Abstract
Serologic tests are one of the available diagnostic tools in COVID–19. Growing literature highlights their role in the clinical management of the disease. Unfortunately, due to the limited availability of commercial tests and the lack of reliable trials establishing the sensitivity and specificity of the diagnostic method, the clinical application of the test needs to be precisely determined. In this paper, we discuss the utility of anti–SARS–CoV–2 serology testing in a clinical setting and propose diagnostic algorithms that include serological tests in patients with confirmed or suspected COVID–19.
Keywords: Serologic tests, COVID-19, Diagnostic algorithms, Public health
1. Introduction
In December 2019, the first reports about the new virus from Coronaviridae family – Severe Acute Respiratory Syndrome Coronavirus 2 (SARS–CoV–2) were published. The initial outbreak was located in Wuhan, China, and followed by the rapid global spread of the infection [1]. On 11 March 2020, WHO declared the outbreak a global pandemic. From this point forward, scientists and clinicians across the globe began to research accurate, rapid, and cost–effective diagnostic methods, as well as safe and effective treatment modalities. Currently, the only molecular test acceptable and recommended by the WHO in confirming the infection relies on capturing and detecting the SARS–CoV–2 genetic material [2]. However, due to its limited sensitivity and specificity, it is critical to complement the diagnostic procedure with additional tests. Specific anti–SARS–CoV–2 serum antibodies have garnered much attention due to their high diagnostic potential. The data indicate that the virus–specific antibodies may serve as a diagnostic tool to increase the sensitivity of molecular tests as well as assist in controlling the spread of the infection in a population [3].
Unfortunately, due to the limited availability of commercial tests and the lack of reliable trials establishing the sensitivity and specificity of the diagnostic method, the clinical utility of the test needs to be determined. The paper aims to discuss the utility of anti–SARS–CoV–2 serology testing in a clinical setting.
2. A comparison of serological and molecular tests
Currently, the only available test for reliable verification of the SARS–CoV–2 infection is real–time Reverse–Transcription Polymerase Chain Reaction (rRT–PCR) [2]. It is a well–established method that enables the identification of genetic material in samples [4]. A positive rRT–PCR result confirms the diagnosis of COVID–19 in a suspected case. However, due to the limited sensitivity and specificity of this assay, the results often require validation. In the case of COVID–19, a molecular test has the highest specificity up to 7 days after contact with the pathogen [3]. After the first week, the number of truly positive results decreases, and the number of false-negative outcomes increases [3]. Additionally, the collection site may interfere with the results. The virus typically localizes in the lower respiratory tract, but routinely, the sample is collected from the throat, which may lead to a higher ratio of false–negative results [2], [5], [6]. A higher number of virions in the upper respiratory tract during the first two weeks of the infection is associated with more severe disease. Interestingly, in the case of a mild form of the disease, the elevated number of virions is observed only in the first week of the infection [7]. Therefore, the number of false–negative results may be associated with the severity of the COVID-19. False–positive outcomes may result from cross–reactions, as well as sample pollution in the laboratory [8]. Moreover, a molecular test result may be unreliable due to untimely collection of the sample, insufficient sample size, virus mutation, or limited replication phase in the host cells [2], [3]. Further limitations involve technical challenges with the analysis, the requirement for adequately prepared and certified laboratory including laboratory personnel, and fairly long waiting time for the results [2], [5]. The limitations, as well as challenges associated with rRT–PCR warrant further research of new diagnostic approaches in COVID–19. Serologic testing appears to be complementary to rRT–PCR. It can detect antibodies directed to the pathogen as well as specific antigens. The comprehensive analysis of the molecular structure of SARS–CoV–2 allowed the researchers to introduce new types of highly sensitive and specific serologic tests [9], [10], [11]. To date, there are several hundred strip tests that enable quick identification of SARS–CoV–2–specific antibodies. These tests include lateral flow immunoassay (LFIA), chemiluminescence immunoassay (CLIA), and enzyme–linked immunosorbent assay (ELISA) [3]. The sensitivity, however, differs significantly among these assays. For example, the results from lateral flow immunoassay are obtained the fastest, but their sensitivity is fairly low [3]. According to the literature, ELISA is the most common assay used to detect specific SARS–CoV–2 antibodies [6], [12], [13], [14]. The primary limitation of serological tests is linked with sparse data regarding their sensitivity and specificity. Therefore, the tests incur the risk of a high number of false–negative and false–positive results. False–negative outcomes may be associated with developing an immune response during early disease and the undetectable level of specific antibodies (window period). Additionally, chronic immunosuppression may result in an impaired immune response and no detectable titer. On the other hand, false–positive outcomes may be a result of molecular similarity in the structure of beta–coronaviruses, which leads to cross–reaction. At present, the WHO recommends SARS–CoV–2 specific antibody analysis in the acute phase of infection and convalescence period, despite the limitations mentioned above. It is particularly recommended to employ highly sensitive and specific serological tests in case of negative rRT–PCR results [2].
3. Anti-SARS–CoV–2 IgM and anti–SARS-CoV–2 IgG
The presence of SARS–CoV–2–specific antibodies strongly correlates with the molecular structure of the virus. Four essential proteins form its structure: envelope protein E, spike protein S, membrane protein M and nucleocapsid protein N. S protein, which consists of two subunits S1 and S2, plays a critical role in the pathogenesis of COVID–19 [9], [10]. The SARS–CoV–2 contains unique end amino acids sequence at the border of S1 and S2 subunits [11]. This finding may contribute to minimizing the risk of cross–reactions and false–positive results. Both subunits of S protein are responsible for penetrating the virus to the infected cell via an ACE2 receptor located in the respiratory, urinary, and digestive tract [15]. Research indicates that subunit S is crucial to specific antibody production [11]. Nucleocapsid protein N also plays a vital role in stimulating the immune response of the infected organism [15]. The data regarding the changes of antibodies titer (both IgM and IgG) throughout the disease is still sparse. However, quantitative detection of antibodies has potential significance for evaluating the severity and prognosis of COVID–19. The current findings indicate that during SARS–CoV–2 infection, IgM antibodies may be detectable between 3. and 42. days, whereas IgG antibodies between 5. and 47. days from the infection [16]. IgM titer may be detectable 10 to 12 days after the first manifestation of the symptoms [12], [13]. Rarely, anti–SARS–CoV–2 IgM detection becomes possible earlier, after 1–7 days following the infection [13]. Anti–SARS–CoV–2 IgG is measurable subsequently to IgM, after 12–14 days from the infection [12], [13]. A study by Havieri et al. examined the changes in anti–SARS–CoV–2 antibodies in 4., 9., 10., and 20., day after the onset of the clinical symptoms. The results found that in the samples from the 4th day, the amount of the antibodies was undetectable. Whereas, in those from the 9th and 20th day, the level raised to 80 and 320 for anti–SARS–CoV–2 IgM and 80 and 1280 for anti–SARS–CoV–2 IgG, respectively [16]. The dynamic changes in serum antibodies titer create a risk of false–negative results, particularly during the initial period of the infection. It is recommended to measure both anti–SARS–CoV–2 IgM and IgG, to increase the serological test sensitivity in the first days of the disease [12], [13], [14]. Since the primary immune response develops during the first 10–14 days of the infection, it is essential to perform the serological tests no sooner than its onset. The accumulating evidence points out that the serological response during COVID–19 resembles the course of other viral infections [13], [14], [17]. The maximum viremia levels are measured during the initial period of the disease. From the 10–14th day forward, the concentration decreases as a consequence of the immune response [18]. These observations justify the therapeutic use of convalescence plasma at an early stage of severe COVID–19 [18].
4. Anti–SARS–CoV–2 antibodies and COVID-19 treatment
The relationship between the levels of anti–SARS–CoV–2 antibodies and the prognosis, as well as the clinical course of COVID–19, is not established. However, the antibodies and the inflammation caused by the immune system's activation by viral proteins are responsible for the symptoms [19], [20]. The rising level of immunoglobulins and pro–inflammatory cytokines leads to organ failure [21]. The findings from in vivo studies reveal that the rise of anti–S–IgG levels are associated with a lower number of detectable virus units. However, higher anti–S–IgG titer amplifies the risk of Acute Lung Injury (ALI), which is a consequence of IgG–mediated stimulation of inflammation agents, such as macrophages and IL–8 [22]. Notably, contradicting evidence regarding the association between higher antibodies levels and severe form of COVID–19 exist [12], [17]. Of note, however, is the significance of neutrophil/lymphocyte ratio (NLR), which allows us to predict the severity of the infection. High NLR correlates with a more severe clinical course of the disease [21], [23].
Nonetheless, other risk factors, such as age or presence of coexisting diseases, can also impact the severity of COVID–19 [17], [23], [24], [25]. Notably, age was qualified as an independent risk factor [17]. The evidence points out that declines in immune system function and higher expression of ACE2 receptors observed in geriatric patients may be responsible for the poor prognosis [26].
5. The role of serological testing in the diagnostic process in confirmed COVID–19
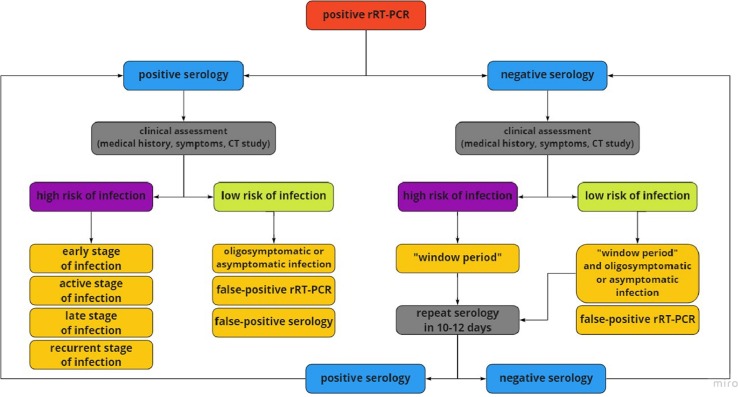
Positive rRT–PCR confirms the presence of active disease [2], [27]. The presence of anti–SARS–CoV–2 antibodies may indicate the phase of COVID–19 disease. High IgM levels are observed in an early stage of the infection, while elevated IgG is characteristic for reinfection or more advanced stage [28]. Fig. 1 presents the suggested approach to serologic testing of patients with positive rRT–PCR results.
Fig. 1.
Suggested approach to serologic testing of patients with positive rRT–PCR results. Assessment of the patient’s clinical condition consists of medical history, symptoms, and CT scan. Contact with suspected COVID–19 case, typical symptoms (fever, dry cough, fatigue) and distinctive CT image (ground-glass opacities, patchy consolidations) suggest a high risk of SARS–CoV–2 infection. For this reason, positive serology can suggest an early, active, or late phase of the disease. It can also suggest recurrent SARS–CoV–2. However, negative serology may result from undetectable concentration of antibodies - “window period”. No contact with suspected COVID–19 case, oligosymptomatic or asymptomatic infection, and nonspecific CT image suggest a low risk of infection. Asymptomatic infection and false-positive rRT–PCR should be considered in positive serology studies whereas asymptomatic infection with “window period” in negative serology studies. In the “window period,” it is advisable to repeat the serological test in 10–12 days. rRT–PCR – real-time Reverse–Transcription Polymerase Chain Reaction, COVID–19 – Coronavirus Disease 2019, CT – Computer Tomography. SARS–CoV–2 – Severe Acute Respiratory Syndrome Coronavirus 2.
6. The role of serological testing in the diagnostic process in suspected COVID–19
The determination of antibody titer can play a complementary role in the diagnostic process of COVID–19. In patients with negative rRT–PCR results and suspected COVID–19, serological tests may help determine the presence or absence of the infection [6], [15]. The variable sensitivity of the tests during the course of the disease strongly influences the results. Zhao et al. demonstrated that in the first seven days of COVID–19, the sensitivity of rRT–PCR and serological tests was 66,7% and 38,3%, respectively. Between the 8th and 14th day from the onset, the sensitivity of rRT–PCR decreased to 54%, whereas the sensitivity of serological tests increased by up to 90%. After the 15th day, the sensitivity of rRT–PCR was 45,5% and serological tests above 90%. Considering these findings, the authors suggest concurrent rRT–PCR and anti–SARS–CoV–2 antibodies testing to increase the diagnostic sensitivity of COVID–19 [12]. In turn, a study by Bin et al. found that the length of virus incubation affects the seroconversion time. In the case of virus incubation of fewer than five days, seroconversion appeared around the 10th day from the beginning of an infection. However, in the case of incubation lasting over five days, seroconversion was detected from the 7th day from the beginning of the disease. Among 80 patients with confirmed SARS–CoV–2 infection, seroconversion was observed in 79 of them [13]. Other findings revealed the rise of antibody titer in more advanced stages of COVID–19, particularly after seven days from the onset [15], [17]. It is recommended to perform both tests to increase the sensitivity of the diagnostic process, particularly rRT–PCR testing in an early stage of the infection and serological testing later in the course of the disease [12]. It is, therefore, advisable to perform serological tests in patients with negative rRT–PCR results in a later stage of COVID–19. The presence of specific antibodies suggests a recent infection or a convalescence period [28]. However, it is critical to emphasize the low sensitivity of rRT–PCR testing in later stages of the disease. Fig. 2, Fig. 3 present the suggested approach to serologic testing of patients with negative rRT–PCR results, with and without past infection.
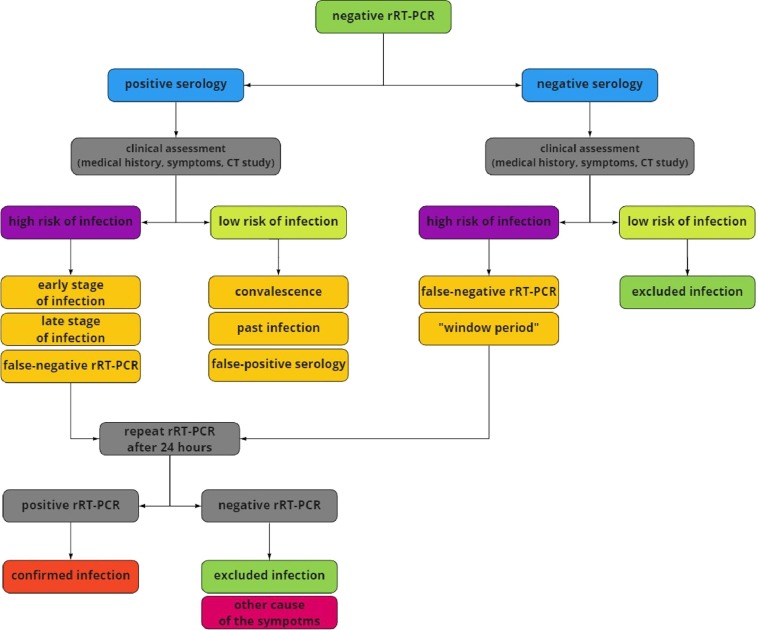
Fig. 2.
Suggested approach to serologic testing of patients with past infection and negative rRT–PCR. Assessment of the patient's clinical condition consists of medical history, symptoms, and CT scan. Contact with suspected COVID–19 case, typical symptoms (fever, dry cough, fatigue), and distinctive CT image (ground-glass opacities, patchy consolidations) suggest a high risk of SARS–CoV–2 infection. For this reason, positive serology can suggest the early or late phase of the disease. It is also necessary to exclude the risk of false-negative rRT–PCR in these cases. No contact with suspected COVID–19 case, oligosymptomatic or asymptomatic infection, and nonspecific CT image suggest a low risk of infection. Convalescence, a past infection, and false-positive serology should be considered in these cases. Due to negative rRT–PCR study and positive serology, it is advisable to repeat the rRT–PCR test. It is also suggested in low-risk cases due to the possibility of asymptomatic COVID–19 in patients with past infection. However, past COVID–19 and negative serology suggest false–negative result and requires repeat testing. rRT–PCR – real-time Reverse–Transcription Polymerase Chain Reaction, COVID–19 – Coronavirus Disease 2019, CT – Computer Tomography. SARS–CoV–2 – Severe Acute Respiratory Syndrome Coronavirus 2.
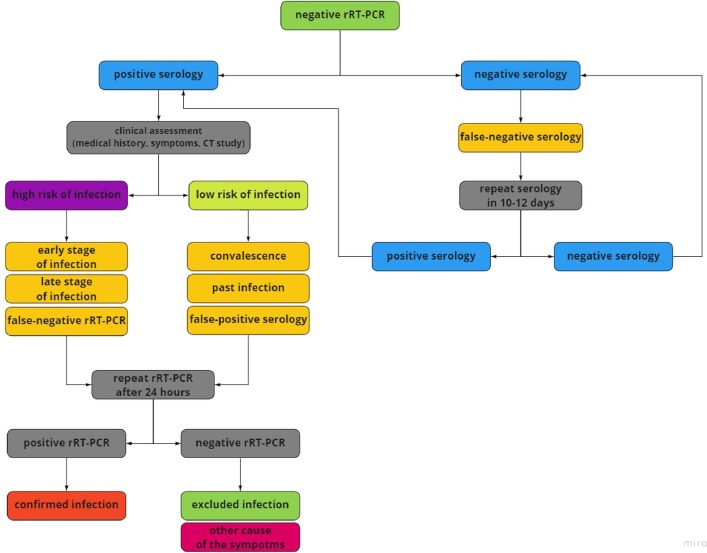
Fig. 3.
Suggested approach to serologic testing of patients without past infection and negative rRT–PCR. Assessment of the patient's clinical condition consists of medical history, symptoms, and CT scan. Contact with suspected COVID–19 case, typical symptoms (fever, dry cough, fatigue) and distinctive CT image (ground-glass opacities, patchy consolidations) suggest a high risk of SARS–CoV–2 infection. For this reason, positive serology can suggest the early or late phase of infection. It is also necessary to exclude the risk of false-negative rRT–PCR study, so repeating the molecular test is advisable. No contact with suspected COVID–19 case, oligosymptomatic or asymptomatic infection, and nonspecific CT image suggest a low risk of infection. Convalescence, a past infection, and false-positive serology should be considered in these cases. Due to no prior disease, negative rRT–PCR, and low risk of infection, it seems unnecessary to perform repeat testing. However, negative rRT–PCR and negative serology indicate the false-negative rRT–PCR and “window period” in cases with high risk of infection. It is advisable to repeat the rRT–PCR test. Insignificant medical history, negative rRT–PCR, negative serology, and low risk of COVID–19 suggest no active and past infection. rRT–PCR – real-time Reverse–Transcription Polymerase Chain Reaction, COVID–19 – Coronavirus Disease 2019, CT – Computer Tomography. SARS–CoV–2 – Severe Acute Respiratory Syndrome Coronavirus 2.
7. Clinical value of serological testing
Serological tests may be utilized to complement the diagnostic approach to suspected or confirmed COVID–19. Moreover, the tests may also be used to limit the spread of the virus. The SARS–CoV–2 virus is primarily transmitted between people through respiratory droplets and contact routes [29]. Serological tests are vital to assess the prevalence of the infection in the targeted population [2], [3], [30]. Fundamentally, it allows the estimation of the number of people infected, as well as tracking the spread of the diseases through the population over time. The surveillance helps public health officials plan for future healthcare needs. Moreover, the tests enable easy identification of oligosymptomatic or asymptomatic individuals and help reach a quarantine decision [30]. The oligosymptomatic or asymptomatic individuals emerge as the primary vector of the transmission that contributes to the uncontrolled spread of the disease [31], [32]. Furthermore, the assessment of the prevalence of infection among healthcare workers is of particular relevance since it helps to estimate and mitigate their exposure and transmission risk. It is suggested that healthcare workers with immunity against SARS–CoV–2 could be directed to the management of COVID–19 patients [30]. The serological tests can be utilized to draw conclusions regarding the effectiveness of various types of personal protective equipment, such as shield, masks, goggles, gowns [30]. Lastly, serology tests may also be used to identify potential convalescent plasma donors and to evaluate the immune response to candidate vaccines [18], [30].
8. Conclusion
Serological testing plays a vital role in the clinical management of COVID–19. Constellations of serological findings, rRT–PCR results, and their clinical interpretation in suspected infection are summarized in Table 1 . Due to the high sensitivity of serological tests after the 10th day from the onset of the disease, it is recommended to utilize qualitative and quantitative anti–SARS–CoV2 IgM and IgG detection in the advanced stage of the infection. Particularly, serologic testing should be advised in patients with negative rRT–PCR results.
Table 1.
Interpretation of molecular, serological and clinical examination.
| rRT–PCR results | Serology results (IgM i/lub IgG) | Risk of infection |
Interpretation | |
|---|---|---|---|---|
| High | Low | |||
| positive | IgM (+) i IgG (−) | + | – | early stage of infection |
| positive | IgM (+) i IgG (+) | + | – | active phase of infection |
| positive | IgM (−) i IgG (+) | + | – | late stage of infection, recurrent stage of infection |
| positive | IgM (+) lub IgG (+) | – | + | oligosymptomatic or asymptomatic infection, false–positive rRT–PCR, false–positive serology |
| positive | IgM (−) i IgG (−) | + | – | “window period” |
| positive | IgM (−) i IgG (−) | – | + | “window period”with oligosymptomatic or asymptomatic infection false–positive rRT–PCR |
| negative | IgM (+) i IgG (−) | + | – | early stage of infection, false–negative rRT–PCR |
| negative | IgM (−) i IgG (+) | + | – | late stage of infection, false–negative rRT–PCR |
| negative | IgM (−) i IgG (+) | – | + | past infection |
| negative | IgM (+) i IgG (+) | – | + | convalescence, false–negative rRT–PCR |
| negative | IgM (−) i IgG (−) | + | – | “window period”, false–negative rRT–PCR |
“+” positive result or present risk; “−” negative result or absent risk.
Funding
Funding by Medical Research Agency, grant number 2020/ABM/COVID19/005.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
N.A.
References
- 1.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID–19) in suspected human cases. Interim guidance: 2 March 2020.
- 3.Rastawicki W., Rokosz-Chudziak N. Characteristics and assessment of the usefulness of serological tests in the diagnostic of infections caused by coronavirus SARS–CoV–2 on the basis of available manufacturer‘s data and literature review. Przegl Epidemiol. 2020;74(1):113–132. doi: 10.32394/pe.74.11. [DOI] [PubMed] [Google Scholar]
- 4.Corman Victor M., Landt Olfert, Kaiser Marco, et al. Detection of 2019 novel coronavirus (2019–nCoV) by real–time RT–PCR. Euro. Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z., Yi Y., Luo X., et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;1–7 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W., Rong-Hui D.u., Li B. Molecular and serological investigation of 2019–nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Y. Yang, M. Yang, C. Shen, et al., Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019–nCoV infections, 2020.
- 8.Andrew N. Cohen, Bruce Kessel, False positives in reverse transcription PCR testing for SARS–CoV–2. MedRxiv preprint, 2020.
- 9.Roujian Lu, Zhao Xiang, Li Juan, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu A., Peng Y., Huang B., et al. Genome composition and divergence of the novel coronavirus (2019–nCoV) originating in China. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walls A.C., Park Y.-J., Tortorici M.A., et al. Structure, function, and antigenicity of the SARS–CoV–2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juanjuan Zhao, Quan Yuan, Haiyan Wang, et al., Antibody responses to SARS–CoV–2 in patients of novel coronavirus disease 2019. MedRxiv preprint 2020. [DOI] [PMC free article] [PubMed]
- 13.Bin Lou, Ting–Dong Li, Shu–Fa Zheng, et al., Serology characteristics of SARS–CoV–2 infection since the exposure and post symptoms onset. MedRxiv preprint, 2020. [DOI] [PMC free article] [PubMed]
- 14.Sun Baoqing, Feng Ying, Mo Xiaoneng, et al. Kinetics of SARS–CoV–2 specific IgM and IgG responses in COVID–19 patients. Emerg. Microbes Infect. 2020 doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Y.-W., Schmitz J.E., Persing D.H., et al. The laboratory diagnosis of COVID–19 infection: current issues and challenges. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haveri Anu, Smura Teemu, Kuivanen Suvi, et al. Serological and molecular findings during SARS–CoV–2 infection: the first case study in Finland, January to February 2020. Euro Surveill. 2020;25(11):2000266. doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.To Kelvin Kai-Wang, Tsang Owen Tak-Yin, Leung Wai-Shing, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS–CoV–2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long Chen, Jing Xiong, Lei Bao, Convalescent plasma as a potential therapy for COVID–19. Lancet Infect Dis, 2020. Comment. [DOI] [PMC free article] [PubMed]
- 19.Maria Infantino, Arianna Damiani, Francesca Li Gobbi, et al., Serological Assays for SARS–CoV–2 Infectious Disease: Benefits, Limitations and Perspectives. IMAJ, vol. 22, April 2020. [PubMed]
- 20.Lina Ling, Lua Lianfeng, Caoa Wei, et al. Hypothesis for potential pathogenesis of SARS–CoV–2 infection–a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 2020;9 doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bicheng Zhang, Xiaoyang Zhou, Chengliang Zhu et al., Immune phenotyping based on neutrophil–to–lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID–19. MedRxiv preprint 2020. [DOI] [PMC free article] [PubMed]
- 22.Liu Li, Wei Qiang, Lin Qingqing, et al. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS–CoV infection. JCI Insight. 2019;4(4):e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin Chuan, Zhou Luoqi, Ziwei Hu, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID–19) in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanga Jing, Zhenga Ya., Goua Xi, et al. Prevalence of comorbidities and its effects in patients infected with SARS–CoV–2: a systematic review and meta–analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan Wei-jie, et al. Comorbidity and its impact on 1590 patients with Covid-19 in China: A Nationwide Analysis. Eur. Respir. J. 2020;55(5) doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Yu., Li Lanjuan. SARS–CoV–2: virus dynamics and host response. Comment. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robin Patel, Esther Babady, Elitza S, et al., Report from the American Society for Microbiology COVID–19 International Summit, 23 March 2020: Value of Diagnostic Testing for SARS–CoV–2/COVID–19. mBio 11: e00722–20. [DOI] [PMC free article] [PubMed]
- 28.Diazyme Laboratories Inc., Covid–19–antibodytests; http://www.diazyme.com/covid–19antibody–tests.
- 29.Morawska L., Cao J. Airborne transmission of SARS–CoV–2: the world should face the reality. Environ. Int. 2020 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter Amy K., Hegde Sonia T. The important role of serology for COVID–19 control. Comment. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothe Camilla, Schunk Mirjam, Sothmann Peter, et al. Transmission of 2019–nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382 doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan Bai, Lingsheng Yao, Tao Wei et al., Presumed Asymptomatic Carrier Transmission of COVID–19. JAMA April 14 2020; Volume 323, Number 14. [DOI] [PMC free article] [PubMed]