Abstract
Many eukaryotes use RNA processing, including alternative splicing, to express multiple gene products from the same gene. The budding yeast Saccharomyces cerevisiae has been successfully used to study the mechanism of splicing and the splicing machinery, but alternative splicing in yeast is relatively rare and has not been extensively studied. Alternative splicing of SKI7/HBS1 is widely conserved, but yeast and a few other eukaryotes have replaced this one alternatively spliced gene with a pair of duplicated, unspliced genes as part of a whole genome doubling (WGD). We show that other examples of alternative splicing known to have functional consequences are widely conserved within Saccharomycotina. A common mechanism by which alternative splicing has disappeared is by replacement of an alternatively spliced gene with duplicate unspliced genes. This loss of alternative splicing does not always take place soon after duplication, but can take place after sufficient time has elapsed for speciation. Saccharomycetaceae that diverged before WGD use alternative splicing more frequently than S. cerevisiae, suggesting that WGD is a major reason for infrequent alternative splicing in yeast. We anticipate that WGDs in other lineages may have had the same effect. Having observed that two functionally distinct splice-isoforms are often replaced by duplicated genes allowed us to reverse the reasoning. We thereby identify several splice isoforms that are likely to produce two functionally distinct proteins because we find them replaced by duplicated genes in related species. We also identify some alternative splicing events that are not conserved in closely related species and unlikely to produce functionally distinct proteins.
Keywords: alternative splicing, yeast, evolution, subfunctionalization
INTRODUCTION
Alternative splicing is thought to be an important mechanism to generate multiple functionally distinct proteins from a single gene. While the majority of mammalian genes are alternatively spliced, other eukaryotes use this process less frequently. Although the budding yeast Saccharomyces cerevisiae has proven to be a powerful tool to understand the mechanism of splicing, only a few genes are thought to encode two functionally distinct proteins through alternative splicing. Even in cases where alternative splicing occurs in S. cerevisiae, the functional significance of both isoforms is incompletely understood.
PTC7 is a well-understood example of an alternatively spliced gene encoding two functionally distinct proteins in S. cerevisiae (see Fig. 1G below). Ptc7 is a protein phosphatase whose localization is altered by splicing. Translation of the unspliced PTC7 mRNA results in the inclusion of a transmembrane helix and insertion of Ptc7 into the nuclear membrane (Juneau et al. 2009). This Ptc7 isoform confers resistance to Latrunculin A (Juneau et al. 2009). Alternatively, when PTC7 mRNA is spliced, the encoded protein is localized in the mitochondria (Juneau et al. 2009). Ptc7 is required to dephosphorylate mitochondrial proteins, which likely reflects that the spliced isoform directly carries out this function (Guo et al. 2017). The spliced and unspliced isoforms also have different effects on mitochondrial function (Awad et al. 2017). Thus, the Ptc7 isoforms seem to perform separate functions.
FIGURE 1.
Alternative splicing of PTC7 orthologs in the Saccharomycotina. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny. Key branches are indicated: the branch leading to the subphylum Saccharomycotina, the branch leading to the family Saccharomycetacea, and the branch leading to the WGD clade. Red species names highlight the WGD clade. Green asterisk indicates the most likely origin of PTC7 intron retention. Red asterisk indicates the most likely replacement of one alternatively spliced gene with duplicate genes. (B–G) Sashimi plots of a representative RNA-seq data set for the indicated species. Each plot indicates in the top left corner the read coverage scale on the y-axis. Arches indicate introns that were spliced out, and the numbers along the arches indicate the number of exon junction reads for that splicing pattern. Below each sashimi plot is the likely RNA structure with boxes indicating coding exons and lines indicating intron. TM indicates a transmembrane helix predicted to be encoded by the retained intron. (B) C. albicans diverged before the likely origin of PTC7 alternative splicing and no alternative splicing was detected. (C) Intron retention in the PTC7 ortholog from C. jadinii. (D) Intron retention in the PTC7 ortholog from T. delbrueckii. (E,F) T. blattae contains two PTC7 orthologs, with one lacking an intron and the other containing a constitutive intron. (G) Intron retention in PTC7 of S. cerevisiae. The box around this panel indicates this panel confirms the previously described alternative splicing.
Similarly, Fes1 is a nucleotide exchange factor for Hsp70 that is targeted to the nucleus when translated from the spliced mRNA and remains cytoplasmic when translated from the unspliced mRNA (see Fig. 7F below; Gowda et al. 2016). The cytoplasmic form has been shown to be important for the degradation of misfolded proteins and is required for growth at high temperature, but the function of nuclear Fes1, if any, is unknown (Gowda et al. 2016).
FIGURE 7.
Alternative splicing of FES1 orthologs in the Saccharomycotina. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. (B) Y. lipolytica diverged before the likely origin of the FES1 retained intron and no splicing was detected. (C) Intron retention in the FES1 ortholog from O. polymorpha. (D) Intron retention in the FES1 ortholog from K. lactis. (E) The retained intron has been lost from the FES1 ortholog in T. blattae. (F) Intron retention in the FES1 ortholog from S. cerevisiae as previously reported. NLS indicates the location of a nuclear localization signal.
A third example is provided by SRC1, which generates two proteins through the use of alternative 5′ splice sites (see Fig. 6F below; Rodriguez-Navarro et al. 2002; Grund et al. 2008). The alternative 5′ splice sites of SRC1 are separated by 4 nt, resulting in the downstream exon being used in two different reading frames. Both the 95 kDa “full-length” and 73 kDa “truncated” proteins are inserted into the nuclear membrane. The lack of the full-length protein causes a growth defect in mutants when the THO complex is also defective, suggesting that the full-length protein has some function that is not shared with the truncated protein (Grund et al. 2008). In contrast, a specific function for the truncated protein has not been identified. The full-length protein is also similar to a paralog that arose as part of a whole genome doubling (WGD) that occurred an estimated 95 million years ago (MYA). The functional differences between this paralog (Heh2) and Src1 are also not well understood.
FIGURE 6.
Alternative splicing of SRC1/HEH2 orthologs in the Saccharomycotina and A. fumigatus. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. The black asterisks indicate a change from alternative 3′ splice sites to alternative 5′ splice sites or vice versa between A. fumigatus and the Saccharomycotina, changes in the position of the minor 5′ splice site in the Candida genus and the Saccharomycetacea, and the addition of an intron retention variant in the ZT clade. (B) Alternative 3′ splice sites in the SRC1/HEH2 ortholog in A. fumigatus. (C) Alternative 5′ splice sites in the SRC1/HEH2 ortholog in Y. lipolytica. (D) Alternative 5′ splice sites in the SRC1/HEH2 ortholog in L. kluyveri. (E) Alternative 5′ splice sites and intron retention in the SRC1/HEH2 ortholog in Z. rouxii. (F,G) Duplicated SRC1 and HEH2 genes in S. cerevisiae. SRC1 uses alternative 5′ splice sites, as previously reported, while HEH2 lacks an intron.
We have recently shown that in Lachancea kluyveri, another member of the Saccharomycetacea, a single gene encodes Ski7 and Hbs1 through alternative splicing, but that this alternatively spliced gene is replaced by separate SKI7 and HBS1 genes in S. cerevisiae (Marshall et al. 2013). Both HBS1 and SKI7 lack introns. Ski7 tethers the Ski complex to the cytoplasmic RNA exosome complex and thereby mediates mRNA degradation (van Hoof et al. 2000; Araki et al. 2001; Kowalinski et al. 2016). Hbs1 instead delivers Dom34 to stalled ribosomes, which allows Dom34 to recycle the ribosomal subunits for subsequent rounds of translation (Shoemaker et al. 2010; Becker et al. 2011; Pisareva et al. 2011). These separable functions of Ski7 and Hbs1 can be assigned to the L. kluyveri splice-isoforms, with proximal 3′ splice site usage resulting in a functional Ski7, and distal 3′ splice site usage in a functional Hbs1. We further showed that this alternative splicing of SKI7/HBS1 is conserved in animals, fungi, and plants (Marshall et al. 2013, 2018).
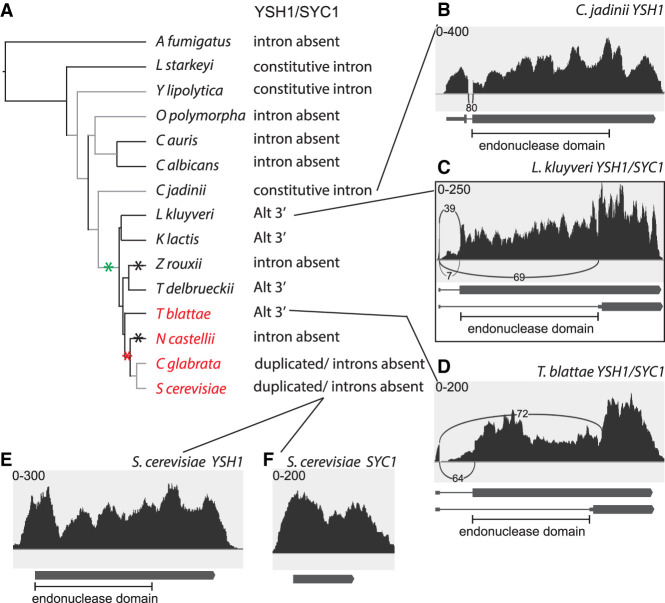
During our studies focused on SKI7/HBS1, we noted anecdotally that two other gene pairs fit the same pattern. First, the S. cerevisiae YSH1 and SYC1 genes arose by duplication of an alternatively spliced YSH1/SYC1 gene that uses alternative 3′ splice sites (see Fig. 3C below; Marshall et al. 2018). In S. cerevisiae, Ysh1 is the endonuclease that is responsible for the 3′ end formation of mRNAs in the cleavage and polyadenylation reaction, while Syc1 is required for cleavage-independent 3′ end formation of some noncoding RNAs (Lidschreiber et al. 2018). Syc1 is similar to the carboxy-terminal domain of Ysh1, and through this domain the two proteins interact with the same partners. However, Syc1 lacks the endonuclease domain (Nedea et al. 2003). Thus it appears that an alternatively spliced YSH1/SYC1 gene in L. kluyveri is replaced by duplicate YSH1 and SYC1 genes that both lack introns (Marshall et al. 2018).
FIGURE 3.
Alternative splicing of YSH1/SYC1 orthologs in the Saccharomycotina. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. Black asterisks indicate that Z. rouxii and N. castellii appear to have lost Syc1. (B) C. jadinii diverged before the likely origin of YSH1/SYC1 alternative splicing and its ortholog contains a constitutive intron. (C) Alternative 3′ splice sites in the YSH1/SYC1 ortholog of L. kluyveri, as previously reported. (D) Alternative 3′ splice sites in the YSH1/SYC1 ortholog of T. blattae. (E,F) Duplicated YSH1 and SYC1 genes in S. cerevisiae lack introns.
The second previously noted example of an alternatively spliced gene being replaced by duplicate genes is PTC7. As noted above, the S. cerevisiae PTC7 gene encodes two protein phosphatases with distinct localization (Juneau et al. 2009). In contrast, Tetrapisispora blattae contains two PTC7 genes, and we have speculated that one contains an intron. This predicted intron contains stop codons that prevent translation of any intron-retained mRNA (Marshall et al. 2013). However, there is no experimental evidence supporting this suggestion and introns in T. blattae have not been carefully annotated. Unlike SKI7/HBS1 alternative splicing, which is conserved in animals, fungi and plants, neither the YSH1/SYC1 nor PTC7 alternative splicing events appeared to be conserved from an ancient ancestor, but when these alternative splicing events arose has not been extensively studied.
Strikingly, the duplication of SKI7 and HBS1, YSH1 and SYC1 and PTC7 each date to the WGD (Kellis et al. 2004; Scannell et al. 2007; Marcet-Houben and Gabaldon 2015). This WGD also gave rise to the SRC1 and HEH2 paralogs mentioned above. These observations suggest that the WGD event in the Saccharomyces lineage may be a contributor to the relatively infrequent use of alternative splicing to diversify its proteome, but the relationship between WGD and loss of alternative splicing is incompletely understood.
Here, we more systematically analyze when previously identified examples of alternative splicing in Saccharomycotina arose, how extensively they are conserved, and whether they were replaced by duplicated genes in any species. The results reveal the generality that most genes that likely encode two distinct functional proteins through alternative splicing in one species of Saccharomycotina are replaced by duplicate unspliced genes in other species. However, this generality does not extend to regulatory alternative splicing, as in the PRP5 gene. In this case, splicing functions in a negative feedback loop to prevent accumulation of the spliced mRNA, and thus the spliced mRNA does not have a separate function (Karaduman et al. 2017). Finally, we use these observations to suggest some alternative splicing events that likely generate a functional gene product, and some that appear less likely to do so.
RESULTS
Study design
The subphylum Saccharomycotina is divided into approximately a dozen clades, which generally correspond to families, and in the current study we included all the clades/families with available genome and RNA-seq data (Supplemental Fig. 1; Shen et al. 2016). We sampled the Saccharomycetaceae family more densely to gain a better understanding of the relationship between WGD and loss of alternative splicing. The Saccharomycetaceae are divided into three clades: the KLE, ZT, and WGD clades (Marcet-Houben and Gabaldon 2015; Shen et al. 2016). The KLE clade is composed of the Kluyveromyces, Lachancea and Eremothecium genera, while the ZT clade is composed of the Zygosaccharomyces and Torulaspora genera (Supplemental Fig. 1). Although the WGD event was initially described as a duplication, more recent analysis (Marcet-Houben and Gabaldon 2015) suggests it was a hybridization between one parent from the KLE clade and another parent from the ZT clade (and we therefore prefer whole genome doubling instead of whole genome duplication to describe the outcome rather than the mechanism). After this single hybridization, the WGD clade diverged into Saccharomyces, Kazachstania, Naumovozyma, Tetrapisispora, and Vanderwaltozyma genera (Supplemental Fig. 1). The original hybridization occurred before the extant species of the KLE and ZT clades diverged from each other and thus, the parents cannot be meaningfully assigned to one of the extant species. We therefore included two representatives from both the KLE and ZT clades, and four from the WGD clade. Within each clade we maximized the diversity analyzed. For example, the first branch within the WGD clade separates the T. blattae lineage from the S. cerevisiae lineage and we therefore included both of these species. In addition to these Saccharomycotina, we analyzed alternative splicing in RNA-seq data from Aspergillus fumigatus, which is a representative of the other Ascomycota.
To find alternatively spliced target genes, we performed extensive literature searches for “alternative splicing” and each species name. We focused on alternative splicing events with some evidence that the alternative splicing event has some functional consequence (to eliminate splicing errors and heterogeneity in splicing without functional consequences). This literature search was supplemented by inspecting RNA-seq data from two species, L. kluyveri and Candida albicans, which identified one novel intron retention event (in the L. kluyveri NUP116/NUP100 gene). Similar inspection of RNA-seq data has previously identified candidate alternative splicing events (Kawashima et al. 2014; Schreiber et al. 2015), and we also included several of the most prominent candidates from those studies. However, other examples from the Schreiber et al. study were not detected in our S. cerevisiae RNA-seq analysis and were only supported by a single exon junction read in the original analysis, suggesting that they may be sequencing or mapping errors rather than robust biological phenomena.
We then mapped RNA-seq reads from each of the species to the genome sequence and inspected each of the genes of interest for evidence of alternative splicing. Although annotating alternative splicing de novo is complicated by difficulties distinguishing errors in splicing, sequencing, or mapping, the previously described functional events were obvious in RNA-seq alignments (see boxed panels in each of the figures below). The use of alternative splice sites was reflected in exon-junction reads where the read starts in one exon and ends in the next exon, precisely defining the splice sites used. Thus, RNA-seq analysis reliably confirmed each of the previously reported alternative splicing events (see boxed panels in Figs. 1–11). As expected from our earlier studies (Marshall et al. 2013, 2018), we detected the use of alternative SKI7/HBS1 3′ splice sites in early diverging Saccharomycotina. Although this confirmed our previous conclusions, it revealed no novel findings, and thus, is not included here.
A conserved alternatively spliced PTC7 gene is replaced by duplicate genes
As described in the introduction, PTC7 is a well-understood example of alternative splicing in S. cerevisiae and encodes two distinct proteins. Ptc7 is a protein phosphatase with distinct functions and localization depending on intron removal or retention (Juneau et al. 2009; Awad et al. 2017; Guo et al. 2017). We detected this intron retention in S. cerevisiae (Fig. 1A and boxed panel 1G). Importantly, we also observed alternative splicing in other pre- and post-WGD species (Fig. 1A,C,D). In each case, the intron length was a multiple of three and lacked stop codons, and a substantial number of reads mapped to within the intron, indicating intron retention. However, this alternative splicing was not detected in any earlier diverging species such as C. albicans (Fig. 1B). Instead, these earlier diverging species lack an intron in the orthologous position (Fig. 1B; Supplemental Fig. 2). Because the divergence time of Cyberlindnera jadinii has not been estimated, we cannot precisely time the emergence of the alternative splicing, but it occurred between 114 MYA and 304 MYA (green asterisk in Fig. 1A). The protein encoded by the unspliced C. albicans gene is localized to the mitochondria (Wang et al. 2007), and thus, the mitochondrial function of the spliced mRNA in S. cerevisiae likely predates the gain of alternative splicing.
We have previously speculated that the PTC7 gene was duplicated in T. blattae and that both duplicates lost alternative splicing (Marshall et al. 2013), but this speculation was not supported by any data at the RNA level. T. blattae is positioned at an interesting juncture, as the split between T. blattae and S. cerevisiae is the earliest known split after WGD (Fig. 1A). We therefore generated RNA-seq data from T. blattae, which confirmed that the PTC7 ortholog predicted to be cytoplasmic is indeed constitutively spliced (Fig. 1F), while the nuclear membrane ortholog lost the capacity to splice (Fig. 1E). Even if there were a low level of intron-retained mRNA for this spliced gene, it would contain a stop codon, preventing the formation of a second protein. Thus, as predicted, the alternatively spliced PTC7 gene is replaced by two genes that have lost the capacity for alternative splicing (red asterisk in Fig. 1A).
Strikingly, in several species with PTC7 intron retention, the intron has splice sites that perfectly match the consensus intron sequence (GUAUGU…UACUAAC…YAG in T. delbrueckii, Z. rouxii and K. lactis). It is therefore not clear why this intron is retained, but sequences outside the core splice sites, secondary structure, and trans-acting factors may all contribute. In contrast, the 5′ splice site in T. blattae does not match perfectly to the consensus sequences (GUAAGU…UACUAAC…UAG). Thus, the disappearance of the intron retention form in T. blattae might be primarily due to the gain of a stop codon within the intron, which is expected to trigger nonsense-mediated decay (Kawashima et al. 2014).
A conserved alternatively spliced MDH gene is replaced by duplicate unspliced genes
It has been reported that exon 3 of a malate dehydrogenase (MDH) gene from Yarrowia lipolytica starts with alternative UAG 3′ splice sites (Kabran et al. 2012). Although the alternative 3′ splice sites are only 4 nt apart (i.e., UAGCUAG), and the two proteins only differ by a single amino acid, the proteins are localized differently. The distal splice site generates a peroxisomal targeting signal (PTS; Supplemental Fig. 3), while the proximal splice site does not create a PTS, and this has been shown to affect protein localization (Kabran et al. 2012). The use of these alternative 3′ splice sites was confirmed by our RNA-seq analysis (Fig. 2A,D).
FIGURE 2.
Alternative splicing of MDH orthologs in the Saccharomycotina and A. fumigatus. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. In addition, the black asterisks indicate a change of splicing pattern. Specifically, the ancestor likely used both intron retention and alternative 3′ splice sites. (*) Loss of alternative 3′ splice sites in L. starkeyi, loss of intron retention in Y. lipolytica, and the conversion of the peroxisomal gene into a pseudogene in O. polymorpha. (B) Intron retention and alternative 3′ splice sites in the A. fumigatus MDH gene. The light gray box indicates a reading frame that is different from the dark gray boxes. PTS indicates a peroxisomal targeting signal. (C) Intron retention (likely coupled with an intronic polyadenylation site) in the L. starkeyi MDH gene. (D) Alternative 3′ splice sites in the Y. lipolytica MDH gene. (E,F) Duplicated MDH genes in S. cerevisiae lack introns. In panels B–D, only the 3′ end of the gene is shown.
We also observed alternative splicing of the orthologous gene in A. fumigatus (Fig. 2B) and Lipomyces starkeyi (Fig. 2C). In A. fumigatus we detected three distinct mRNAs: The most abundant and annotated mRNA retained the final intron. A second mRNA uses a proximal 3′ splice site (AAG) for a novel intron and a third mRNA uses a distal 3′ splice site (UAG) for the same intron. As in Y. lipolytica the 3′ splice sites are 4 nt apart. Use of the proximal 3′ splice site results in the exact same protein sequence as intron retention (replacing only the wobble base of the last sense codon and the stop codon; Supplemental Fig. 3). In contrast, as in Y. lipolytica, the distal 3′ splice site adds a single amino acid and generates a putative PTS1.
Similarly, in L. starkeyi the MDH ortholog either retains the final intron and encodes a likely cytoplasmic protein, or splices the final intron generating a PTS1 signal (Fig. 2C; Supplemental Fig. 3). The most parsimonious explanation of these observations is that the A. fumigatus gene structure represents the ancestral state, with Y. lipolytica having lost the intron retention and L. starkeyi having lost the proximal 3′ splice site due to a single nucleotide change (AAG to AAU; black asterisk in Fig. 2A). This implies that the alternative splicing event is conserved from a common ancestor of all Saccharomycotina and A. fumigatus that lived >590 MYA.
In all other Saccharomycotina we detected two orthologs for this gene (Fig. 2A,E,F; Supplemental Fig. 3). None of these orthologs had an intron in the same position as the Y. lipolytica gene. However, in each species one ortholog encodes a protein that ends with a PTS1 signal, while the other gene lacked a similar sequence (Supplemental Fig. 3). We conclude that a single alternatively spliced gene was replaced by two unspliced genes. Note that “unspliced” here and in other cases below indicates unspliced at this position. In some cases other introns are present. The common ancestor where this duplication and loss of splicing occurred lived >304 MYA (red asterisk in Fig. 2A).
A conserved alternatively spliced YSH1/SYC1 gene is replaced by duplicate unspliced genes
We have previously reported that the L. kluyveri YSH1/SYC1 gene uses alternative 3′ splice sites (Marshall et al. 2018) and this was confirmed by RNA-seq (Fig. 3A,C). This gene is the ortholog of both YSH1 (CPSF73 in mammals) and SYC1 in S. cerevisiae, which arose as part of the WGD. Ysh1 and Syc1 share a similar carboxy-terminal domain. In addition, Ysh1 has an endonuclease domain that is absent from Syc1. Alternative splicing in L. kluyveri generates Ysh1-like and Syc1-like proteins from a single gene (Fig. 3C; Supplemental Fig. 4). The orthologs in L. kluyveri, Kluyveromyces lactis, Torulaspora delbrueckii, and T. blattae each contain an intron that uses alternative 3′ splice sites. The 5′ splice site for this intron is in the 5′ UTR with a proximal 3′ splice site and AUG codon upstream of the endonuclease domain and a distal 3′ splice site and AUG codon internal to the ORF that would skip the endonuclease domain (Fig. 3A,C,D). None of the earlier diverging species contains an orthologous alternatively spliced intron. Thus, alternative splicing likely arose >304 MYA. However, some earlier diverging species (C. jadinii, Y. lipolytica, and L. starkeyi) contain an intron in what appears likely the orthologous position, but this intron is constitutively spliced (Fig. 3B). This suggests that alternative splicing arose by adding a distal 3′ splice site to a preexisting intron.
In the four WGD species, the duplicated orthologs followed three different paths (Fig. 3A). T. blattae maintained one alternatively spliced gene and lost the duplicate copy (Fig. 3D). In the lineage leading to S. cerevisiae and Candida glabrata, both genes lost the intron and one gene expresses Ysh1, and the other only Syc1 without either one being spliced (Fig. 3E,F). Finally, Naumovozyma castellii contains only one copy of this gene, which is not alternatively spliced. Thus, N. castellii appears to have lost Syc1 as a distinct splice isoform. It is possible that it expresses Syc1 by some other mechanism such as alternative start codon use and/or alternative transcription start sites.
A conserved alternatively spliced NUP116/NUP100 gene is replaced by duplicate unspliced genes
While identifying novel alternative splicing events in L. kluyveri, we discovered a novel potential intron that is retained some of the time in the NUP116/NUP100 gene (Fig. 4A,C). Intron retention results in the annotated protein, while splicing generates a protein that lacks 127 internal amino acids. We observed the same intron retention pattern in orthologs from other species of the KLE and ZT clades (all pre-WGD Saccharomycetaceae) (Fig. 4A,C,D). The splice sites and branchpoint sequences (GUAUGU … UACUAAC … UAG in all four species) are perfect matches for the consensus splicing signals in those species (Neuveglise et al. 2011) and it is not clear why this intron is sometimes retained. This conservation suggests that this alternative splicing event arose in a common ancestor of the Saccharomycetaceae that lived about 114 MYA.
FIGURE 4.
Alternative splicing of NUP116/NUP100 orthologs in the Saccharomycotina. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. (B) C. jadinii diverged before the likely origin of NUP116/NUP100 alternative splicing and its ortholog lacks an intron. The C. jadinii ortholog contains a sequence that is homologous to the sequence in Nup116 that binds to Gle2. (C) Intron retention in the NUP116/NUP100 ortholog of L. kluyveri. (D) Intron retention in the NUP116/NUP100 ortholog of T. delbrueckii. (E,F) Duplicated NUP116 and NUP100 genes in S. cerevisiae lack introns and only Nup116 contains the Gle2 binding site.
The NUP116/NUP100 gene is the ortholog of both NUP116 and NUP100, two S. cerevisiae nucleoporin-encoding genes that resulted from the WGD. The functional difference between Nup116 and Nup100 is a Gle2 binding site that is present in Nup116 but absent in Nup100 (Fig. 4E,F; Supplemental Fig. 5; Bailer et al. 1998). This sequence is conserved in the retained intron of the KLE and ZT clades (Fig. 4C,D; Supplemental Fig. 5). This suggests that the spliced mRNA encodes a protein that has the Nup100 function, while the intron retained mRNA encodes a protein with Nup116 function. This Gle2 binding site is also clearly present in orthologs that diverged earlier (Fig. 4B; Supplemental Fig. 5). This suggests that the intron arose by converting coding sequence to an intron by gaining splicing sites.
The NUP116/NUP100 intron retention event was not detectable in the WGD clade, which suggests that this alternative splicing event was lost after WGD >69 MYA (Fig. 4A,E,F). The low similarity of the protein level, and the repetitive nature of nucleoporin sequences, limited the ability to generate a high confidence alignment of the Nup100 proteins from the WGD clade (Supplemental Fig. 5). Nevertheless, the alignment is consistent with the intron being precisely deleted in NUP100, possibly by recombination with a cDNA (Fink 1987). The Nup116 proteins aligned better, allowing us to identify where the splice sites used to be. This revealed that all three splice signals are mutated in the WGD species (e.g., to GAAUGU …UACCAAU … UGG in S. cerevisiae). It is impossible to determine which change originally caused loss of alternative splicing and which occurred subsequently. Thus, an alternative spliced NUP116/100 gene appears to be replaced in WGD species by duplicated unspliced genes.
Conserved alternative splicing of a GND gene was lost but not replaced by duplicate genes
It has previously been reported that the C. albicans 6-phosphogluconate dehydrogenase (GND) gene uses alternative 3′ splice sites (Strijbis et al. 2012). In this case, use of the proximal 3′ splice site causes inclusion of a PTS2 signal and targeting to the peroxisome, while use of the distal 3′ splice site results in a cytoplasmic protein (Strijbis et al. 2012). Although it was reported that the peroxisomal splice-isoform accounts for only ∼0.1% of the total mRNA, we were able to detect exon junction reads in two different RNA-seq data sets (one of which is shown in Fig. 5D). The evidence for alternative splicing was more robust in RNA-seq data from L. starkeyi, Y. lipolytica, and C. jadinii (Fig. 5A–C). In each case, a minor splice isoform included a putative PTS2 (Supplemental Fig. 6). Although the peroxisomal isoform appears to be minor, in these species we detected dozens of exon junction reads. Thus this alternative splicing appears to have arisen in a common ancestor >332 MYA. The orthologs from A. fumigatus and Candida auris have a potential proximal 3′ splice site that would cause inclusion of a PTS2 (Supplemental Fig. 6), but we did not detect exon junction reads for this splicing pattern. Whether this splice site is used infrequently or under specific conditions remains to be determined, and thus this alternative splicing may have arisen even earlier than >332 MYA.
FIGURE 5.
Alternative splicing of GND orthologs in the Saccharomycotina and A. fumigatus. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. The alt 3′? indicates that although a potential alternative 3′ splice site is conserved, we did not detect any exon junction reads indicating its usage under the specific conditions analyzed. The black asterisk indicates a change from alternative 3′ splice site to exon skipping. (B) Alternative 3′ splice sites in the GND ortholog in L. starkeyi. (C) Exon skipping in the GND ortholog in Y. lipolytica. (D) Alternative 3′ splice sites in the GND ortholog in C. albicans, as previously reported. (E) The T. delbrueckii ortholog contains a constitutive intron in the orthologous position.
The alternative splicing of the GND gene appears to have been lost twice, in Ogataea polymorpha and in the Saccharomycetaceae (both >114 MYA). In these species, we failed to detect any RNA-seq reads suggesting alternative splicing, nor was there a candidate alternative 3′ splice site that would cause inclusion of a PTS2. After WGD, S. cerevisiae retained both copies of the GND gene (GND1 and GND2), but these do not appear to be related to the loss of alternative splicing as neither gene has a PTS (Supplemental Fig. 6), and neither has been localized to peroxisomes (Huh et al. 2003).
A conserved alternatively spliced SRC1/HEH2 gene is twice replaced by duplicate genes
As mentioned in the introduction, S. cerevisiae uses alternative 5′ splice sites in SRC1 to express a full-length and a truncated protein. This alternative splicing is clearly visible in RNA-seq data (Fig. 6F). Genetic evidence indicates that the full-length protein is functional, but whether the truncated protein has a distinct function is not entirely clear.
Analyzing RNA-seq data from various species showed that alternative splicing of SRC1 is ancient, with alternative 5′ splice sites present in many different Saccharomycotina (Fig. 6A–E). Thus, the use of alternative 5′ splice sites is conserved from >332 MYA. Even the A. fumigatus ortholog is alternatively spliced (Fig. 6B), although it uses alternative 3′ splice sites, pushing the likely origin of alternative splicing to >590 MYA. In each of these orthologs, the alternative splice sites cause the downstream exon to be used in two different reading frames, with one splice pattern generating a full-length protein, and the other splice pattern a truncated protein. The full-length protein has been shown to be functional, and consistent with that, the encoded protein in this reading frame is conserved (Supplemental Fig. 7).
Interestingly, instead of the single alternatively spliced gene, O. polymorpha has two unspliced orthologs, with one being full-length, and the other truncated (Fig. 6A; Supplemental Fig. 7). The observation that the alternatively spliced gene is replaced by two unspliced genes in O. polymorpha suggests that both proteins are functional.
The functionality of the truncated protein is further supported by amino acid conservation patterns (Supplemental Fig. 7). The exon downstream from the alternatively spliced intron is translated in two different reading frames and the amino acid sequence in both reading frames is conserved. In the first ∼150 nt of the exon, two reading frames are used and the encoded amino acid sequence conservation is higher for the truncated protein (Supplemental Fig. 7). The second part of the exon forms part of the 3′ UTR in the truncated mRNA, but encodes conserved amino acids in the full-length protein (Supplemental Fig. 7). This pattern of sequence conservation in both reading frames strongly suggests that both proteins are functional.
More support for the functionality of the truncated protein comes from changes in the position of the alternative 5′ splice sites. In the early diverging species Y. lipolytica, the 5′ splice site for the truncated form is 5 nt distal of the full-length 5′ splice site (Fig. 6C). The same pattern occurs in C. jadinii. Interestingly, this spacing has changed twice: In the Saccharomycetacea (including S. cerevisiae and L. kluyveri; Fig. 6D–F), the truncating 5′ splice site is moved from +5 to −4 relative to the full-length encoding splice site. This same change from +5 to −4 occurred in Candida albicans. The observation that these shifts in the truncating 5′ splice sites each time occur in a multiple of three suggests that the sequence of the truncating protein is important, not just the fact that the alternative form is truncated. Thus, duplication in O. polymorpha, conservation of sequence, and conservation of reading frame all suggest that both splice isoforms are functional.
In each of the Saccharomycotina, both 5′ splice sites in SRC1 are poor matches to the consensus. Specifically, the splice site for full-length Src1 is GUgaGU in all Saccharomycetacea, while the truncating 5′ splice site at −4 is always GcAaGU (deviations from the consensus GUAUGU are in lower case). Furthermore, C. auris and C. jadinii both use the alternative 5′ splice site at +5, and the GcAaGU motif is accordingly shifted from −4 to +5. Thus the shift in the alternative 5′ splice site correlates with the shift in GcAaGU.
We noted one additional change in alternative splicing pattern that may be related to why both SRC1 and HEH2 were maintained after WGD. The RNA-seq data from both species of the ZT clade indicate that three different protein encoding mRNAs are expressed from a single SRC1/HEH2 gene (Fig. 6E). In addition to the use of alternative 5′ splice sites, all in frame stop codons were lost from the intron, such that an intron-retained mRNA encodes a third protein. Because the WGD arose after hybridization between the KLE clade and the ZT clade, the common ancestor of the WGD clade likely inherited one gene from the KLE clade that expressed two different SRC1/HEH2 proteins plus one gene from the ZT clade that expressed three different SRC1/HEH2 proteins, for a total of five proteins. If the three proteins from the ZT clade each were functionally distinct, the WGD species might have to maintain expression of all three proteins. This could be accomplished by completely losing the gene inherited from the KLE clade, but that is not what appears to have happened. Instead the three isoform gene inherited from the ZT clade and the two isoform gene inherited from the KLE clade likely each lost one splice isoform. This reduced the total number of proteins from the gene pair from five to three.
Conserved alternative splicing of a FES1 gene was lost but not replaced by duplicate genes
FES1 encodes a nucleotide exchange factor for Hsp70. It has previously been reported that an intron can be spliced out near the 3′ end. The second exon of FES1 adds a 16 amino acid Lys and Arg rich sequence that functions as a nuclear localization sequence (NLS) (Gowda et al. 2016). Alternatively, the intron can be retained. The retained intron contains four sense codons, a stop codon and a poly(A) site. This splicing pattern was readily apparent in our RNA-seq analysis (Fig. 7F), confirming the previous report. We detected intron retention in FES1 orthologs of eight other species (Fig. 7A,C,D). In each case the retained intron is in the same position in the gene. The species that is most diverged and showed intron retention is O. polymorpha (Fig. 7C). Thus, FES1 intron retention is conserved between species, and arose >304 MYA.
In S. cerevisiae the second exon of FES1 has been shown to add an NLS (Gowda et al. 2016). In these other species the second exon is similarly short (14–17 AA) and Lys/Arg-rich, suggesting that they also encode an NLS (Supplemental Fig. 8). Alternatively, when the intron is retained translation stops four to seven codons into the intron. This suggests that the alternative localization of Fes1 splice isoforms is likely also conserved.
Contrary to the genes discussed so far, the alternatively spliced FES1 gene was not replaced by duplicate genes in any of the species we examined (Fig. 7A). We did note that T. blattae, C. albicans, and C. auris have lost the second exon. Thus either they have lost the nuclear isoform of Fes1, or they use some other signal to import Fes1 into the nucleus.
Conserved inefficient splicing of PRP5 has not been replaced with duplicated genes
The alternative splicing events discussed so far all result in the production of two different proteins that are both likely to be functional, and whose function appear to differ. Instead of producing multiple functional proteins, in some cases alternative splicing is used to modulate the amount of one protein. A prime example of this is PRP5, which encodes an RNA helicase required for splicing (Vijayraghavan et al. 1989). Interestingly, PRP5 is regulated by feedback inhibition such that when Prp5 activity is high, it causes splicing of its own mRNA, removing the start codon and reducing its own expression (Karaduman et al. 2017). We detected this inefficient splicing/intron retention in RNA-seq data from S. cerevisiae (Fig. 8E). Importantly, we also detected inefficient splicing of the orthologous intron in a number of other Saccharomycotina, and even in A. fumigatus (Fig. 8A,B,D). In each case, splicing removed the start codon of the PRP5 ORF. This suggests that in each of these species splicing functions to reduce the expression of Prp5, presumably in response to Prp5 abundance. This feedback inhibition splicing event therefore appears to be conserved from >590 MYA.
FIGURE 8.
Inefficient splicing of PRP5 orthologs in the Saccharomycotina and A. fumigatus disrupts the ORF and likely serves to feedback inhibit PRP5 expression. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. N.D. indicates that splicing pattern could not be determined because of the low expression level in the conditions analyzed. (B) Splicing of the first intron of the PRP5 ortholog in A. fumigatus disrupts the ORF by removing the start codon. (C) The inefficiently spliced intron has been lost from the PRP5 ortholog in L. starkeyi. (D) Splicing of the first intron of the PRP5 ortholog in C. jadinii disrupts the ORF by removing the start codon. (E) Splicing of the first intron of PRP5 in S. cerevisiae disrupts the ORF by removing the start codon, as previously reported. In panels B–D, only the 5′ end of the gene is shown.
Similar to other genes, the inefficiently spliced intron of PRP5 appears to have been lost (e.g., in L. starkeyi; Fig. 8C). However, even though this inefficient splicing is ancient and predates WGD, we did not find PRP5 duplicated with loss of alternative splicing in any species. This characteristic is expected for an alternative splicing event that regulates the amount of protein, rather than produces multiple proteins, because a feedback inhibition splicing event could not operate in trans (see Discussion).
Alternative splice site usage in MTR2, GCR1, and APE2 are not well conserved
We next investigated some other genes that have been reported to be alternatively spliced in S. cerevisiae, but where the functional consequences were not clear. MTR2, GCR1, and APE2 all have been reported to use alternative 5′ and/or 3′ splice sites in S. cerevisiae (Davis et al. 2000; Parenteau et al. 2008; Meyer et al. 2011; Hossain et al. 2016), and in each case these were readily detectable in RNA-seq data (Figs. 9D, 10D, 11D). However, the orthologs, even within the family Saccharomycetaceae did not use alternative splice sites.
FIGURE 9.
The alternatively spliced intron in MTR2 is not well conserved. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. (B) The K. lactis MTR2 ortholog does not contain an intron. (C) The C. glabrata MTR2 ortholog does not contain an intron. (D) Splicing of MTR2 in S. cerevisiae is inefficient and uses multiple 5′ and 3′ splice sites, as previously reported.
FIGURE 10.
Alternative splicing of GCR1 orthologs in the Saccharomycetacea. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. A clear ortholog of GCR1 is not present outside the Saccharomycetacea. (B) The K. lactis GCR1 ortholog does not contain an intron. (C) The C. glabrata GCR1 ortholog contains an intron, but no alternative 5′ or 3′ splice sites. Translation from an AUG codon in the intron the predicted to produce an alternate isoform. (D) Splicing of GCR1 in S. cerevisiae is inefficient and uses alternative 3′ splice sites, as previously reported. It may also use alternative 5′ splice sites at low frequency. Translation from an AUG codon in the intron produces an alternate isoform.
In the case of MTR2, the intron was absent from all other Saccharomycetaceae (Fig. 9A,B), confirming that this intron recently arose in the Saccharomyces genus (Talkish et al. 2019). Remarkably, deletion of the MTR2 intron is lethal (Parenteau et al. 2008). Given the recent origin of the intron and its inefficient splicing we suggest that the DNA sequence that was deleted in the previous study may have some essential function other than serving as an intron.
In contrast, the orthologous intron is conserved in GCR1 and APE2 orthologs from other Saccharomycetaceae but uses only one 5′ and one 3′ splice site (Figs. 10, 11). To the best of our knowledge, it has not been shown that the distinct spliced mRNAs in these genes have distinct functions, and the absence of conservation of multiple splice sites suggests that the choice of splice site may not be functionally important. The apparent origin of these alternative splice sites after WGD also precludes them being replaced by duplicate genes as part of the WGD.
FIGURE 11.
Alternative splicing of APE2 and AAP1 orthologs in the Saccharomycetacea. (A) Splicing pattern is shown for each of the species analyzed, along with the species phylogeny as in Figure 1A. A clear ortholog of APE2 could not be identified outside the Saccharomycetacea. (B) The K. lactis APE2/AAP1 ortholog contains an intron, but no alternative 5′ or 3′ splice sites. Translation from an AUG codon in the intron is predicted to produce an alternate isoform. (C,D) Duplicated APE2 and AAP1 genes in S. cerevisiae. APE2 uses alternative 3′ splice sites, as previously reported, while AAP1 lacks the intron. In addition, there may be a transcription start site within the intron and translation from an AUG codon in the intron is predicted to produce an alternate isoform. Mito indicates the presence of a predicted mitochondrial targeting sequence in the optional first exon.
One common feature between MTR2, APE2, and GCR1 is that splicing these genes appeared inefficient with a significant number of reads mapping within the intron. For all three genes, either the intron retained mRNA or an mRNA with a transcription start site within the annotated intron appeared to be abundant (Figs. 9D, 10D, 11D). Interestingly, it has been shown that GCR1 produces two distinct proteins by the use of start codons in exon 1 and in the intron, respectively (Hossain et al. 2016). Furthermore, both proteins are required for optimal growth (Hossain et al. 2016). This inefficient splicing and the intronic AUG were conserved in GCR1 from some but not all Saccharomycetacea (Fig. 10; Supplemental Fig. 9). Thus, the inefficient splicing of GCR1 and the potential to encode two proteins is conserved from an ancestor >97 MYA, but the use of multiple splice sites is a more recent addition in Saccharomyces. Orthologs outside the Saccharomycetacea could not be identified unambiguously, preventing us from tracing the intron evolution further back in time.
Similarly, for APE2, the RNA-seq data suggested that an alternative protein could be encoded starting at an AUG codon near the end of the annotated intron. In this case, this alternate AUG codon appears to be present as part of an mRNA that starts at an alternate transcription start site within the intron (Fig. 11D). This potential to use an alternative start codon and an alternative transcription start site was conserved in all Saccharomycetacea (Supplemental Fig. 10), thereby dating back to a common ancestor >114 MYA. This conservation pattern strongly suggests that the start site within the annotated intron is functionally important. Ape2 encodes an aminopeptidase that has been shown to localize to both the cytoplasm and mitochondria (Huh et al. 2003). Interestingly, translation starting from the AUG in exon 1 encodes a predicted mitochondrial protein, while translation starting within the annotated intron encodes a predicted cytoplasmic protein (Fig. 11; Supplemental Fig. 10). Overall this suggests that APE2 uses alternative translation start sites, and possibly alternative transcription start sites, to encode two functionally distinct proteins.
We also investigated whether MTR2, GCR1, and APE2 were replaced by duplicate genes, and found only one species with duplicated GCR1 and three species with duplicated APE2, all resulting from the WGD. T. blattae encodes two orthologs of GCR1 (Supplemental Fig. 9). One of these retains the inefficiently spliced intron, while the other lacks an intron. Due to the low sequence conservation it is unclear whether the protein from the second T. blattae gene corresponds to the spliced or unspliced isoform of its single gene orthologs (Supplemental Fig. 9).
APE2 was present in duplicates in three species from the WGD clade, with the paralog named AAP1 in S. cerevisiae. The fourth WGD species (T. blattae) appears to have lost both copies of APE2/AAP1 after WGD. In S. cerevisiae, Aap1 has been localized to the cytoplasm and nucleus (Huh et al. 2003), but the basis for the difference in localization between Ape2 and Aap1 is unknown. In the other two WGD species with duplicated APE2/AAP1 genes, one gene of the pair (AAP1) lacks an intron and only appears to encode the cytoplasmic/nuclear protein, while the other gene (APE2) uses alternative start sites to encode predicted cytoplasmic and mitochondrial isoforms (Fig. 11; Supplemental Fig. 10). Thus in this case, after WGD, AAP1 appears to have lost a conserved alternative start site.
DISCUSSION
Here, we survey the evolution of alternative splicing in the Saccharomycotina during their 330 million year divergence, especially as it relates to genome and gene duplication. Although alternative splicing is rarer in the Saccharomycotina than in Metazoa, all 14 Saccharomycotina species studied here use alternative splicing to functionally diversify their proteome.
Alternative splicing events in Saccharomycotina are conserved
By analyzing RNA-seq data from diverse Saccharomycotina we have shown that well-characterized alternative splicing events are conserved for hundreds of millions of years. Several alternative splicing events were even conserved with A. fumigatus, which we analyzed as a representative Ascomycete outside the Saccharomycotina. Thus these most anciently conserved alternative splicing events are conserved from >590 MYA, and include SKI7/HBS1, MDH, GND, SRC1/HEH2, and PRP5. Other splicing events arose later, with FES1 alternative splicing arising >304 MYA, and PTC7, NUP100/NUP116, and YSH1 alternative splicing both arising >114 MYA. In contrast, the alternative 5′ and/or 3′ splice site usage of MTR2, GCR1, and APE2 are more recent additions specifically in the Saccharomyces genus. Although alternative 5′ and 3′ splice site usage in GCR1 and APE2 is a recent addition, both genes do appear to use alternative start codons, that are potentially linked to intron retention and/or the use of alternative transcription start sites. This conservation during >100 million years indicates that alternative splicing in the Saccharomycotina is functionally important and deserves further studies.
We also describe several changes in alternative splicing events, where the use of alternative 5′ sites, alternative 3′ sites, intron retention and/or exon skipping replace each other while maintaining the functional consequences at the protein level. The observation that alternative splicing itself is more conserved than the exact mechanism (or cis-acting elements) suggests that the production of functionally distinct proteins is critical, but multiple means that accomplish this are functional.
These conservation times are similar to the divergence of vertebrates from each other. For example, human and fish diverged an estimated 435 MYA, while humans diverged from marsupials 159 MYA and from mice 90 MYA. Although, we describe several examples of conservation, our analysis is not exhaustive, and we expect there to be additional alternative splicing events that are conserved from >100 MYA.
Loss of alternative splicing events often follows gene duplication
Our RNA-seq analysis revealed six instances where an alternative splicing event was lost in the Saccharomycotina and replaced by duplicated genes. The parents of the WGD event appear to have used alternative splicing in eight different genes. Four of these eight genes are replaced by a pair of duplicated unspliced genes in at least some WGD species (SKI7/HBS1, PTC7, NUP116/NUP100, YSH1/SYC1). Some of these losses of alternative splicing are shared with all WGD species, and thus may have occurred soon after WGD (before Tetrapisispora and Saccharomyces diverged). However, the PTC7 and YSH1/SYC1 loss of alternative slicing must have occurred after the T. blattae lineage diverged from the S. cerevisiae lineage since alternative splicing is maintained in one species but replaced by duplicated genes in the other species. These different outcomes in different WGD species suggest that the loss of alternative splicing does not always take place soon after duplication, but can take place after sufficient time has elapsed for speciation.
Interestingly, loss of alternative splicing is more frequent in WGD species than other Saccharomycotina. Specifically, the WGD clade diverged at about the same time that C. albicans diverged from C. auris, K. lactis from L. kluyveri, and Zygosaccharomyces rouxii from T. delbrueckii (Fig. 1A), but only one loss of an alternative splicing event occurred in any of these six species (YSH1/SYC1 in Z. rouxii).
While loss of alternative splicing is more common after WGD, we also detected two losses that followed small-scale duplications: a single alternatively spliced SRC1/HEH2 gene was replaced by duplicated unspliced genes in O. polymorpha, and a single alternatively spliced MDH gene was replaced by duplicated unspliced genes in a common ancestor of Saccharomyces, Candida, and Ogataea.
In the majority of cases, loss of alternative splicing was accomplished by loss of the intron, but in a minority of cases the alternatively spliced intron was converted to a constitutive intron (e.g., one of the T. blattae PTC7 duplicates). The exact nucleotide changes that caused the loss of alternative splicing in many cases could not be identified. One contributing factor was that many of the splicing events occurred in relatively poorly conserved regions of the genes. Additionally, over the long times examined here many nucleotide changes occurred, and the initial cause of loss of alternative splicing is lost among additional changes.
Our observation that loss of alternative splicing often occurs after gene duplication suggests that gene duplication may be the initiating and rate-limiting event. The observation that alternative splicing was replaced with duplicated genes multiple times does not imply an evolutionary advantage for such a replacement. Instead, we consider neutral events sufficient to explain our observations. Although we repeatedly found loss of alternative splicing, we found many species that have maintained it and neither species has gone extinct. Thus, both mechanisms to express two different proteins have lead to evolutionary success in each of these genes. If there is any advantage, it must be specific for specific genes/species rather than a general rule of one being advantageous over the other. Instead, a series of neutral events can lead to loss of alternative splicing: After duplication, one paralog might randomly lose one splice isoform. This should not reduce fitness since a functionally redundant splice isoform is still produced by the other paralog. Later, the other paralog might lose the second splice isoform also without a change in fitness. Thus, fitness neutral events are sufficient to explain the reproducible loss of alternative splicing events we observed.
The replacement of bifunctional genes by duplicated genes is probably not specific for alternative splicing, and we suspect that genes that make distinct functional products by other RNA or protein processing events could also be replaced by duplicated genes. This would include genes using multiple start codons, edited mRNAs, and even genes that encode both a protein and an ncRNA such as a snoRNA. One example of this appears to be that before WGD APE2/AAP1 used alternative transcription and translation start sites to encode mitochondrial and cytoplasmic proteins, and the upstream start site and mitochondrial form appear to have been lost from AAP1. Another example is that it has been suggested that the snR38 snoRNA and TEF4 mRNA are processed from one primary transcript in pre-WGD species but replaced by duplicated genes that produce only the snoRNA and only the mRNA, respectively, in some post-WGD species (Hooks et al. 2014). Furthermore, we anticipate that other small-scale duplication and WGD events, including the two rounds of WGD in the human lineage, had similar effects.
Alternative splicing arose by intronization and exonization
Alternative splicing can arise by generation of an intron from previously exonic events (intronization), or by generating an exon out of previously intronic sequences (exonization). NUP116/NUP100 and YSH1/SYC1 provide clear examples of intronization: in both cases conserved amino acid sequences are encoded in (alternative) introns, and the sequence conservation predates the origin of the intron. Thus, preexisting conserved exon sequence must have been converted into alternative intron sequence, presumably by the point mutations that introduce splice sites. Because these introns arose in an ancient ancestor we are unable to determine the exact sequence changes that occurred.
We detected one example of exonization: the conversion of intronic sequences to exonic sequences in the SRC1 gene of the ZT clade by loss of all the stop codons from the intron.
In contrast to these clear examples of intronization in YSH1/SYC1 and NUP116/NUP100 and exonization in SRC1/HEH2, we were not able to clearly establish the origin of other alternative splicing events, nor is it possible to establish what nucleotide changes caused the initial gain or loss of splicing. Many of the alternative splicing events affect the extreme amino or carboxyl termini (or in YSH1/SYC1 the 5′ UTR), which are difficult to align over long evolutionary times. Furthermore, given the long time since these splicing events originated and disappeared, many additional sequence changes occurred obscuring the initial change that affected splicing patterns.
Distinguishing functionally distinct splice isoforms, regulatory splicing, and unproductive splicing from patterns of evolution
Individual alternative splicing events can have a variety of functions, or not have functional consequences at all. Broadly speaking, alternative splicing events can be functional because they produce multiple proteins with distinct functions or because they regulate the amount of protein being produced by the gene. In addition to these functional splicing events, some heterogeneity in splice sites may be splicing errors (where one splice product is nonfunctional) or splicing inaccuracies (where the proteins may differ by a few amino acids, but are interchangeable). Finally, RNA-seq alignment algorithms are not fool proof and can artifactually suggest exon junctions that do not exist in vivo. We suggest that examination of RNA-seq in related species is a facile way to help distinguish between these. Alternative splicing that is functionally important should be conserved, and we do indeed find that the case for several genes. On the other hand, the alternative splicing events that are not conserved such as MTR2 are most likely nonfunctional noise of the splicing machinery, or in rare instances may be recent species-specific evolutionary innovations. Furthermore, we suggest that alternative splicing that produces functionally distinct proteins can be expected to be replaced with duplicate unspliced genes in related species (e.g., PTC7, MDH, GND, SKI7/HBS1, YSH1/SYC1, and NUP116/NUP100), while alternative splicing that regulates the protein amount should not be easily replaced by unspliced duplicated genes (e.g., PRP5). In the case of the PRP5 example, the feedback inhibition mechanism cannot be replaced by an unspliced gene that always produces a functional mRNA and a constitutively spliced gene that always produces the nonproductive RNA.
Using this logic, conservation patterns suggest the NLS added to Fes1 by alternative splicing may not be critical for two reasons: The NLS-containing isoform is lost from C. albicans, C. auris, and T. blattae and this alternative splicing is not replaced by duplicated unspliced genes in any of the species examined. It remains to be determined whether these species have lost nuclear Fes1 or they use an alternative mechanism to target this Hsp70 nucleotide exchange factor to the nucleus. On the other hand, our observation that the SRC1 alternatively spliced gene is replaced by duplicated genes in O. polymorpha suggests that both protein products have a critical function.
MATERIALS AND METHODS
The times of species divergence cited are from timetree.org and represent a consensus of available data
We performed extensive literature searches for “alternative splicing” and each of our target species. We included all examples with some evidence that the alternative splicing event resulted in functionally distinct proteins. The previously predicted alternative splicing of PGK in Y. lipolytica (Freitag et al. 2012) could not be detected in RNA-seq data from that species (data not shown) and thus was not included. We also excluded some cases of regulated splicing where only the amount of the encoded protein is affected because this would require quantitative RNA-seq analysis from many different conditions (e.g., C. albicans DUR31) (Donovan et al. 2018). This literature search was supplemented by inspecting RNA-seq data from two strategically placed species, L. kluyveri and C. albicans, which identified one novel intron retention event (in the L. kluyveri NUP116/NUP100 ortholog).
For most species, raw RNA-seq reads from previous studies were downloaded from EBI (https://www.ebi.ac.uk/ena). Because there was no publicly available RNA-seq data from the Tetrapisispora/Vanderwaltozyma branch we generated RNA-seq data from T. blattae (deposited in SRA under project PRJNA613484). As explained above, this branch diverged soon after WGD and thus was critical in understanding the fate of alternative splicing after WGD. We obtained T. blattae from the United States Department of Agriculture, Agricultural Research Service Culture Collection (https://nrrl.ncaur.usda.gov/), grew duplicate cultures in YPD medium at 30°C, isolated RNA by a hot phenol method (He et al. 2008), and generated paired-end 150 nt RNA-seq data (25,338,942 and 30,978,862 pairs for the biological replicates).
Both downloaded and newly generated RNA-seq reads were adaptor and quality trimmed with TrimGalore when needed (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), and aligned to the genome downloaded from Genbank (https://www.ncbi.nlm.nih.gov/genome/) using TopHat2 (Kim et al. 2013). Several of the data sets were also aligned with HiSat2 (Kim et al. 2015) and RNA STAR (Dobin et al. 2013) with identical results. At least two data sets from each species were analyzed (Supplemental Table 1). If more than two data sets were available, preference was given to data sets generated in two independent studies of the same species, paired-end reads, 150 nt reads, data sets with 20–30 million reads, data sets from the same wild-type strain as that used for whole genome sequencing and data sets from standard growth conditions. These preferences were all designed to ease the detection of alternative splicing. TopHat2 settings were adjusted to allow introns from 30 nucleotides to 10 kb in size. The resulting alignment files were manually inspected in IGV (https://software.broadinstitute.org/software/igv/), and Sashimi plots were generated in IGV.
A gene was considered to use alternative splice sites if the number of exon junction reads for the minor splicing patterns was at least 10% of that for the major splicing pattern detected. An exception was made for GND1/GND2 alternative splicing because it has previously been shown that the PTS2-included form accounts for only ∼0.1% of the mRNA, yet is functionally important (Strijbis et al. 2012). This low level of alternative splicing could be reliably detected because of the very high expression of this gene (Fig. 5B–D). The PTC7 and NUP100/NUP116 intron was considered retained if the read coverage within the intron was at least 10% of the exon junction reads, if the intron length was a multiple of three, and if the intron contained no in frame stop codons. For MDH2/MDH3 and FES1 the intron was considered to be retained if read coverage within the intron was at least 10% of the exon junction reads, and extended past the first in-frame stop codon.
To understand the functional divergence of splice isoforms and duplicate genes, multiple sequence alignments were generated with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) after correcting some of the protein sequences based on the observed splicing patterns. PSORT II (https://psort.hgc.jp/form2.html) was used to aid in the prediction of protein localization, and TMHMM2.0 was used to predict transmembrane helices in PTC7 http://www.cbs.dtu.dk/services/TMHMM/.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) grant R01GM099790 to A.v.H. We thank the reviewers for thought provoking comments that improved the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.075655.120.
REFERENCES
- Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J 20: 4684–4693. 10.1093/emboj/20.17.4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad AM, Venkataramanan S, Nag A, Galivanche AR, Bradley MC, Neves LT, Douglass S, Clarke CF, Johnson TL. 2017. Chromatin-remodeling SWI/SNF complex regulates coenzyme Q6 synthesis and a metabolic shift to respiration in yeast. J Biol Chem 292: 14851–14866. 10.1074/jbc.M117.798397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. 1998. Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO J 17: 1107–1119. 10.1093/emboj/17.4.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. 2011. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol 18: 715–720. 10.1038/nsmb.2057 [DOI] [PubMed] [Google Scholar]
- Davis CA, Grate L, Spingola M, Ares M Jr. 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res 28: 1700–1706. 10.1093/nar/28.8.1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan PD, Holland LM, Lombardi L, Coughlan AY, Higgins DG, Wolfe KH, Butler G. 2018. TPP riboswitch-dependent regulation of an ancient thiamin transporter in Candida. PLoS Genet 14: e1007429 10.1371/journal.pgen.1007429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink GR. 1987. Pseudogenes in yeast? Cell 49: 5–6. 10.1016/0092-8674(87)90746-X [DOI] [PubMed] [Google Scholar]
- Freitag J, Ast J, Bolker M. 2012. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485: 522–525. 10.1038/nature11051 [DOI] [PubMed] [Google Scholar]
- Gowda NK, Kaimal JM, Masser AE, Kang W, Friedlander MR, Andreasson C. 2016. Cytosolic splice isoform of Hsp70 nucleotide exchange factor Fes1 is required for the degradation of misfolded proteins in yeast. Mol Biol Cell 27: 1210–1219. 10.1091/mbc.E15-10-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund SE, Fischer T, Cabal GG, Antunez O, Perez-Ortin JE, Hurt E. 2008. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J Cell Biol 182: 897–910. 10.1083/jcb.200803098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Niemi NM, Hutchins PD, Condon SGF, Jochem A, Ulbrich A, Higbee AJ, Russell JD, Senes A, Coon JJ, et al. 2017. Ptc7p dephosphorylates select mitochondrial proteins to enhance metabolic function. Cell Rep 18: 307–313. 10.1016/j.celrep.2016.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Amrani N, Johansson MJ, Jacobson A. 2008. Chapter 6. Qualitative and quantitative assessment of the activity of the yeast nonsense-mediated mRNA decay pathway. Methods Enzymol 449: 127–147. 10.1016/S0076-6879(08)02406-3 [DOI] [PubMed] [Google Scholar]
- Hooks KB, Delneri D, Griffiths-Jones S. 2014. Intron evolution in Saccharomycetaceae. Genome Biol Evol 6: 2543–2556. 10.1093/gbe/evu196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Claggett JM, Edwards SR, Shi A, Pennebaker SL, Cheng MY, Hasty J, Johnson TL. 2016. Posttranscriptional regulation of Gcr1 expression and activity is crucial for metabolic adjustment in response to glucose availability. Mol Cell 62: 346–358. 10.1016/j.molcel.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Juneau K, Nislow C, Davis RW. 2009. Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics 183: 185–194. 10.1534/genetics.109.105155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabran P, Rossignol T, Gaillardin C, Nicaud JM, Neuveglise C. 2012. Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA Res 19: 231–244. 10.1093/dnares/dss007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaduman R, Chanarat S, Pfander B, Jentsch S. 2017. Error-prone splicing controlled by the ubiquitin relative Hub1. Mol Cell 67: 423–432 e424. 10.1016/j.molcel.2017.06.021 [DOI] [PubMed] [Google Scholar]
- Kawashima T, Douglass S, Gabunilas J, Pellegrini M, Chanfreau GF. 2014. Widespread use of non-productive alternative splice sites in Saccharomyces cerevisiae. PLoS Genet 10: e1004249 10.1371/journal.pgen.1004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624. 10.1038/nature02424 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Kogel A, Ebert J, Reichelt P, Stegmann E, Habermann B, Conti E. 2016. Structure of a cytoplasmic 11-subunit RNA exosome complex. Mol Cell 63: 125–134. 10.1016/j.molcel.2016.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidschreiber M, Easter AD, Battaglia S, Rodriguez-Molina JB, Casanal A, Carminati M, Baejen C, Grzechnik P, Maier KC, Cramer P, et al. 2018. The APT complex is involved in non-coding RNA transcription and is distinct from CPF. Nucleic Acids Res 46: 11528–11538. 10.1093/nar/gky845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldon T. 2015. Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the Baker's yeast lineage. PLoS Biol 13: e1002220 10.1371/journal.pbio.1002220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall AN, Montealegre MC, Jimenez-Lopez C, Lorenz MC, van Hoof A. 2013. Alternative splicing and subfunctionalization generates functional diversity in fungal proteomes. PLoS Genet 9: e1003376 10.1371/journal.pgen.1003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall AN, Han J, Kim M, van Hoof A. 2018. Conservation of mRNA quality control factor Ski7 and its diversification through changes in alternative splicing and gene duplication. Proc Natl Acad Sci 115: E6808–E6816. 10.1073/pnas.1801997115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Plass M, Perez-Valle J, Eyras E, Vilardell J. 2011. Deciphering 3′ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol Cell 43: 1033–1039. 10.1016/j.molcel.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem 278: 33000–33010. 10.1074/jbc.M304454200 [DOI] [PubMed] [Google Scholar]
- Neuveglise C, Marck C, Gaillardin C. 2011. The intronome of budding yeasts. C R Biol 334: 662–670. 10.1016/j.crvi.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Parenteau J, Durand M, Veronneau S, Lacombe AA, Morin G, Guerin V, Cecez B, Gervais-Bird J, Koh CS, Brunelle D, et al. 2008. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol Biol Cell 19: 1932–1941. 10.1091/mbc.e07-12-1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. 2011. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 30: 1804–1817. 10.1038/emboj.2011.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Igual JC, Perez-Ortin JE. 2002. SRC1: an intron-containing yeast gene involved in sister chromatid segregation. Yeast 19: 43–54. 10.1002/yea.803 [DOI] [PubMed] [Google Scholar]
- Scannell DR, Frank AC, Conant GC, Byrne KP, Woolfit M, Wolfe KH. 2007. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci 104: 8397–8402. 10.1073/pnas.0608218104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber K, Csaba G, Haslbeck M, Zimmer R. 2015. Alternative splicing in next generation sequencing data of Saccharomyces cerevisiae. PLoS One 10: e0140487 10.1371/journal.pone.0140487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XX, Zhou X, Kominek J, Kurtzman CP, Hittinger CT, Rokas A. 2016. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 (Bethesda) 6: 3927–3939. 10.1534/g3.116.034744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. 2010. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330: 369–372. 10.1126/science.1192430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbis K, van den Burg J, Visser WF, van den Berg M, Distel B. 2012. Alternative splicing directs dual localization of Candida albicans 6-phosphogluconate dehydrogenase to cytosol and peroxisomes. FEMS Yeast Res 12: 61–68. 10.1111/j.1567-1364.2011.00761.x [DOI] [PubMed] [Google Scholar]
- Talkish J, Igel H, Perriman RJ, Shiue L, Katzman S, Munding EM, Shelansky R, Donohue JP, Ares M. 2019. Rapidly evolving protointrons in Saccharomyces genomes revealed by a hungry spliceosome. PLOS Genet 15: e1008249 10.1371/journal.pgen.1008249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Staples RR, Baker RE, Parker R. 2000. Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol Cell Biol 20: 8230–8243. 10.1128/MCB.20.21.8230-8243.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Company M, Abelson J. 1989. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev 3: 1206–1216. 10.1101/gad.3.8.1206 [DOI] [PubMed] [Google Scholar]
- Wang J, Yan Z, Shen SH, Whiteway M, Jiang L. 2007. Expression of CaPTC7 is developmentally regulated during serum-induced morphogenesis in the human fungal pathogen Candida albicans. Can J Microbiol 53: 237–244. 10.1139/w06-125 [DOI] [PubMed] [Google Scholar]