Abstract
Human CD4+ T cells are often subdivided into distinct subtypes, including Th1, Th2, Th17, and Treg cells, that are thought to carry out distinct functions in the body. Typically, these T-cell subpopulations are defined by the expression of distinct gene repertoires; however, there is variability between studies regarding the methods used for isolation and the markers used to define each T-cell subtype. Therefore, how reliably studies can be compared to one another remains an open question. Moreover, previous analysis of gene expression in CD4+ T-cell subsets has largely focused on gene expression rather than alternative splicing. Here we take a meta-analysis approach, comparing eleven independent RNA-seq studies of human Th1, Th2, Th17, and/or Treg cells to determine the consistency in gene expression and splicing within each subtype across studies. We find that known master-regulators are consistently enriched in the appropriate subtype; however, cytokines and other genes often used as markers are more variable. Importantly, we also identify previously unknown transcriptomic markers that appear to consistently differentiate between subsets, including a few Treg-specific splicing patterns. Together this work highlights the heterogeneity in gene expression between samples designated as the same subtype, but also suggests additional markers that can be used to define functional groupings.
Keywords: T cells, Th1, Th2, Th17, Treg, transcriptomics, alternative splicing
INTRODUCTION
The adaptive immune response relies on the ability of T cells to detect foreign antigen and respond by carrying out appropriate functions, such as the secretion of cytotoxins or cytokines. Importantly, T cells are not a uniform cell type, rather it is now recognized that multiple subtypes of T cells are generated during development and/or an immune response based on the nature of the foreign antigen and/or the context in which the antigen engages with T cells (DuPage and Bluestone 2016). In particular, subtypes of CD4+ T cells differ in the antigens they engage and in the nature of their response to antigen, resulting in the optimal functional response to various types of immune challenge. However, there is abundant variation in the field in how CD4+ T subtypes are isolated and defined (DuPage and Bluestone 2016; Stockinger and Omenetti 2017). While this variability is widely acknowledged, its impact on the nature of the molecular characteristics of the cells studied has not been analyzed in detail.
Three of the most widely studied T-cell subtypes are the T-helper 1 (Th1), T-helper 2 (Th2), and T-helper 17 (Th17) subsets of CD4+ T cells, each of which have been defined as expressing signature cytokines. Th1 cells secrete the cytokine interferon gamma (IFNγ) to promote an innate immune response against viruses or cancer (Tran et al. 2014), while parasite-fighting Th2 cells secrete the cytokines interleukin (IL)-4, IL-5, and IL-13 (Pulendran and Artis 2012). Th17 cells secrete IL-17 and IL-23 to fight fungal infections (Harrington et al. 2005) or cancer. Importantly, an inappropriate balance of T-cell subtypes not only reduces effectiveness of fighting pathogens but can also cause disease. Overabundance or activity of Th1 cells contribute to colitis (Harbour et al. 2015), Th2 cells contribute to asthma and allergies (Venkayya et al. 2002), and Th17 cells contribute to multiple sclerosis and other autoimmune diseases (Vaknin-Dembinsky et al. 2006).
T-cell subsets have historically been defined based on the expression of a lineage-defining master transcription factor (Shih et al. 2014). The Th-specific master regulatory transcription factors include: T-box transcription factor TBX21 (T-bet) for Th1 (Szabo et al. 2002), GATA-binding protein 3 (GATA3) for Th2 (Zhang et al. 1997; Zheng and Flavell 1997), and RAR-related orphan receptor gamma (RORγ) for Th17 (Ivanov et al. 2006). However, using these two key factors, a master regulator and signature cytokines, to define T-cell subsets is increasingly appreciated to be too simple to adequately explain the breadth and plasticity that has been observed for T-cell populations (DuPage and Bluestone 2016). For example, another common T-cell subtype are regulatory T cells (Treg), which suppress immune responses. Treg suppressive function was found to depend on the master regulator Forkhead Box P3 (FOXP3) (Zheng and Rudensky 2007), but there are no well-defined signature cytokines for Treg cells. Treg cells produce TGFB, IL-10, or IL-35, which are critical anti-inflammatory cytokines, but not all of these cytokines are produced by all Treg cells (Shih et al. 2014). Moreover, core signature cytokines and master regulators such as IFNγ and T-bet have been observed in Th17, Th22 and other hematopoietic cells (Shih et al. 2014).
Another definition used to discriminate T-cell subsets is their ability to sense and migrate to specific chemokines through expression of distinct chemokine receptors, including: CXCR3 for Th1, CCR4 for Th2, CCR6 for Th17 and IL2RA for Tregs (DuPage and Bluestone 2016). However, transitionary T cells exist that simultaneously express chemokine receptors from two subsets (Cohen et al. 2011). Moreover, based on all of the above definitions, T cells have shown remarkable plasticity in their ability to polarize between distinct T-cell subtypes (Bending et al. 2009; DuPage and Bluestone 2016). Therefore, it is clear that better descriptions are needed of the molecular differences that define functionally distinct T-cell subpopulations.
A complication to the study of CD4+ subtypes is that methods to isolate populations vary widely across the field. One approach to purifying T-cell subsets is the use of antibodies to chemokine receptors or other extracellular marker proteins to isolate specific T-cell subsets directly from blood using flow cytometry or magnetic beads. However, these methods are limited by the above-mentioned variability and overlap in expression of these marker proteins. In contrast, an alternate method to enrich for T-cell subsets is polarizing naïve CD4+ T cells toward distinct phenotypes in vitro with various cytokine cocktails. While these cytokine cocktails are meant to mimic the environment that promotes the development of each subtype, the conditions used vary from one laboratory to another, thereby also inducing variability between studies. Importantly, while the field acknowledges that distinct methods of isolation of CD4+ subtypes likely generate functionally different cells (DuPage and Bluestone 2016), the extent to which these cell populations differ has not been thoroughly examined. Moreover, the use of the same designation (i.e., Th1) for cells purified from blood or polarized in culture complicates the literature and begs the question of whether or not it is appropriate to compare cell populations from distinct studies (DuPage and Bluestone 2016; Zhu 2018).
Here we seek to better understand the variability and or similarities between T-cell subsets generated by distinct methodologies by taking a meta-analysis approach to compare gene expression across a range of T-cell subsets that have been isolated and defined by a variety of methods. By comparing RNA-seq data across eleven independent studies we identify a set of ∼20–50 genes whose expression is well-correlated with distinct T-cell subsets. Notably, these include some, but not all, of the genes encoding the cytokines and receptors typically used to define T-cell subsets, as well as some additional genes that have not previously been considered indicators of cell fate. In addition, we also investigated the extent to which alternative splicing contributes to transcriptomic variation between subsets. We indeed find a small set of genes for which splicing patterns at least somewhat correlate with cell subtype; however, splicing seems to be less definitively regulated in a subtype-specific manner than transcription. Together, our findings are consistent with models arguing for more elaborate and nuanced definitions of T-cell populations (Shih et al. 2014). Moreover, our data provide important information as to the underlying biologic differences between CD4+ subsets and suggest new molecular markers that may also be used to define these populations.
RESULTS
Selection of data sets and RNA-seq analysis pipeline
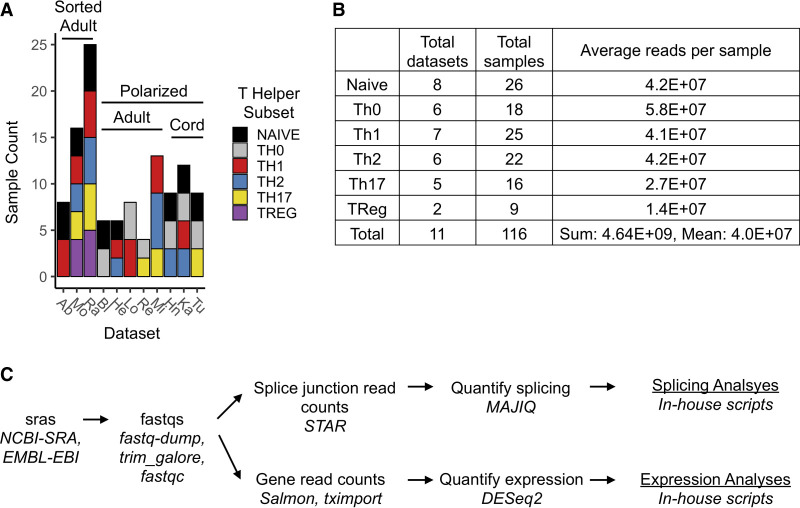
Given the extensive variability in the methods used in the field to generate and define CD4+ T-cell subsets we were interested in determining how much molecular variation exists in the resulting cell populations and whether there are transcriptomic signatures that robustly identify T-cell subsets regardless of experimental conditions. To carry out a meta-analysis of transcriptomic variation in T-cell subtypes, we identified RNA-seq experiments in the NCBI-GEO and EMBL-EBI-ArrayExpress databases that were from poly(A)-selected RNA samples derived from CD4+ T cells purified from the blood of healthy humans and had at least two out of three of the following types of samples: naïve (no stimulation), Th0 (stimulated in the absence of polarizing cytokines), or specific T-cell subsets. We differentiate between naïve and Th0 cells, as stimulation through the T-cell receptor alone (as in Th0) has been shown by us and others to dramatically impact transcriptome expression compared to naïve cells (Martinez et al. 2012, 2015). We also confirmed the quality of the data sets by requiring that all samples had >60% uniquely mapped reads and no nonhuman overrepresented sequences. Lastly, we performed principle component analysis of samples by gene expression to confirm that in each study samples clustered by cell type (Supplemental Fig. S1). In total, 10 publicly available data sets, plus one in-house data set (Bl, GSE135118), met these inclusion and quality control criteria (Fig. 1A; Supplemental Table S1). These 11 data sets include multiple replicate samples of naïve and Th0 CD4+ cells, as well as Th1, Th2, Th17, and Treg subpopulations (Fig. 1A,B). Importantly, these data sets encompassed samples of specific T-cell subsets obtained using one of two general approaches: (i) sorting cells from whole blood using known extracellular protein markers, or (ii) in vitro polarization of naïve CD4+ T cells toward Th1, Th2, or Th17 cell fates with specific cytokine cocktails. Data sets that generated T-cell populations by in vitro polarization obtained naïve precursors from either adult whole blood or neonatal cord blood and used distinct cytokine combinations (Fig. 1A). A detailed description of the experimental conditions and RNA-seq technical specifications for each data set is shown in Supplemental Table S1.
FIGURE 1.
Data sets and analysis pipeline used for meta-analysis. (A) Data sets used in this study and the breakdown of samples and isolation methods for each data set. Sorted: samples sorted from blood; Polarized: samples in vitro polarized; Adult or Cord: cells derived from adult or cord blood. Ab (Abadier et al. 2017), Mo (Monaco et al. 2019), Ra (Ranzani et al. 2015), Bl (this study), He (Hertweck et al. 2016), Lo (Locci et al. 2016), Re (Revu et al. 2018), Mi (Micossé et al. 2019), Hn (Henriksson et al. 2019), Ka (Kanduri et al. 2015), Tu (Tuomela et al. 2016). For further detail, see Supplemental Table S1. (B) Total number and quality of data sets for each T-cell subtype used in this study. (C) Pipelines used to process RNA-seq data and quantify gene expression and splicing variation.
To begin to look for common patterns of gene and isoform expression in the above data sets, raw RNA-seq data from all samples were uniformly processed by trimming low quality base calls and adaptor sequences and aligning reads to the hg38 genome (Fig. 1C). Gene expression was then quantified with the Salmon and DESeq2 algorithms, while local splicing variations were quantified with the MAJIQ algorithm (Fig. 1C), which is optimized to detect complex and unannotated splicing events (Vaquero-Garcia et al. 2016). Importantly, all but two of these data sets used a donor-paired experimental design (i.e., multiple cell types were isolated or derived from each given donor), allowing us to directly compare transcriptome profiles within T-cell subsets from an individual donor. RNA-seq-based genotyping was used to confirm all sample pairings, as well as to identify sample pairings in those studies in which such information was not given (see Materials and Methods). For expression analysis we used the asinh transformation. The asinh transformation is a commonly used method for stabilization of variance in TPM in gene expression analysis (Huber et al. 2002; SEQC/MAQC-III Consortium 2014; Francesconi et al. 2019).
Master regulators are consistently expressed in a subtype-specific manner, while other common markers are not
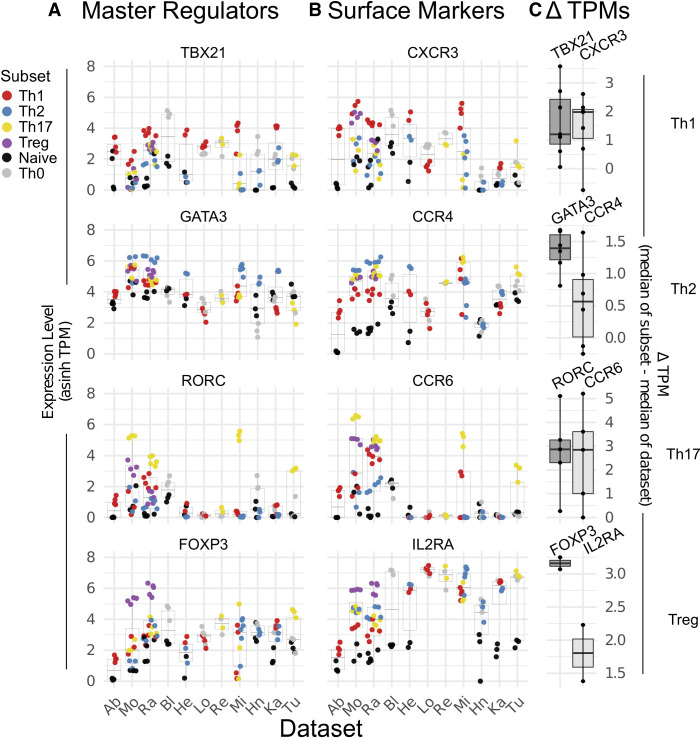
In order to determine the validity and robustness of both the data sets and our analysis pipeline, we first assessed the expression of well-accepted signature genes of Th1, Th2, Th17, and Treg cell populations across the various subpopulations. Although the absolute expression of previously described “master regulators” for Th1 (TBX21), Th2 (GATA3), Th17 (RORC), and Treg (FOXP3) varied from study to study, within any given study the majority of the Th1, Th2, Th17, and Treg samples expressed their respectively known master regulators more highly than any other cell type (Fig. 2A). For example, 25 of the 25 Th1 populations across all seven Th1-containing data sets, expressed the Th1 master regulator TBX21, as or more highly than other cells in the same study (Fig. 2A, top panel). Similarly, 21 of 22 Th2 samples highly expressed the Th2 master regulator GATA3 (Fig. 2A, second panel), and all of the Th17 and Treg samples were enriched for their respective master regulators RORC and FOXP3 (Fig. 2A, bottom two panels). As emphasized above, each study analyzed here used different protocols to obtain T Helper cell RNA-seq samples. Therefore, these enrichment data demonstrate that master regulator gene expression clearly segregates Th1, Th2, Th17, and Treg populations independent of experimental method.
FIGURE 2.
Expression of classical master regulators and cell surface markers across T-cell subsets. Expression of signature genes typically used to delineate T Helper cells, including genes encoding (A) master transcription regulators and (B) cell-surface proteins, across all data sets. Each subplot represents a distinct gene, while each column in a specific subplot is a distinct study. Studies are listed at bottom and ordered as in Figure 1A. Cell type listed on right is the subtype expected to be enriched for the two genes plotted on that row. For each study, the mean and distribution of the given gene is displayed as a box plot, with the values for each individual sample represented as a dot. Dots are colored by cell subtype. Note that not all studies contain all cell types, as described in Figure 1A. Expression values shown as the inverse hyperbolic sine (asinh) of transcripts per million (TPM). (C) Comparison of the median TPM in the expression of the indicated genes in the corresponding cell subtype relative to all others in the data set. Dots are values for individual data sets, and the boxplot shows the range and median for each gene. In each case, the darker color is the master regulator and the lighter color is the surface marker.
Interestingly, unlike the clear expression patterns of master regulators, several extracellular markers commonly used to isolate T Helper cell subpopulations showed less consistent Th1, Th2, Th17, and Treg-specific expression patterns (Fig. 2B,C). The Th1 extracellular marker CXCR3 was preferentially expressed six out of seven data sets; however, in one data set (Lo), Th1 cells express less CXCR3 than Th0 cells. This variability in expression cannot be explained by the method of cell isolation as Lo used similar methods as other studies that do show Th1-specific expression of CXCR3 (in vitro polarized, He and Si). Similarly, CCR4 only was enriched in Th2 cells in only three of the six data sets that include these cells (Mo, Ra, and He), while the Th17 extracellular marker CCR6 showed Th17-specific expression patterns in only 9/16 samples across four out of five data sets. Finally, while the Treg extracellular marker IL2RA was highly expressed in 9/9 Treg samples from both data sets with Treg samples, its difference in expression compared to other T-cell subsets is markedly lower than that observed for FoxP3 (Fig. 2C).
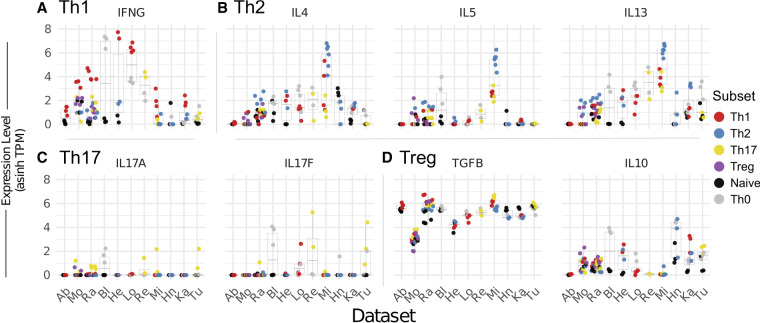
We also find significant variability in the extent to which T-cell subtype samples expressed their expected signature cytokines (Fig. 3). Almost all of the Th1 samples (22/25) do express the Th1-specific cytokine IFNG more highly than other cell types in the same study. In contrast, we observe no enrichment of the Treg-associated cytokines TGFB or IL10 in Treg cells relative to others (Fig. 3). Th17 and Th2 cells show some bias in expression of their associated cytokines, IL4/5/13 and IL17A/F, respectively, over other cell types, but only approximately half of the individual Th17 or Th2 cell samples express higher levels of these cytokines than other cells in the same studies (Fig. 3). Taken together, the above analysis of genes previously associated with Th1, Th2, Th17, and Treg populations reveals significant variability of all but the “master regulator” genes. This raises concerns about the validity of using mRNA levels of cytokines and cell surface receptors as consistent and reliable identifiers of T-cell subtypes, although it is possible that cytokine protein expression does correlate well with T-cell subtype as many cytokines are regulated at a posttranscriptional level (Anderson 2008). In addition, the data we analyze here does not rule out the possibility that there is more synchrony of gene expression at other time points during T-cell differentiation.
FIGURE 3.
Expression of cytokines across T-cell subsets. Expression of cytokines associated with (A) Th1, (B) Th2, (C) Th17, or (D) Treg cells, across all data sets. Each plot represents a distinct gene, while each column is a distinct study. Studies are listed at bottom and ordered as in Figure 1A. For each study, the mean and distribution of the given gene is displayed as a box plot, with the values for each individual sample represented as a dot. Dots are colored by cell subtype. Note that not all studies contain all cell types, as described in Figure 1A. Expression values shown as the inverse hyperbolic sine (asinh) of transcripts per million (TPM).
Th1, Th2, Th17, and Treg populations highly express core sets of genes
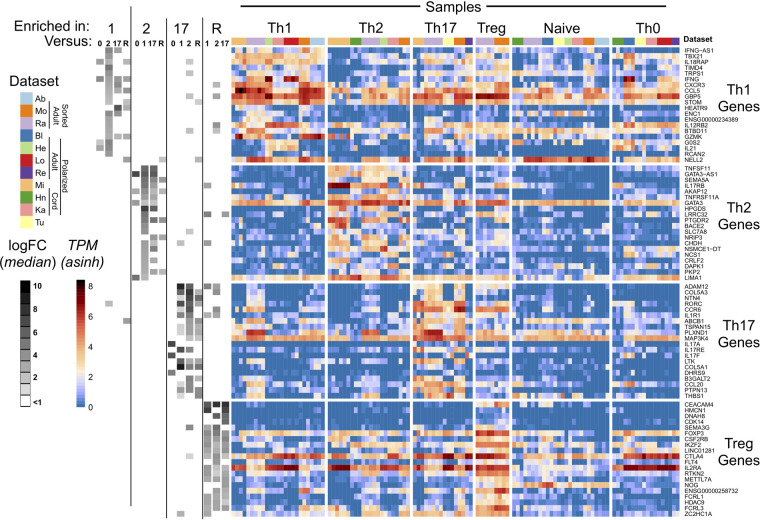
Given the relatively limited consistency of expression of genes expected to correlate with T-cell subtypes, we next asked if other genes might be consistently enriched in Th1, Th2, Th17, and/or Treg cells regardless of variation in culture conditions and cellular sources. Specifically we mined the full gene expression data as determined by DESeq2 (Fig. 1C) for genes “reliably expressed” in a given T-cell subset, which we define as genes that are significantly more highly expressed in a given subset relative to others, in at least two of the studies analyzed (see Materials and Methods). 555 unique genes were reliably higher in at least one T-cell subset over another across multiple data sets (Supplemental Table S2). These 555 subset-associated genes were then ranked by the number of data sets in which they were enriched, followed by the fold difference in expression between the subset analyzed versus others (Supplemental Fig. S2).
Notably, by this ranking, the top 20 most associated genes for each T Helper subset (Fig. 4) include the “master regulators” TBX21 for Th1, GATA3 for Th2, RORC for Th17, and FOXP3 for Treg. Moreover, consistent with the analysis of specific cytokines and chemokine receptors in Figures 2 and 3, the top Th1-associated genes include CXCR3 and IFNG, the top 20 Th17-associated genes include IL17A/F and CCR6, and IL2RA is among the top 20 Treg-associated genes (Fig. 4). In contrast, none of the Th2 signature cytokines or receptors genes (IL-4/5/13 and CCR4) are significantly enriched in Th2 cells compared to the other populations surveyed (Fig. 4).
FIGURE 4.
T cells consistently express core set of genes in a subset-specific manner. Reliably expressed T Helper genes (as defined in Materials and Methods), ranked by number of supporting data sets and log2FC differences. The heatmap shows inverse hyperbolic sine (asinh)-transformed TPMs. Each column is a sample, and samples are grouped by cell type and author. Gene names are on the right. Grayscale boxes in the leftmost segment indicate whether a given core T Helper gene was reliably more highly expressed in a given subset over others. The median logFC between the given subtype over others is quantified by the grayscale of the boxes (darker indicates higher logFC). All differential expression analyses were performed between samples from the same data set. The data sets with Treg samples lacked Th0 samples, so there is no “enriched in Treg vs. Th0” column.
In addition to the master regulators, cytokines, and chemokine receptors specific to distinct CD4+ T-cell subtypes, many of the core genes enriched in Th1, Th2, Th17, and Treg cells have been implicated in the biology of these cells (see Discussion). However, at least two of the top 20 Th1 core genes (TRPS1 and STOM) and three of the top 20 Th2 core genes (TNFSF11, TNFRSF11A, and LRRC32) have not previously been implicated in Th1 and Th2 biology, respectively, and may represent potential new markers of CD4+ subtypes (see Discussion). Therefore, our identification of subset-associated genes highlights additional genes that may contribute to the function of particular T-cell subsets and/or be useful as markers for subpopulations. Importantly, the expression of many of these core genes in CD4+ T-cell subtypes is independent, at least at steady state, from the expression of master regulators, as indicated by limited correlation in the expression levels of the genes in Figure 4 with the corresponding master regulators (Supplemental Fig. S3).
Analysis of local splicing variations reveals Treg-biased isoform expression
Given our success in identifying genes whose expression is highly associated with specific T-cell subsets, we next asked the question of whether particular splicing patterns (i.e., mRNA isoforms) of certain genes was also correlated with T-cell subtype. Alternative splicing is a ubiquitous mRNA processing step that arises from differential inclusion of exons, introns or portions thereof. Such splicing variations often result in different protein isoforms, which can have disparate functions (Braunschweig et al. 2013). Recent studies have revealed widespread and co-regulated alternative splicing early in CD4+ T-cell activation in the absence of polarizing cytokines (Ip et al. 2007; Martinez and Lynch 2013; Martinez et al. 2015). While a few studies have reported differential splicing or expression of splicing regulatory proteins in particular T-cell subsets in mice (Stubbington et al. 2015; Middleton et al. 2017), a comprehensive comparison across human T-cell subsets has not been reported.
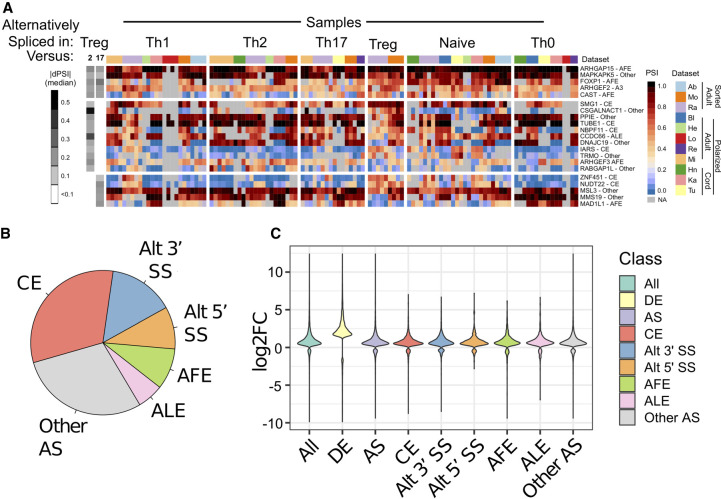
In contrast to the results with gene expression, we find very few instances in which we observe consistent differences in splicing patterns in a T-cell subtype-specific manner (Fig. 5A; Supplemental Table S3). The few instances of subset-specific splicing that we can detect across data sets are cases of modest differential isoform expression in Treg cells versus Th2 or Th17 cells (Fig. 5A). These splicing events represent all standard classes of splicing patterns (Fig. 5B) and occur in genes that do not display any differences in overall expression (Fig. 5C; Supplemental Table S2). Therefore, the isoform differences that do exist between T-cell subtypes are not readily detected in typical gene expression profiling.
FIGURE 5.
Only a limited number of splicing events are consistently regulated in a subset-specific manner in T cells. (A) Heatmap of PSI value for splicing events that show reproducible differences between Treg cells and Th2 and/or Th17 cells. Each column is a sample, and samples are grouped by cell type and author. Gene names and splicing event are on the right. Gray boxes in the heatmap indicate splicing events that lacked sufficient RNA-seq read depth to accurately quantify PSI. Grayscale boxes in the leftmost segment indicate comparisons that met significance threshold. The median dPSI of significant differences between the given subtype over others is quantified by the grayscale of the boxes (darker indicates higher dPSI). Significant differences are based on comparison between samples from the same data set (Ra and Mo have Treg), but PSI values are shown for all samples. (B) Pie chart of splicing events identified as differentially spliced between all T Helper subset comparisons. Categories include cassette exons (CE), alternative first exons (AFE), alternative 3′ss (Alt3′ss), alternative last exon (ALE), or alternative 5′ss (Alt5′ss), or other nondefined patterns of splicing (Other AS). (C) Change in expression (Log2FC) in all genes that are differentially expressed (DE) or alternatively spliced (AS) between two cell subtypes studies as compared to all genes (ALL). The change in expression of each type of splicing event is also shown as for B.
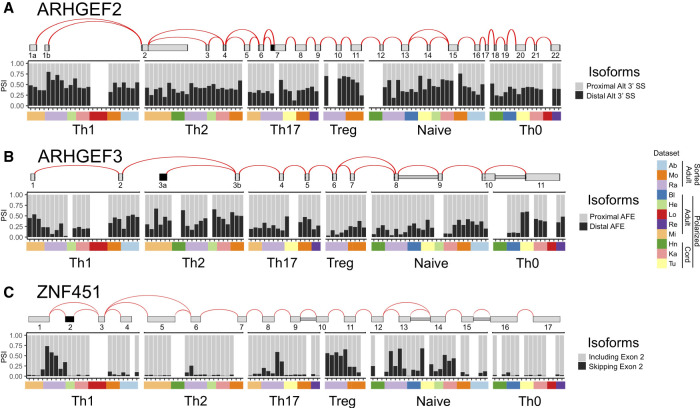
Overall, the data in Figure 5 suggests that alternative splicing is not a general determinant of T-cell identity. However, we do note that some of the observed Treg-biased splicing events are in genes linked to cytokine expression and immune function and thus are potentially of interest for future studies. For example, ARHGEF2 exhibits differential use of alternative 3′ splice sites at the beginning of exon 7 in Treg cells versus Th2 and Th17 cells (Figs. 5A, 6A). ARHGEF2 encodes GEF-H1, a Rho guanine nucleotide exchange factor (Rho-GEF) that is involved in the response to intracellular pathogens and is required for expression of IL1β and IL6 (Wang et al. 2017). Tregs generally express more of the isoform that uses the distal 3′ splice site than Th2 or Th17 cells (Figs. 5A, 6A). Use of this distal 3′ splice site results in the removal of a single alanine residue in the linker between the microtubule binding domain and the enzymatic Dbl-homology domain and has been shown to correlate with loss of RhoA-enhancing activity by GEF-H1 (Chen et al. 2019). A related Rho-GEF, ARHGEF3, is also somewhat differentially spliced between Treg and Th2 cells in that a proximal alternative first exon is favored in Tregs versus Th2 cells, thus altering the first 32–38 amino acids of the encoded protein (Fig. 6B). While functional differences have not been identified between the isoforms with distinct amino-termini, ARHGEF3 has been linked to activation of RhoA in myeloid development (D'Amato et al. 2015).
FIGURE 6.
Treg-biased splicing events are predicted to alter protein function. Details of the splicing patterns for genes (A) ARHGEF2, (B) ARHGEF3, and (C) ZNF451, that exhibit some Treg bias (top) and the quantification of the variable events across samples (bottom).
Finally, a particularly interesting case is ZNF451, which exhibits preferentially skipping of an in-frame exon 2 in Treg cells as compared to Th17 cells (and perhaps also Th2 cells, although not scored as significant due to limited read count, Fig. 6C). ZNF451 is a SUMO E3 ligase and has also been shown to physically interact with Smad4 to repress its activation of TGF-β signaling (Feng et al. 2014; Cappadocia et al. 2015). Notably, exon 2, encodes part of the domain required to recruit SUMO to substrates (Cappadocia et al. 2015). While the role of the E3 ligase activity of ZNF451 to TGF-β signaling has not been defined, the differential expression of exon 2 in Treg cells suggests that this splicing difference may be a mechanism to differentially regulate TGF-β signaling in T-cell subsets.
DISCUSSION
Much variability exists in how T-cell subsets are differentiated and defined. To investigate how much variability exists in the gene expression profiles of nominally similar but experimentally distinct T-cell populations, we took a meta-analysis approach comparing RNA-seq data collected across distinct laboratories and methods. Importantly, while our transcriptomic analysis does confirm enriched expression of subtype-specific master regulators, we find that many other genes markers commonly associated with Th1, Th2, Th17, or Treg cells show limited predictive power to differentiate subtypes, at least for the range of time points and conditions encompassed here. On the other hand, our analysis uncovers a handful of genes previously unknown to be regulated in a cell-type specific manner that show significant enrichment in one T-cell subset compared to others. Finally, we show that while alternative splicing appears to play a limited role in shaping T-cell differentiation into subsets, a few exceptions to this rule may exist, especially in Treg cells.
Using a pair-wise comparison method we were able to identify ∼860 genes that exhibit differential expression between at least two T-cell subsets (Fig. 4; Supplemental Table S3). We emphasize that the genes highlighted in Figure 4 are consistently enriched in a CD4+ T-cell subset regardless of purification method or cell source, and thus are likely genes that are intimately tied to the biology of each subset. For example, the top ten Th1 reliably expressed genes were mostly previously known to be important for Th1 biology, including the master regulator TBX21 (Szabo et al. 2002), the Th1-associated cytokines IFNG and CXCR3 (DuPage and Bluestone 2016), the IFNG enhancer IFNG-AS1 (Collier et al. 2014), as well as IL18RAP (Jenner et al. 2009), TIMD4 (Nakajima et al. 2005), GBP5 (Lund et al. 2007), and CCL5 (Shadidi et al. 2003).
Similarly, the top ten reliably expressed genes in Th2, Th17 and Treg cells include those implicated in relevant biology. For example, the top Th2 core genes includes the Th2 master regulator GATA3 and other genes previously implicated in Th2 biology such as GATA3-AS1, PTGDR2 (aka CRTH2), SEMA5A, IL17RB, AKAP12, and HPGDS (Angkasekwinai et al. 2007; Lund et al. 2007; Wang et al. 2007; Zhang et al. 2013; Mitson-Salazar et al. 2016); while the top ten Th17 reliably expressed genes includes Th17 master regulator RORC, as well as genes known to be important for Th17 biology including ADAM12 (Zhou et al. 2013), COL5A3 (Castro et al. 2017), CCR6, IL1R1 (Hu et al. 2011), ABCB1 (Ramesh et al. 2014), PLXND1 (Guo et al. 2016), and MAP3K4 (Cleret-Buhot et al. 2015). Finally, the top ten Treg reliably expressed genes includes Treg master regulator FOXP3 and additional genes known to be important for Treg biology including CEACAM4 (Hua et al. 2015), HMCN1 (Sadlon et al. 2010), DNAH8 (Regateiro et al. 2012), SEMA3G, CSF2RB (Bhairavabhotla et al. 2016), IKZF2 (Thornton et al. 2010), LINC01281 (Ranzani et al. 2015), and CTLA4 (Takahashi et al. 2000).
Importantly, beyond confirming genes known to be involved in CD4+ T-cell identity, our analysis also revealed genes that may represent novel biology or new markers for specific CD4+ T-cell subsets. For example, Th1 identified core genes CCL5 and TIMD4 encode for cell-surface-exposed proteins, so it would be especially interesting if these proteins could be used for isolating or quantifying Th1 cell populations. In addition, one of the top 10 Th1-reliable genes, TRPS1, is not known to be important for Th1 cell populations specifically but is implicated in Th17 (Yosef et al. 2013). STOM was also reliably expressed in Th1 samples, but it is not known what role, if any, STOM plays in T-cell biology. Similarly, Th2 identified core genes TNFSF11 and SEMA5A encode for cell-surface-exposed proteins and are more consistent Th2 cell markers at the mRNA level than CCR4 or IL4/5/13 (Fig. 4), suggesting that TNFSF11 and SEMA5A proteins might have utility in isolation of Th2 cells by flow cytometry, while the enrichment of TNFSF11A and LLRC32 among the highly Th2-associated genes, suggest new Th2 biology. Th17 identified core genes ADAM12, COL5A3, NTN4, and TSPAN15 are as at least as consistently expressed in a Th17-specific manner than the commonly used genes RORC, CCR6, or IL17A/IL17F (Fig. 4). ADAM12 and TSPAN15 encode for cell-surface-exposed proteins, so could be used for isolating or quantifying Th17 cell populations. Treg identified core genes CEACAM4, CSF2RB, IKZF2, and LINC01281 are as good or better markers at the mRNA level for Treg cells than the commonly used genes FOXP3, IL2RA, IL10, or TGFB. CEACAM4 and CSF2RB encode for cell-surface-exposed proteins, so may have utility for isolating or quantifying Treg cell populations. Taken together we conclude that surveying a combination of differentially expressed genes may be more informative for defining and studying T-cell subsets than simply relying on one or two markers.
Finally, a major question we sought to answer in this study is whether cytokines, or distinct CD4+ T-cell differentiation programs, also impact alternative splicing. Surprisingly, we find little evidence for widespread coordinated changes in splicing that correlate strongly with T-cell subset identity. This could reflect the fact that splicing represents a fine-tuning of T subset function rather than a major determinant, or is regulated at different times during differentiation than gene expression. Alternatively, splicing may be more sensitive than gene expression to variations in the methods used to isolate subpopulations of CD4+ T cells or the depth of sequencing, as RNA-seq-based splicing quantifications is known to require more read-depth than gene expression as only a subset of reads report on differential isoforms (Vaquero-Garcia et al. 2016). Regardless, we did identify a few genes for which splicing might contribute to differential function of Treg cells, such as AHRGEF2 and ZNF451. Similar to the unappreciated subset-biased gene expression programs mentioned above, these splicing events represent potentially new biology that we anticipate will motivate further study. We also do not test here the possibility that other forms of posttranscriptional gene regulation, such as 3′ end processing or translational control, could impact differential protein expression in CD4+ T-cell subsets. Such investigations would be an interesting goal of future analyses.
MATERIALS AND METHODS
Experimental model and subject details
For the in-house Bl data set, CD4+ human peripheral blood mononuclear cells were obtained via apheresis from de-identified healthy blood donors after informed consent by the University of Pennsylvania Human Immunology Core. Samples were collected from three donors, ND307 (age: 46, sex: male), ND523 (age: 26, sex: female), and ND535 (age: 32, sex: male).
Primary T-cell isolation and in vitro culturing of naïve CD4+ T cells into Th0 cells
From the CD4+ T cells apheresis, naïve CD4+ T cells were negatively selected for with MACS Miltenyi CD45RO microbeads (130-046-001). Six-well plates were coated for 3 h with 2.5 µg anti-CD3 (555336) at 37°C then washed with PBS. 10 × 106 Naïve CD4+ T cells were then cultured in complete RPMI in the anti-CD3-coated six well plates with 2.5 µg soluble anti-CD28 (348040) and 10 IU of IL-2. Cells were harvested after 48 h.
RNA-sequencing of primary T cells
RNA was isolated with RNA Bee (Tel-Test Inc.), according to the manufacturer's protocol, from bulk naïve CD4+ T cells (cultured for 0 h) and Th0 cells (cultured for 48 h with anti-CD3 and anti-CD28). The RNA integrity number (RIN) was measured with a bioanalyzer, and all samples had a RIN > 8.0 (Supplemental Fig. S4). RNA-sequencing libraries were generated by and sequenced by GeneWiz. The libraries were poly(A) selected (nonstranded) and paired-end sequenced at a 150 bp read length.
Selection of data sets and RNA-seq data processing
The following search terms were used to find appropriate data sets on EMBL-EBI-ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) and NCBI-GEO (https://www.ncbi.nlm.nih.gov/geo/): “T Helper,” “Naïve,” “CD4,” “Th0,” “Th1,” “Th2,” “Th17,” and “T Regulatory.” Data sets that had at least two of the following types of samples were retained for further analysis: Naïve, Th0, or Th1/Th2/Th17/Treg.
SRA files for the publicly available data sets were downloaded from the NCBI Sequence Read Archive (Supplemental Table S1). SRA files were converted to fastqs with fastq-dump (sratoolkit v2.9.2; Leinonen et al. 2011) using the following commands: –split-3 –gzip. Sequencing adaptors and low quality base calls were trimmed from reads using trim_galore (v0.5.0, downloaded from www.bioinformatics.babraham.ac.uk/ projects/trim_galore/) using the following commands: –stringency 5 –length 35 -q 20. For gene expression analyses, transcript level counts were obtained using Salmon (v0.11.3; Patro et al. 2017) in mapping-based mode with default settings. The Salmon transcriptome indices were prepared with the GRCh38 genome and Ensembl GRCh38.94 transcript database. Transcript level counts were collapsed into gene level counts with tximport (v1.12.0; Soneson et al. 2015). For alternative splicing analyses, fastq reads were aligned with STAR (2.5.2a) to the GRCh38 genome supplemented with the Ensembl GRCh38.94 transcript database using the following commands: –outSAMattributes All –alignSJoverhangMin 8 –readFilesCommand zcat –outSAMunmapped Within. The aligned bam files were then quantified for alternative splicing analyses with MAJIQ (v2.1-59f0404; Vaquero-Garcia et al. 2016), using MAJIQ build with the following additional command: –simplify 0.01.
Identifying and confirming which samples derived from the same human donors
All but one data set (Ra) used a paired-donor experimental design, meaning two or more samples representing different T-cell subsets derived from the same human donor. To determine which samples derived from the same donor, single nucleotide variants were identified for each RNA-seq sample, and then the proportion of shared genomic variation between samples was calculated. To identify and call the single nucleotide variants from the genome-aligned bam files, bcftools (v1.9; Li 2011) was used. The command used was bcftools mpileup -Ou –f <genome.fasta> <bam file> | bcftools call -mv -Ob –output <bcf file>. The bcf files were indexed, and then merged (–output-type z) with bcftools into a vcf file. The merged vcf file was then filtered with bcftools view –min-af 0.25 –output-type z, then the vcf was normalized and converted back to a bcf with bcftools norm -m-any | bcftools norm -Ob –check-ref w –f <genome.fastq>. The resulting bcf was indexed with bcftools, and then processed with plink (v1.90b6.7; Purcell et al. 2007) using the following commands: –bcf <bcf file> –const-fid 0 –allow-extra-chr 0 –recode –out <working directory>. To quantify the proportion of shared genomic variation between samples (identify by descent or IBD), plink was run with the following command: –file <working directory> –genome –out <working directory>. Groups of samples were determined to derive from the same donor if IBD scores were greater within the group than outside the group. Reassuringly, this analysis confirmed which samples derived from the same donor in data sets that provided donor information, so we feel confident in in our determination of sample donor pairs for data sets from which donor information was not provided.
Quality control checks
The trimmed fastqs were analyzed by FastQC (v0.11.2 downloaded from www.bioinformatics.babraham.ac.uk/projects/fastqc/). FastqQC results were used to confirm each sample did not have any nonhuman overrepresented sequences. One publicly available data set relevant to this study (but not used for further analyses) failed this quality control check because some samples had overrepresented bacterial genome sequences possibly indicating a bacterial contamination during cell culture. To confirm samples from the same donor were convincingly differentiated or sorted into different T-cell subtypes, PCA analysis was carried out on the gene-level counts using DESeq2::plotPCA (v1.22.1; Love et al. 2014). One publicly available data set relevant to this study (but not used for further analyses) failed this quality control check because the T-cell subtype explained <10% of the variance in gene expression across samples in the data set (at least 57% of the variance in gene expression was attributed to the identity of the donor, suggesting inefficient cell sorting). The final quality control check was to confirm a sufficient percentage of reads in the fastqs aligned to the human genome uniquely and unambiguously at a rate of at least 60% according to the STAR logs.
Differential expression analyses
DESeq2 (v1.22.1) in R (v3.5.1) was used to quantify differential expression between T-cell subtypes for each data set (see bitbucket repository for commands used). For each test for differential expression between two T-cell subtypes, samples were only ever compared from the same data set (see Fig. 1A). For example, Bl Naïve vs. Bl Th0. Before testing for differentially expressed genes, genes were filtered to only retain genes with total counts greater than 10 in at least two samples in at least one subtype. Mitochondrial and ribosomal genes were also filtered out. For all but the Ra and Mi data sets, the donor-aware model matrix supplied to DESeq2 controlled for gene expression variation due to donor by using “design = ∼Donor + Subtype.” The Ra and Mi data set design matrices were simply “design = ∼Subtype.” To control for noisy estimates of log2 fold-change in lowly expressed genes and genes with a high coefficient of variation, a log fold-shrinkage was applied to the DESeq2 differential expression results: “DESeq2::fcShrink(<deseq_obj>, <coefficient>, type=apelgm, lfcThreshold=1.5, svale=True”).
Identifying consistently differentially expressed genes between T-cell subtypes
To identify core sets of genes highly expressed in each CD4+ T-cell subtype, we performed differential expression analyses between all pairs of subtypes, controlling for the human donor source of the sample (i.e., Th0 vs. Th1, Th2 vs. Th1, Th17 vs. Th1, TReg vs. Th1). Differential expression analyses were done with DESeq2, and the resulting log2 fold changes (log2FC) and the s-values (the probability that the sign of the log2FC is wrong) were used to identify core genes. For example, core Th1 genes were defined as genes more highly expressed (log2FC > 1) in Th1 samples than Th0, Th2, Th17, or TReg samples; Th1 core genes needed to be consistently higher in Th1 than Th0, Th2, Th17, or TReg in the data sets that included both Th1 and Th0 samples (genes higher in two out of two data sets), Th1 and Th2 samples (genes higher in at least four out of five data sets), Th1 and Th17 samples (genes higher in three out of three data sets), or Th1 and TReg samples (genes higher in two out of two data sets). Th1 core genes were further filtered out if none of the differential expression comparisons showed the gene ever having an s-value <0.001. Core genes were then identified for Th2, Th17, and TReg populations. The resulting core genes are represented in Supplemental Table S3, whereby each core gene can be represented by multiple rows: Each row summarizes the differential expression results for the gene from a given CD4+ T-cell comparison (Th0 vs. Th1, Th2 vs. Th1, etc.). In many cases, core genes were identified for multiple T-cell subtypes. For example, CXCR3 is a core gene for Th1 (mean log2FC 2.95 over Th2 in five data sets, log2FC 4.2 over Th17 in three data sets) and CXCR3 is also a core gene for TRegs (log2FC 2.36 over Th2 in two data sets).
To better visualize these T-cell subtype core genes, the core genes table was filtered, per gene, to select which T-cell comparison (i.e., Th0 vs. Th1) had the greatest number of data sets showing higher expression with s-value <0.001 (ties decided by the greatest mean log2FC across the data sets). After the above filtering, each core gene was represented by a single T-cell subtype comparison. For each CD4+ T-cell subtype comparison, core genes were sorted by log2FC to rank genes by the greatest differential expression in favor of the given subtype comparison. Figure 4 shows these sorted genes’ asinh for transcripts per million (TPM) levels. Asinh(x) is calculated as ln[x + sqrt(x^2 + 1)].
Splicing analyses
To look for genes exhibiting consistent differences in splicing between T-cell subtypes, the MAJIQ algorithm (v2.1-59f0404; Vaquero-Garcia et al. 2016) was used to quantify alternative splicing from the genome-aligned bam files (see bitbucket repository for details). With the exception of the Ra and Mi data sets, differential splicing was quantified between all pairs of samples from the same donor (e.g., Ka Th1 donor 1 vs. Ka Th2 donor 2). For Ra and Mi, samples were compared in bulk versus each other (e.g., all Th1 vs. all Th2 in Ra). MAJIQ deltapsi quantifies the difference in percent splice included for every splice junction (e.g., GeneA junctionX shows a 20% difference in inclusion between Ka Th1 donor 1 vs. Ka Th2 donor 2 or 20% difference in inclusion between Ra Th1 samples and Ra Th2 samples). MAJIQ also quantifies the probability that a difference in splice inclusion is above some threshold, and we used a threshold of 20%. Significant differences in splicing were those identified as having a 95% probability of being greater than a 20% difference in splicing.
Identifying consistently differentially spliced genes between T-cell subtypes
To identify consistently differentially spliced splice junctions, for each junction and for each T-cell subtype comparison, we first identified the splice junctions that showed a significant difference in splicing in at least one donor from at least one data set. Next, we filtered out junctions for which any other donors disagreed on the direction of the change in splicing. We next filtered out junctions for which two data sets disagreed on the direction of the change in splicing. Next, we filtered out splicing changes where the number of data sets that agreed the splicing change was at least 10% in the same direction was fewer than two. Forty-three genes passed these filters, and these genes are listed in Supplemental Table S4.
DATA DEPOSITION
The new RNA-seq from naïve and Th0 cells generated for this study is available in GEO (GSE135118). Accessions for the other data sets used in this study include: Hn (E-MTAB-6300), Mi (E-MTAB-5739), Ra (E-MTAB-2319), Tu (GSE52260), Ka (GSE71645), Ab (GSE107981), Re (GSE110097), He (GSE62484), Mo (GSE107011), and Lo (GSE78276). All scripts used to analyze data for this study are made publicly available here: https://bitbucket.org/cradens/t_cell_meta_anlaysis/.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
K.W.L. is supported by R35 GM118048 (National Institute of General Medical Sciences), Y.B. is supported by R01 GM128096 and R01 GM128096, C.M.R. was supported in part by T32 GM008216, and D.B. was supported by a supplement R35 GM118048-S1.
Author contributions: C.M.R., Y.B., and K.W.L. designed the study and analysis approach, C.M.R. identified data sets and carried out all the analysis, D.B. isolated cell subsets and generated RNA-seq libraries, P.J. developed several analysis algorithms used in the study. C.M.R., Y.B., and K.W.L. wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.075929.120.
REFERENCES
- Abadier M, Pramod AB, McArdle S, Marki A, Fan Z, Gutierrez E, Groisman A, Ley K. 2017. Effector and regulatory T cells roll at high shear stress by inducible tether and sling formation. Cell Rep 21: 3885–3899. 10.1016/j.celrep.2017.11.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. 2008. Post-transcriptional control of cytokine production. Nat Immunol 9: 353–359. 10.1038/ni1584 [DOI] [PubMed] [Google Scholar]
- Angkasekwinai P, Park H, Wang Y-H, Wang Y-H, Chang SH, Corry DB, Liu Y-J, Zhu Z, Dong C. 2007. Interleukin 25 promotes the initiation of proallergic type 2 responses. J Exp Med 204: 1509–1517. 10.1084/jem.20061675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bending D, De la Peña H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, Cooke A. 2009. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest 119: 565–572. 10.1172/JCI37865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhairavabhotla R, Kim YC, Glass DD, Escobar TM, Patel MC, Zahr R, Nguyen CK, Kilaru GK, Muljo SA, Shevach EM. 2016. Transcriptome profiling of human FoxP3+ regulatory T cells. Hum Immunol 77: 201 10.1016/j.humimm.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. 2013. Dynamic integration of splicing within gene regulatory pathways. Cell 152: 1252–1269. 10.1016/j.cell.2013.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L, Pichler A, Lima CD. 2015. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat Struct Mol Biol 22: 968–975. 10.1038/nsmb.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro G, Liu X, Ngo K, De Leon-Tabaldo A, Zhao S, Luna-Roman R, Yu J, Cao T, Kuhn R, Wilkinson P, et al. 2017. RORγt and RORα signature genes in human Th17 cells. PLoS One 12: e0181868 10.1371/journal.pone.0181868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gao F, He M, Ding XF, Wong AM, Sze SC, Yu AC, Sun T, Chan AW, Wang X, et al. 2019. Long-read RNA sequencing identifies alternative splice variants in hepatocellular carcinoma and tumor-specific isoforms. Hepatology 70: 1011–1025. 10.1002/hep.30500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleret-Buhot A, Zhang Y, Planas D, Goulet J-P, Monteiro P, Gosselin A, Wacleche VS, Tremblay CL, Jenabian M-A, Routy J-P, et al. 2015. Identification of novel HIV-1 dependency factors in primary CCR4+CCR6+Th17 cells via a genome-wide transcriptional approach. Retrovirology 12: 102 10.1186/s12977-015-0226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Crome SQ, MacDonald KG, Dai EL, Mager DL, Levings MK. 2011. Human Th1 and Th17 cells exhibit epigenetic stability at signature cytokine and transcription factor loci. J Immunol 187: 5615–5626. 10.4049/jimmunol.1101058 [DOI] [PubMed] [Google Scholar]
- Collier SP, Henderson MA, Tossberg JT, Aune TM. 2014. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J Immunol 193: 3959–3965. 10.4049/jimmunol.1401099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amato L, Dell'Aversana C, Conte M, Ciotta A, Scisciola L, Carissimo A, Nebbioso A, Altucci L. 2015. ARHGEF3 controls HDACi-induced differentiation via RhoA-dependent pathways in acute myeloid leukemias. Epigenetics 10: 6–18. 10.4161/15592294.2014.988035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Bluestone JA. 2016. Harnessing the plasticity of CD4+ T cells to treat immune-mediated disease. Nat Rev Immunol 16: 149–163. 10.1038/nri.2015.18 [DOI] [PubMed] [Google Scholar]
- Feng Y, Wu H, Xu Y, Zhang Z, Liu T, Lin X, Feng XH. 2014. Zinc finger protein 451 is a novel Smad corepressor in transforming growth factor-β signaling. J Biol Chem 289: 2072–2083. 10.1074/jbc.M113.526905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi M, Di Stefano B, Berenguer C, de Andres-Aguayo L, Plana-Carmona M, Mendez-Lago M, Guillaumet-Adkins A, Rodriguez-Esteban G, Gut M, Gut IG, et al. 2019. Single cell RNA-seq identifies the origins of heterogeneity in efficient cell transdifferentiation and reprogramming. Elife 8: e41627 10.7554/eLife.41627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, MacIsaac KD, Chen Y, Miller RJ, Jain R, Joyce-Shaikh B, Ferguson H, Wang I-M, Cristescu R, Mudgett J, et al. 2016. Inhibition of RORγT skews TCRα gene rearrangement and limits T cell repertoire diversity. Cell Rep 17: 3206–3218. 10.1016/j.celrep.2016.11.073 [DOI] [PubMed] [Google Scholar]
- Harbour SN, Maynard CL, Zindl CL, Schoeb TR, Weaver CT. 2015. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc Natl Acad Sci 112: 7061–7066. 10.1073/pnas.1415675112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6: 1123–1132. 10.1038/ni1254 [DOI] [PubMed] [Google Scholar]
- Henriksson J, Chen X, Gomes T, Ullah U, Meyer KB, Miragaia R, Duddy G, Pramanik J, Yusa K, Lahesmaa R, et al. 2019. Genome-wide CRISPR screens in T helper cells reveal pervasive crosstalk between activation and differentiation. Cell 176: 882–896.e818. 10.1016/j.cell.2018.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck A, Evans CM, Eskandarpour M, Lau JCH, Oleinika K, Jackson I, Kelly A, Ambrose J, Adamson P, Cousins DJ, et al. 2016. T-bet activates Th1 genes through mediator and the super elongation complex. Cell Rep 15: 2756–2770. 10.1016/j.celrep.2016.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Troutman TD, Edukulla R, Pasare C. 2011. Priming microenvironments dictate cytokine requirements for T helper 17 cell lineage commitment. Immunity 35: 1010–1022. 10.1016/j.immuni.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Davis SP, Hill JA, Yamagata T. 2015. Diverse gene expression in human regulatory T cell subsets uncovers connection between regulatory T cell genes and suppressive function. J Immunol 195: 3642–3653. 10.4049/jimmunol.1500349 [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18: S96–S104. 10.1093/bioinformatics/18.suppl_1.S96 [DOI] [PubMed] [Google Scholar]
- Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. 2007. Global analysis of alternative splicing during T-cell activation. RNA 13: 563–572. 10.1261/rna.457207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. 2006. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126: 1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, Glimcher LH, Lord GM. 2009. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Natl Acad Sci 106: 17876–17881. 10.1073/pnas.0909357106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri K, Tripathi S, Larjo A, Mannerström H, Ullah U, Lund R, Hawkins RD, Ren B, Lähdesmäki H, Lahesmaa R. 2015. Identification of global regulators of T-helper cell lineage specification. Genome Med 7: 122 10.1186/s13073-015-0237-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M. International Nucleotide Sequence Database Consortium. 2011. The sequence read archive. Nucleic Acids Res 39: D19–D21. 10.1093/nar/gkq1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M, Wu JE, Arumemi F, Mikulski Z, Dahlberg C, Miller AT, Crotty S. 2016. Activin A programs the differentiation of human TFH cells. Nat Immunol 17: 976–984. 10.1038/ni.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund RJ, Löytömäki M, Naumanen T, Dixon C, Chen Z, Ahlfors H, Tuomela S, Tahvanainen J, Scheinin J, Henttinen T, et al. 2007. Genome-wide identification of novel genes involved in early Th1 and Th2 cell differentiation. J Immunol 178: 3648–3660. 10.4049/jimmunol.178.6.3648 [DOI] [PubMed] [Google Scholar]
- Martinez NM, Lynch KW. 2013. Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol Rev 253: 216–236. 10.1111/imr.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NM, Pan Q, Cole BS, Yarosh CA, Babcock GA, Heyd F, Zhu W, Ajith S, Blencowe BJ, Lynch KW. 2012. Alternative splicing networks regulated by signaling in human T cells. RNA 18: 1029–1040. 10.1261/rna.032243.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NM, Agosto L, Qiu J, Mallory MJ, Gazzara MR, Barash Y, Fu XD, Lynch KW. 2015. Widespread JNK-dependent alternative splicing induces a positive feedback loop through CELF2-mediated regulation of MKK7 during T-cell activation. Genes Dev 29: 2054–2066. 10.1101/gad.267245.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micossé C, von Meyenn L, Steck O, Kipfer E, Adam C, Simillion C, Seyed Jafari SM, Olah P, Yawlkar N, Simon D, et al. 2019. Human “TH9” cells are a subpopulation of PPAR-γ+ TH2 cells. Sci Immunol 4: eaat5943 10.1126/sciimmunol.aat5943 [DOI] [PubMed] [Google Scholar]
- Middleton R, Gao D, Thomas A, Singh B, Au A, Wong JJ-L, Bomane A, Cosson B, Eyras E, Rasko JEJ, et al. 2017. IRFinder: assessing the impact of intron retention on mammalian gene expression. Genome Biol 18: 51 10.1186/s13059-017-1184-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitson-Salazar A, Yin Y, Wansley DL, Young M, Bolan H, Arceo S, Ho N, Koh C, Milner JD, Stone KD, et al. 2016. Hematopoietic prostaglandin D synthase defines a proeosinophilic pathogenic effector human TH2 cell subpopulation with enhanced function. J Allergy Clin Immunol 137: 907–918.e909. 10.1016/j.jaci.2015.08.007 [DOI] [PubMed] [Google Scholar]
- Monaco G, Lee B, Xu W, Mustafah S, Hwang YY, Carré C, Burdin N, Visan L, Ceccarelli M, Poidinger M, et al. 2019. RNA-seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep 26: 1627–1640.e1627. 10.1016/j.celrep.2019.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Wooding S, Satta Y, Jinnai N, Goto S, Hayasaka I, Saitou N, Guan-Jun J, Tokunaga K, Jorde LB, et al. 2005. Evidence for natural selection in the HAVCR1 gene: high degree of amino-acid variability in the mucin domain of human HAVCR1 protein. Genes Immunity 6: 398–406. 10.1038/sj.gene.6364215 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Artis D. 2012. New paradigms in type 2 immunity. Science 337: 431–435. 10.1126/science.1221064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh R, Kozhaya L, McKevitt K, Djuretic IM, Carlson TJ, Quintero MA, McCauley JL, Abreu MT, Unutmaz D, Sundrud MS. 2014. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J Exp Med 211: 89–104. 10.1084/jem.20130301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M, et al. 2015. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol 16: 318–325. 10.1038/ni.3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regateiro FS, Chen Y, Kendal AR, Hilbrands R, Adams E, Cobbold SP, Ma J, Andersen KG, Betz AG, Zhang M, et al. 2012. Foxp3 expression is required for the induction of therapeutic tissue tolerance. J Immunol 189: 3947–3956. 10.4049/jimmunol.1200449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revu S, Wu J, Henkel M, Rittenhouse N, Menk A, Delgoffe GM, Poholek AC, McGeachy MJ. 2018. IL-23 and IL-1β drive human Th17 cell differentiation and metabolic reprogramming in absence of CD28 costimulation. Cell Rep 22: 2642–2653. 10.1016/j.celrep.2018.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlon TJ, Wilkinson BG, Pederson S, Brown CY, Bresatz S, Gargett T, Melville EL, Peng K, D'Andrea RJ, Glonek GG, et al. 2010. Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J Immunol 185: 1071–1081. 10.4049/jimmunol.1000082 [DOI] [PubMed] [Google Scholar]
- SEQC/MAQC-III Consortium. 2014. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol 32: 903–914. 10.1038/nbt.2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadidi KR, Aarvak T, Henriksen JE, Natvig JB, Thompson KM. 2003. The chemokines CCL5, CCL2 and CXCL12 play significant roles in the migration of Th1 cells into rheumatoid synovial tissue. Scand J Immunol 57: 192–198. 10.1046/j.1365-3083.2003.01214.x [DOI] [PubMed] [Google Scholar]
- Shih H-Y, Sciumè G, Poholek AC, Vahedi G, Hirahara K, Villarino AV, Bonelli M, Bosselut R, Kanno Y, Muljo SA, et al. 2014. Transcriptional and epigenetic networks of helper T and innate lymphoid cells. Immunol Rev 261: 23–49. 10.1111/imr.12208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love M, Robinson M. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences [version 1; peer review: 2 approved]. F1000Res 4: 1521 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B, Omenetti S. 2017. The dichotomous nature of T helper 17 cells. Nat Rev Immunol 17: 535–544. 10.1038/nri.2017.50 [DOI] [PubMed] [Google Scholar]
- Stubbington MJ, Mahata B, Svensson V, Deonarine A, Nissen JK, Betz AG, Teichmann SA. 2015. An atlas of mouse CD4+ T cell transcriptomes. Biol Direct 10: 14 10.1186/s13062-015-0045-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. 2002. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science 295: 338–342. 10.1126/science.1065543 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Tagami T, Yamazaki S, Uede T, Shimizu J, Sakaguchi N, Mak TW, Sakaguchi S. 2000. Immunologic self-tolerance maintained by CD25+CD4+ regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J Exp Med 192: 303–310. 10.1084/jem.192.2.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. 2010. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol 184: 3433–3441. 10.4049/jimmunol.0904028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. 2014. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344: 641–645. 10.1126/science.1251102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomela S, Rautio S, Ahlfors H, Öling V, Salo V, Ullah U, Chen Z, Hämälistö S, Tripathi SK, Äijö T, et al. 2016. Comparative analysis of human and mouse transcriptomes of Th17 cell priming. Oncotarget 7: 13416–13428. 10.18632/oncotarget.7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin-Dembinsky A, Balashov K, Weiner HL. 2006. IL-23 is increased in dendritic cells in multiple sclerosis and down-regulation of IL-23 by antisense oligos increases dendritic cell IL-10 production. J Immunol 176: 7768–7774. 10.4049/jimmunol.176.12.7768 [DOI] [PubMed] [Google Scholar]
- Vaquero-Garcia J, Barrera A, Gazzara MR, Gonzalez-Vallinas J, Lahens NF, Hogenesch JB, Lynch KW, Barash Y. 2016. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife 5: e11752 10.7554/eLife.11752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkayya R, Lam M, Willkom M, Grünig G, Corry DB, Erle DJ. 2002. The Th2 lymphocyte products IL-4 and IL-13 rapidly induce airway hyperresponsiveness through direct effects on resident airway cells. Am J Respir Cell Mol Biol 26: 202–208. 10.1165/ajrcmb.26.2.4600 [DOI] [PubMed] [Google Scholar]
- Wang Y-H, Angkasekwinai P, Lu N, Voo KS, Arima K, Hanabuchi S, Hippe A, Corrigan CJ, Dong C, Homey B, et al. 2007. IL-25 augments type 2 immune responses by enhancing the expansion and functions of TSLP-DC-activated Th2 memory cells. J Exp Med 204: 1837–1847. 10.1084/jem.20070406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang J, Yang J, Yang X, He J, Wang R, Liu S, Zhou L, Ma L. 2017. Guanine nucleotide exchange factor -H1 promotes inflammatory cytokine production and intracellular mycobacterial elimination in macrophages. Cell Cycle 16: 1695–1704. 10.1080/15384101.2017.1347739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. 2013. Dynamic regulatory network controlling TH17 cell differentiation. Nature 496: 461–468. 10.1038/nature11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. 1997. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem 272: 21597–21603. 10.1074/jbc.272.34.21597 [DOI] [PubMed] [Google Scholar]
- Zhang H, Nestor CE, Zhao S, Lentini A, Bohle B, Benson M, Wang H. 2013. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1. J Allergy Clin Immunol 132: 1005–1008. 10.1016/j.jaci.2013.05.033 [DOI] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89: 587–596. 10.1016/S0092-8674(00)80240-8 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Rudensky AY. 2007. Foxp3 in control of the regulatory T cell lineage. Nat Immunol 8: 457–462. 10.1038/ni1455 [DOI] [PubMed] [Google Scholar]
- Zhou AX, Hed AE, Mercer F, Kozhaya L, Unutmaz D. 2013. The metalloprotease ADAM12 regulates the effector function of human Th17 cells. PLoS One 8: e81146 10.1371/journal.pone.0081146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. 2018. T helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harb Perspect Biol 10: a030338 10.1101/cshperspect.a030338 [DOI] [PMC free article] [PubMed] [Google Scholar]