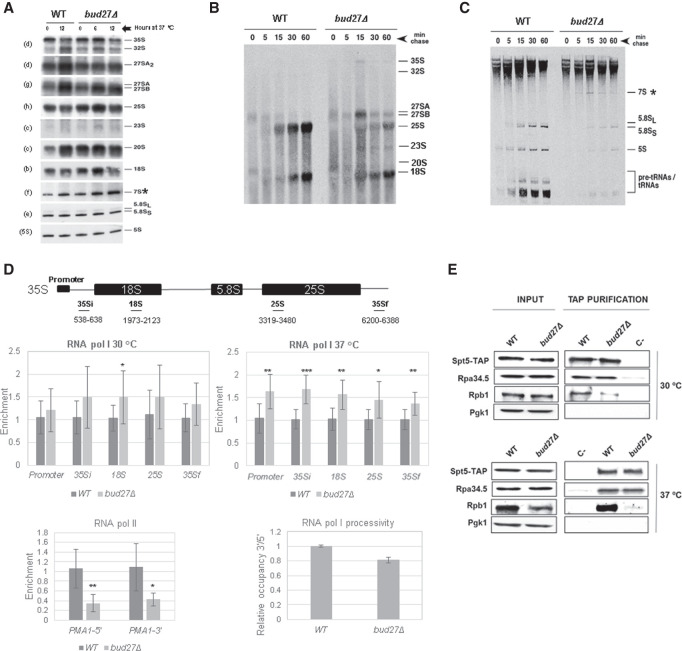
FIGURE 1.
Bud27 is required for the optimal synthesis and maturation of the rRNAs transcribed by RNA polymerase I. (A) Northern blot analysis of the pre- and mature rRNAs of the wild-type and bud27Δ strains. Cells were grown in YPD medium at 30°C and shifted to 37°C for 6 or 12 h. Total RNA was extracted and equal amounts (5 µg) were subjected to northern hybridization. Probes, in brackets, are described in Supplemental Table S2 and Supplemental Figure S2. 7S pre-rRNA is indicated by an asterisk. (B,C) The wild-type and bud27Δ strains were transformed with YCplac33 (CEN, URA3), grown at 30°C in SD-Ura to the mid-log phase and shifted to 37°C for 12 h. Cells were pulse-labeled with [5,6-3H]uracil for 2 min, followed by a chase with unlabeled uracil largely in excess for the indicated times. Total RNA was extracted from each sample and 20,000 c.p.m. were loaded and separated on 1.2% agarose-6% formaldehyde gel (B) or on 7% polyacrylamide-8 M urea gel (C) before being transferred to nylon membranes and visualized by fluorography. The positions of the different pre- and mature rRNAs are indicated. We assume that 7S pre-rRNAs correspond to the species labeled by an asterisk, which were only evident in bud27Δ cells. (D) RNA Pol I and Pol II occupancy (Rpb8-TAP occupancy) was analyzed by chromatin immunoprecipitation (ChIP) in the wild-type and bud27Δ cells containing a functional TAP-tagged version of Rpb8, which is a common subunit to the three RNA pols. Cells were grown in YPD at 30°C or shifted to 37°C for 12 h. RNA pols were precipitated using Dynabeads Pan Mouse IgG as described in the Materials and Methods. The occupancy on the rDNA unit was analyzed for the indicated amplicons. The values found for the immunoprecipitated PCR products were compared to those of the total input, and the ratio of each PCR product of transcribed genes to a nontranscribed region of chromosome V was calculated. The average and standard deviations of three biological replicates are shown. RNA Pol II occupancy on PMA1 gene (5′ and 3′ regions) was analyzed at 30°C. Statistical significance, by t-Student. (*) P < 0.05, (**) P < 0.005, (***) P < 001. The lowest right panel shows the processivity analysis of the data from RNA Pol I occupancy, at 37°C. (E) Wild-type and bud27Δ cells containing a functional TAP-tagged version of Spt5 were grown in YPD medium at 30°C or shifted to 37°C for 12 h. Whole-cell extracts were performed and subjected to pull-down with an anti-TAP resin. The obtained immunoprecipitates were analyzed by western blot with antibodies against TAP (Spt5), Rpa34.5 (RNA Pol I), Rpb1 (RNA Pol II), and Pgk1 as a control.