SUMMARY
Human pluripotent stem cells differentiated to insulin-secreting β Cells (SC-β Cells) in islet organoids could provide an unlimited cell source for diabetes cell replacement therapy. However, current SC-β Cells generated in vitro are transcriptionally and functionally immature compared to native adult β Cells. Here, we use single-cell transcriptomic profiling to catalog changes that occur in transplanted SC-β Cells. We find that transplanted SC-β Cells exhibit drastic transcriptional changes and mature to more closely resemble adult β Cells. Insulin and IAPP protein secretions increase upon transplantation, along with expression of maturation genes lacking with differentiation in vitro, including INS, MAFA, CHGB, and G6PC2. Other differentiated cell types, such as SC-α and SC-enterochromaffin (SC-EC) cells, also exhibit large transcriptional changes. This study provides a comprehensive resource for understanding human islet cell maturation and provides important insights into maturation of SC-β Cells and other SC-islet cell types to enable future differentiation strategy improvements.
In Brief
Augsornworawat et al. demonstrate that in vitro human stem-cell-derived islets have immature transcriptome profiles when compared to native human islets. After transplantation, immature SC-islet cells undergo drastic changes in transcriptome and acquire mature gene expression. Grafted SC-β Cells express mature β Cell genes, including MAFA and G6PC2, resembling native human islets.
Graphical Abstract
INTRODUCTION
Diabetes is a global disease caused by death or dysfunction of pancreatic insulin (INS)-producing β Cells. Current conventional therapeutic strategies, such as exogenous INS treatment, are insufficient in recapitulating the role of healthy endogenous β Cells for many patients (Atkinson et al., 2014). Transplantation of replacement human islets (HIs) is a promising alternative (Latres et al., 2019; Shapiro et al., 2006, 2000). However, this therapy faces the challenge of limited availability from cadaveric donors. This is compounded by the need of multiple donors to treat one diabetic patient in most cases. Human pluripotent stem cells (hPSCs) differentiated into functional islets (SC-islets) containing β Cells (SC-β Cells) would provide a renewable cell source for cell replacement therapy (Aguayo-Mazzucato and Bonner-Weir, 2010). Recently, differentiation strategies have been developed to successfully specify fate selection of differentiating hPSCs into SC-islets (Pagliuca et al., 2014; Rezania et al., 2014). Further refinements to these protocols have improved function and gene expression (Ghazizadeh et al., 2017; Hogrebe et al., 2020; Millman et al., 2016; Nair et al., 2019; Velazco-Cruz et al., 2020b, 2019). Nonetheless, SC-islets generated in vitro remain unequal to cadaveric islets both functionally and transcriptionally.
Single-cell RNA sequencing (scRNA-seq) and other assays of global transcription have been used to rigorously study and uncover pancreatic identities (Augsornworawat and Millman, 2020), including assessment of SC-β Cell populations in diabetic models (Balboa et al., 2018; Maxwell et al., 2020), analysis of SC-islets from in vitro differentiation (Pagliuca et al., 2014; Veres et al., 2019), and studying islets or islet development (Baron et al., 2016; Byrnes et al., 2018; Enge et al., 2017; Hogrebe et al., 2020; Muraro et al., 2016; Russell et al., 2020; Scavuzzo et al., 2018; Segerstolpe et al., 2016; Velazco-Cruz et al., 2020a; Xin et al., 2018). When compared to β Cells from cadaveric islets, these technologies revealed that in-vitro-derived SC-β Cells lack or have low expression of many important maturation genes, such as MAFA and G6PC2 (Hogrebe et al., 2020; Pagliuca et al., 2014; Veres et al., 2019). However, after transplantation, the function and amount of INS secreted by SC-β Cells increases (Balboa et al., 2018; Hogrebe et al., 2020; Nair et al., 2019; Pagliuca et al., 2014; Rezania et al., 2014; Vegas et al., 2016; Velazco-Cruz et al., 2019). The cause of this increase is unknown and may be due to differentiation of accompanying progenitors, remodeling of the graft, and the maturation state of cells not being set by the in vitro conditions under which they were derived. However, further assessment of these transplanted cells has been limited to immunostaining and functional assessment of ex vivo grafts (Balboa et al., 2018; Bruin et al., 2015; Pepper et al., 2017; Robert et al., 2019; Velazco-Cruz et al., 2019).
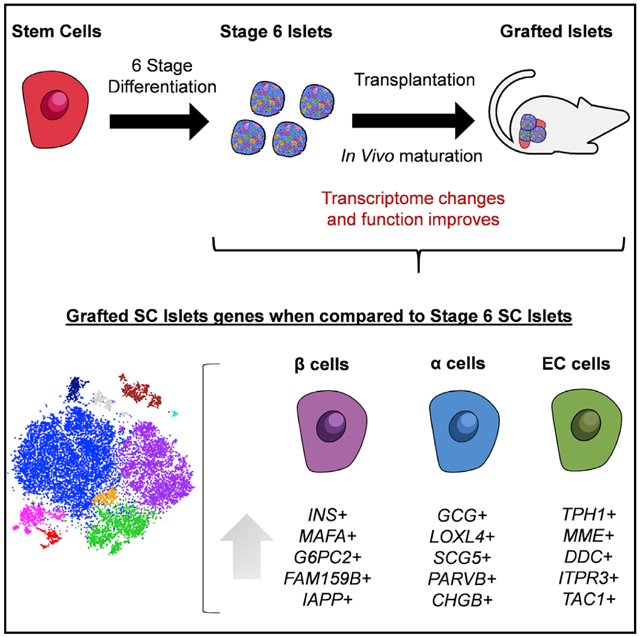
Here, we utilize scRNA-seq to elucidate the cellular identity, maturation, and changes in gene expression of transplanted (grafted) SC-islets compared to in-vitro-generated (stage 6) SC-islets and primary cadaveric islets. We demonstrate that β and α cells differentiated from both human embryonic stem cell (hESC) and human induced pluripotent stem cell (hiPSC) lines undergo functional and drastic transcriptional changes after transplantation to closely resemble β and α cells from cadaveric islets. Specifically, for SC-β Cells, we find that transplantation increases expression of many missing or lowly expressed β Cell maturation and identity genes, including MAFA, G6PC2, MNX1, and INS, while decreasing GCG and SST. In addition, we chart the transcriptional changes of stem-cell-derived enterochromaffin (SC-EC) cells that were recently described to be generated during SC-islet differentiation (Veres et al., 2019). This is a critical resource for understanding and advancing current directed differentiation technologies to develop novel cell therapies for diabetes.
RESULTS
Retrieval and Identification of SC-Islet Cell Populations after Transplantation
We differentiated a hESC and hiPSC line with our six-stage differentiation protocol into SC-islets (Hogrebe et al., 2020) (Figures S1A and S1B). Differentiated SC-islets were then transplanted into the kidney capsule of streptozotocin (STZ)-induced diabetic mice (Figure 1A). Glucose tolerance test (GTT) was performed in transplanted diabetic mice, showing improvements in blood glucose regulation after 4 weeks and further reversal of diabetes at 6 months (Figure 1B). Moreover, we detected increasing glucose stimulated human C-peptide (CP) secretion in serum of transplanted mice (Figure 1C). After 6 months, we harvested the kidneys from mice to extract transplanted SC-islets. These grafts display endocrine cell protein markers including chromogranin A (CHGA), Homeobox protein Nkx-6.1 (NKX6.1), CP, and glucagon (GCG) (Figure 1D). The extracted SC-islet grafts were processed for ex vivo analysis by single-cell dispersing the excised kidney with collagenase D and enriching grafted cells with a magnetic bead mouse cell depletion kit. After library preparation with the 10x Genomics Chromium platform, we sequenced 45,674 cells from isogenic matched stage 6 (in vitro) and grafted (in vivo) cells with the NovaSeq 6000 (Figures 1E and 1F). Filtering was performed to remove cells with high mitochondrial content or high gene number, indicative of apoptotic cells and doublets, respectively. To remove residual mouse kidney cells, we performed unsupervised clustering of the grafted samples to generate t-distributed Stochastic Neighbor Embedding (tSNE) plots (Figure S1C). We identified a cluster of cells in each sample containing low human-mapped gene counts, which marked predominantly mouse cells (Figure S1D). TTC36, a kidney gene marker that aligns to both the mouse and human genome, is represented in each cluster with low gene counts in all graft samples. To verify, we generated violin plots to show upregulated expression of TTC36 in the identified low gene count population (Figure S1E; Table S1) (Zhou et al., 2016). For all grafted samples, we removed all cells enriched with TTC36 as a method to distinguish mouse cells and remove them from subsequent analysis.
Figure 1. Single-Cell Transcriptional Characterization of Stage 6 and Grafted hPSC-Islets Identifies Endocrine and Non-Endocrine Cell Populations.
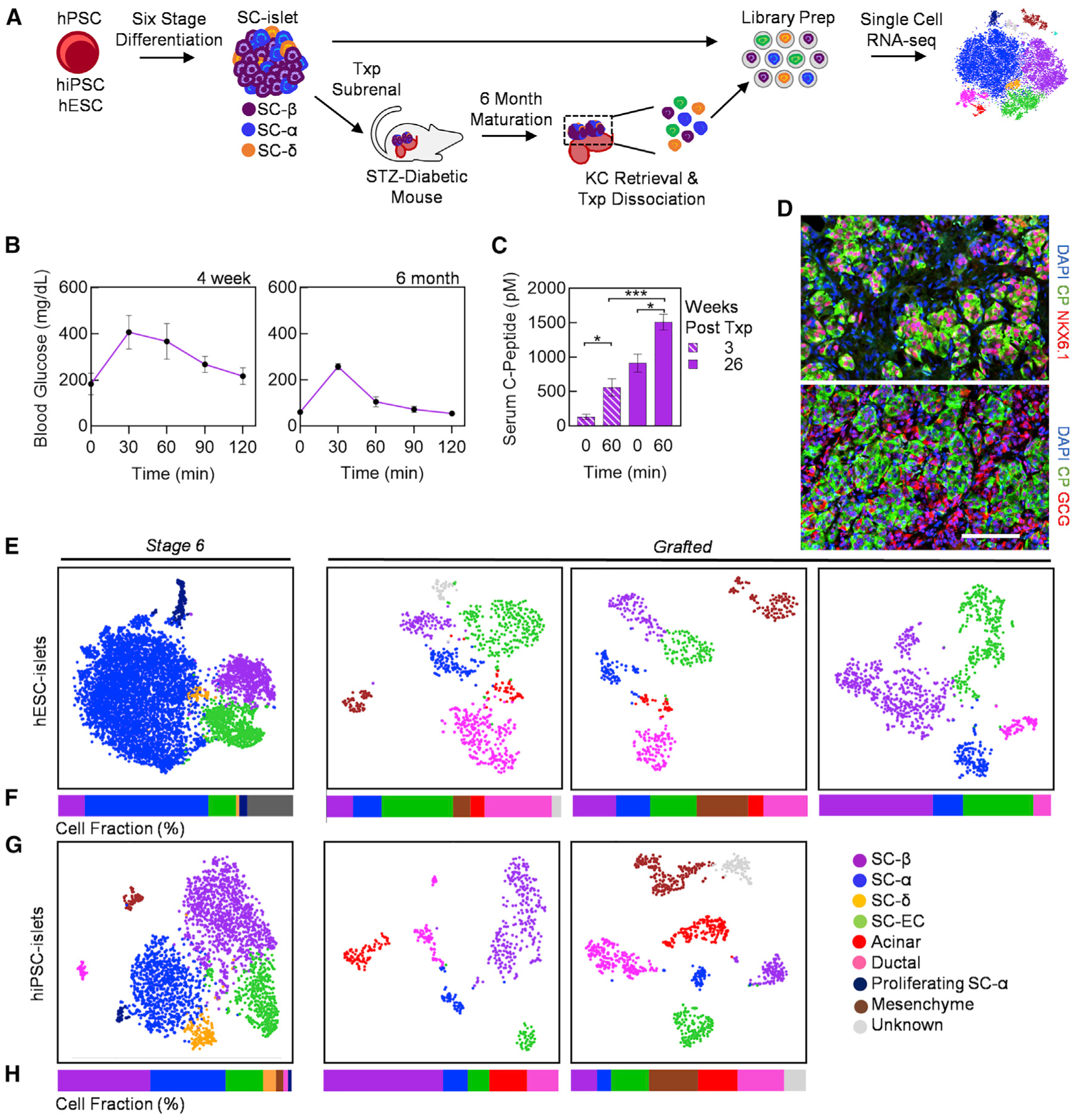
(A) Schematic summary of single-cell analysis of stage 6 SC-islets and grafted SC-islets following 6 months of in vivo maturation in diabetic mice.
(B) Glucose tolerance test (GTT) at 4 weeks and 6 months after transplantation of SC-islets (n = 5).
(C) In vivo glucose-stimulated CP secretion at 0 min, before, and 60 min after glucose injection at 3 and 26 weeks post-transplantation (n = 5). *p < 0.05, ***p < 0.001 by two-way unpaired t test.
(D) Immunostaining of kidney graft explanted form SC-islet transplanted mice at 6 months. Scale bar, 100 μm.
(E–H) Individual tSNE projection (E and G) and cell population fractions (F and H) from unsupervised cluster of stage 6 and grafted hESC- and hiPSC-islets. CHGA, chromogranin A; CP, C-peptide; EC, enterochromaffin; GCG, glucagon. Error bars represent SEM. See also Figure S1 and Table S2.
After filtering, we had 15,914 cells for analysis from stage 6 hiPSC-islets, stage 6 hESC-islets, hESC-islet grafts from three mice, and hiPSC-islet grafts from two mice. Single-cell datasets of the same condition, three hESC-islet grafts and two hiPSC-islet grafts, were pooled to obtain sufficient cell numbers for subsequent analyses, comparable to previously published literature (Enge et al., 2017; Segerstolpe et al., 2016; Wang et al., 2016; Xin et al., 2016). Single-cell analysis revealed the majority of cell types to be pancreatic in nature for both stage 6 and grafted cells. Within the individual sample tSNE projections, we identified key pancreatic endocrine cell populations, including SC-β, stem-cell-derived α (SC-α), and stem-cell-derived δ (SC-δ) cells. The SC-β Cells identified in the sequencing data appear to match prior reports that defined SC-β Cells by the expression of CP and NKX6.1, while the polyhormonal cells identified in prior reports appear to be within the SC-α and SC-δ cell populations in the sequencing data (Pagliuca et al., 2014; Velazco-Cruz et al., 2019). Other cell types, including exocrine, also persist to some extent, as found in previous transplantation studies (Velazco-Cruz et al., 2019) (Figures 1E–1H and S1F). The fate of these exocrine cells is hard to determine with our dataset and has been similarly hard in other datasets (Korbutt et al., 1996). Our cluster-matched cell types were assigned based on cell-type-specific genes found in published scRNA-seq literature for cadaveric and SC-islets (Table S2) (Muraro et al., 2016; Veres et al., 2019). The fractional yields and defining genes of each cell type can be found in Table S2. SC-β and SC-α cells are present in both stage 6 and grafted populations. We also identify populations of proliferating SC-α cells (GCG, ARX, CRYBA2, MKI67, PCNA, and MCM2) in stage 6 SC-islets (Figure S2A), similar to as previously reported (Balboa et al., 2018). These proliferative genes are not expressed in other endocrine cell types. Similarly, identifiable SC-δ cell populations are only present in stage 6 SC-islets (Figures 1E–1H). While SST+/HHEX+ cells are found in the grafted samples, their overall gene expression signatures are not sufficiently different from SC-α cells to cluster separately. Recently, SC-EC cells were found to be an off-target cell type generated from SC-islet differentiation protocols and have been implicated as an inhibitory cell type for SC-islet in vitro function (Veres et al., 2019). These cells synthesize and secrete serotonin (TPH1, SLC18A1) and are typically found in the gastrointestinal tract or gastric glands (Grün et al., 2015). We identify SC-EC cells in stage 6 and grafted SC-islets (Figures 1E–1H). Our analysis shows that many cell types generated in vitro by our six-stage differentiation protocol persist in grafted SC-islets.
SC-β Cells Mature within Transplanted Grafts
To study gene expression changes that occur in SC-β Cells after transplantation, we projected individually sequenced stage 6 and grafted cells onto a combined tSNE, separated by cell line, and quantified the percentage of each cell type in the combined plots (Figure 2A). It is important to note that combined plots are used for subsequent gene and gene set analysis, as the dimensional reduction provided by tSNE plots depends on the samples used, and absolute distances should not be compared across different plots. Similarly, we designated cell types based on literature of pancreatic cell types matched to upregulated genes within each cluster (Figure S2A; Table S3) (Muraro et al., 2016; Veres et al., 2019). We detected 1,671 and 1,683 β Cells derived from hESC and hiPSC lines, respectively. Focusing on the SC-β Cell population (Figure 2B), we identify 5,867 genes with differential expression (adjusted p value <0.05) in grafted compared to stage 6 SC-β Cells (Table S4). Despite some variations among SC-β Cells from hiPSC and hESC sources (Figures S2B and S2C; Table S3), which we expected due to differences in genetic background (Kim et al., 2009; Wang et al., 2016), we observe substantial reductions in transcriptome variations among SC-β Cells after transplantation when compared between the two lines (Figures S2D and S2E; Table S3). Drastic and congruent transcriptome changes are observed in transplanted SC-β Cells when segmented by the original derivation line (Figures 2C, 2D, S2F, and S2G; Table S4).
Figure 2. Comparison of Stage 6 and Grafted hiPSC-β and hESC-β Cell Transcriptomes.
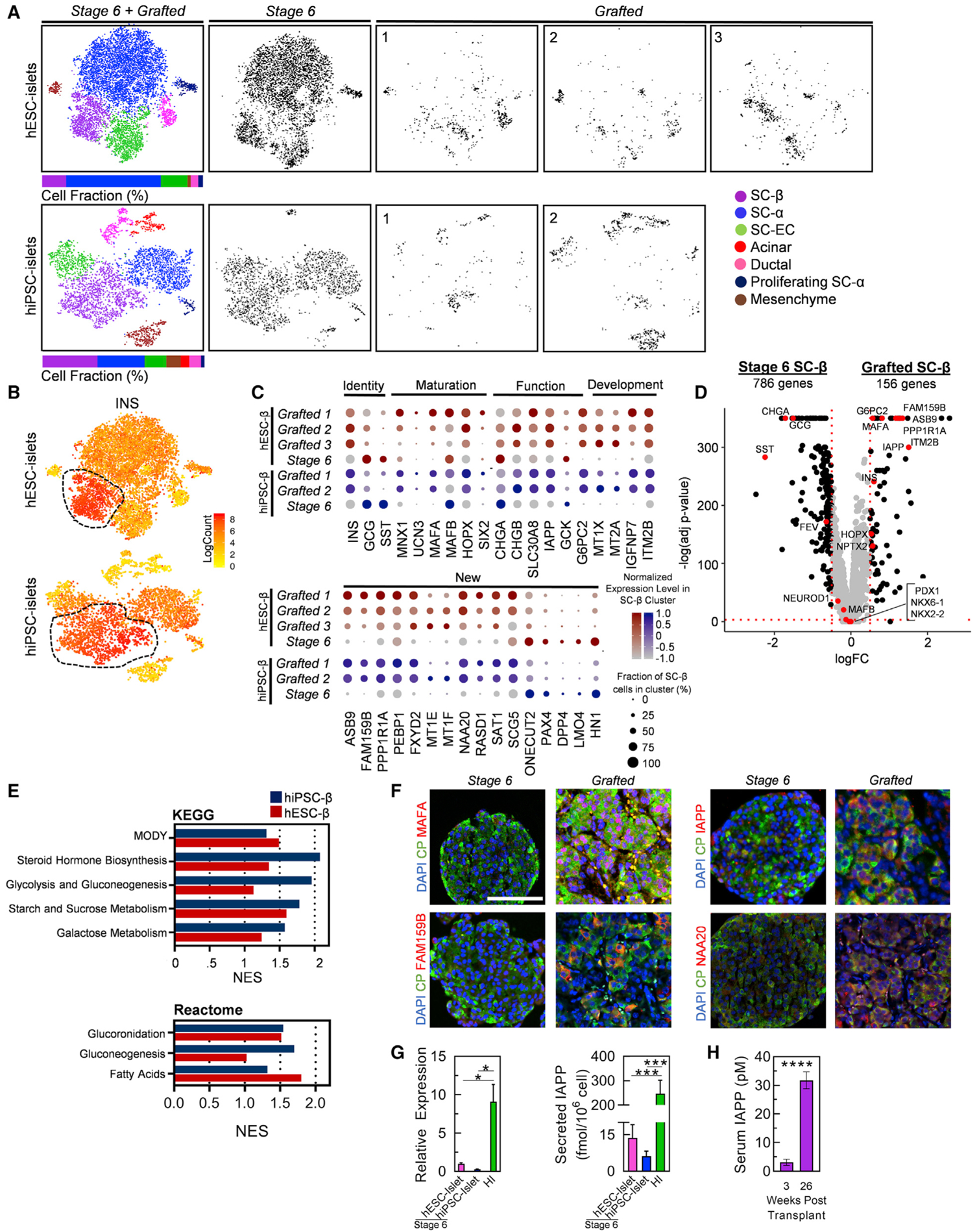
(A) Unsupervised clustering tSNE projections of hESC-islets and hiPSC-islets with islet cell-type identification and cell proportions for each individual sample (gray).
(B) Heatmap of INS gene expression to distinguish SC-β Cell population for both hESC-islet and hiPSC-islet samples.
(C) Normalized expression values for key β Cell identity, maturation, functional, developmental, and newly identified gene sets compared between stage 6 and grafted SC-β Cells for both hiPSC-β and hESC-β datasets. Percentage of cells positive for designated gene is represented by circle size, and saturation indicates low (gray) or high (blue/red) expression of the gene.
(D) Volcano plot displaying fold change (FC) differences of β Cell genes between stage 6 and grafted SC-β Cells of all hESC- and hiPSC-derived samples combined. Dashed lines are drawn to define restriction of log FC value of 0.25 and −log of adjusted p value 0.001. Key β Cell genes are labeled and colored red.
(E) KEGG and Reactome GSEA, quantified by the normalized enrichment score (NES), for pathways upregulated in grafted hESC-β (red) and hiPSC-β (blue) compared to stage 6 hESC-β or hiPSC-β Cells, respectively. NES values and gene lists are available in Table S4.
(F) Immunostaining of stage 6 and 6-month kidney grafts containing SC-islets for identified β Cell maturation markers (red) co-stained with CP (green). Scale bar, 100 μm.
(G) Analysis of IAPP gene expression with real-time PCR (left; n = 4) and secreted IAPP protein (right; n = 11, n = 8, n = 9 for stage 6 hESC-islet, stage 6 hiPSC-islet, and HIs, respectively) in vitro. *p < 0.05, ***p < 0.001 by two-way unpaired t test.
(H) IAPP protein in serum of mice transplanted with SC-islets for 3 and 26 weeks (n = 5). ****p < 0.0001 by two-way unpaired t test.
INS, insulin; MODY, maturity onset diabetes of the young; CHGB, chromogranin B; IAPP, islet amyloid polypeptide; HI, human islet. Error bars represent SEM. See also Figures S1 and S2 and Table S3.
Of particular interest, studied maturation genes such as INS, MAFA, SIX2, MNX1, and G6PC2 are now more highly expressed in a greater fraction of cells within the grafted SC-β Cells, whereas they were previously missing or had lower expression in SC-β Cells in vitro (Figures 2C and 2D; Table S4). Moreover, many additional genes associated with mature β Cells are found more highly expressed in grafted compared to stage 6 SC-β Cells. UCN3, recognized as a β Cell maturation marker from mouse studies (Blum et al., 2012), is increased. KLF9 and HOPX, which were identified to be upregulated in adult β Cells when compared to fetal primary β Cells, are increased (Hrvatin et al., 2014). FAM159B, ITM2B, and IGFBP7, which are regulators of physiological function expressed in cadaveric β Cells (Camunas-Soler et al., 2020; Segerstolpe et al., 2016; Tyler et al., 2019), are found upregulated in our grafted SC-β Cells. Gene set enrichment analysis (GSEA) reveals higher expression of KEGG and REACTOME pathways pertaining to β Cell identity and processes in the grafted SC-β Cells (Figure 2E and S2H; Table S4).
To verify our identified SC-β Cell maturation genes, we performed histology stains of in-vitro-derived and transplanted SC-islets. We stained the graft of transplanted cells for maturation markers including MAFA, FAM159B, NAA20, and islet amyloid polypeptide (IAPP) (Figure 2F). Transplanted SC-β Cells show marked expression of protein products from these upregulated genes. Because IAPP encodes a hormone that is also secreted by β Cells, we confirmed that the transcript was lower using real-time PCR and detected lower secretion of the protein in stage 6 SC-β Cells compared to primary islets (Figure 2G). To test if secreted IAPP protein could be a useful biomarker correlating with maturation, we measured IAPP in the serum of transplanted mice 3 and 26 weeks after transplantation (Figure 2H). We found that IAPP secretion increased 10.1 ± 2.63, correlating with the scRNA-seq results of increased IAPP transcript (Figure 2C). This is also a greater dynamic range than what we observed with CP, which only increased 2.7 ± 0.5× during the same time period (Figure 1C). The protein analysis here is consistent with the scRNA-seq data.
Other misexpressed genes have lower expression in SC-β Cells after transplantation (Figures 2C and 2D; Table S4). We and others previously identified several genes (CHGA, MAFB, GCK, and GLUT1) that were overexpressed in stage 6 SC-islets compared to cadaveric islets (Augsornworawat et al., 2019; Hogrebe et al., 2020; Nair et al., 2019; Velazco-Cruz et al., 2019; Veres et al., 2019) and here find that they have reduced expression in grafted compared to stage 6 SC-β Cells. We observe reductions in GCG and SST gene expression and other genes expressed by α and δ cells (ALDH1A1, FTL, and CRYBA2) with transplantation. Genes enriched in non-functional stem-cell-derived INS-expressing endocrine compared to adult cadaveric β Cells (FOXA1, NR2F1, FEV, SOX11, ONECUT2, and TCF3) (Hrvatin et al., 2014) are lowly expressed in grafted SC-β Cells.
Next, we reconstructed the single-cell data from stage 6 and grafted SC-β Cells into developmental trajectories to obtain a greater understanding of the dynamic changes in β Cell maturation transcripts. Stage 6 SC-β Cells from hESC and hiPSC lines form separate branches at the initial pseudo-time state and merge to a single trajectory that consists mainly of grafted SC-β Cells (Figures 3A and 3B), suggesting that the transcriptomes among different cell lines become more similar upon maturation. Reduction of GCG and SST is also observed with pseudo-time, thus verifying our observation of β Cell fate commitment after transplantation (Figure 3C). Using the top 100 most differentially expressed genes, we were able to observe a multitude of genes that trend similarly to GCG and SST including ADLH1A1, MAP1B, and CRYBA2 (Figure 3D; Table S6). Conversely, key β Cell maturation genes, specifically MAFA and G6PC2, increase in expression with pseudo-time (Figures 3E and 3F). Of note, we identified additional regulatory and identity genes including FXYD2, IAPP, MT1X, and SAT1 to increase in concert with MAFA and G6PC2. When evaluating the most differentially expressed genes, JUN, NEAT1, PPP1R1A, SLC30A1, and ASB9 are upregulated at the terminal end of the trajectory (Figure 3D), indicating further β Cell development.
Figure 3. Pseudo-Time Analysis Projects Stage 6 SC-β Cell Maturation into Grafted SC-β Cells after Transplantation.
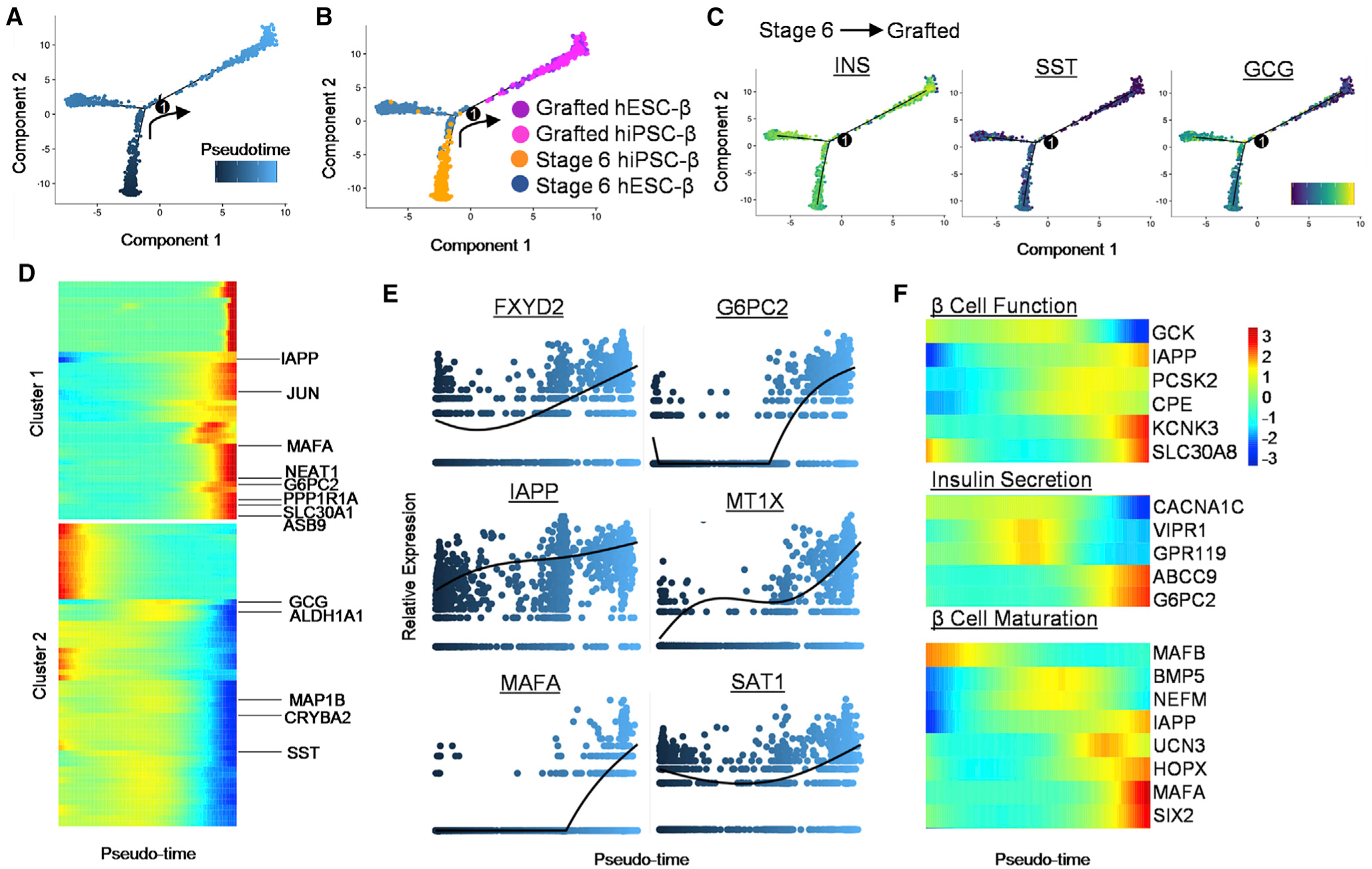
(A) Pseudo-time trajectory of stage 6 and grafted SC-β Cells with one branching point.
(B) Distribution of SC-β Cells from grafted hESC-β (purple), grafted hiPSC-β (pink), stage 6 hiPSC-β (orange), and stage 6 hESC-β Cells (blue). (C) Changes in gene expression of endocrine hormones—INS, somatostatin (SST), and GCG—along pseudo-time projection for SC-β Cells. Scale: relative expression.
(D) Analysis of top 100 branch-dependent genes determined based on clusters identified through pseudo-time analysis of SC-β Cells, found in Table S4. Scale: relative expression.
(E) Relative expression level of selected β Cell genes along SC-β Cell pseudo-time.
(F) Gene expression heatmap of select β Cell genes focused on function and maturation along pseudo-time. Scale: normalized expression. See also Figure S3 and Table S4.
We also analyze additional gene sets pertaining to β Cell function and maturation. We see upregulation of KCNK3, SLC30A8, and IAPP, known to support β Cell function (Figures 3E, 3F, and S3) (Camunas-Soler et al., 2020; Velazco-Cruz et al., 2019). CACNA1C, a calcium channel subunit important for INS secretion, lessens with maturation; VIPR1 and GPR119, potent GSIS activators (Ohishi and Yoshida, 2012; Tsutsumi et al., 2002), have intermediate activation; and ABCC9 and G6PC2, expressed in functional β Cells, are present at the final state in grafted SC-β Cells (Figure 3F). Consistent with previous HI and SC-β Cell comparison studies, we also observe reduced expression of GCK in the grafted SC-β Cells. Looking into identified β Cell maturation gene sets (Veres et al., 2019), we observe all transcripts, with the exception of MAFB, to increase with pseudo-time (Figures 3F and S3). Though categorized as maturation genes, the expression pattern of MAFB trends similar to primary human β Cells (Hrvatin et al., 2014; Veres et al., 2019). Altogether, our clustering and pseudo-time ordering analyses suggest that transplanted SC-β Cells display dynamic gene patterns leading to β Cell maturity.
Grafted SC-β Cells Closely Resemble Primary Human β Cells
Previous studies show that SC-β Cells are markedly different from native adult β Cells, possessing key β Cell identity genes but short of many other genes by global perspective (Pagliuca et al., 2014). To explore if our grafted SC-β Cells acquired gene expression that better resemble native adult β Cells, we performed subsequent comparisons by incorporating scRNA-seq data from three healthy cadaveric donor HIs (Xin et al., 2018) into a new combined tSNE projection. We pooled 3,368 SC-β Cells (1,765 from hESC and 1,603 hiPSC) with cadaveric HI β Cells, making a total of 4,696 β Cells for our analysis. With a total of 10 single-cell sequencing samples, we detected all cell populations identified previously in Figure 1, which includes pancreatic endocrine and exocrine cell types (Figures 4A, 4B, and S4A; Table S5) (Muraro et al., 2016; Veres et al., 2019). With the addition of primary islet data, we now identify endothelial cells marked by high expression of PECAM1, CD34, and ENTPD1. Notably, we do not detect SC-EC or proliferating α cells within cadaveric HIs (Figures 4A and 4B).
Figure 4. Grafted SC-β Cells Resemble Primary Human β Cells Compared to Stage 6 SC-β Cells.
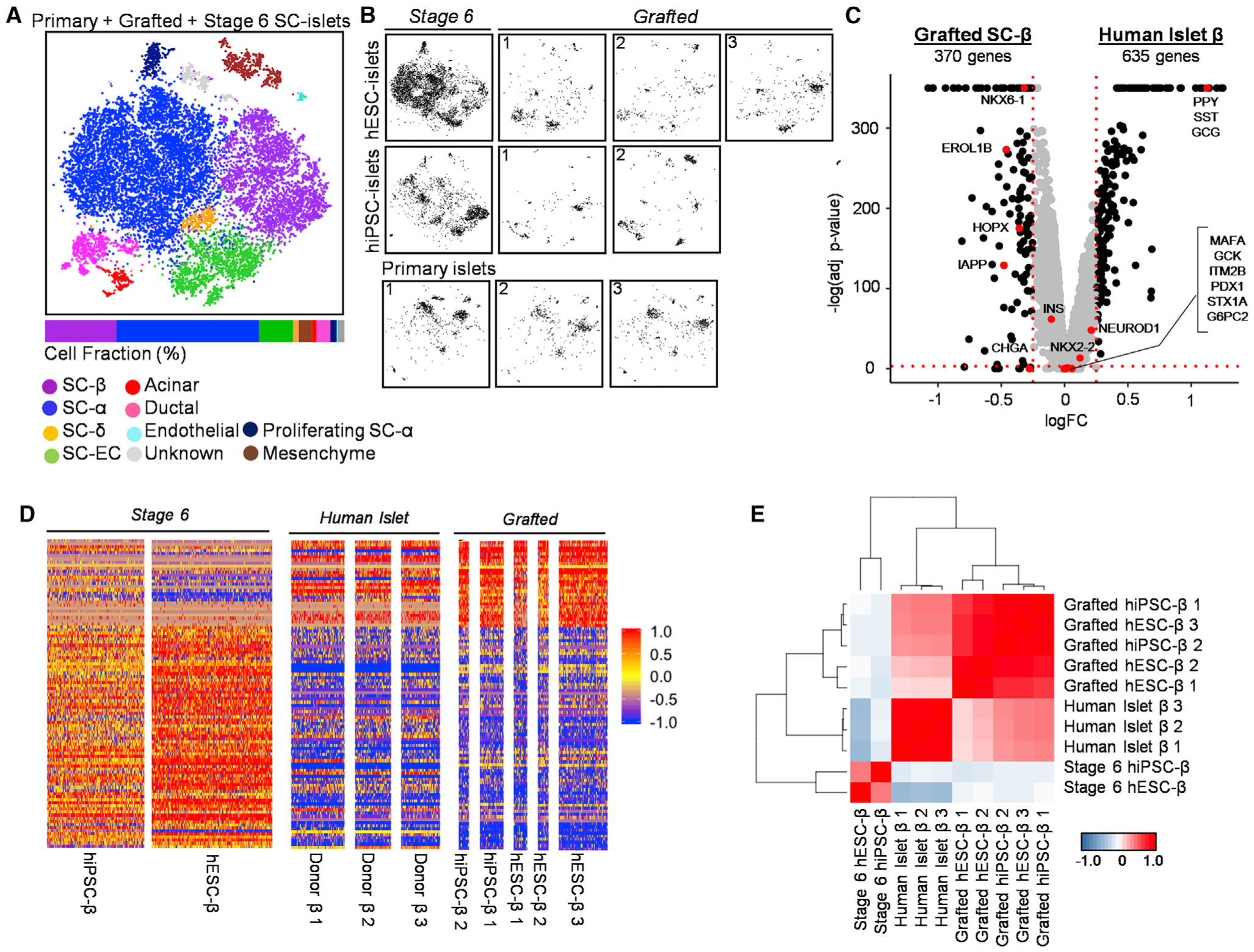
(A) Unsupervised tSNE projection from 10 islet datasets including stage 6 hiPSC- and hESC-islets, three grafted hESC-islets, two grafted hiPSC-islets, and three primary HIs.
(B) Individual location of different samples within combined tSNE projection.
(C) Volcano plot displaying FC differences of β Cell genes between grafted SC-β (left) and HI β Cells (right). Dashed lines are drawn to define restriction of log FC value of 0.25 and −log of adjusted p value 0.001. Key β Cell genes are labeled and colored red.
(D) Heatmap showing gene expression values for stage 6, grafted SC-β, and primary HI (donor) β Cells of the 100 most differentially expressed genes between stage 6 and grafted SC-β Cells listed in Table S6. Scale: normalized expression.
(E) Pearson correlation matrix and hierarchical clustering to identify most similar populations among all β Cell samples using top 430 variable genes. Pearson correlation values detailed in Table S6. Scale: correlation coefficient.
We performed differential gene expression analysis of β Cells from SC-islet grafts and native HIs. β Cell identity genes (INS, NEUROD1, and NKX2–2), maturation genes (MAFA and G6PC2), and other notable genes (ITM2B, PDX1, GCK, and STX1A) are strikingly similar across the two conditions (Figures 4C, S4B, and S4C; Table S6). Furthermore, β Cells from native islets have similar transcriptome profiles when compared to grafted SC-β Cells (Figure 4D). Quantitatively, Pearson correlation shows that native β Cells are more similar to grafted SC-β Cells when compared across all samples (Figure 4E; Table S6). These results suggest that SC-β Cells have the ability to acquire native adult β Cell genes when placed in an in vivo setting.
We also observe genes that are misexpressed in our grafted SC-β Cells when compared to native adult β Cells. Some key functional and β Cell identity genes we identified as reduced in our grafted SC-β Cells, including NKX6–1, CHGA, IAPP, HOPX, and EROL1B, continue to have higher expression when compared to native islet β Cells (Figures 4C, S4B, and S4C; Table S6). Conversely, β Cells from native islets have higher expression of multiple endocrine hormone genes, specifically PPY, SST, and GCG (Figures 4C, S4B, and S4C; Table S6). We also observe Human Leukocyte Antigen (HLA) genes (B2M, HLA-B, and HLA-A) and ribosomal protein small (RPS) and other large (RPL) genes to be upregulated in native adult β Cells (Table S6). Of note, no major β Cell identity genes were under-expressed in our grafted SC-β Cells when compared to native adult β Cells.
In addition to these marked differences, we refined our differentially expressed list of genes upregulated in both grafted SC-β Cells and native adult β Cells (Tables S4 and S6) and established a curated list of genes that are shared across the two conditions (Table S6). In our list, metallothionein (MT1E, MT1F, MT1X, and MT2A) and β Cell function regulatory (FAM159B, G6PC2, and IAPP) genes are highlighted. Notably, IAPP, FAM159B, and MT1X were previously associated with adult HIs and are now confirmed as maturation genes (Camunas-Soler et al., 2020; Muraro et al., 2016). Additional genes including PEBP1, MT2A, FXYD2, NAA20, RASD1, and SAT1 are also upregulated. Pseudo-time ordering analysis also verified these identified genes to be upregulated at similar expression values for both grafted SC-β Cells and native adult β Cells (Figures S4D–S4F). We validated some of our identified maturation genes using qPCR (Figure S4G). Furthermore, we observed that a very small fraction (2.09% and 1.20%) of stage 6 SC-β Cells expressed MAFA but still lacked expression of other important genes, such as G6PC2 (Figures S4H and S4I). Collectively, the results support that our grafted SC-β Cells have transcriptional profiles that more closely resemble native β Cells in cadaveric HIs compared to stage 6 cells.
SC-α Cells Mature within Transplanted Grafts
SC-α cells are generated from our six-stage SC-islet differentiation protocol and account for an average of 51.8% and 11.1% cell proportions in stage 6 and grafted SC-islets, respectively (Table S2). While we see some variation in stage 6 SC-α cells from different genetic backgrounds (Kim et al., 2009; Wang et al., 2016), there is considerable convergence of grafted transcriptome profiles after transplantation (Figure S5A; Table S3). Further analysis of the SC-α cell population (Figure 5A) indicates grafted SC-α cells undergo significant transcriptome changes with greater expression of specific maturation genes compared to stage 6 SC-α cells (Figures 5B–5E; Table S6). However, the grafted SC-α cells are not as similar to HI α cells as SC-β Cells are to HI β Cells (Figures 4E and 5D; Table S6). A marked difference is the reduced level of GCG and increased expression of INS in the grafted SC-α cells compared to native islet α cells (Figures 5C, 5E, and S5B). In fact, this degree of polyhormonality and this higher expression of FEV, an immature α cell marker (Veres et al., 2019), are key characteristics reported in precursor α cells, found in juvenile HIs (Ramond et al., 2018; Veres et al., 2019). Of note, ARX, α key α cell marker, remains elevated in both stage 6 and grafted SC-α cells compared to HI α cells (Figures 5E and S5B). The expression of proliferating genes (MKI67, PCNA, and MCM2) are also found predominantly in the stage 6 SC-α cell population (Figure S2A), but we did not extensively study this population here. This population is not detected in grafted SC-islets (Figures 1E–1H and S4A) or adult HIs and is not detectable in other HI α cell studies (Baron et al., 2016; Muraro et al., 2016). Despite these differences, differential gene expression analysis indicates stage 6 SC-α cells are maturing to a transcriptional state similar to primary islet α cells. For further examination of the SC-α cell maturation state, we curated a maturation gene list (Table S6) from upregulated genes in grafted (Table S4) and HI α cells (Table S6) compared to stage 6 SC-α cells.
Figure 5. Single-Cell Transcriptome and Pseudo-Time Analysis Indicates Stage 6 SC-α Cells Mature during Transplantation to More Closely Resemble HI α cells.
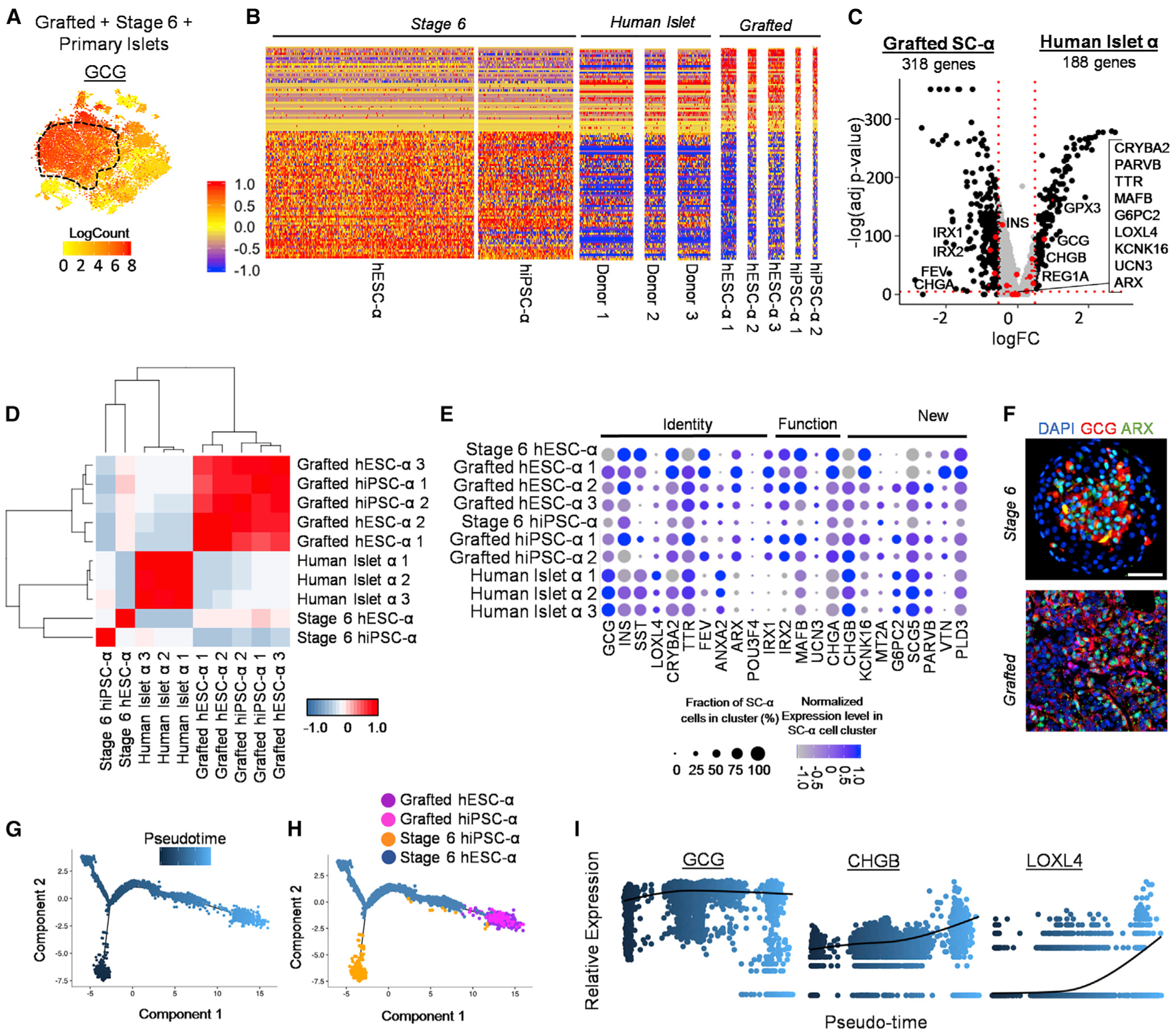
(A) Heatmap of GCG gene expression to distinguish SC-α cell populations within tSNE plot from Figure 4A.
(B) Normalized gene expression values for two stage 6 SC-α, five grafted SC-α, and three primary HI (donor) α cells of the 100 most differentially expressed genes between stage 6 and grafted SC-α cells listed in Table S6. Scale: normalized expression.
(C) Volcano plot displaying FC differences of α cell genes between grafted SC-α (left) and HI α cells (right). Dashed lines are drawn to define restriction of log FC value of 0.25 and −log of adjusted p value 0.001. Key α cell genes are labeled and colored red.
(D) Pearson correlation matrix and hierarchical clustering to identify most similar populations among all α cell samples using top 430 variable genes. Pearson correlation values detailed in Table S6. Scale: correlation coefficient.
(E) Normalized expression values for key α cell identity, function, and newly identified genes among stage 6 SC-α, grafted SC-α, and primary human α cells. Percentage of cells positive for designated gene is represented by circle size, and saturation indicates low (gray) or high (blue) expression of the gene.
(F) Immunostaining of stage 6 and 6-month kidney graft containing SC-islets for α cell markers. Scale bar, 50 μm.
(G and H) Pseudotime trajectory (G) and distribution (H) of SC-α cells from seven hPSC-α samples.
(I) Relative expression level of selected α cell genes along SC-α cell pseudo-time. Scale: relative expression. Each dot represents a different cell. See also Figure S5 and Tables S3, S4, and S6.
To dynamically visualize transcriptional changes, we reconstructed our stage 6 and grafted SC-α cell data into a pseudo-time trajectory (Figures 5G and 5H). Similar to SC-β Cells, the hiPSC-α and the hESC-α cells formed separate branches at the initial pseudo-time. hESC-α cells, however, are more representative across the bulk of the trajectory. These branches merged to form a population that consists mainly of grafted hiPSC-α and hESC-α cells at the terminal pseudo-time, as was observed in our β Cell analysis. Of particular note, well-known (CHGB, GCG, and LOXL4) and newly identified (SCG5 and PARVB) genes from our α cell maturation gene list increase with pseudo-time (Figures 5I and S5C). On the other hand, expression of CHGA and FTL reduces with pseudo-time, following trends observed when incorporating primary islet α cells into the analysis (Figures S5C–S5G; Table S4). Altogether, our results support that transplantation of SC-β and SC-α cells induce drastic transcriptome changes to more closely resemble their respective islet endocrine cell type.
Transcriptional Changes of EC Cells within Transplanted Grafts
We identify SC-EC cells, an off-target of SC-islet and SC-β Cell differentiation protocols, in all stem-cell-derived samples. However, we did not detect EC cells in cadaveric HI samples (Figures 6A and S6A), consistent with prior reports (Veres et al., 2019). We performed a comparison of the SC-EC cells before and after transplantation. Several transcriptional changes occur after transplantation (Figure 6B), and interestingly, transcriptome similarity between the grafted SC-EC cells has the highest Pearson correlation compared to other interrogated cell types (Figure 6C; Table S4). Following transplantation, EC cell genes, including MME, DDC, ITPR3, TAC1, CHGA, SLC18A1, and CHGB, are up-regulated, and TPH1 protein is expressed (Figures 6D and 6E; Table S4) (Grün et al., 2015; Veres et al., 2019). We also identify new EC cell genes that increase after transplantation, such as DUSP1 and ZFP36. Conversely, islet endocrine genes (INS, GCG and SST) are reduced, suggesting further commitment to the EC cell fate.
Figure 6. Analysis of Pseudo-Time and Single-Cell Transcriptomes of Stage 6 hPSC-ECs and Grafted hPSC-EC Cells Following Trans plantation.
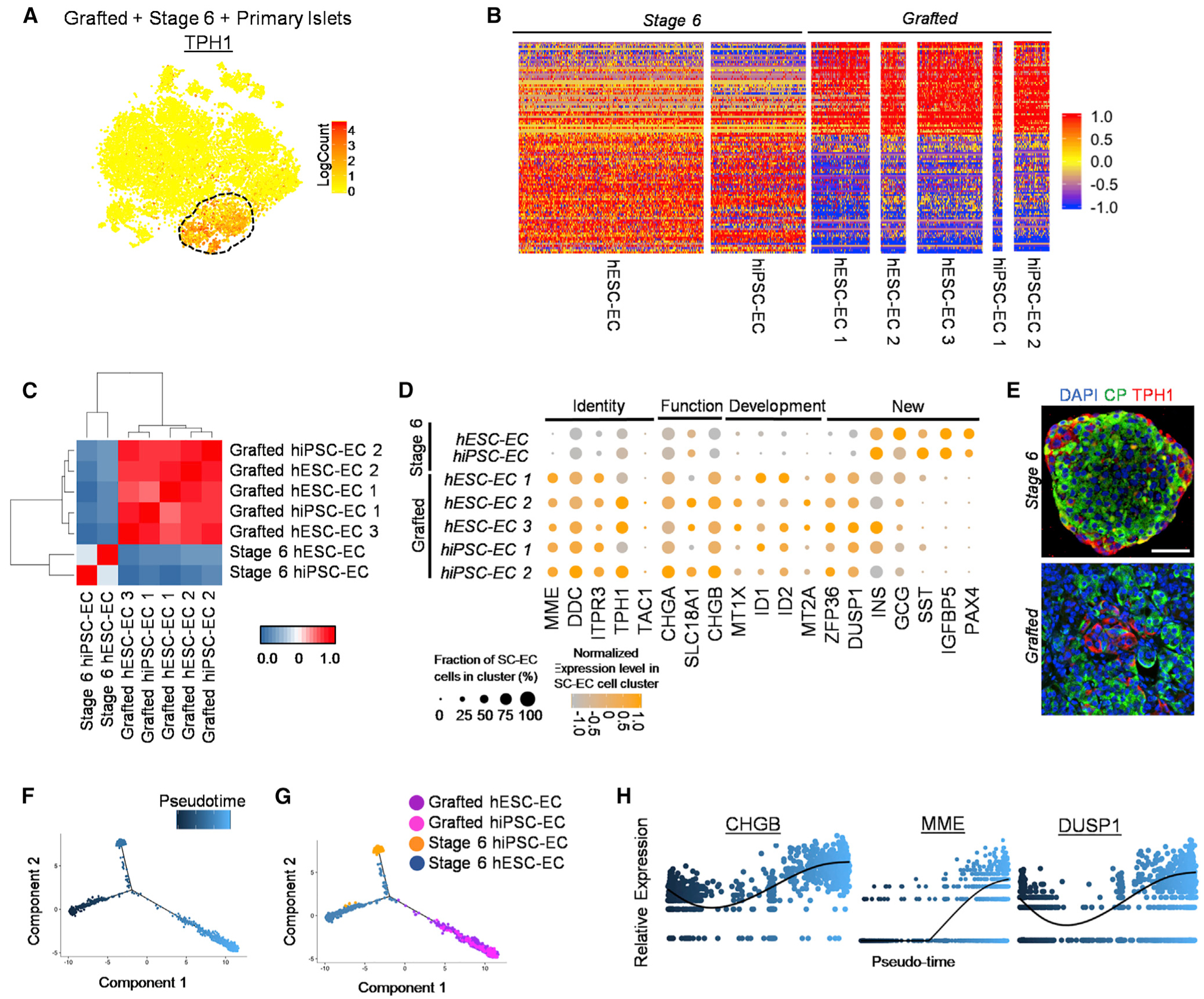
(A) Heatmap of TPH1 gene expression to distinguish SC-EC cell population within tSNE plot from Figure 4A.
(B) Gene expression values for two stage 6 SC-EC cells and five grafted SC-EC cells for the 100 most differentially expressed genes between stage 6 and grafted SC-EC cells listed in Table S6. Scale: normalized expression.
(C) Pearson correlation matrix and hierarchical clustering to identify most similar populations among all EC cell samples using top 430 variable genes. Scale: correlation coefficient.
(D) Expression values for key EC cell identity, function, development, and newly identified genes between stage 6 and grafted SC-EC cells.
(E) Immunostaining of stage 6 and 6-month kidney graft containing SC-islets for EC cell marker TPH1. Scale bar, 50 μm.
(F and G) Distribution (F) of SC-EC cells along pseudo-time trajectory (G) from seven SC-EC samples.
(H) Relative expression of select EC cell genes along SC-EC cell pseudo-time. Scale: relative expression. Each dot represents a different cell.
TPH1, tryptophan hydroxylase 1. See also Figure S6 and Table S4.
Pseudo-time ordering analysis reveals similar reconstruction patterns observed with SC-α cells and SC-β Cells (Figures 6F and 6G). The grafted SC-EC cell populations continue to overlap at the terminal point in pseudo-time, suggesting a uniform commitment to EC cell fate. Genes previously found upregulated in SC-EC cells following transplantation from differential gene expression analysis are also increased with pseudo-time (Figures 6H, S6B, and S6C; Table S4). ZFP36 and MME are initially lowly expressed in a few cells. However, along the pseudo-time, they are upregulated in a greater number of cells. Elevated genes in the grafted SC-EC cells (Table S4) may serve as maturation markers. Yet, we are unable to validate this list, as HIs do not have EC cell populations, and there are limited EC cell primary data available. Overall, we establish SC-EC cells are present in our differentiated SC-islets, and following transplantation, they continue to exist and change gene expression in vivo.
DISCUSSION
The ability to robustly generate pancreatic islets from hPSCs marks an important accomplishment in developmental science and bioregenerative medicine. Nonetheless, SC-β Cells generated from current protocols have transcriptome and functional attributes that resemble juvenile, immature β Cells rather than adult mature β Cells (Hogrebe et al., 2020; Pagliuca et al., 2014; Velazco-Cruz et al., 2019). Herein, we demonstrate that SC-β Cells, derived from both ES and iPS cell lines, can develop into mature β Cells after transplantation. Previous publications display enhanced INS secretion with prolonged transplantation times, which we replicate in our study. Herein, we use scRNA-seq to characterize and explore the transcriptome changes within these functionally improved cells, which has not previously been explored. We performed scRNA-seq on in-vitro-generated stage 6 and grafted SC-islets following 6 months transplantation in diabetic mice. In our SC-islets, we identify key pancreatic and islet cell types, including SC-β, SC-α, SC-δ, and exocrine cells. After transplantation, the grafted SC-β cell population acquired expression of adult HI β Cell genes, expressing known maturation markers, including MAFA and G6PC2.
The expression of genes important to β Cell phenotype changed with transplantation of SC-β Cells. MAFA is widely considered a marker of β Cell maturation, but function in SC-β Cells in vitro has been achieved with little to no MAFA (Hogrebe et al., 2020; Liu and Hebrok, 2017; Nair et al., 2019; Velazco-Cruz et al., 2019). Thus, the precise role of MAFA in β Cell maturation is unclear. As we show SC-β Cells can further mature to express MAFA in vivo, our platform could be used in the future to interrogate transcriptional changes in this and other genes. We also observe reduction of endocrine genes overexpressed in SC-β Cells in vitro (CHGA, MAFB, GCK, and GLUT1) to be more consistent with primary HIs and reduction of GCG and SST gene expression. An interesting observation was that GCG and SST gene expression was actually lower in grafted SC-β Cells compared to primary HIs, likely due in part to isolation of these primary cells (Teo et al., 2018). While prior reports have demonstrated GCG and SST transcript in β Cells from primary HIs (Segerstolpe et al., 2016; Teo et al., 2018; Tyler et al., 2019), their molecular role in primary β Cells and SC-β Cells is unclear. Furthermore, in our study, we have also provided a new list of genes corresponding with SC-β Cell maturation in vivo. We validated protein expression of some new and pre-defined β Cell maturation genes. A limitation of this study and opportunities for further investigation are further validation of other β Cell and newly identified α and EC cell markers. Another limitation of this study is that we did not trace individual cells across different comparisons through the report.
While the focus of this study is on β Cells, α and EC cells are also explored, as they constitute major endocrine cell populations with current differentiation methodologies. Stage 6 SC-α cells mature after transplantation, indicated by increased α cell maturation marker expression. However, the SC-α cells do not resemble human primary α cells as closely as SC-β Cells do to their primary counterparts. This may be a result of our six-stage differentiation protocol targeting the generation of SC-β, rather than SC-α, cells. Nonetheless, we have generated a list of α cell maturation markers that provides insight on enhancing α cell function and targeting further maturation in SC-islet differentiations. EC cells are an intestinal, serotonin-secreting endocrine off-target cell type generated during endocrine differentiation (Veres et al., 2019). They are present in both the stage 6 and grafted SC-islet populations, but their effects on SC-islet function and maturation is unknown.
GSEA and pseudo-time ordering analysis revealed multiple transcriptional genes and gene sets enriched in grafted SC-islet subpopulations. A shared observation in grafted SC-β, SC-α, and SC-EC cells is the increased expression of metallothionein and FOS/JUN genes. Enrichment of these pathways indicates further development and terminal differentiation of these cell types. Interestingly, several identified SC-β Cell maturation genes are highly expressed prior to MAFA expression in pseudo-time, possibly indicating they regulate MAFA expression. Potentially, prolonged exposure to factors associated with these signaling components may lead to SC-islet, namely, SC-β and SC-α cell, maturity in vitro. Targeting these pathways could result in improved in vitro differentiation protocols.
The approach in this study involved the transplantation SC-β Cells into mice with pre-existing STZ-induced diabetes. However, the conditions of optimal maturation, such as transplantation time, diabetic model, and STZ induction before or after transplant, are unknown and would be an interesting study. Cells in an in vivo setting, particularly when not in their natural organ, may undergo de-differentiation or trans-differentiation to become other cell types (Teo et al., 2018; Ye et al., 2015). Because we did not observe progenitor populations in our sequencing data, we believe that the majority of our transplanted populations are retained to their identity. However, it may be possible that some SC-α cell or exocrine cell populations may trans-differentiate into INS-producing cells as demand for blood INS and GCG changes overtime (Ye et al., 2015), and this study did not utilize lineage tracing. An additional limitation of this study was the yields of SC-β Cells we recovered from individual mice, ranging from 111 to 560 SC-β Cells. This prompted us to pool the results from five mice for most of the analysis in this study, but improvements, such as better purification, additional mice, and more differentiations, would better enable robust studies. Furthermore, incorporating the primary HI datasets caused small variations in identified cell types, although overall gene expression differences were not meaningfully affected. An additional limitation is the technical processing of scRNA-seq, which conceivably may yield free-floating mRNA transcripts yielding false positives (Sachs et al., 2020). However, our analysis here of the cell-defining and hormone transcripts comparisons among cell types was significantly different between endocrine and non-endocrine cell types, as we have shown previously (Maxwell et al., 2020), making this possibility unlikely.
In conclusion, we provide a resource thoroughly detailing the maturation of SC-β Cells through in vivo transplantation. SC-β Cells derived from both hiPSC and hESC lines are capable of developing into cells transcriptionally similar to primary human β Cells. This dataset provides valuable information on potential genes and target pathways that may contribute to understanding human β Cell maturation following transplantation. More importantly, it can lead to further developments in SC-β Cell generation for cell replacement therapies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeffrey R. Millman (jmillman@wustl.edu).
Materials Availability
HUES8 cell line is available through Harvard Stem Cell Institute (HSCI). The WS4corr cell line is available upon request to Lead Contact with required MTA.
Data and Code Availability
The single cell RNA sequencing data from this study is made available at the Gene Expression Omnibus (GEO). The accession number for the raw and processed Stage 6 hESC-islet and all Grafted SC-islet data is GEO: GSE151117. The accession number for the raw and processed Stage 6 hiPSC-islet data is GEO: GSE139535. The accession number for the raw and processed human islet data is GEO: GSE114297.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal models
Male, 7 wk old, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratories; stock no. 005557) were used in this study. Mice were fed on chow diet and on a 12 hour light/dark cycle. Animals were randomly assigned to experimental groups. All animal studies performed in this research were in accordance with Washington University International Care and Use Committee (IACUC) regulations.
Human cell lines
Two human PSC lines were used in this study including an hESC line (HUES8) and an hiPSC (WS4corr) line. Both cell lines are authenticated and have normal karyotype. The sex of HUES8 and WS4corr are male and female, respectively. The cells were cultured in mTeSR1 (StemCell Technologies; 05850) in a 5% CO2 incubator at 37°C. Cells were single cell dispersed with TrypLE (Life Technologies; 12-604-039) and passaged every 3–4 days. A Vi-Cell XR (Beckman Coulter) counted viable single cells and we seeded 1.1×105 cells/cm2 cells onto tissue culture-treated flasks, coated with DMEM-diluted (GIBCO, 11965–084) Matrigel-coated (Fisher, 356230) for propagation in mTeSR1 with 10μM Y27532 (Abcam; ab120129). All work for this study was performed in accordance with the Embryonic Stem Cell Research Oversight (ESCRO) committee through Washington University in St. Louis.
Primary cell culture
Cadaveric human islets were purchased from Prodo Labs and cultured in islet media (CMRL Supplemented (Mediatech; 99–603-CV) with 10% Fetal Bovine Serum (GE Healthcare; SH3007003)). For RNA, donor islets were from a 54 year-old Hispanic female. Details for donor islets for sequencing comparison are as follows (1) Caucasian, 32 years, female, (2) Hispanic, 23 years, male, (3) Caucasian, 49 years, male.
Stem cell-derived β Cell differentiation
The differentiation is based on our previous publication (Hogrebe etal., 2020).Undifferentiated hPSCs were single cell dispersed, counted, and seeded onto tissue culture plastic treated flasks with mTeSR1 and 10μM Y27532 at 5.2×105 cells/cm2. After 24 hours, differentiation began using medias and added factors. The differentiation protocol including base media formulation and differentiation factors is available in Table S1. 7 days into stage 6, cells were dispersed with TrypLE and seeded ~5×106 cells into an individual well of a 6-well plate with 4 mL of ESFM media on an OrbiShaker (Benchmark) at 100 RPM. Cells were used for transplantation between day 13–16 in stage 6.
METHOD DETAILS
Mouse Transplants
All animal studies performed in this research were in accordance with Washington University International Care and Use Committee (IACUC) regulations. Prior to transplantation of cells, male, 7 wk old, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratories; stock no. 005557) were treated with 45 mg/kg streptozotocin (STZ; R&D; 1621500) in 0.9% sterile saline (Moltox; 51–405022.052) for 5 consecutive days. Mice were diabetic (blood glucose > 500 mg/dL) 8 days after the final STZ injection. Surgery and assessments were performed by unblinded individuals and mice were randomly assigned to experimental groups. For SC-islet transplantation, mice were anesthetized with isoflurane and injected with ~5×106 SC-islet cells under the kidney capsule, from either hiPSC (n = 3) or hESC (n = 2) derived SC-β Cells. Mice were monitored for 6 months after transplantation before removal of the transplanted kidney for SC-islet retrieval. For glucose tolerance tests (GTT) and in vivo glucose stimulated c-peptide secretion, mice were fasted for 4 hr and injected with 2 g glucose/kg body weight. For GTT, blood glucose was measured every 30 min from 0 to 120 min with a Contour Blood Glucose Monitoring System (Bayer; 9545C). For in vivo glucose stimulated c-peptide secretion, serum was collected before and 1 hr after glucose stimulation. Hormones were quantified by collecting serum and measuring C-peptide and IAPP with the Human Ultrasensitive C-Peptide ELISA kit (Mercodia; 10-1141-01) and Human Amylin ELISA kit (Miltenyi; EZHA-52K), respectively.
Immunostaining
Stage 6 clusters and excised transplant kidneys were fixed overnight in 4% paraformaldehyde (PFA) (Electron Microscopy Science; 15714) at 4°C. Fixed kidneys and Stage 6 clusters, suspended in Histogel (Thermo Scientific; hg-4000–012), were further processed by the Division of Comparative Medicine (DCM) Research Animal Diagnostic Laboratory Core at Washington University. Histoclear (Electron Microscopy Sicences; 64111–04) removed paraffin from slides containing both Stage 6 and kidney sections. Sections were then rehydrated and antigens were retrieved in a pressure cooker (Proteogenix; 2100 Retriever) with 0.05 M EDTA (Ambion; AM9261). Slides were blocked and stained with primary antibodies diluted in immunostaining solution (PBS (Fisher; MT21040CV) with 0.1% Triton X (Acros Organics; 327371000) and 5% donkey serum (Jackson Immunoresearch; 017000–121)) overnight at 4°C, stained with diluted secondary antibodies for 2 hr at room temperature, and stained for DAPI (SouthernBiotech; 0100–20). The immunostained cells and grafts were imaged with a Nikon A1Rsi confocal microscope. Antibody details and dilutions are available in Table S7.
Tissue and Cell Preparation for Sequencing
Stage 6 SC-islet preparation.
At stage 6 day 14, SC-islets were washed with PBS and single cell dispersed with TrypLE at 37°C for ~20 minutes. The dispersed cells were centrifuged, counted, and resuspended in DMEM for library prep.
Grafted SC-islet cell retrieval.
Transplanted mice were euthanized through cervical dislocation and the kidney transplanted with SC-islets were removed. Excess scar tissue and fat was removed from around the kidney. Images were taken of the kidneys to visualize the transplanted cells. The kidney was sliced into several small pieces with a razor blade and placed into a solution of 2mg/mL collagenase D (Sigma; 11088858001) in RPMI (GIBCO; 11875–085). The tissue was incubated at 37°C for 45 min. PBS was added to the solution and additional breakup was enforced with a pipette before filtering the cells through a 70mm cell strainer (Corning; 431751). Collected flow through was centrifuged, supernatant was removed, and resuspended with MACS buffer (0.05% BAS in PBS). A Miltenyi mouse cell isolation kit (kit, 130-104-694; LS column, 130-042-401) was used to remove excess mouse cells from the cell solution. Cells in flow through were collected, centrifuged, counted, and resuspended in DMEM for library prep.
Single-cell RNA sequencing preparation
Single cell dispersed cells were delivered to Washington University in St. Louis Genome Technology Access Center (GTAC) for library preparation and sequencing. For library preparations, the Chromium Single Cell 3′ Library and Gel Bead Kit v2 was used following the provided protocol. In summary, a microfluidic platform is used to profile digital gene expression for individual cells by creating an emulsion for each cell with a unique barcoded cDNA, breaking the emulsion, and amplifying the cDNA. The library was sequenced with a NovaSeq 6000 System (Illumina) at the recommended 26×98bp. Gene expression levels found in violin plots and heatmaps are displayed in Log(normalized read count) and normalized average gene expression, respectively.
Single-cell RNA sequencing data analysis: Raw Data Processing
Seurat v2.3.4 was used for single cell analyses and comparison between all groups (Butler et al., 2018). Monocle v2.12.0 was used for pseudotime analysis between Stage 6 and grafted SC-β Cells (Qiu et al., 2017; Trapnell et al., 2014) and WS4corr Stage 6 data is from Maxwell et al. (https://doi.org/10.1126/scitranslmed.aax9106) SC-islet samples were aligned to the human genome and filtered out low gene counts (< 500 for grafted and < 3000 for Stage 6) and high mitochondrial percentages (> 20%) for further data analysis, indicating doublets, debris, and dying cells. This filtering removed several mouse cells present in the grafted SC-islet samples. The minimum number of genes is lower for the grafted compared to Stage 6 SC-islet samples to filter out the mouse cells, because our datasets are aligned to the human genome, therefore the mouse cells have low gene counts. Therefore, we clustered the grafted SC-islet samples and identified the clusters with low gene counts. We identified TTC36, a kidney gene which is present in both mouse and human genomes, to also be highly expressed in the low gene number clusters. We removed all cells with TTC36 log-normalized expression > 3, to remove all remaining mouse cells from further analysis. The genes defining the cell cluster that was identified as mouse cells can be found in Table S1. Each sample dataset was normalized with global-scaling normalization. FindVariableGenes function removed outlier genes using a scaled z-score dispersion.
Clustering
For individual tSNE plots, cells with similar gene expression patterns were plotted near each other based on PCA using the FindClusters function. We used a resolution of 0.2 and 5 dimensions to generate tSNE plots for each individual sample (Stage 6 iPS SC-islets, Stage 6 ES SC-islets, 3 Grafted ES SC-islets, 2 Grafted iPS SC-islets). The top 50 genes that separated each cluster within the individual samples were identified with FindAllMarkers and these gene sets were used to designate the different SC-islet cell types including beta, alpha, and delta cells. These gene lists can be found in Table S2. The FeaturePlot function was used to visualize expression of specific genes across the different clusters, represented in tSNE plots. Violin plots were generated to compare the expression of specific genes for the SC-β Cell populations within the SC-islets for each sample.
DE between Stage 6 and Grafted SC-islets
We combine datasets for the same hPSC source using canonical correlation analysis (CCA) with RunMultiCCA (num.ccs = 10). The CCA subspaces are aligned with AlignSubspace. Unsurpervised tSNE plots were generated for the combined dataset with RunTSNE using 17 dimensions. Clusters were designated specific cell types based on differentially expressed genes in each cluster compared to all other clusters, identified with FindMarkers. These gene lists can be found in Table S3. For further analyses, we compare Stage 6 and Grafted SC-β Cells by only comparing the most enriched INS β Cell cluster. Additionally, we do this for SC-α and SC-EC cells. We are able to identify all differentially expressed genes for SC-β, SC-α, and SC-EC cells between conditions using FindMarkers with a log fold change threshold of 0.25; these genes can be found in Table S4. We generate a heatmap for the top differentially expressed genes.
Pathway Enrichment
Genes defining branch points in pseudotime analysis and between sample groups were analyzed for KEGG and GO pathway enrichment and differential expression of HALLMARK gene sets by GSEA (4.0.2). Top enriched genes for the identified pathways and gene sets, with NES scores above 1.0 or below −1.0 and FDR q-value < 0.25 for either or both of the derived cell line conditions, were considered in our analysis.
Pseudotime Trajectory with Monocle
Filtered scRNA-seq data containing only β, α, or EC cells from Seurat was transferred for subsequent pseudotime trajectory analysis using Monocle v 2.12.0 (Trapnell et al., 2014). The data was reanalyzed by clustering the datasets using clusterCells to identify Stage 6 and Grafted populations. Differentially expressed genes between the two identities were calculated using differentialGeneTest. We used the top 800, 1000, 1500 genes by lowest q-value to reduce dimensions using DDRTree method for β, α, or EC cells, respectively. Cells were ordered using the orderCells function and pseutdotime was defined by selecting states representative of Stage 6 cells being the initial state. The pseudotime heatmap and gene plots were generated using plot_pseudotime_heatmap and plot_genes_in_pseudotime, respectively. The top differentially expressed genes between branches of pseudotime were defined for β, α, and EC cell populations. The list of genes can be found in Table S4.
Comparison to human islets
Next, we compare our Stage 6 and Grafted SC-islets, and specifically SC-β and SC-α cells, to primary human islets from cadaveric donors. The human islet datasets come from Xin et al., 2018 (https://doi.org/10.2337/db18-0365) and can be found at Gene Expression Omnibus database with accession number GEO: GSE114297. We selected three human islet patients for our comparison analysis. As previously described, 10 datasets (Stage 6 SC-islets (iPSC and ESC), grafted SC-islets (iPSC (n = 2) and ESC (n = 3)), human islet donors (n = 3)) were normalized and subspaces were aligned using CCA to generate unsupervised tSNE plots with resolution of 0.1 and 30 dimensions. Clusters were designated specific cell types based on differentially expressed genes in each cluster compared to all other clusters, identified with FindMarkers. These gene lists can be found in Table S5. The combined tSNE plot is used to specifically compare β, α, and EC cells among samples, by selecting cells containing the INS, GCG, or TPH1 gene (low.thresholds = c(0.1)). In addition we analyzed MAFA+ β Cells from within the β Cell enriched cluster (MAFA log-normalized expression > 0.2). Differential expression of the β, MAFA+ β, and α cells from each sample (Stage 6, Grafted, Human Islet) were identified. A heatmap and/or correlation matrix was generated reflecting each sample’s expression of the differentially expressed genes or transcriptome similarity, respectively. A list of the genes differentially expressed for a specific cell type, between human islet and Stage 6 or human islet and Grafted stem cell-derived cells can be found in Table S6. For correlation matrix, the average expression levels of the most differentially expressed genes were used and generated by Pearson method using cor. Values for the correlation matrix can be found in Table S6. Pseudotime trajectory construction with monocle was also used for β and α cells in combination with human islet data to generate branching trajectories and gene plots in pseudo-time.
Real-Time PCR
RNA from human islets and Stage 6 hiPSC-β and hESC-β Cells was extracted using an RNeasy Mini Kit (QIAGEN; 74016) and DNase treatment (QIAGEN; 79254). cDNA was synthesized using the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems; 4368814) and a thermocycler (Applied Biosystems; A37028). PowerUp SYBR Green Master Mix (Applied Biosystems; A25741) generated real-time PCR reactions in a StepOne Plus (Applied Biosystems). Data was analyzed with the ΔΔCt methodology. TBP and GUSB were used for normalization and primer sequences are available in Table S1.
Software
Data was analyzed using Rstudio version 3.6.1 with Seurat version 2.3.4 (Butler et al., 2018) and Monocle version 2.12.0 (Qiu et al., 2017; Trapnell et al., 2014).
IAPP Secretion in vitro
To quantify IAPP protein secretion in vitro, the cells were cultured on transwells in a solution of 128 mM NaCl, 5 mM KCl, 2.7 mM CaCl2, 1.2 mM MgSO4, 1 mM Na2HPO4, 1.2 mM KH2PO4, 5 mM NaHCO3, 20 mM glucose, 10 mM HEPES (GIBCO; 15630–080) and 0.1% bovine serum albumin (Proliant; 68700) for 1 hr. The solution was collected and IAPP quantified with the Human Amylin ELISA kit.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism 8.2.1 was used to calculate two-sided unpaired and paired t tests. p < 0.05 was considered statistically significant. Data is displayed as mean ± s.e.m. Sample size, n, represents total biological replicates. Tests used for individual panels, sample size (n), and statistical parameters are detailed in figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti C-peptide | DSHB | Cat#GN-ID4; RRID:AB_2255626 |
| Mouse anti NKX6–1 | DSHB | Cat#F55A12; RRID:AB_532379 |
| Rabbit anti Glucagon | Cell Marque | Cat#259A-18; RRID:AB_1158356 |
| Goat anti PDX1 | R&D Systems | Cat#AF2419; RRID:AB_355257 |
| Rabbit anti CHGA | ABCAM | Cat#ab15160; RRID:AB_301704 |
| Rabbit anti MAFA | LifeSpan Bioscience | Cat#LP9872; RRID:AB_2665528 |
| Rabbit anti FAM159B | Invitrogen | Cat#PA5–52855; RRID:AB_2641287 |
| Rabbit anti NAA20 | Invitrogen | Cat#PA5–66574; RRID:AB_2663038 |
| Rabbit anti IAPP | Sigma Aldrich | Cat#PA5–84142; RRID:AB_2791294 |
| Sheep anti ARX | Invitrogen | Cat#PA5–47896; RRID:AB_2605931 |
| Sheep anti TPH1 | EMD Millipore | Cat#AB1541; RRID:AB_90754 |
| Sheep anti Trypsin | R&D | Cat#AF3586; RRID:AB_2846900 |
| Mouse anti KRT19 | Dako | Cat#M0888; RRID:AB_2234418 |
| anti-rat-alexa fluor 488 | Invitrogen | Cat#a21208; RRID:AB_141709 |
| anti-rabbit-alexa fluor 488 | Invitrogen | Cat#a21206; RRID:AB_2535792 |
| anti-rat-alexa fluor 488 | Invitrogen | Cat#a21208; RRID:AB_141709 |
| anti-mouse-alexa fluor 594 | Invitrogen | Cat#a21203; RRID:AB_141633 |
| anti-rabbit-alexa fluor 594 | Invitrogen | Cat#a21207; RRID:AB_141637 |
| anti-goat-alexa fluor 594 | Invitrogen | Cat#a11058; RRID:AB_2534105 |
| anit-sheep-alexa fluor 594 | Invitrogen | Cat#a11016; RRID:AB_10562537 |
| Biological Samples | ||
| Healthy Human Islets | Prodo Labs | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| mTeSR1 | StemCell Technologies | Cat#05850 |
| Y27632 | Abcam | Cat#ab120129 |
| Matrigel | Corning | Cat#356230 |
| TrypLE | Life Technologies | Cat#12-604-039 |
| DNase | QIAGEN | Cat#79254 |
| HEPES | GIBCO | Cat#15630–080 |
| Tris | MilliporeSigma | Cat#T6066 |
| EDTA | Ambion | Cat#327371000 |
| Paraformaldehyde | Electron Microscopy Science | Cat#15714 |
| Donkey Serum | Jackson Immunoresearch | Cat#017-000-121 |
| Triton X-100 | Acros Organics | Cat#327371000 |
| DMEM | MilliporeSigma | Cat#D6429 |
| Fetal Bovine Serum | MilliporeSigma | Cat#F4135 |
| Histo-clear II | Electron Microscopy Sciences | Cat#64111–04 |
| Histogel | Thermo Scientific | Cat#Hg-4000–012 |
| 0.5 M EDTA | Ambion | Cat#AM9261 |
| 200 proof ethanol | Decon | Cat#2716 |
| STZ | R&D | Cat#1621500 |
| Critical Commercial Assays | ||
| Ultrasensitive C-Peptide Elisa | Mercodia | Cat#10-1141-01 |
| Human Amylin Elisa | Miltenyi | Cat#EZHA-52K |
| RNeasy Mini Kit | QIAGEN | Cat#74016 |
| High Capacity cDNA Reverse Transcriptase Kit | Applied Biosystems | Cat#4368814 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | Cat#A25741 |
| Chromium Single Cell 3’ Library &Gel Bead Kit | 10X Genomics | Cat#PN-120267 |
| Deposited Data | ||
| raw and processed single cell RNA seq data - Stage 6 hESC-islet, 3 Grafted hESC-islet, 2 Grafted hiPSC-islet | This Paper | GEO: GSE151117 |
| raw and processed single cell RNA seq data - Stage 6 hiPSC-islet | Maxwell et al., 2020 | GEO: GSE139535 |
| raw and processed single cell RNA seq data - Human Islets | Xin etal., 2018 | GEO: GSE114297 |
| Experimental Models: Cell Lines | ||
| Human: HUES8 hESC | HSCI | hES Cell Line: HUES-8 |
| Human: WS4corr | Maxwell et al., 2020 | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: NOD.Cg-Prkdcscid Il2rgtm1Wj/SzJ (NSG) | The Jackson Laboratories | JAX: 005557; RRID:IMSR_JAX:005557 |
| Oligonucleotides | ||
| Primers for this study, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| Fiji ImageJ | ImageJ public freeware | https://imagej.net/Fiji/Downloads; RRID:SCR_003070 |
| GraphPad Prism 8.2.1 | GraphPad |
https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| FLOWJO | FLOWJO | https://www.flowjo.com/solutions/flowjo/downloads; RRID:SCR_008520 |
| GSEA 4.0.2 | GSEA | https://www.gsea-msigdb.org/gsea/index.jsp; RRID:SCR_003199 |
| RStudio | RStudio | https://rstudio.com/products/rstudio/download/; RRID:SCR_000432 |
| R | R | https://cran.r-project.org/src/base/R-4/ |
| Seurat v2.3.4 | Butler et al., 2018 | https://satijalab.org/seurat/install.html; RRID:SCR_007322 |
| Monocle v2.12.0 | Qiu et al., 2017 | http://cole-trapnell-lab.github.io/monocle-release/docs/#installing-monocle |
| Other | ||
| Vi-Cell XR | Beckman Coulter | Cat#Vi-Cell XR |
| Tanswells | Corning | Cat#431752 |
| #1.5 glass bottom 96 well plate | Cellvis | Cat#963–1.5H-N |
| 8-channel peristaltic pump | ISMATEC | Cat#ISM931C |
| Inlet/outlet two-stop tubing | ISMATEC | Cat#070602–04i-ND |
| Dispensing nozzle | BioRep | Cat#Peri-Nozzle |
| Connection tubbing | BioRep | Cat#Peri-TUB-040 |
| Bio-Gel P-4 | Bio-Rad | Cat#150–4124 |
| Cell Chamber | BioRep | Cat#Peri-Chamber |
| Pressure cooker | Electron Microscopy Sciences | Model#Retriever 2100 |
| DAPI Fluoromount-G | SouthernBiotech | Cat#0100–20 |
| Phosphate-Buffered Saline (PBS) | Corning | Cat#21–040-CV |
| micro coverslip | VWR | Cat#48393–081 |
Highlights
SC-islets show improved insulin and IAPP release with time in transplanted diabetic mice
SC-islet cells undergo drastic transcriptome changes after 6 months in vivo
Grafted SC-β Cells mature and express MAFA, CHGB, G6PC2, and FAM159B in vivo
Differentially expressed genes upon transplantation resemble native human islet cells
ACKNOWLEDGMENTS
This work was supported by the NIH (5R01DK114233), a JDRF Career Development Award (5-CDA-2017-391-A-N), Washington University-Centene Personalized Medicine Initiative, and startup funds from Washington University School of Medicine Department of Medicine. Further support was provided by a grant jointly funded by Washington University (Departments of Surgery, Medicine, and Pediatrics), Mid-America Transplant Services, and The Foundation for Barnes-Jewish Hospital. P.A. was supported by the David and Deborah Winston Fellowship in Diabetes Research. K.G.M. was supported by the NIH (T32DK108742). Sequencing work was performed by the Washington University Genome Technology Access Center in the Department of Genetics (NIH P30CA91842 and UL1TR000448). We thank Dr. Kevin D’Amour (ViaCyte, Inc.) for the MAFA antibody.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108067.
DECLARATION OF INTERESTS
L.V.-C. and J.R.M. are inventors on patent applications related to SC-β Cell technology described in this manuscript.
REFERENCES
- Aguayo-Mazzucato C, and Bonner-Weir S (2010). Stem cell therapy for type 1 diabetes mellitus. Nat. Rev. Endocrinol 6, 139–148. [DOI] [PubMed] [Google Scholar]
- Atkinson MA, Eisenbarth GS, and Michels AW (2014). Type 1 diabetes. Lancet 383, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsornworawat P, and Millman JR (2020). Single-cell RNA sequencing for engineering and studying human islets. Curr. Opin. Biomed. Eng 16, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsornworawat P, Velazco-Cruz L, Song J, and Millman JR (2019). A hydrogel platform for in vitro three dimensional assembly of human stem cell-derived islet cells and endothelial cells. Acta Biomater. 97, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa D, Saarimäki-Vire J, Borshagovski D, Survila M, Lindholm P, Galli E, Eurola S, Ustinov J, Grym H, Huopio H, et al. (2018). Insulin mutations impair beta-cell development in a patient-derived iPSC model of neonatal diabetes. eLife 7, e38519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, Ryu JH, Wagner BK, Shen-Orr SS, Klein AM, et al. (2016). A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst. 3, 346–360.e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Hrvatin S, Schuetz C, Bonal C, Rezania A, and Melton DA (2012). Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat. Biotechnol 30, 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruin JE, Asadi A, Fox JK, Erener S, Rezania A, and Kieffer TJ (2015). Accelerated Maturation of Human Stem Cell-Derived Pancreatic Progenitor Cells into Insulin-Secreting Cells in Immunodeficient Rats Relative to Mice. Stem Cell Reports 5, 1081–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes LE, Wong DM, Subramaniam M, Meyer NP, Gilchrist CL, Knox SM, Tward AD, Ye CJ, and Sneddon JB (2018). Lineage dynamics of murine pancreatic development at single-cell resolution. Nat. Commun 9, 3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camunas-Soler J, Dai XQ, Hang Y, Bautista A, Lyon J, Suzuki K, Kim SK, Quake SR, and MacDonald PE (2020). Patch-Seq Links Single-Cell Transcriptomes to Human Islet Dysfunction in Diabetes. Cell Metab. 31, 1017–1031.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enge M, Arda HE, Mignardi M, Beausang J, Bottino R, Kim SK, and Quake SR (2017). Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell 171, 321–330.e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazizadeh Z, Kao DI, Amin S, Cook B, Rao S, Zhou T, Zhang T, Xiang Z, Kenyon R, Kaymakcalan O, et al. (2017). ROCKII inhibition promotes the maturation of human pancreatic beta-like cells. Nat. Commun 8, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, and van Oudenaarden A (2015). Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255. [DOI] [PubMed] [Google Scholar]
- Hogrebe NJ, Augsornworawat P, Maxwell KG, Velazco-Cruz L, and Millman JR (2020). Targeting the cytoskeleton to direct pancreatic differentiation of human pluripotent stem cells. Nat. Biotechnol 38, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, O’Donnell CW, Deng F, Millman JR, Pagliuca FW, DiIorio P, Rezania A, Gifford DK, and Melton DA (2014). Differentiated human stem cells resemble fetal, not adult, β Cells. Proc. Natl. Acad. Sci. USA 111, 3038–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Miller K, Jo J, Kilimnik G, Wojcik P, and Hara M (2009). Islet architecture: A comparative study. Islets 1, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbutt GS, Elliott JF, Ao Z, Smith DK, Warnock GL, and Rajotte RV (1996). Large scale isolation, growth, and function of porcine neonatal islet cells. J. Clin. Invest 97, 2119–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latres E, Finan DA, Greenstein JL, Kowalski A, and Kieffer TJ (2019). Navigating Two Roads to Glucose Normalization in Diabetes: Automated Insulin Delivery Devices and Cell Therapy. Cell Metab. 29, 545–563. [DOI] [PubMed] [Google Scholar]
- Liu JS, and Hebrok M (2017). All mixed up: defining roles for β-cell subtypes in mature islets. Genes Dev. 31, 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KG, Augsornworawat P, Velazco-Cruz L, Kim MH, Asada R, Hogrebe NJ, Morikawa S, Urano F, and Millman JR (2020). Gene-edited human stem cell-derived β Cells from a patient with monogenic diabetes reverse preexisting diabetes in mice. Sci. Transl. Med 12, eaax9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman JR, Xie C, Van Dervort A, Gürtler M, Pagliuca FW, and Melton DA (2016). Generation of stem cell-derived b-cells from patients with type 1 diabetes. Nat. Commun 7, 11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro MJ, Dharmadhikari G, Grun D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJ, et al. (2016). A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 3, 385–394.e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair GG, Liu JS, Russ HA, Tran S, Saxton MS, Chen R, Juang C, Li ML, Nguyen VQ, Giacometti S, et al. (2019). Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β Cells. Nat. Cell Biol 21, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi T, and Yoshida S (2012). The therapeutic potential of GPR119 agonists for type 2 diabetes. Expert Opin. Investig. Drugs 21, 321–328. [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, and Melton DA (2014). Generation of functional human pancreatic β Cells in vitro. Cell 159, 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper AR, Pawlick R, Bruni A, Wink J, Rafiei Y, O’Gorman D, Yan-Do R, Gala-Lopez B, Kin T, MacDonald PE, and Shapiro AMJ (2017). Transplantation of Human Pancreatic Endoderm Cells Reverses Diabetes Post Transplantation in a Prevascularized Subcutaneous Site. Stem Cell Reports 8, 1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C (2017). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramond C, Beydag-Tasöz BS, Azad A, van de Bunt M, Petersen MBK, Beer NL, Glaser N, Berthault C, Gloyn AL, Hansson M, et al. (2018). Understanding human fetal pancreas development using subpopulation sorting, RNA sequencing and single-cell profiling. Development 145, dev165480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. (2014). Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol 32, 1121–1133. [DOI] [PubMed] [Google Scholar]
- Robert T, De Mesmaeker I, Van Hulle FO, Suenens KG, Stangé GM, Ling Z, Haller C, Bouche N, Keymeulen B, Kraus MRC, and Pipeleers DG (2019). Cell Mass Increase Associated with Formation of Glucose-Controlling b-Cell Mass in Device-Encapsulated Implants of hiPS-Derived Pancreatic Endoderm. Stem Cells Transl. Med 8, 1296–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R, Carnese PP, Hennings TG, Walker EM, Russ HA, Liu JS, Giacometti S, Stein R, and Hebrok M (2020). Loss of the transcription factor MAFB limits b-cell derivation from human PSCs. Nat. Commun 11, 2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs S, Bastidas-Ponce A, Tritschler S, Bakhti M, Böttcher A, Sánchez-Garrido MA, Tarquis-Medina M, Kleinert M, Fischer K, Jall S, et al. (2020). Targeted pharmacological therapy restores β-cell function for diabetes remission. Nat. Metab 2, 192–209. [DOI] [PubMed] [Google Scholar]
- Scavuzzo MA, Hill MC, Chmielowiec J, Yang D, Teaw J, Sheng K, Kong Y, Bettini M, Zong C, Martin JF, and Borowiak M (2018). Endocrine lineage biases arise in temporally distinct endocrine progenitors during pancreatic morphogenesis. Nat. Commun 9, 3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstolpe Å, Palasantza A, Eliasson P, Andersson EM, Andréasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, et al. (2016). Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab. 24, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro AMJ, Lakey JRT, Ryan EA, Korbutt GS, Toth E, Warnock GL, Kneteman NM, and Rajotte RV (2000). Islet Transplantation in Seven Patients with Type 1 Diabetes Mellitus Using a Glucocorticoid-Free Immunosuppressive Regimen. N. Engl. J. Med 343, 230–238. [DOI] [PubMed] [Google Scholar]
- Shapiro AM, Ricordi C, Hering BJ, Auchincloss H, Lindblad R, Robertson RP, Secchi A, Brendel MD, Berney T, Brennan DC, et al. (2006). International trial of the Edmonton protocol for islet transplantation. N. Engl. J. Med 355, 1318–1330. [DOI] [PubMed] [Google Scholar]
- Teo AKK, Lim CS, Cheow LF, Kin T, Shapiro JA, Kang NY, Burkholder W, and Lau HH (2018). Single-cell analyses of human islet cells reveal de-differentiation signatures. Cell Death Discov. 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi M, Claus TH, Liang Y, Li Y, Yang L, Zhu J, Dela Cruz F, Peng X, Chen H, Yung SL, et al. (2002). A potent and highly selective VPAC2 agonist enhances glucose-induced insulin release and glucose disposal: a potential therapy for type 2 diabetes. Diabetes 51, 1453–1460. [DOI] [PubMed] [Google Scholar]
- Tyler SR, Rotti PG, Sun X, Yi Y, Xie W, Winter MC, Flamme-Wiese MJ, Tucker BA, Mullins RF, Norris AW, et al. (2019). PyMINEr Finds Gene and Autocrine-Paracrine Networks from Human Islet scRNA-Seq. Cell Rep. 26, 1951–1964.e1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegas AJ, Veiseh O, Gürtler M, Millman JR, Pagliuca FW, Bader AR, Doloff JC, Li J, Chen M, Olejnik K, et al. (2016). Long-term glycemic control using polymer-encapsulated human stem cell-derived betα cells in immune-competent mice. Nat. Med 22, 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazco-Cruz L, Song J, Maxwell KG, Goedegebuure MM, Augsornworawat P, Hogrebe NJ, and Millman JR (2019). Acquisition of Dynamic Function in Human Stem Cell-Derived β Cells. Stem Cell Reports 12, 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazco-Cruz L, Goedegebuure MM, Maxwell KG, Augsornworawat P, Hogrebe NJ, and Millman JR (2020a). SIX2 regulates human β Cell differentiation from stem cells and functional maturation in vitro. Cell Rep. 31, 107687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazco-Cruz L, Goedegebuure MM, and Millman JR (2020b). Advances Toward Engineering Functionally Mature Human Pluripotent Stem Cell-Derived β Cells. Front. Bioeng. Biotechnol 8, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres A, Faust AL, Bushnell HL, Engquist EN, Kenty JH, Harb G, Poh YC, Sintov E, Gürtler M, Pagliuca FW, et al. (2019). Charting cellular identity during human in vitro β-cell differentiation. Nature 569, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Schug J, Won K-J, Liu C, Naji A, Avrahami D, Golson ML, and Kaestner KH (2016). Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65, 3028–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y, Kim J, Okamoto H, Ni M, Wei Y, Adler C, Murphy AJ, Yancopoulos GD, Lin C, and Gromada J (2016). RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 24, 608–615. [DOI] [PubMed] [Google Scholar]
- Xin Y, Dominguez Gutierrez G, Okamoto H, Kim J, Lee AH, Adler C, Ni M, Yancopoulos GD, Murphy AJ, and Gromada J (2018). Pseudotime Ordering of Single Human β-Cells Reveals States of Insulin Production and Unfolded Protein Response. Diabetes 67, 1783–1794. [DOI] [PubMed] [Google Scholar]
- Ye L, Robertson MA, Hesselson D, Stainier DY, and Anderson RM (2015). Glucagon is essential for alpha cell transdifferentiation and beta cell neogenesis. Development 142, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, He Q, Chen J, Liu Y, Mao Z, Lyu Z, Ni D, Long Y, Ju P, Liu J, et al. (2016). The expression patterns of Tetratricopeptide repeat domain 36 (Ttc36). Gene Expr. Patterns 22, 37–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single cell RNA sequencing data from this study is made available at the Gene Expression Omnibus (GEO). The accession number for the raw and processed Stage 6 hESC-islet and all Grafted SC-islet data is GEO: GSE151117. The accession number for the raw and processed Stage 6 hiPSC-islet data is GEO: GSE139535. The accession number for the raw and processed human islet data is GEO: GSE114297.


