that AhR expression was abolished in the KBM7Rep cells transduced with the AhR sgRNA by immunoblot, while it was clearly present in the ctrl Cas9 population (Figure 4a). We confirmed that editing the AhR gene in the KBM7 reporter cells led to a reduced capacity to respond to poly(I:C) in terms of GFP and IL-8 expression (Figure 4b and c). In the absence of AhR, the IL-8 response to TNF-α was decreased (by sixfold), while the IP-10 one was increased (by 7.4-fold; Figure 4c and d). Of note, while AhR gene edition led to a significant increase in IP-10 production when cells were exposed to a range of doses of poly(I:C) (considering the entire dose range for the statistical test; Figure 3d), this effect was maintained but not significant considering a single dose tested of 25 µg/ml (Figure 4d).
Figure 4.
Cells edited for AhR exhibit a decreased IL-8 and an increased IP-10 response to poly(I:C). (a) Immunoblots of cell lysates of KBM7Rep cells KO for AhR or ctrl Cas9 cells revealed with an anti-AhR mAb or with an anti-actin mAb. (b) NF-κB activity or (c) IL-8 and (d) IP-10 were measured by FACS or CBA, respectively, under the various conditions and cells as indicated, in response to poly(I:C; 25 μg/ml) or TNF-α (10 ng/ml; n = 4 independent experiments). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 (two-way ANOVA followed by Sidak’s multiple comparisons test for each panel).
Agonist and antagonist of AhR modulate IL-8 production by TLR3-stimulated cells
To evaluate whether AhR was involved in the TLR3 pathway by an independent approach, we used well-described AhR agonists and antagonists.47,48 The KBM7Rep cells edited for AhR and the ctrl Cas9 cell population were exposed to poly(I:C; 25 µg/ml) or to TNF-α (10 ng/ml) and to increasing doses of agonist (FICZ) or antagonist (SR-1). We observed that FICZ increased the NF-κB and IL-8 responses to poly(I:C), while SR-1 inhibited them (Figure 5a and b). Importantly, these effects were abolished in the absence of AhR. Regarding the IP-10 responses, SR-1 and FICZ had almost no effect on the response to poly(I:C; Figure 5c). The effects of the AhR ligands on the response to TNF-α were not detected, with the exception of a non-statistically significant inhibition of the NF-κB reporter expression by SR-1 exposure (Figure 5a). Thus, the AhR agonist and the antagonist impact the TLR3 response in an AhR-dependent manner, suggesting that the activity of AhR can impact the TLR3 response.
Figure 5.
AhR agonist and antagonist affect IL-8 response to poly(I:C). (a) GFP expression, (b) IL-8 and (c) IP-10 productions were determined in control or AHR KO cells in response to poly(I:C; 25 μg/ml) or TNF-α (10 ng/ml) in the presence of increasing doses of AhR agonist (FICZ) and antagonist (SR-1; n = 4 independent experiments). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 (for each curve vs. the appropriate DMSO control, two-way ANOVA).
AhR pathway is involved in the TLR3-mediated response in primary human macrophages
To extend our findings to primary cells naturally expressing TLR3, we repeated similar experiments in primary human monocyte-derived macrophages (MDMs). Here, we used a fixed concentration of AhR ligands (5 µM) and increasing doses of poly(I:C) and TNF-α. Interestingly, the agonist FICZ significantly increased the IL-8 response to TLR3 stimulation, while the antagonist SR-1 had the opposite effect (Figure 6a). Similar but lower effects of the AhR ligands were observed in response to TNF-α (Figure 6a). The IP-10 response to poly(I:C) was inhibited by FICZ but also to a lesser extent by SR-1 (Figure 6b). In contrast, both AhR ligands exhibited no clear effect on the IP-10 response to TNF-α (Figure 6b).
Figure 6.
Primary myeloid cells response to poly(I:C) is affected by AhR agonist and antagonist. (a) and (b) AhR modulators impact human monocyte derived macrophages (MDMs) response to poly(I:C) and TNF-α. MDMs were treated with FICZ (5 µM), SR-1 (5 µM) or vehicle (DMSO) for 1 h before stimulation with increasing doses of poly(I:C) or TNF-α. (a) IL-8 and (b) IP-10 were measured by CBA (n = 5 independent donors). *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001 (for each curve vs. the appropriate DMSO control, two-way ANOVA). (c) AhR activity was enhanced by the agonist and inhibited by the antagonist. CYP1A1 mRNA expression of MDM treated with FICZ (5 μM) or SR-1 (5 μM) for 17 h was quantified by quantitative RT-PCR (n = 5 independent donors). *P ≤ 0.05; ****P ≤ 0.0001 (Kruskal–Wallis test with Dunn’s multiple comparisons test).
As a control of the activity of the AhR ligands on MDMs, we measured their effects on the expression of the CYP1A1 mRNA by quantitative RT-PCR.46 As expected, FICZ dramatically increased the CYP1A1 expression (by 36-fold), while SR-1 reduced these levels (by twofold; Figure 6c).
Discussion
Here, we applied a genome-wide CRISPR screen to identify regulators of the TLR3 pathway using reporter cells expressing GFP upon NF-κB activation. Following mutagenesis and enrichment of cells with reduced GFP expression after poly(I:C) exposure, several genes were identified. TLR3 itself and UNC93B1 grouped together as the genes with the highest enrichment of corresponding sgRNAs, confirming the validity of our screening process. The ER resident chaperone UNC93B1 is essential for the functioning of all the NAS TLRs.16,18 This multi-pass transmembrane protein controls their trafficking from the ER to the endosomes and remains associated with the TLRs through post-Golgi steps.12,13 Although the details of the mechanism by which TLR3 trafficking is controlled by UNC93B1 remain elusive, it was reassuring to identify this key protein as a strong hit in our genetic screen.
We also identified the TICAM1/TRIF adaptor (Supplemental Figure S2), which is recruited by TLR3 at the level of its cytosolic tail upon ligand binding to its ectodomain in endosomes.49 TICAM1/TRIF is specific of the TLR3 pathway, as the other NAS TLRs use MyD88 instead, with the exception of TLR4 which can use both.50 Two additional genes related to the NF-κB pathway were also identified by the screen. First, CHUK, also named IKKα, encodes a serine kinase part of the IKK complex.51 CHUK phosphorylates IκB and thus triggers its degradation via the ubiquitination pathway, thereby activating the NF-κB transcription factor.51,52 Second, the RELA gene product is complexed with the p50 subunit of NF-κB and is required for its activities on transcription.52 Taken together, these results indicated that our screen strategy was suitable to identify molecular players involved at different levels in the regulation of the TLR3 pathway.
Among the newly identified hits by the screen, we focused on DLX1, SOD1 and AhR to validate our finding using independent read-outs. We observed that editing of each of these three genes led to a strong inhibition of the IL-8 response to poly(I:C) in addition to a diminished NF-κB activity as measured by GFP expression in the reporter cells. However, compared to the DLX1 and SOD1 sgRNAs, the AhR sgRNA appeared to have the highest positive impact on the IP-10 response to poly(I:C). Moreover, DLX1 is mostly expressed during development53 and thus might have been identified in our screen as the result of the particular cell line used, that is, the near-haploid KBM7 cell which was derived from a chronic myeloid leukaemia.54 Superoxide dismutase 1 (SOD1) is a cytosolic enzyme able to convert free superoxide radicals into hydrogen peroxide H2O2 and oxygen.55 The absence of SOD1 may thus affect the finely tuned concentrations of cytosolic ROS that may impact the TLR3 pathway.
We further focused on AhR, an environmental sensor involved in the differentiation of several T-cell types such as CD4+ Th17 and Treg42 and implicated in the induced tolerance to LPS.56 We found that AhR editing had a dual effect on the TLR3 response: abrogation of the IL-8 production and enhancement of the IP-10 response. AhR can accommodate different ligands, including the carcinogenic pollutant 2,3,7,8-tétrachlorodibenzo-p-dioxine,57 or tryptophan metabolites, as well as ligands derived from food.58,59 In the absence of ligand, AhR is complexed in the cytosol with several chaperones.60,61 Ligand binding induces AhR dissociation from its chaperones and its nuclear translocation, where it regulates gene expression with its canonical partner ARNT.61 RelA and RelB, two subunits of the NF-κB complex, have been shown to bind AhR independently of ARNT.62–64 AhR has recently attracted attention due to its close association with NF-κB members that appears crucial in pathological responses induced by environmental insults.65 In macrophages stimulated by LPS, AhR, NF-κB and STAT1 form a complex that negatively regulate the production of IL-6. From these studies, AhR emerges as a key regulator of inflammatory responses through epigenetic mechanisms that remain in large part elusive.65,66
Interestingly, RelB-AhR response elements are present in the IL-8 promotor and are required for proper IL-8 expression in U937 macrophages.63 This may explain that TLR3-mediated production of IL-8 is AhR-dependent. In contrast, in cells edited for AhR, we observed increased expression of IP-10 in response to TLR3, suggesting that AhR can repress IP-10 production. Similarly, in a recent study performed with human dendritic cells, TLR4 and TLR3-mediated activation combined with AhR agonists also led to dual effects, increasing some cytokine productions and decreasing others.67 The authors proposed that this was due to the presence of RelB-AhR response elements in the promoter of the up-regulated cytokines and to the AhR-induced repressor down-regulating the others.
Finally, in primary human macrophages, TLR3-mediated production of IL-8 was increased by AhR agonist and decreased by AhR antagonist. However, IP-10 response to poly(I:C) was strongly inhibited by AhR agonist and to some extent by AhR antagonist. The inconsistency of the impact of AhR stimulation on IP-10 response to TLR3 observed between KBM7 and primary macrophages supports cell-type-specific regulatory mechanism(s) that remain obscure.
Future work should establish how AhR is involved in the different TLR pathways in different cell types. Our study is in line with the idea that AhR is a powerful modulator and environmental sensor involved in the regulation of the TLR3 pathway.
Supplemental Material
Supplemental material, INI915507 Supplemental Material1 for A genome-wide CRISPR screen identifies regulation factors of the TLR3 signalling pathway by Laurent Zablocki-Thomas, Sam A Menzies, Paul J Lehner, Nicolas Manel and Philippe Benaroch in Innate Immunity
Supplemental material, INI915507 Supplemental Material2 for A genome-wide CRISPR screen identifies regulation factors of the TLR3 signalling pathway by Laurent Zablocki-Thomas, Sam A Menzies, Paul J Lehner, Nicolas Manel and Philippe Benaroch in Innate Immunity
Acknowledgements
The authors would like to thank Francesca Graziano, François-Xavier Gobert and Elodie Segura from the Institut Curie, as well as Xavier Coumoul from Université Paris Descartes, for helpful discussions. Vasco Rodrigues and Francesca Graziano are thanked for their critical reading of the manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: This work was supported by Institut Curie, INSERM, CNRS and by grants from the ANR-10-IDEX-0001-02 PSL and ANR-11-LABX-0043 to N.M. and P.B.; INCA 40832, LNCC PARIS – RS19/75-1, ARC PJA 20181208045, Sidaction 2018-1-AEQ-11984, ANRS and ANR 15-CE13-0009-01 to P.B.; and Sidaction VIH2016126002, ANRS ECTZ25472, ANRS ECTZ36691, ANR-14-CE14-0004-02 and ANR-17-CE15-0025-01 to N.M. L.Z.-T. was supported by a doctoral fellowship from École normale supérieure de Lyon, followed by another one from the Association de la Recherche contre le Cancer (ARC).
ORCID iD
Laurent Zablocki-Thomas https://orcid.org/0000-0001-9917-7815
Supplemental material
Supplemental material for this article is available online.
References
- 1.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol 2014; 5: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasius AL, Beutler B. Intracellular Toll-like receptors. Immunity 2010; 32: 305–315. [DOI] [PubMed] [Google Scholar]
- 3.Alexopoulou L, Holt AC, Medzhitov R, et al. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001; 413: 732–738. [DOI] [PubMed] [Google Scholar]
- 4.Zhang SY, Jouanguy E, Ugolini S, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science 2007; 317: 1522–1527. [DOI] [PubMed] [Google Scholar]
- 5.Zhang SY, Herman M, Ciancanelli MJ, et al. TLR3 immunity to infection in mice and humans. Curr Opin Immunol 2013; 25: 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto M, Sato S, Mori K, et al. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-Like receptor signaling. J Immunol 2002; 169: 6668–6672. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, Sato S, Hemmi H, et al. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 2003; 301: 640–643. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Z, Mak TW, Sen G, et al. Toll-like receptor 3-mediated activation of NF-kappaB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-beta. Proc Natl Acad Sci U S A 2004; 101: 3533–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brentano F, Schorr O, Gay RE, et al. RNA released from necrotic synovial fluid cells activates rheumatoid arthritis synovial fibroblasts via Toll-like receptor 3. Arthritis Rheum 2005; 52: 2656–2665. [DOI] [PubMed] [Google Scholar]
- 10.Lee S-Y, Yoon B-Y, Kim J-I, et al. Interleukin-17 increases the expression of Toll-like receptor 3 via the STAT3 pathway in rheumatoid arthritis fibroblast-like synoviocytes. Immunology 2014; 141: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng L, Zhu W, Jiang C, et al. Toll-like receptor 3 upregulation in macrophages participates in the initiation and maintenance of pristane-induced arthritis in rats. Arthritis Res Ther 2010; 12: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee BL, Moon JE, Shu JH, et al. UNC93B1 mediates differential trafficking of endosomal TLRs. Elife 2013; 2: e00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YM, Brinkmann MM, Paquet ME, et al. UNC93B1 delivers nucleotide-sensing Toll-like receptors to endolysosomes. Nature 2008; 452: 234–238. [DOI] [PubMed] [Google Scholar]
- 14.Pelka K, Bertheloot D, Reimer E, et al. The chaperone UNC93B1 regulates Toll-like receptor stability independently of endosomal TLR transport. Immunity 2018; 48: 911–922.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiya T, Kajita E, Miwa S, et al. TLR3 and TLR7 are targeted to the same intracellular compartments by distinct regulatory elements. J Biol Chem 2005; 280: 37107–3717. [DOI] [PubMed] [Google Scholar]
- 16.Tabeta K, Hoebe K, Janssen EM, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol 2006; 7: 156–164. [DOI] [PubMed] [Google Scholar]
- 17.Brinkmann MM, Spooner E, Hoebe K, et al. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol 2007; 177: 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casrouge A, Zhang SY, Eidenschenk C, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 2006; 314: 308–312. [DOI] [PubMed] [Google Scholar]
- 19.Itoh H, Tatematsu M, Watanabe A, et al. UNC93B1 physically associates with human TLR8 and regulates TLR8-mediated signaling. PLoS One 2011; 6: e28500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewald SE, Engel A, Lee J, et al. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med 2011; 208: 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi R, Singh D, Kao CC. Proteolytic processing regulates Toll-like receptor 3 stability and endosomal localization. J Biol Chem 2012; 287: 32617–32629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Cattaneo A, Gobert F-X, Muller M, et al. Cleavage of Toll-like receptor 3 by cathepsins B and H is essential for signaling. Proc Natl Acad Sci U S A 2012; 109: 9053–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnsen IB, Nguyen TT, Ringdal M, et al. Toll-like receptor 3 associates with c-Src tyrosine kinase on endosomes to initiate antiviral signaling. EMBO J 2006; 25: 3335–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chow J, Franz KM, Kagan JC. PRRs are watching you: Localization of innate sensing and signaling regulators. Virology 2015; 479–480: 104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol 2009; 9: 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol 2004; 4: 499–511. [DOI] [PubMed] [Google Scholar]
- 27.Sato S, Sugiyama M, Yamamoto M, et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN- β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor. J Immunol 2003; 171: 4304–4310. [DOI] [PubMed] [Google Scholar]
- 28.Cusson-Hermance N, Khurana S, Lee TH, et al. Rip1 mediates the TRIF-dependent Toll-like receptor 3- and 4-induced NF-κB activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem 2005; 280: 36560–36566. [DOI] [PubMed] [Google Scholar]
- 29.Oganesyan G, Saha SK, Guo B, et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 2006; 439: 208–211. [DOI] [PubMed] [Google Scholar]
- 30.Doyle SE, Vaidya SA, O’Connell R, et al. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 2002; 17: 251–263. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhu S, Cai C, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 2014; 509: 487–491. [DOI] [PubMed] [Google Scholar]
- 32.Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014; 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Decalf J, Desdouits M, Rodrigues V, et al. Sensing of HIV-1 entry triggers a type I interferon response in human primary macrophages. J Virol 2017; 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yilla M, Tan A, Ito K, et al. Involvement of the vacuolar H(+)-ATPases in the secretory pathway of HepG2 cells. J Biol Chem 1993; 268: 19092–19100. [PubMed] [Google Scholar]
- 35.Lee CC, Carette JE, Brummelkamp TR, et al. A reporter screen in a human haploid cell line identifies CYLD as a constitutive inhibitor of NF-κB. PLoS One 2013; 8: e70339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014; 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart SE, Menzies SA, Popa SJ, et al. A genome-wide CRISPR screen reconciles the role of N-linked glycosylation in galectin-3 transport to the cell surface. J Cell Sci 2017; 130: 3234–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timms RT, Menzies SA, Tchasovnikarova IA, et al. Genetic dissection of mammalian ERAD through comparative haploid and CRISPR forward genetic screens. Nat Commun 2016; 7: 11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silvin A, Yu CI, Lahaye X, et al. Constitutive resistance to viral infection in human CD141+ dendritic cells. Sci Immunol 2017; 2: eaai8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Bouteiller O, Merck E, Hasan UA, et al. Recognition of double-stranded RNA by human Toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J Biol Chem 2005; 280: 38133–38145. [DOI] [PubMed] [Google Scholar]
- 41.Burr SP, Costa ASH, Grice GL, et al. Mitochondrial protein lipoylation and the 2-oxoglutarate dehydrogenase complex controls HIF1α stability in aerobic conditions. Cell Metab 2016; 24: 740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutiérrez-Vázquez C, Quintana FJ. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity 2018; 48: 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer BK, Perdew GH. Characterization of the AhR-hsp90-XAP2 core complex and the role of the immunophilin-related protein XAP2 in AhR stabilization. Biochemistry 1999; 38: 8907–8917. [DOI] [PubMed] [Google Scholar]
- 44.Perdew GH. Association of the Ah receptor with the 90-kDa heat shock protein. J Biol Chem 1988; 263: 13802–13806. [PubMed] [Google Scholar]
- 45.Ikuta T, Eguchi H, Tachibana T, et al. Nuclear localization and export signals of the human aryl hydrocarbon receptor. J Biol Chem 1998; 273: 2895–2904. [DOI] [PubMed] [Google Scholar]
- 46.Nebert DW, Roe AL, Dieter MZ, et al. Role of the aromatic hydrocarbon receptor and (Ah) gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol 2000; 59: 65–85. [DOI] [PubMed] [Google Scholar]
- 47.Rannug U, Rannug A, Sjöberg U, et al. Structure elucidation of two tryptophan-derived, high affinity Ah receptor ligands. Chem Biol 1995; 2: 841–845. [DOI] [PubMed] [Google Scholar]
- 48.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 2010; 329: 1345–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita M, Chattopadhyay S, Fensterl V, et al. Epidermal growth factor receptor is essential for Toll-like receptor 3 signaling. Sci Signal 2012; 5: ra50–ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Neill LAJ, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 2007; 7: 353–364. [DOI] [PubMed] [Google Scholar]
- 51.Häcker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE 2006; 2006: re13. [DOI] [PubMed] [Google Scholar]
- 52.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 2012; 26: 203–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merlo GR, Zerega B, Paleari L, et al. Multiple functions of Dlx genes. Int J Dev Biol 2000; 44: 619–626. [PubMed] [Google Scholar]
- 54.Kotecki M, Reddy PS, Cochran BH. Isolation and characterization of a near-haploid human cell line. Exp Cell Res 1999; 252: 273–280. [DOI] [PubMed] [Google Scholar]
- 55.Dimayuga FO, Wang C, Clark JM, et al. SOD1 overexpression alters ROS production and reduces neurotoxic inflammatory signaling in microglial cells. J Neuroimmunol 2007; 182: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bessede A, Gargaro M, Pallotta MT, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature 2014; 511: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guyton KZ, Rusyn I, Chiu WA, et al. Application of the key characteristics of carcinogens in cancer hazard identification. Carcinogenesis 2018; 39: 614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veldhoen M, Hirota K, Christensen J, et al. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med 2009; 206: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H, Wei Y, Yu D. Control of lymphocyte homeostasis and effector function by the aryl hydrocarbon receptor. Int Immunopharmacol 2015; 28: 818–824. [DOI] [PubMed] [Google Scholar]
- 60.Stockinger B, Di Meglio P, Gialitakis M, et al. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol 2014; 32: 403–432. [DOI] [PubMed] [Google Scholar]
- 61.Larigot L, Juricek L, Dairou J, et al. AhR signaling pathways and regulatory functions. Biochim Open 2018; 7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen PH, Chang H, Chang JT, et al. Aryl hydrocarbon receptor in association with RelA modulates IL-6 expression in non-smoking lung cancer. Oncogene 2012; 31: 2555–2565. [DOI] [PubMed] [Google Scholar]
- 63.Vogel CFA, Sciullo E, Li W, et al. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol 2007; 21: 2941–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogel CFA, Sciullo E, Matsumura F. Involvement of RelB in aryl hydrocarbon receptor-mediated induction of chemokines. Biochem Biophys Res Commun 2007; 363: 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian Y. Ah receptor and NF-κB interplay on the stage of epigenome. Biochem Pharmacol 2009; 77: 670–680. [DOI] [PubMed] [Google Scholar]
- 66.Kimura A, Naka T, Nakahama T, et al. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med 2009; 206: 2027–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kado S, Chang WLW, Chi AN, et al. Aryl hydrocarbon receptor signaling modifies Toll-like receptor-regulated responses in human dendritic cells. Arch Toxicol 2017; 91: 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, INI915507 Supplemental Material1 for A genome-wide CRISPR screen identifies regulation factors of the TLR3 signalling pathway by Laurent Zablocki-Thomas, Sam A Menzies, Paul J Lehner, Nicolas Manel and Philippe Benaroch in Innate Immunity
Supplemental material, INI915507 Supplemental Material2 for A genome-wide CRISPR screen identifies regulation factors of the TLR3 signalling pathway by Laurent Zablocki-Thomas, Sam A Menzies, Paul J Lehner, Nicolas Manel and Philippe Benaroch in Innate Immunity