Abstract
Over the past 10 years, the field of immunometabolism made great strides to unveil the crucial role of intracellular metabolism in regulating immune cell function. Emerging insights into how systemic inflammation and metabolism influence each other provide a critical additional dimension on the organismal level. Here, we discuss the concept of systemic immunometabolism and review the current understanding of the communication circuits that underlie the reciprocal impact of systemic inflammation and metabolism across organs in inflammatory and infectious diseases, as well as how these mechanisms apply to homeostasis. We present current challenges of systemic immunometabolic research, and in this context, highlight opportunities and put forward ideas to effectively explore organismal physiological complexity in both health and disease.
Lercher at al. review the current understanding of the communication circuits that underlie systemic immunometabolism and how these mechanisms contribute to homeostasis and disease. They discuss current challenges of systemic immunometabolic research and approaches to effectively explore organismal physiological complexity.
Introduction
In a seminal experiment, Urey and Miller demonstrated that simple metabolite precursors and conditions resembling the atmosphere of the early Earth can generate amino acids and RNA nucleobases (Ferus et al., 2017; Miller, 1953). Metabolites are biochemical intermediates or end-products of cellular metabolic pathways, represent cornerstones of life, and, as molecular basis of metabolism, are considered a driving force of evolution. Anabolic processes build up biomass, whereas catabolic processes generate energy and redox equivalents to catalyze thermodynamically unfavorable reactions. Next to their bioenergetic and biosynthetic roles, metabolites are key communication signals that are evolutionarily conserved from prokaryotes, including quorum sensing between bacteria, to complex multicellular eukaryotic organisms. Extracellular cues may shift the metabolic homeostasis of cells under certain environmental conditions. Such signals are sensed by central pathways like mTOR, which integrate available environmental information including nutrients and other stimuli to coordinate metabolic adaptations and proliferation of cells (Saxton and Sabatini, 2017). These environment-driven metabolic adaptations play particularly important roles for the immune system. The organism is estimated to spend up to 30% of its basal metabolic rate for the activation of the adaptive immune system alone (Muehlenbein et al., 2010; Straub et al., 2010). This demonstrates its evolutionary value for multicellular organisms to control pathogens, tissue repair, and many other vital functions (Bantug et al., 2018; Pearce and Everts, 2015).
The chemist Emil Fischer made fundamental discoveries in the field of carbohydrates and published the structures of glucose and its isomers in 1891 (Fischer, 1891). Three decades later, biochemist Hans Krebs described the first metabolic cycle in 1932, while physiologist Otto Warburg made seminal findings about the peculiar metabolism of cancer cells (Krebs and Henesleit, 1932; Warburg, 1956; Warburg et al., 1927). Around two decades later, Otto Warburg noted that increased cellular glycolysis was not only a hallmark of tumor cell development, but also for leukocyte activation (Warburg et al., 1958). Half a century of seminal discoveries about the immune system later (Doherty and Robertson, 2004), immunologists rediscovered metabolic adaptations as critical initial steps for immune responses (Febbraio et al., 2004; Hotamisligil et al., 1993; Pearce et al., 2009; Wieman et al., 2007). This eventually resulted in the establishment of the thriving research field of immunometabolism. Research in this field studies the metabolic control of the immune-regulated processes including infectious or tumorigenic diseases, tissue regeneration, and how these immune responses influence metabolic organs (Buck et al., 2017; Eming et al., 2017; Norata et al., 2015). Initial in vitro studies were instrumental to unravel cytokine-driven reprogramming of the metabolic network and the resulting switch from the reliance on mitochondrial respiration toward glycolysis in activated macrophages and dendritic cells (Jha et al., 2015; Pearce and Everts, 2015; Pearce and Pearce, 2013; Russell et al., 2019). Similarly, engagement of cognate antigen and T cell receptor (TCR) signaling alters metabolism in naive T cells, which is a prerequisite for subsequent rapid clonal expansion and effector function acquisition (Bantug et al., 2018; Sinclair et al., 2013). In vitro studies showed that T cell proliferation relies on increased oxidative phosphorylation (OXPHOS), glycolysis, and Warburg metabolism (Bantug et al., 2018; Chapman et al., 2020; Warburg et al., 1927). Glycolytic reprogramming of T cells also directly affects effector cytokine production (Chang et al., 2013), and multiple studies have explored the contribution of OXPHOS, fatty acid oxidation (FAO), and glycolysis on T cell memory formation (Klein Geltink et al., 2017; Pearce et al., 2009; Phan et al., 2016; Raud et al., 2018).
Cell culture systems are indispensable for breaking new mechanistic grounds, but they cannot recapitulate the vast complexity of the organism. This is reflected by a recent study highlighting the differences between metabolic reprogramming of in vitro-activated T cells compared to their infection-activated counterparts in vivo (Ma et al., 2019). Metabolic parameters such as organ-specific pH, partial pressure of oxygen, nutrient gradients, and disease-dependent changes of the metabolic environment are not considered by standard cell culture conditions, but clearly influence T cell activation (Buck et al., 2017; Geiger et al., 2016; Johnson et al., 2018; Lercher et al., 2019; Ma et al., 2017; Murray, 2016; Nakaya et al., 2014; Roy et al., 2020; Sinclair et al., 2013). Similarly, tumor-associated fibroblasts have a high affinity for glucose and deplete the tumor microenvironment from glucose, resulting in metabolic competition that impairs the function of infiltrating tumor-specific T cells (Chang et al., 2015). Next to glucose, numerous other extracellular metabolites, including amino acids, may modulate T cell function in the tumor microenvironment or during chronic viral infection (Geiger et al., 2016; Lercher et al., 2019; Murray et al., 2015).
In this review, we will discuss recent advances and technological approaches to study immune-metabolic crosstalk at the organismal level, in between organs, and within tissues and provide examples of how these communication networks are shaped by pathogenic and/or commensal microbes. We conclude by providing examples of how research of systemic immunometabolism revealed therapeutic opportunities to target essential mechanisms at the tissue and organismal level.
The Many Components of Systemic Immunometabolism
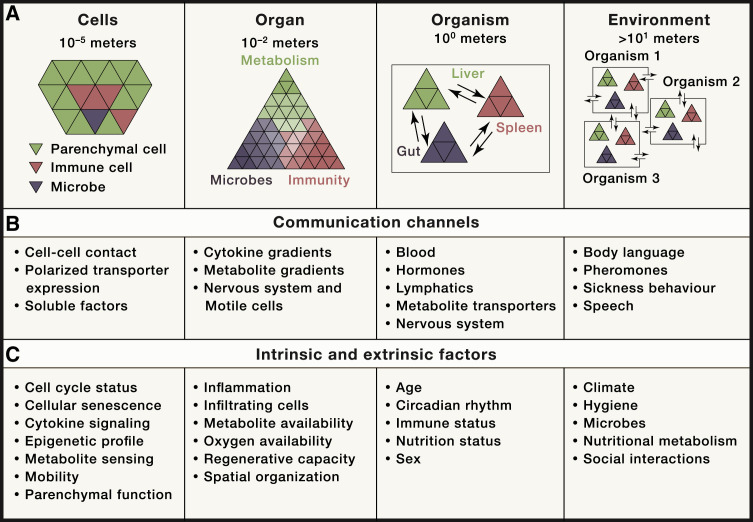
Organs and specialized tissue-resident cells evolved to compartmentalize metabolic processes. This was accompanied by co-evolution of communication systems to coordinate metabolic processes across organs and with the microbiota (Figure 1 A). The crosstalk between immune and metabolic processes is conserved across the animal kingdom. A good example is the fat body in Drosophila that coordinates processes that are mirrored by the crosstalk between the immune system, adipose tissue, and liver in vertebrates (Beller et al., 2010; Hotamisligil, 2017; Padmanabha and Baker, 2014). This supports the notion that most organs, even if specialized on metabolic tasks, may critically impact systemic immune responses.
Figure 1.
Communication Levels of Systemic Immunometabolism
(A) Organizational units of higher organisms ranging from the cellular level, to organs, to the whole organism, up to the environment and inter-individual relationships.
(B) Communication channels that are critically involved for communication within and/or between the individual organizational units.
(C) Intrinsic and extrinsic factors shaping the identity of the described organizational units.
Recent advances in single-cell sequencing technology vastly expanded our understanding of cellular heterogeneity of organs and how this correlates with organ-specific immune responses and composition (Aizarani et al., 2019; Han et al., 2020; Jaitin et al., 2019; Krausgruber et al., 2020; Stubbington et al., 2017; Zhao et al., 2020). For inter-organ coordination, mammals evolved blood, lymph, and nervous systems as communication channels (Figure 1B). Distal communication is achieved by tissue-specific sensing and production of soluble messenger molecules including hormones, neurotrophic peptides, cytokines, and metabolites (Chu et al., 2020; Hotamisligil, 2017; Krauss et al., 2012; Perry et al., 2015; Priest and Tontonoz, 2019). On a cellular level, metabolic processes are spatially organized, and intracellular nutrient distribution is important for organelle communication and function (Pareek et al., 2020; Sdelci et al., 2019). Tissue-specific and context-dependent expression of metabolite transporters and receptors form an additional versatile layer of inter-organ communication. While we are yet to de-orphanize many of these proteins, it seems clear that this will open a new era of increased understanding of how pathophysiological processes involving multiple organs are coordinated across cellular membranes (César-Razquin et al., 2015). Thus, such communication networks orchestrate and coordinate responses to environmental cues or immunological challenges such as infection-associated sickness behavior with symptoms of fever, lethargy, fatigue, social withdrawal, and/or anorexia (Hart, 1988). Organism-wide response programs are largely conserved across species, albeit there are differences such as febrile responses to viral infection between mice and humans (Dybing et al., 2000; Hart, 1988). These immune-metabolic programs are driven by cytokine signaling in the brain and impact pathogen clearance and disease-tolerance mechanisms (Ayres, 2020a; Dantzer et al., 2008; Hart, 1988). In case of bacterial infection, reduced food intake and infection-associated anorexia can be beneficial and promote survival, whereas it aggravates pathology during influenza infection (Wang et al., 2016). A recently identified regulator of anorectic behavior is the neurotrophic peptide GDF15, which signals through its receptor GFRAL in the hindbrain (Borner et al., 2020; Johnen et al., 2007; Tsai et al., 2016, 2018). GDF15 is expressed by a wide range of cells, including macrophages (Fairlie et al., 1999; Hsu et al., 2017; Patel et al., 2019), and was shown to modulate systemic metabolism (Luan et al., 2019; Santos et al., 2019). Conversely, bacteria may interfere with anorexia-related response programs to promote host survival and ensure prolonged pathogen dissemination (Rao et al., 2017). The underlying endogenous regulation of feeding behavior during infection is thought to redistribute energy and balance the energetic demands of the activated immune system with the organism's core functions of maintenance, growth, and reproduction programs, known as the “life history programs” (Ganeshan et al., 2019; Muehlenbein et al., 2010; Schieber and Ayres, 2016; Straub et al., 2010). Related evolutionarily conserved mechanisms have been reviewed elsewhere (Wang et al., 2019).
Nutritional metabolism is a key determinant of body composition, systemic metabolism, and immune responses and linked to numerous diseases. Unbalanced diet leads to inflammation and influences systemic immune responses and co-morbidities (Cornier et al., 2008; Marcos et al., 2003). This is probably most extensively studied in obesity-associated chronic inflammation, metabolic syndrome, and type 2 diabetes (T2D) (Hotamisligil, 2017). Poor metabolic health is a risk factor for developing a more severe course during infectious diseases. Concurringly, diabetes and hypertension are among the most important risk factors for coronavirus disease 2019 (COVID-19) in the global SARS-CoV-2 pandemic (Ayres, 2020b; Lu et al., 2020; Shen et al., 2020; Zhou et al., 2020).
The adipose tissue is an important immune cell niche during homeostasis and an important immune-metabolic communication hub in metabolic syndrome (Hotamisligil, 2017; Schipper et al., 2012). Diet-induced obesity leads to the accumulation of a broad range of immune cells in the adipose tissue that can contribute to systemic metabolism (Aouadi et al., 2013; Jaitin et al., 2019). Particularly, the recruitment of macrophages in the adipose tissue augments systemic glucose intolerance via pro-inflammatory cytokines like TNFα (Aouadi et al., 2013; Weisberg et al., 2003). Adipocytes instruct tissue-resident macrophages toward intracellular lipid accumulation and degradation and thus contribute to overall adipose tissue lipolysis (Xu et al., 2013). Recent single-cell analyses highlighted a role of Trem2 + lipid-associated macrophages in counteracting adipocyte hypertrophy during obesity. Yet, it remains elusive whether macrophage-intrinsic Trem2 is necessary and sufficient to ameliorate obesity-associated metabolic syndrome (Jaitin et al., 2019).
Beyond the adipose tissue, nutritional metabolism broadly affects immune responses in malignant and infectious diseases. Diet-associated obesity and increased availability of extracellular lipids rewires cellular metabolism of natural killer cells, leading to impaired anti-tumoral activity in animals and patients (Michelet et al., 2018). Ketogenic diet improves survival in a model of influenza infection in the presence of γδ T cells in the lung, boosting protective mucus production by airway epithelial cells (Goldberg et al., 2019). Perturbing nutritional metabolism by intermitted fasting promotes localization of pro-inflammatory monocytes (Jordan et al., 2019) and memory T cells (Collins et al., 2019) to the bone marrow, a nutrient-rich immune cell niche, and increases disease tolerance during infection (Ganeshan et al., 2019). Dietary restriction of the amino acid methionine proved beneficial in a model of autoimmune disease by repressing epigenetic remodeling and effector functions of TH17 via intracellular S-adenosyl-methionine (SAM) levels (Roy et al., 2020). Additionally, dietary deficiency of glycine and serine blunts cytotoxic CD8 T cell responses, likely via reduced cell-intrinsic one carbon metabolism and de novo nucleotide synthesis (Ma et al., 2017). This corroborates previous in vitro studies that highlighted increased amino acid dependencies of activated T cells (Geiger et al., 2016; Murray, 2016; Van de Velde et al., 2016). Of note, pathogens themselves may trigger systemic metabolite changes and by that effect, immune responses (Balmer et al., 2016; Qiu et al., 2019), and vice versa, circulating immune cells can actively shape systemic metabolite levels (Fan et al., 2019; Miyajima et al., 2017). These cases exemplify that nutritional metabolism is an important regulator of homeostasis of systemic metabolism that shapes immunity and vice versa. Yet, the cellular responders and molecular basis for diet-induced effects, particularly during infectious diseases, remain to be fully understood.
Generally, studies of systemic immunometabolism in model organisms like mice are influenced by several fundamental metabolic cross-species variances. These include differences in surface-area-to-volume ratio, resting heart rate, relative food intake, or basal metabolic rate between mice and humans (Speakman, 2013). Additional confounding factors, which may be controlled for in experimental models, include sex, age, dietary habits, circadian rhythm, and the microbiome (Figure 1C) (Druzd et al., 2017; Dyar et al., 2018; Ferraro et al., 1992; Houtkooper et al., 2011; Lazzer et al., 2010; Rothschild et al., 2018; Weger et al., 2019).
Next, we will focus on selected metabolically active organs such as the liver, adipose tissue, and the gut to showcase how inflammation-induced local metabolic changes can affect systemic immunometabolism.
Immunometabolism between Organs
Adipose tissue is connective tissue primarily formed by adipocytes and serves as a major energy store for the organism (Spallanzani et al., 2019). Adipocytes contain large lipid droplets and are actively involved in many systemic metabolic pathways including lipid metabolism and the regulation of glucose homeostasis. They communicate to other organs including the liver and muscle and contribute to systemic metabolism via secretion of fatty acids, adipokines, lipokines, and cytokines (Cao et al., 2008; Cawthorn and Sethi, 2008; Scheja and Heeren, 2019). A major research breakthrough was the revelation that adipose tissue itself can secrete the pro-inflammatory cytokine TNFα to modulate systemic glucose metabolism (Hotamisligil et al., 1993). This was also one of the first molecular studies to link metabolic and immunological processes, providing a pillar for the emerging research field of immunometabolism.
The adipose tissue is centrally involved in cachexia, a multifactorial syndrome leading to uncontrolled loss of body mass that is accompanied by severe prognostic implications for patients suffering from cancer, chronic infections, and inflammatory diseases (Baracos et al., 2018). In this syndrome, weight loss is only partially mediated through anorexia (Ayres and Schneider, 2009; Murray and Murray, 1979; Wang et al., 2016). In general, pro-inflammatory signaling pathways trigger metabolic changes, resulting in a negative energy balance and a shift toward catabolism in a broad range of organs, including the adipose tissue. Specifically, cytokines like TNFα, IL-6, and interferons are associated with cancer-associated cachexia, whereas pathogen-specific CD8 T cells are essential for cachexia in a model of chronic viral infection (Arner and Langin, 2014; Baazim et al., 2019; Cawthorn and Sethi, 2008; Fearon et al., 2012; Patel and Patel, 2017). Adipose tissue depletion is accompanied by release of energy in the form of increased free fatty acids (FFAs) into the circulation. FFAs directly contribute to energy distribution when they are taken up by other organs and fuel ATP production via oxidation and the tricarboxylic acid (TCA) cycle, or they may be re-esterified and stored as lipid droplets. FFAs can also instruct metabolic reprogramming in receiving tissues like the liver, where they facilitate a shift of glucose utilization and by that, shape systemic metabolism (Rosen and Spiegelman, 2006). During diet-induced T2D, macrophage-derived IL-6 boosts lipolysis in adipose tissue, which leads to increased glucose production in the liver via increased FFA-derived acetyl-CoA in hepatocytes (Perry et al., 2015). Apart from FFAs, the adipose tissue can communicate with distal organs via exosomal miRNAs, and, for example, modulates production of the hepatokine FGF21, a well-known regulator of systemic glucose metabolism (Thomou et al., 2017).
Such metabolic crosstalk between adipose tissue and liver highlights the communication networks to facilitate labor distribution across the organs within an organism. The liver evolved as a major hub for glucose, fatty acid, and amino acid metabolism and, as such, controls serum levels and homeostasis of the respective metabolites (van den Berghe, 1991; Trefts et al., 2017). Owed to its anatomical location, there is also extensive communication between the liver and intestine via the portal vein (Kubes and Jenne, 2018; Tripathi et al., 2018). Hepatocytes are the functional unit of the liver parenchyma and account for approximately 80% of total liver cells (Racanelli and Rehermann, 2006). At the tissue level, hepatocytes are organized in hexagonal lobules with a central vein at the center and the portal triads, consisting of hepatic artery, hepatic portal vein, and bile duct, at the edges. The directed blood flow from the edges to the center, together with bile flow from center to the periphery, establishes nutrient and oxygen gradients that drive metabolic zonation. This phenomenon, whereby hepatocytes assume specific metabolic tasks depending on their anatomical position within a liver lobule, is best described in the liver (Ben-Moshe and Itzkovitz, 2019; Jungermann and Sasse, 1978; Kietzmann, 2017), but it is likely that metabolic zonation exists in most organs to varying extents. Single-cell transcriptome (single-cell RNA sequencing [scRNA-seq]) analyses combined with mathematical modeling and single-molecule fluorescence in situ hybridization (smFISH) identified marker genes of liver zonation during homeostasis (Halpern et al., 2017). Yet, so far it remains elusive how pathogens and/or immune responses influence metabolic zonation of the liver. Ongoing and future studies will aim to assess the robustness and plasticity of metabolic zonation in liver pathologies. Next to their metabolic core functions, hepatocytes serve as immune signaling platforms during inflammation (Bénéchet et al., 2019; Crispe, 2016; Kubes and Jenne, 2018; Lercher et al., 2019; Rehermann and Nascimbeni, 2005; Zhou et al., 2016). Systemic pathogens and hepatotropic viruses enter the liver primarily via the hepatic artery and are thought to infect hepatocytes close to the blood vessel (Kubes and Jenne, 2018; Protzer et al., 2012; Robinson et al., 2016). Immune cell recruitment and pro-inflammatory cytokine signaling might lead to the establishment of an immunological gradient across the liver lobule. Virus-induced innate immune responses are dominated by the antiviral cytokine type I interferon (IFN-I). IFN-I induces an antiviral state in a broad range of target cells via interferon-stimulated genes (ISGs), but is also a modulator of cellular metabolism (Fritsch and Weichhart, 2016; McNab et al., 2015; Schoggins et al., 2011). IFN-I rewires cellular metabolism of innate immune cells to license optimal antigen presentation or boost the production of immune-modulatory metabolites like itaconate (Mills et al., 2016; O’Neill and Artyomov, 2019; Pantel et al., 2014; Wu et al., 2016). Similarly, IFN-I modulates intracellular cholesterol homeostasis in non-immune cells to increase antiviral responses (York et al., 2015). In a model of metabolic syndrome, IFN-I shapes hepatic glucose metabolism via infiltrating T cells, corroborating data from non-alcoholic fatty liver disease (NAFLD) patients (Ghazarian et al., 2017). Apart from inducing a robust antiviral immune response, IFN-I modulates cellular redox homeostasis and central metabolic pathways in hepatocytes (Bhattacharya et al., 2015; Lercher et al., 2019; Schoggins et al., 2011). This initiates an endogenous regulatory circuit to modulate adaptive antiviral T cell responses and tissue damage via circulating metabolites in a murine infection that is similar to serum metabolite changes in patients with severe COVID-19 (Lercher et al., 2019; Shen et al., 2020). Conversely, inflammation-induced metabolic reprogramming of hepatocytes can influence systemic energy metabolism (Okin and Medzhitov, 2016). This highlights the important role of the liver as central modulator of systemic immunometabolism (Figure 1).
Metabolic Communication across Kingdoms: Immunometabolism and the Microbiota
Eukaryotes live with trillions of bacteria and, depending on the context, their relationship can be pathogenic, commensal, or symbiotic (Hooper and Gordon, 2001). The gut represents a large barrier surface, where a single layer of epithelial cells is a major mediator of crosstalk between gut microbes in the lumen and host cells, including immune cells in the lamina propria. Metabolism is at the center of many of these inter-kingdom interactions. Microbes harbor distinct metabolic pathways and thus can synthesize a vast diversity of small molecules that can be further metabolized (Visconti et al., 2019). Recent chemical and forward genetic screening approaches further highlighted the vast potential of microbiome-derived metabolites to initiate distinct signaling pathways in host cells (Chen et al., 2019; Colosimo et al., 2019; Visconti et al., 2019). Immune responses in the gut are coordinated by epithelial cells and intraepithelial lymphocytes (IELs). They are tightly regulated to adequately respond to pathogens, while avoiding excessive tissue damage and detrimental responses to commensals or food antigens (Figure 1) (Olivares-Villagómez and Van Kaer, 2018).
From an evolutionary perspective, the metabolic labor division and crosstalk between eukaryotic host and prokaryotic microbiome appears highly advantageous (Figure 1A). Gut bacteria metabolize otherwise indigestible food components and by that, fuel metabolic processes of the host and contribute to serum metabolite levels (Cummings et al., 1987; McNeil, 1984; Uchimura et al., 2018; Weger et al., 2019; Wikoff et al., 2009). Yet, the metabolic flexibility of commensal bacteria can also be detrimental, as they can interfere with drug metabolism and associated pathologies (Pryor et al., 2019; Zimmermann et al., 2019). The microbiome affects local and systemic immune responses through the production of immune-reactive metabolites (Abt et al., 2012; Belkaid and Hand, 2014). Prime examples are short-chain fatty acids (SCFAs), which have a local effect in the gut to dampen immune responses toward commensal bacteria (Chang et al., 2014). Increased dietary fiber leads to elevated levels of systemic SCFA via the gut microbiome and is associated with altered bone marrow hematopoiesis. This is accompanied by ameliorated tissue damage in a model of influenza virus infection via increased lung infiltration of alternatively activated macrophages (Trompette et al., 2018). In addition, SCFAs facilitate the development of regulatory T cells (Treg) in the periphery (Arpaia et al., 2013; Haghikia et al., 2015). Contrarily, gut-derived long-chain fatty acids (LCFAs) increase pro-inflammatory T cell responses and ameliorate autoimmune pathologies (Haghikia et al., 2015).
Non-classical T cells, including natural killer T (NKT) or mucosal-associated invariant T (MAIT) cells, specifically recognize microbe-derived metabolites via their TCRs. Microbe-derived metabolite antigens shape development, activation, and maintenance of these cells and are critical in mediating initial immune responses and tissue repair at barrier surfaces (An et al., 2014; Brennan et al., 2013; Constantinides et al., 2019; Legoux et al., 2019). During early life, gut microbe-derived riboflavins are critical for adequate MAIT cell development and skin wound healing in adult animals, yet the communication networks between gut and skin in humans remain elusive (Constantinides et al., 2019). These non-classical T cells represent yet another communication hub between commensal bacteria, the immune system, and individual organs. Of note, microbiome-related studies in laboratory mice are confounded by the controlled hygienic standards of animal facilities and inter-cage diversity, as well as inter-individual variability of the microbiome (Almeida et al., 2019; Pasolli et al., 2019). The microbial diversity is influenced by the genetic background of the host and, to a great extent, by environmental factors (Rothschild et al., 2018). In line, the microbiome of laboratory animals vastly differs from their feral counterparts (Rosshart et al., 2017). Not only is wild microbiome more resilient, but also profoundly affects the immune system, which appears closer to human in wild animals. This can be recapitulated by co-housing or fecal transplant experiments from wild to laboratory animals (Beura et al., 2016; Rosshart et al., 2017, 2019). Embryo transfer of laboratory mice into wild mothers results in mouse colonies that unify well-controlled genetics and a highly resilient microbiome and may, thus, provide a highly relevant model for translational research (Rosshart et al., 2019). This showcases that microbiome-related studies warrant high standardization, meticulous documentation, and disclosure of information about microbiome composition by methods such as 16S RNA sequencing. Undoubtedly, germ-free or antibiotic-treated animals are important experimental models, but show altered immune responses toward pathogens and might not be ideal for studying immune-metabolic relationships (Abt et al., 2012; Kennedy et al., 2018). Reconstitution with a defined microbial community and working with gnotobiotic or isobiotic mice is a laborious alternative, but would allow even better experimental standardization across research groups (Hooper et al., 2012; Wymore Brand et al., 2015). Guidelines for optimization of microbiome-related study design have been reviewed elsewhere (Laukens et al., 2016).
At the tissue level, there are close interactions between IELs and epithelial cells, which themselves are immune signaling platforms that locally modulate IEL function. For example, innate immune signaling pathways in gut epithelial cells are critical for metabolic adaptation and function of IELs in bacterial infection (Hoytema van Konijnenburg et al., 2017). Further, single-cell transcriptome profiling of intestinal epithelial cells unraveled location-dependent heterogeneity and plasticity in homeostasis and following bacterial infection (Haber et al., 2017). Another related immune-regulatory mechanism is the proximal-to-distal gradient of T cell responses along the gut. Mesenteric lymph nodes (mLNs) drain anatomically distinct segments of the gut and by that, distribute inflammatory and tolerogenic immune responses to antigens (Esterházy et al., 2019). The mLNs are surrounded by white adipose tissue (WAT), which contributes to immune responses in the gut and is a reservoir for potent tissue-resident memory T cells (Han et al., 2017). Structural damage to the lymphatics disrupts the communication between microbiome and immune cells and microbes accumulating in the WAT lead to chronic inflammatory responses that cannot be resolved (Fonseca et al., 2015). The compartmentalization of pro- and anti-inflammatory responses along the gut correlates with distinct expression of metabolic genes in migratory dendritic cells, respective to the draining lymph node (Esterházy et al., 2019). Thus, in analogy to metabolic zonation of liver lobules, there is evidence of metabolic zonation along the gut (Quinn et al., 2020).
Technological Approaches to Study Systemic Immunometabolism
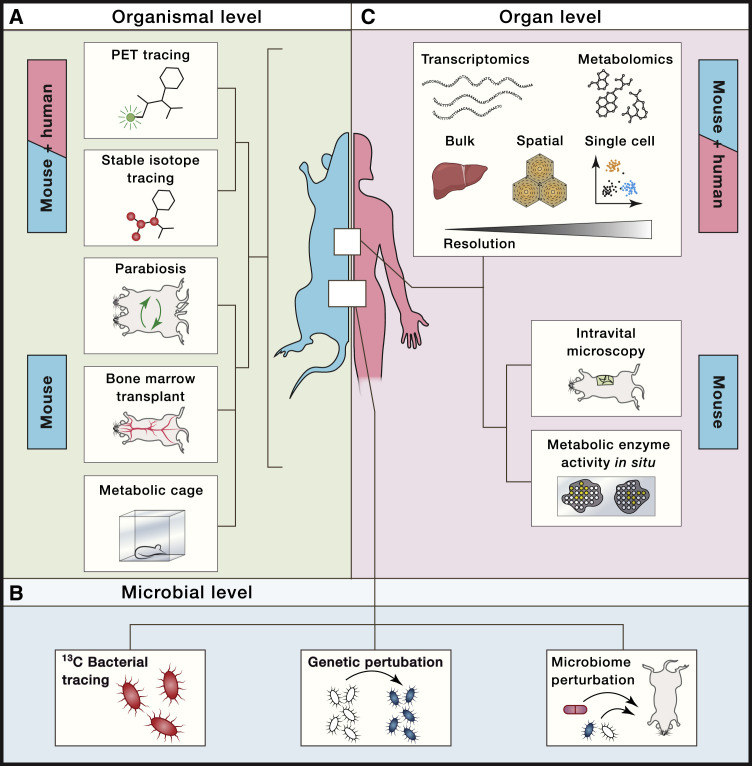
Experimental dissection of immunometabolic relationships at the organismal level during homeostasis and disease remains challenging. Experimental animals can be housed in metabolic cages to longitudinally monitor energy expenditure, physical activity, indirect calorimetry based on oxygen consumption and carbon dioxide production, and food and water intake (Speakman, 2013; Tschöp et al., 2011). Time-resolved imaging techniques may provide strong complementary evidence for immunometabolic alterations within an organism. A functional imaging readout well suited to address such questions is positron emission tomography (PET). This non-invasive whole-body imaging technique is based on radioactively labeled tracers like 18F-labeled deoxy-glucose (FDG) to identify tissues with high glucose demands in both animal models and patients (Lammertsma, 2017; Vaquero and Kinahan, 2015; Zhu et al., 2011). Complementary, heavy isotope tracing of 13C-labeled metabolites by mass spectrometry, together with computational metabolic flux modeling, offers yet another powerful technique to assess de novo host metabolite synthesis and turnover in vivo (Jang et al., 2018). By transplanting 13C heavy isotope-labeled bacteria and subsequent metabolite tracing, this methodological approach further allows dissection of microbiome- versus host-derived metabolites (Figure 2 A) (Jang et al., 2018; Uchimura et al., 2018). Additionally, 13C tracing of bacterial metabolites by mass spectrometry allows identification of unknown bacterial metabolites. Complementary, metabolic pathways of bacteria of the microbiome may be genetically perturbed in situ by engineering a donor strain with broad phylogenetic recipient range to harness horizontal gene transfer (Figure 2B) (Ronda et al., 2019).
Figure 2.
Technical Approaches to Study Systemic Immunometabolism
(A–C) Schematic depiction of selected complementary technical approaches to dissect immune-metabolic crosstalk and relationships in experimental model systems and patients at the (A) organismal (e.g., PET tracing, stable isotope tracing, parabiosis, bone marrow transplant experiments, or metabolic cages), (B) microbial (e.g., 13C bacterial tracing experiments, genetic or microbime perturbations), and (C) organ (e.g., bulk, spatial or single-cell transcriptomics and metabolomics, intravital microscopy, or measuring metabolic enzyme activity in situ) levels.
To dissect immune-metabolic relationships at the cell and tissue level, imaging techniques visualizing cells via defined markers combined with functional well-established metabolic readouts to quantify, for example, mitochondria, reactive oxygen species, lipid droplets, or glucose, are widely used. This includes the quantification of metabolic enzyme activity in situ for research focused on certain metabolic pathways (Miller et al., 2017). Spatiotemporal resolution of cell-cell interactions and migration dynamics of immune cells within tissues in vivo could be achieved via intravital microscopy (Figure 2C) (Karreman et al., 2016).
A critical question for investigating communication networks in vivo relates to the specific sender-respective responder cell types and tissue-specific metabolic pathways. Widely used genetic systems like the Cre-loxP system are valuable genetic systems to specifically ablate genes in a cell-type-specific manner, but come with certain limitations as discussed elsewhere (Becher et al., 2018; Kurachi et al., 2019). Recombinant adeno-associated viral (AAV) vectors pose a versatile alternative to specifically manipulate gene expression in a wide range of tissues in vivo (Srivastava, 2016). Adoptive cell transfers and bone marrow transplants from genetically modified animals to wild--type animals or vice versa are another versatile tool to address the contribution of the hematopoietic immune cell compartment to an observed metabolic phenotype. Parabiosis experiments may augment such approaches to examine the contribution of migrating immune cells and/or circulatory factors to systemic metabolism in homeostasis and disease (Han et al., 2017). These and other technologies, particularly when integrated with single-cell sequencing and methods providing spatial resolution (Stark et al., 2019), are expected to offer unprecedented new mechanistic insights into systemic metabolism and inflammation across and between both lymphoid and non-lymphoid organs.
Development and application of methods with spatial resolution gain increased momentum as exemplified by current advances toward spatial in situ transcriptomics (Moor and Itzkovitz, 2017; Stark et al., 2019). Multiplexed error-robust FISH (MERFISH) is an advanced imaging-based method with single-cell-transcript-level resolution that may be combined with single-cell sequencing (Figure 2B) (Chen et al., 2015; Moffitt et al., 2018). Other approaches are based on direct hybridization of RNA from tissue sections of interest to spatially barcoded probes immobilized either on a plane surface with the prospect of providing single-cell or even sub-cellular resolution (Rodriques et al., 2019; Ståhl et al., 2016; Vickovic et al., 2019). Alternatively, multiplexed immunohistochemistry using DNA-barcoded antibodies enable detection of protein abundances within tissues combined with a high throughput sequencing readout (Stoeckius et al., 2017). Multi-dimensional in situ imaging at high resolution is also achievable by using heavy metal-labeled antibodies for protein detection and subsequent tissue ablation via a laser and mass-cytometry-based (CyTOF) analyses of the liberated particles (Hartmann and Bendall, 2020).
To ultimately address spatiotemporal dynamics of immunometabolic processes at the tissue level, advances in spatial metabolomics will be instrumental (Miura et al., 2012). Methods for in situ spatial single-cell metabolomics integrating matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry (IMS) with bright field and fluorescence microscopy are under development (Alexandrov, 2020). These methods deliver spatially resolved single-cell metabolite profiles and allow visualization of tissue sections via their metabolite distribution (Miura et al., 2012; Rappez et al., 2019; Sun et al., 2019). A combination of spatial scRNA-seq and metabolomics is bound to open new perspectives for the immunometabolic relationships within organs and contribute to mapping inter-organ communication within an organism.
In summary, it is evident that studying systemic immunometabolism requires a broad complementary technological portfolio to investigate utilization, distribution, and immune-metabolic communication networks in space and time. The integration of spatial reorganization of cells within tissues and metabolic phenotype changes of an organ or organism will be of paramount importance to elevate our knowledge on systemic immunometabolism.
Immunometabolism in the Clinics
The discovery of so-called antimetabolite drugs and the subsequent development of the folate pathway inhibitor methotrexate by Sidney Farber is considered the beginning of modern chemotherapy (Farber and Diamond, 1948; Miller, 2006). This and similar discoveries put metabolic pathways as attractive targets in the spotlight early on. Many chemotherapeutic drugs used in the clinics nowadays exploit the metabolic vulnerabilities of highly proliferative cancer cells (DeVita and Chu, 2008).
A common metabolic feature of proliferating cancer and activated immune cells is increased energy demand. This demand is generally met by upregulation of glycolysis and OXPHOS, but also depends on extracellular metabolite availability and import. A currently explored antitumoral strategy is the inhibition of glycolysis via 2-deoxy-glucose (2-DG), a competitive inhibitor of glycolysis (Wick et al., 1957). 2-DG is well tolerated in patients and showed beneficial effects in solid tumors (Raez et al., 2013; Stein et al., 2010). Recent studies also suggested a beneficial effect of 2-DG treatment in systemic lupus erythematosus (SLE) and autoimmune disease mediated by glycolytically highly active T cells (Choi et al., 2018; Yin et al., 2015). Cancers and immune cells can upregulate PKM2, an inactive isoform of the glycolysis gene PKM, to activate hypoxic signaling, angiogenesis, and cytokine production (Luo et al., 2011; Palsson-McDermott et al., 2015). The small molecule TEPP-46 inhibits this process by restoring enzymatic activity of PKM2 and yielded promising results in experimental models of cancer and infectious diseases (Anastasiou et al., 2012; Palsson-McDermott et al., 2015). Glycolysis produces pyruvate that is conjugated to coenzyme A (CoA) to be fed into the TCA cycle as acetyl-CoA. Reprogramming of the TCA cycle and associated genes and metabolites are linked to inflammatory processes or malignant transformation (Lampropoulou et al., 2016; Mills et al., 2016; O’Neill and Artyomov, 2019; Yen et al., 2010). The TCA cycle gene SDH facilitates inflammatory processes by boosting OXPHOS and the accumulation of reactive oxygen species (ROS) (Mills et al., 2016). Dimethyl-malonate and 4-octylitaconate are small molecules targeting SDH and ameliorate inflammation in several experimental models (Chouchani et al., 2014; Mills et al., 2018). An isoform of isocitrate dehydrogenase, IDH2, is an oncogene and targeting of the mutant form with the drug Enasidenib appears as promising treatment option in acute myeloid leukemia patients (Stein et al., 2019; Yen et al., 2017). Dimethyl-fumarate is a derivate of the TCA metabolite fumarate, thought to interfere with the NRF2 pathway and glycolysis, and is currently used as an immunomodulatory drug to treat multiple sclerosis and psoriasis in patients (Gross et al., 2015; Kornberg et al., 2018; Mrowietz et al., 2017). These findings support the idea that cell-type-specific metabolic dependencies are an attractive therapeutic target to selectively modulate activity of any cell type of interest. An example thereof is the possibility to boost anti-tumoral activity of chimeric antigen receptor (CAR) T cell responses to tumor by genetic or pharmacological perturbation of CPT1A, the rate-limiting enzyme of FAO (Klein Geltink et al., 2017).
Research in the field of systemic immunometabolism helped to unveil metabolic dependencies of highly proliferating cells on the metabolic environment. This is particularly evident in rapidly proliferating T cells or malignant cells that are sensitive toward extracellular amino acid scarcity. This opens an opportunity to tune immune responses or interfere with cancer cell progression and growth by modulating amino acid availability. A prime example is the IDO1-mediated depletion of the essential amino acid tryptophan and accumulation of the immunosuppressive metabolite kynurenine (Prendergast et al., 2017). Several clinical studies explored IDO1 as a target to re-activate T cells and, after setbacks, inhibitors like Epacadostat and BMS986205 might be most effective in combination therapy against cancer (Van den Eynde et al., 2020; Komiya and Huang, 2018). A second example is the semi-essential amino acid arginine, whose extracellular availability is critical for both cancer and immune cells (Brunner et al., 2020; Feun et al., 2012; Geiger et al., 2016). The therapeutic potential of arginine depletion via recombinant enzymes, including arginine deiminase (ADI-PEG20) and recombinant arginase 1 (recARG1), was evaluated in clinical trials (Abou-Alfa et al., 2018; Ensor et al., 2002; De Santo et al., 2018). Since arginine-depletion seems to reduce pro-inflammatory immune cell activity, it could also provide a strategy to ameliorate inflammatory diseases. Analogously, the essential amino acid methionine is another limiting factor of cancer cell metabolism and malignant transformation and, concurringly, dietary methionine restriction reduces cancer progression in humans (Gao et al., 2019; Hoffman and Erbe, 1976). Complementarily, the high glucose demand of tumors can be exploited by adhering to a ketogenic diet, which reduces circulating glucose availability (Champ et al., 2014). Preclinical patient data and small cohort trials suggest a beneficial effect of ketogenic diet and the associated blood glucose in several tumors (Fine et al., 2012; Weber et al., 2018). Apart from cancer, recent studies in animal models show that extracellular availability of amino acids or glucose influences immune responses in autoinflammatory or infectious diseases (Brunner et al., 2020; Goldberg et al., 2019; Lercher et al., 2019; Ma et al., 2017; Roy et al., 2020; Zhang et al., 2019). Specifically, extracellular amino acids are important regulators of the protective properties of Treg cells and thus modulate pathophysiology of autoimmune diseases (Do et al., 2020; Ikeda et al., 2017; Shi et al., 2019). This shows a potentially broad applicability of the immune-metabolic intervention strategies that are currently predominantly explored in cancer. Further reviews discussing the potential of metabolic drugs and nutritional metabolism for the treatment of inflammatory diseases and cancer are provided elsewhere (Kanarek et al., 2020; Pålsson-McDermott and O’Neill, 2020).
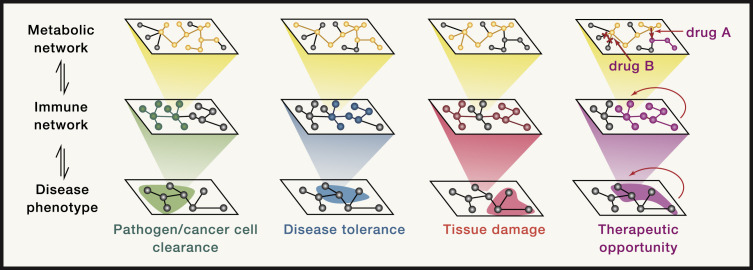
Altogether, disease pathologies are the result of the interplay of multiple molecular processes (Menche et al., 2015). Similarly, cells and signaling molecules of the immune system are organized in complex network-like structures whose activity is modulated by the underlying metabolic program (Jha et al., 2015; Rieckmann et al., 2017). Targeted intervention with the metabolic networks bears the potential to tailor immune responses to ameliorate and/or prevent disease (Figure 3 ). As we keep discovering the whole breadth of crosstalk between immune-metabolic networks and disease pathophysiology, therapeutic interventions targeting specific organs and/or cell types are becoming increasingly in reach.
Figure 3.
Reprogramming of Systemic Immunometabolism as Therapeutic Intervention Strategy
Schematic depiction of the hierarchical order of metabolic, immune, and disease pathology networks. The metabolic network of a cell or organism is the basal regulator of immune responses in different tissues or organs. The quality of the immune response, immune cell activation state, and cytokine signaling in turn shapes beneficial or detrimental disease pathophysiology. These outcomes include removal of the perturbation (e.g., pathogen or cancer cell) and disease tolerance (e.g., pathogen-host co-adaptations), but also extensive tissue damage (e.g., viral hepatitis) that can be life threatening to the host. Specific therapeutic interventions at the level of the metabolic network bears the potential to effectively modulate immune-related disease pathologies.
Concluding Remarks
Immune-metabolic relationships form the basis of life and are at the center of many diseases including cancer, immune-mediated diseases, and metabolic syndrome. The field of immunometabolism is one of the most exciting interdisciplinary research areas. We expect fundamental new insights for our understanding of how the intricate regulation of the immune system interdepends on metabolism. Research will be propelled by the integration of sophisticated in vitro systems, disease-relevant animal models, and patient-derived data, exploiting the respective strengths of the approach while considering the existing limitations. Technological advances on all levels including imaging, single-cell analysis, and spatially resolved techniques will empower the immunometabolism research community in their quest. Conceptually, we are convinced to see an even stronger and more profound consideration of physiological processes, such as the core functions of parenchyma and connective tissue, spatial zonation of organs, and drainage by blood and lymph. This will open new dimensions that eventually result in a truly holistic understanding of immunology on the cellular, organ, and organism level. Systemic immunometabolism will be at the forefront to integrate hitherto rather neglected roles of non-immune cells and non-lymphoid organs, thereby filling important gaps in our understanding of how immune cells communicate with the organism to execute their diverse tasks most effectively.
There is already a surge in novel therapeutic strategies based on the exploitation of metabolic vulnerabilities and targeting of specific metabolic processes. Systemic immunometabolism will improve our mechanistic understanding of the inner workings of the human organism, promising to contribute to a stream of innovative (immuno-) therapeutic approaches for a great range of diseases.
Acknowledgments
We thank Jakob-Wendelin Genger and Henrique Colaço for valuable feedback. A.L. was supported by a DOC fellowship of the Austrian Academy of Sciences. A.B. received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement 677006; “CMIL” awarded to A.B.). The authors apologize for not being able to cite all related studies due to space constraints.
References
- Abou-Alfa G.K., Qin S., Ryoo B.-Y., Lu S.-N., Yen C.-J., Feng Y.-H., Lim H.Y., Izzo F., Colombo M., Sarker D., et al. Phase III randomized study of second line ADI-PEG 20 plus best supportive care versus placebo plus best supportive care in patients with advanced hepatocellular carcinoma. Ann. Oncol. 2018;29:1402–1408. doi: 10.1093/annonc/mdy101. [DOI] [PubMed] [Google Scholar]
- Abt M.C., Osborne L.C., Monticelli L.A., Doering T.A., Alenghat T., Sonnenberg G.F., Paley M.A., Antenus M., Williams K.L., Erikson J., et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizarani N., Saviano A., Sagar, Mailly L., Durand S., Herman J.S., Pessaux P., Baumert T.F., Grün D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov T. Spatial Metabolomics and Imaging Mass Spectrometry in the Age of Artificial Intelligence. Annual Review of Biomedical Data Science. 2020;3 doi: 10.1146/annurev-biodatasci-011420-031537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A., Lawley T.D., Finn R.D. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D., Oh S.F., Olszak T., Neves J.F., Avci F.Y., Erturk-Hasdemir D., Lu X., Zeissig S., Blumberg R.S., Kasper D.L. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D., Yu Y., Israelsen W.J., Jiang J.-K., Boxer M.B., Hong B.S., Tempel W., Dimov S., Shen M., Jha A., et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat. Chem. Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouadi M., Tencerova M., Vangala P., Yawe J.C., Nicoloro S.M., Amano S.U., Cohen J.L., Czech M.P. Gene silencing in adipose tissue macrophages regulates whole-body metabolism in obese mice. Proc. Natl. Acad. Sci. USA. 2013;110:8278–8283. doi: 10.1073/pnas.1300492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner P., Langin D. Lipolysis in lipid turnover, cancer cachexia, and obesity-induced insulin resistance. Trends Endocrinol. Metab. 2014;25:255–262. doi: 10.1016/j.tem.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P., Liu H., Cross J.R., Pfeffer K., Coffer P.J., Rudensky A.Y. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J.S. The Biology of Physiological Health. Cell. 2020;181:250–269. doi: 10.1016/j.cell.2020.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J.S. A metabolic handbook for the COVID-19 pandemic. Nature Metabolism. 2020;2:572–585. doi: 10.1038/s42255-020-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J.S., Schneider D.S. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 2009;7:e1000150. doi: 10.1371/journal.pbio.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baazim H., Schweiger M., Moschinger M., Xu H., Scherer T., Popa A., Gallage S., Ali A., Khamina K., Kosack L., et al. CD8+ T cells induce cachexia during chronic viral infection. Nat. Immunol. 2019;20:701–710. doi: 10.1038/s41590-019-0397-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer M.L., Ma E.H., Bantug G.R., Grählert J., Pfister S., Glatter T., Jauch A., Dimeloe S., Slack E., Dehio P., et al. Memory CD8(+) T Cells Require Increased Concentrations of Acetate Induced by Stress for Optimal Function. Immunity. 2016;44:1312–1324. doi: 10.1016/j.immuni.2016.03.016. [DOI] [PubMed] [Google Scholar]
- Bantug G.R., Galluzzi L., Kroemer G., Hess C. The spectrum of T cell metabolism in health and disease. Nat. Rev. Immunol. 2018;18:19–34. doi: 10.1038/nri.2017.99. [DOI] [PubMed] [Google Scholar]
- Baracos V.E., Martin L., Korc M., Guttridge D.C., Fearon K.C.H. Macmillan Publishers Limited; 2018. Cancer-associated cachexia. [DOI] [PubMed] [Google Scholar]
- Becher B., Waisman A., Lu L.F. Conditional Gene-Targeting in Mice: Problems and Solutions. Immunity. 2018;48:835–836. doi: 10.1016/j.immuni.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M., Bulankina A.V., Hsiao H.H., Urlaub H., Jäckle H., Kühnlein R.P. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 2010;12:521–532. doi: 10.1016/j.cmet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Ben-Moshe S., Itzkovitz S. Spatial heterogeneity in the mammalian liver. Nat. Rev. Gastroenterol. Hepatol. 2019;16:395–410. doi: 10.1038/s41575-019-0134-x. [DOI] [PubMed] [Google Scholar]
- Bénéchet A.P., De Simone G., Di Lucia P., Cilenti F., Barbiera G., Le Bert N., Fumagalli V., Lusito E., Moalli F., Bianchessi V., et al. Dynamics and genomic landscape of CD8+ T cells undergoing hepatic priming. Nature. 2019;574:200–205. doi: 10.1038/s41586-019-1620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Hamilton S.E., Bi K., Schenkel J.M., Odumade O.A., Casey K.A., Thompson E.A., Fraser K.A., Rosato P.C., Filali-Mouhim A., et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532:512–516. doi: 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A., Hegazy A.N., Deigendesch N., Kosack L., Cupovic J., Kandasamy R.K., Hildebrandt A., Merkler D., Kühl A.A., Vilagos B., et al. Superoxide Dismutase 1 Protects Hepatocytes from Type I Interferon-Driven Oxidative Damage. Immunity. 2015;43:974–986. doi: 10.1016/j.immuni.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner T., Shaulson E.D., Ghidewon M.Y., Barnett A.B., Horn C.C., Doyle R.P., Grill H.J., Hayes M.R., De Jonghe B.C. GDF15 Induces Anorexia through Nausea and Emesis. Cell Metab. 2020;31:351–362.e5. doi: 10.1016/j.cmet.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan P.J., Brigl M., Brenner M.B. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- Brunner J.S., Vulliard L., Hofmann M., Kieler M., Lercher A., Vogel A., Russier M., Brüggenthies J.B., Kerndl M., Saferding V., et al. Environmental arginine controls multinuclear giant cell metabolism and formation. Nat. Commun. 2020;11:431. doi: 10.1038/s41467-020-14285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.D., Sowell R.T., Kaech S.M., Pearce E.L. Elsevier Inc.; 2017. Metabolic Instruction of Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Gerhold K., Mayers J.R., Wiest M.M., Watkins S.M., Hotamisligil G.S. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthorn W.P., Sethi J.K. TNF-α and adipocyte biology. FEBS Lett. 2008;582:117–131. doi: 10.1016/j.febslet.2007.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- César-Razquin A., Snijder B., Frappier-Brinton T., Isserlin R., Gyimesi G., Bai X., Reithmeier R.A., Hepworth D., Hediger M.A., Edwards A.M., Superti-Furga G. A Call for Systematic Research on Solute Carriers. Cell. 2015;162:478–487. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Champ C.E., Palmer J.D., Volek J.S., Werner-Wasik M., Andrews D.W., Evans J.J., Glass J., Kim L., Shi W. Targeting metabolism with a ketogenic diet during the treatment of glioblastoma multiforme. J. Neurooncol. 2014;117:125–131. doi: 10.1007/s11060-014-1362-0. [DOI] [PubMed] [Google Scholar]
- Chang C.H., Curtis J.D., Maggi L.B., Jr., Faubert B., Villarino A.V., O’Sullivan D., Huang S.C.C., van der Windt G.J.W., Blagih J., Qiu J., et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P.V., Hao L., Offermanns S., Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.H., Qiu J., O’Sullivan D., Buck M.D., Noguchi T., Curtis J.D., Chen Q., Gindin M., Gubin M.M., van der Windt G.J.W., et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman N.M., Boothby M.R., Chi H. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 2020;20:55–70. doi: 10.1038/s41577-019-0203-y. [DOI] [PubMed] [Google Scholar]
- Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:1360–1363. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Nwe P.-K., Yang Y., Rosen C.E., Bielecka A.A., Kuchroo M., Cline G.W., Kruse A.C., Ring A.M., Crawford J.M., Palm N.W. A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell. 2019;177:1217–1231.e18. doi: 10.1016/j.cell.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.C., Titov A.A., Abboud G., Seay H.R., Brusko T.M., Roopenian D.C., Salek-Ardakani S., Morel L. Inhibition of glucose metabolism selectively targets autoreactive follicular helper T cells. Nat. Commun. 2018;9:4369. doi: 10.1038/s41467-018-06686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani E.T., Pell V.R., Gaude E., Aksentijević D., Sundier S.Y., Robb E.L., Logan A., Nadtochiy S.M., Ord E.N.J., Smith A.C., et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C., Artis D., Chiu I.M. Neuro-immune Interactions in the Tissues. Immunity. 2020;52:464–474. doi: 10.1016/j.immuni.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N., Han S.-J.J., Enamorado M., Link V.M., Huang B., Moseman E.A., Kishton R.J., Shannon J.P., Dixit D., Schwab S.R., et al. The Bone Marrow Protects and Optimizes Immunological Memory during Dietary Restriction. Cell. 2019;178:1088–1101.e15. doi: 10.1016/j.cell.2019.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo D.A., Kohn J.A., Luo P.M., Piscotta F.J., Han S.M., Pickard A.J., Rao A., Cross J.R., Cohen L.J., Brady S.F. Mapping Interactions of Microbial Metabolites with Human G-Protein-Coupled Receptors. Cell Host Microbe. 2019;26:273–282.e7. doi: 10.1016/j.chom.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides M.G., Link V.M., Tamoutounour S., Wong A.C., Perez-Chaparro P.J., Han S.-J., Chen Y.E., Li K., Farhat S., Weckel A., et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019;366 doi: 10.1126/science.aax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornier M.A., Dabelea D., Hernandez T.L., Lindstrom R.C., Steig A.J., Stob N.R., Van Pelt R.E., Wang H., Eckel R.H. The metabolic syndrome. Endocr. Rev. 2008;29:777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispe I.N. Hepatocytes as Immunological Agents. J. Immunol. 2016;196:17–21. doi: 10.4049/jimmunol.1501668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings J.H., Pomare E.W., Branch W.J., Naylor C.P., Macfarlane G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R., O’Connor J.C., Freund G.G., Johnson R.W., Kelley K.W. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santo C., Cheng P., Beggs A., Egan S., Bessudo A., Mussai F. Metabolic therapy with PEG-arginase induces a sustained complete remission in immunotherapy-resistant melanoma. J. Hematol. Oncol. 2018;11:1–5. doi: 10.1186/s13045-018-0612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVita V.T., Jr., Chu E. A history of cancer chemotherapy. Cancer Res. 2008;68:8643–8653. doi: 10.1158/0008-5472.CAN-07-6611. [DOI] [PubMed] [Google Scholar]
- Do M.H., Wang X., Zhang X., Chou C., Nixon B.G., Capistrano K.J., Peng M., Efeyan A., Sabatini D.M., Li M.O. Nutrient mTORC1 signaling underpins regulatory T cell control of immune tolerance. J. Exp. Med. 2020;217 doi: 10.1084/jem.20190848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty M., Robertson M.J. Some early Trends in Immunology. Trends Immunol. 2004;25:623–631. doi: 10.1016/j.it.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Druzd D., Matveeva O., Ince L., Harrison U., He W., Schmal C., Herzel H., Tsang A.H., Kawakami N., Leliavski A., et al. Lymphocyte Circadian Clocks Control Lymph Node Trafficking and Adaptive Immune Responses. Immunity. 2017;46:120–132. doi: 10.1016/j.immuni.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyar K.A., Lutter D., Artati A., Ceglia N.J., Liu Y., Armenta D., Jastroch M., Schneider S., de Mateo S., Cervantes M., et al. Atlas of Circadian Metabolism Reveals System-wide Coordination and Communication between Clocks. Cell. 2018;174:1571–1585.e11. doi: 10.1016/j.cell.2018.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybing J.K., Schultz-Cherry S., Swayne D.E., Suarez D.L., Perdue M.L. Distinct pathogenesis of hong kong-origin H5N1 viruses in mice compared to that of other highly pathogenic H5 avian influenza viruses. J. Virol. 2000;74:1443–1450. doi: 10.1128/jvi.74.3.1443-1450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming S.A., Wynn T.A., Martin P. Inflammation and metabolism in tissue repair and regeneration. Science. 2017;356:1026–1030. doi: 10.1126/science.aam7928. [DOI] [PubMed] [Google Scholar]
- Ensor C.M., Holtsberg F.W., Bomalaski J.S., Clark M.A. Pegylated arginine deiminase (ADI-SS PEG20,000 mw) inhibits human melanomas and hepatocellular carcinomas in vitro and in vivo. Cancer Res. 2002;62:5443–5450. [PubMed] [Google Scholar]
- Esterházy D., Canesso M.C.C., Mesin L., Muller P.A., de Castro T.B.R., Lockhart A., ElJalby M., Faria A.M.C., Mucida D. Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature. 2019;569:126–130. doi: 10.1038/s41586-019-1125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairlie W.D., Moore A.G., Bauskin A.R., Russell P.K., Zhang H.P., Breit S.N. MIC-1 is a novel TGF-beta superfamily cytokine associated with macrophage activation. J. Leukoc. Biol. 1999;65:2–5. doi: 10.1002/jlb.65.1.2. [DOI] [PubMed] [Google Scholar]
- Fan K.Q., Li Y.Y., Wang H.L., Mao X.T., Guo J.X., Wang F., Huang L.J., Li Y.N., Ma X.Y., Gao Z.J., et al. Stress-Induced Metabolic Disorder in Peripheral CD4+ T Cells Leads to Anxiety-like Behavior. Cell. 2019;179:864–879.e19. doi: 10.1016/j.cell.2019.10.001. [DOI] [PubMed] [Google Scholar]
- Farber S., Diamond L.K. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- Fearon K.C.H., Glass D.J., Guttridge D.C. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metab. 2012;16:153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Febbraio M.A., Hiscock N., Sacchetti M., Fischer C.P., Pedersen B.K. Interleukin-6 is a novel factor mediating glucose homeostasis during skeletal muscle contraction. Diabetes. 2004;53:1643–1648. doi: 10.2337/diabetes.53.7.1643. [DOI] [PubMed] [Google Scholar]
- Ferraro R., Lillioja S., Fontvieille A.M., Rising R., Bogardus C., Ravussin E. Lower sedentary metabolic rate in women compared with men. J. Clin. Invest. 1992;90:780–784. doi: 10.1172/JCI115951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferus M., Pietrucci F., Saitta A.M., Knížek A., Kubelík P., Ivanek O., Shestivska V., Civiš S. Formation of nucleobases in a Miller-Urey reducing atmosphere. Proc. Natl. Acad. Sci. USA. 2017;114:4306–4311. doi: 10.1073/pnas.1700010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feun L.G., Marini A., Walker G., Elgart G., Moffat F., Rodgers S.E., Wu C.J., You M., Wangpaichitr M., Kuo M.T., et al. Negative argininosuccinate synthetase expression in melanoma tumours may predict clinical benefit from arginine-depleting therapy with pegylated arginine deiminase. Br. J. Cancer. 2012;106:1481–1485. doi: 10.1038/bjc.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine E.J., Segal-Isaacson C.J., Feinman R.D., Herszkopf S., Romano M.C., Tomuta N., Bontempo A.F., Negassa A., Sparano J.A. Targeting insulin inhibition as a metabolic therapy in advanced cancer: a pilot safety and feasibility dietary trial in 10 patients. Nutrition. 2012;28:1028–1035. doi: 10.1016/j.nut.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Fischer E. Vol. 24. 1891. Ueber die Configuration des Traubenzuckers und seiner Isomeren. Berichte der deutschen chemischen Gesellschaft; pp. 1836–1845. [Google Scholar]
- Fonseca D.M., Hand T.W., Han S.-J., Gerner M.Y., Glatman Zaretsky A., Byrd A.L., Harrison O.J., Ortiz A.M., Quinones M., Trinchieri G., et al. Microbiota-Dependent Sequelae of Acute Infection Compromise Tissue-Specific Immunity. Cell. 2015;163:354–366. doi: 10.1016/j.cell.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch S.D., Weichhart T. Effects of interferons and viruses on metabolism. Front. Immunol. 2016;7:1–13. doi: 10.3389/fimmu.2016.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshan K., Nikkanen J., Man K., Leong Y.A., Sogawa Y., Maschek J.A., Van Ry T., Chagwedera D.N., Cox J.E., Chawla A. Energetic Trade-Offs and Hypometabolic States Promote Disease Tolerance. Cell. 2019;177:399–413.e12. doi: 10.1016/j.cell.2019.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Sanderson S.M., Dai Z., Reid M.A., Cooper D.E., Lu M., Richie J.P., Jr., Ciccarella A., Calcagnotto A., Mikhael P.G., et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature. 2019;572:397–401. doi: 10.1038/s41586-019-1437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R., Rieckmann J.C., Wolf T., Basso C., Feng Y., Fuhrer T., Kogadeeva M., Picotti P., Meissner F., Mann M., et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell. 2016;167:829–842.e13. doi: 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazarian M., Revelo X.S., Nøhr M.K., Luck H., Zeng K., Lei H., Tsai S., Schroer S.A., Park Y.J., Chng M.H.Y., et al. Type I interferon responses drive intrahepatic T cells to promote metabolic syndrome. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aai7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg E.L., Molony R.D., Kudo E., Sidorov S., Kong Y., Dixit V.D., Iwasaki A. Ketogenic diet activates protective γδ T cell responses against influenza virus infection. Sci. Immunol. 2019;4:eaav2026. doi: 10.1126/sciimmunol.aav2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.C., Schulte-Mecklenbeck A., Klinsing S., Posevitz-Fejfár A., Wiendl H., Klotz L. Dimethyl fumarate treatment alters circulating T helper cell subsets in multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2015;3:e183. doi: 10.1212/NXI.0000000000000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghikia A., Jörg S., Duscha A., Berg J., Manzel A., Waschbisch A., Hammer A., Lee D.-H., May C., Wilck N., et al. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity. 2015;43:817–829. doi: 10.1016/j.immuni.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Halpern K.B., Shenhav R., Matcovitch-Natan O., Tóth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E., et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.-J., Glatman Zaretsky A., Andrade-Oliveira V., Collins N., Dzutsev A., Shaik J., Morais da Fonseca D., Harrison O.J., Tamoutounour S., Byrd A.L., et al. White Adipose Tissue Is a Reservoir for Memory T Cells and Promotes Protective Memory Responses to Infection. Immunity. 2017;47:1154–1168.e6. doi: 10.1016/j.immuni.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W., et al. Construction of a human cell landscape at single-cell level. Nature. 2020;581:303–309. doi: 10.1038/s41586-020-2157-4. [DOI] [PubMed] [Google Scholar]
- Hart B.L. Biological basis of the behavior of sick animals. Neurosci. Biobehav. Rev. 1988;12:123–137. doi: 10.1016/s0149-7634(88)80004-6. [DOI] [PubMed] [Google Scholar]
- Hartmann F.J., Bendall S.C. Immune monitoring using mass cytometry and related high-dimensional imaging approaches. Nat. Rev. Rheumatol. 2020;16:87–99. doi: 10.1038/s41584-019-0338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R.M., Erbe R.W. High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc. Natl. Acad. Sci. USA. 1976;73:1523–1527. doi: 10.1073/pnas.73.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., Gordon J.I. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G.S. Nature Publishing Group; 2017. Inflammation, metaflammation and immunometabolic disorders. [DOI] [PubMed] [Google Scholar]
- Hotamisligil G.S., Shargill N.S., Spiegelman B.M. Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- Houtkooper R.H., Argmann C., Houten S.M., Cantó C., Jeninga E.H., Andreux P.A., Thomas C., Doenlen R., Schoonjans K., Auwerx J. The metabolic footprint of aging in mice. Sci. Rep. 2011;1:134. doi: 10.1038/srep00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoytema van Konijnenburg D.P., Reis B.S., Pedicord V.A., Farache J., Victora G.D., Mucida D. Intestinal Epithelial and Intraepithelial T Cell Crosstalk Mediates a Dynamic Response to Infection. Cell. 2017;171:783–794.e13. doi: 10.1016/j.cell.2017.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.Y., Crawley S., Chen M., Ayupova D.A., Lindhout D.A., Higbee J., Kutach A., Joo W., Gao Z., Fu D., et al. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature. 2017;550:255–259. doi: 10.1038/nature24042. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Kinoshita M., Kayama H., Nagamori S., Kongpracha P., Umemoto E., Okumura R., Kurakawa T., Murakami M., Mikami N., et al. Slc3a2 Mediates Branched-Chain Amino-Acid-Dependent Maintenance of Regulatory T Cells. Cell Rep. 2017;21:1824–1838. doi: 10.1016/j.celrep.2017.10.082. [DOI] [PubMed] [Google Scholar]
- Jaitin D.A., Adlung L., Thaiss C.A., Weiner A., Li B., Descamps H., Lundgren P., Bleriot C., Liu Z., Deczkowska A., et al. Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell. 2019;178:686–698.e14. doi: 10.1016/j.cell.2019.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C., Chen L., Rabinowitz J.D. Metabolomics and Isotope Tracing. Cell. 2018;173:822–837. doi: 10.1016/j.cell.2018.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A.K., Huang S.C.C., Sergushichev A., Lampropoulou V., Ivanova Y., Loginicheva E., Chmielewski K., Stewart K.M., Ashall J., Everts B., et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Johnen H., Lin S., Kuffner T., Brown D.A., Tsai V.W.-W., Bauskin A.R., Wu L., Pankhurst G., Jiang L., Junankar S., et al. Tumor-induced anorexia and weight loss are mediated by the TGF-beta superfamily cytokine MIC-1. Nat. Med. 2007;13:1333–1340. doi: 10.1038/nm1677. [DOI] [PubMed] [Google Scholar]
- Johnson M.O., Wolf M.M., Madden M.Z., Andrejeva G., Sugiura A., Contreras D.C., Maseda D., Liberti M.V., Paz K., Kishton R.J., et al. Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism. Cell. 2018;175:1780–1795.e19. doi: 10.1016/j.cell.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S., Tung N., Casanova-Acebes M., Chang C., Cantoni C., Zhang D., Wirtz T.H., Naik S., Rose S.A., Brocker C.N., et al. Dietary Intake Regulates the Circulating Inflammatory Monocyte Pool. Cell. 2019;178:1102–1114.e17. doi: 10.1016/j.cell.2019.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungermann K., Sasse D. Heterogeneity of liver parenchymal cells. Trends Biochem. Sci. 1978;3:198–202. [Google Scholar]
- Kanarek N., Petrova B., Sabatini D.M. Dietary modifications for enhanced cancer therapy. Nature. 2020;579:507–517. doi: 10.1038/s41586-020-2124-0. [DOI] [PubMed] [Google Scholar]
- Karreman M.A., Hyenne V., Schwab Y., Goetz J.G. Intravital Correlative Microscopy: Imaging Life at the Nanoscale. Trends Cell Biol. 2016;26:848–863. doi: 10.1016/j.tcb.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Kennedy E.A., King K.Y., Baldridge M.T. Mouse microbiota models: Comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 2018;9:1534. doi: 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietzmann T. Metabolic zonation of the liver: The oxygen gradient revisited. Redox Biol. 2017;11:622–630. doi: 10.1016/j.redox.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Geltink R.I., O’Sullivan D., Corrado M., Bremser A., Buck M.D., Buescher J.M., Firat E., Zhu X., Niedermann G., Caputa G., et al. Mitochondrial Priming by CD28. Cell. 2017;171:385–397.e11. doi: 10.1016/j.cell.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T., Huang C.H. Updates in the Clinical Development of Epacadostat and Other Indoleamine 2,3-Dioxygenase 1 Inhibitors (IDO1) for Human Cancers. Front. Oncol. 2018;8:423. doi: 10.3389/fonc.2018.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg M.D., Bhargava P., Kim P.M., Putluri V., Snowman A.M., Putluri N., Calabresi P.A., Snyder S.H. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science. 2018;360:449–453. doi: 10.1126/science.aan4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausgruber T., Fortelny N., Fife-Gernedl V., Senekowitsch M., Schuster L.C., Lercher A., Nemc A., Schmidl C., Rendeiro A.F., Bergthaler A., Bock C. Structural cells are key regulators of organ-specific immune responses. Nature. 2020;583:296–302. doi: 10.1038/s41586-020-2424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M., Schaller S., Borchers S., Findeisen R., Lippert J., Kuepfer L. Integrating cellular metabolism into a multiscale whole-body model. PLoS Comput. Biol. 2012;8:e1002750. doi: 10.1371/journal.pcbi.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs H.A., Henesleit K. Vol. 11. 1932. Untersuchungen ueber die Harnstoffbildung Tierkoerper Klinische Wochenschrift; pp. 757–759. [Google Scholar]
- Kubes P., Jenne C. Immune Responses in the Liver. Annu. Rev. Immunol. 2018;36:247–277. doi: 10.1146/annurev-immunol-051116-052415. [DOI] [PubMed] [Google Scholar]
- Kurachi M., Ngiow S.F., Kurachi J., Chen Z., Wherry E.J. Hidden Caveat of Inducible Cre Recombinase. Immunity. 2019;51:591–592. doi: 10.1016/j.immuni.2019.09.010. [DOI] [PubMed] [Google Scholar]
- Lammertsma A.A. Forward to the past: The case for quantitative PET imaging. J. Nucl. Med. 2017;58:1019–1024. doi: 10.2967/jnumed.116.188029. [DOI] [PubMed] [Google Scholar]
- Lampropoulou V., Sergushichev A., Bambouskova M., Nair S., Vincent E.E., Loginicheva E., Cervantes-Barragan L., Ma X., Huang S.C.C., Griss T., et al. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 2016;24:158–166. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukens D., Brinkman B.M., Raes J., De Vos M., Vandenabeele P. Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol. Rev. 2016;40:117–132. doi: 10.1093/femsre/fuv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzer S., Bedogni G., Lafortuna C.L., Marazzi N., Busti C., Galli R., De Col A., Agosti F., Sartorio A. Relationship between basal metabolic rate, gender, age, and body composition in 8,780 white obese subjects. Obesity (Silver Spring) 2010;18:71–78. doi: 10.1038/oby.2009.162. [DOI] [PubMed] [Google Scholar]
- Legoux F., Bellet D., Daviaud C., El Morr Y., Darbois A., Niort K., Procopio E., Salou M., Gilet J., Ryffel B., et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 2019;366:494–499. doi: 10.1126/science.aaw2719. [DOI] [PubMed] [Google Scholar]
- Lercher A., Bhattacharya A., Popa A.M., Caldera M., Schlapansky M.F., Baazim H., Agerer B., Gürtl B., Kosack L., Májek P., et al. Type I Interferon Signaling Disrupts the Hepatic Urea Cycle and Alters Systemic Metabolism to Suppress T Cell Function. Immunity. 2019;51:1074–1087.e9. doi: 10.1016/j.immuni.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H.H., Wang A., Hilliard B.K., Carvalho F., Rosen C.E., Ahasic A.M., Herzog E.L., Kang I., Pisani M.A., Yu S., et al. GDF15 Is an Inflammation-Induced Central Mediator of Tissue Tolerance. Cell. 2019;178:1231–1244.e11. doi: 10.1016/j.cell.2019.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Hu H., Chang R., Zhong J., Knabel M., O’Meally R., Cole R.N., Pandey A., Semenza G.L. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E.H., Bantug G., Griss T., Condotta S., Johnson R.M., Samborska B., Mainolfi N., Suri V., Guak H., Balmer M.L., et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab. 2017;25:345–357. doi: 10.1016/j.cmet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Ma E.H., Verway M.J., Johnson R.M., Roy D.G., Steadman M., Hayes S., Williams K.S., Sheldon R.D., Samborska B., Kosinski P.A., et al. Metabolic Profiling Using Stable Isotope Tracing Reveals Distinct Patterns of Glucose Utilization by Physiologically Activated CD8+ T Cells. Immunity. 2019;51:856–870.e5. doi: 10.1016/j.immuni.2019.09.003. [DOI] [PubMed] [Google Scholar]
- Marcos A., Nova E., Montero A. Changes in the immune system are conditioned by nutrition. Eur. J. Clin. Nutr. 2003;57(Suppl 1):S66–S69. doi: 10.1038/sj.ejcn.1601819. [DOI] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil N.I. The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr. 1984;39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., Barabási A.L. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347 doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet X., Dyck L., Hogan A., Loftus R.M., Duquette D., Wei K., Beyaz S., Tavakkoli A., Foley C., Donnelly R., et al. Metabolic reprogramming of natural killer cells in obesity limits antitumor responses. Nat. Immunol. 2018;19:1330–1340. doi: 10.1038/s41590-018-0251-7. [DOI] [PubMed] [Google Scholar]
- Miller S.L. A production of amino acids under possible primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- Miller D.R. A tribute to Sidney Farber-- the father of modern chemotherapy. Br. J. Haematol. 2006;134:20–26. doi: 10.1111/j.1365-2141.2006.06119.x. [DOI] [PubMed] [Google Scholar]
- Miller A., Nagy C., Knapp B., Laengle J., Ponweiser E., Groeger M., Starkl P., Bergmann M., Wagner O., Haschemi A. Exploring Metabolic Configurations of Single Cells within Complex Tissue Microenvironments. Cell Metab. 2017;26:788–800.e6. doi: 10.1016/j.cmet.2017.08.014. [DOI] [PubMed] [Google Scholar]
- Mills E.L., Kelly B., Logan A., Costa A.S.H., Varma M., Bryant C.E., Tourlomousis P., Däbritz J.H.M., Gottlieb E., Latorre I., et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell. 2016;167:457–470.e13. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.L., Ryan D.G., Prag H.A., Dikovskaya D., Menon D., Zaslona Z., Jedrychowski M.P., Costa A.S.H., Higgins M., Hams E., et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura D., Fujimura Y., Wariishi H. In situ metabolomic mass spectrometry imaging: recent advances and difficulties. J. Proteomics. 2012;75:5052–5060. doi: 10.1016/j.jprot.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Miyajima M., Zhang B., Sugiura Y., Sonomura K., Guerrini M.M., Tsutsui Y., Maruya M., Vogelzang A., Chamoto K., Honda K., et al. Metabolic shift induced by systemic activation of T cells in PD-1-deficient mice perturbs brain monoamines and emotional behavior. Nat. Immunol. 2017;18:1342–1352. doi: 10.1038/ni.3867. [DOI] [PubMed] [Google Scholar]
- Moffitt J.R., Bambah-Mukku D., Eichhorn S.W., Vaughn E., Shekhar K., Perez J.D., Rubinstein N.D., Hao J., Regev A., Dulac C., et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362 doi: 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor A.E., Itzkovitz S. Spatial transcriptomics: paving the way for tissue-level systems biology. Curr. Opin. Biotechnol. 2017;46:126–133. doi: 10.1016/j.copbio.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Mrowietz U., Szepietowski J.C., Loewe R., van de Kerkhof P., Lamarca R., Ocker W.G., Tebbs V.M., Pau-Charles I. Efficacy and safety of LAS41008 (dimethyl fumarate) in adults with moderate-to-severe chronic plaque psoriasis: a randomized, double-blind, Fumaderm® - and placebo-controlled trial (BRIDGE) Br. J. Dermatol. 2017;176:615–623. doi: 10.1111/bjd.14947. [DOI] [PubMed] [Google Scholar]
- Muehlenbein M.P., Hirschtick J.L., Bonner J.Z., Swartz A.M. Toward quantifying the usage costs of human immunity: Altered metabolic rates and hormone levels during acute immune activation in men. Am. J. Hum. Biol. 2010;22:546–556. doi: 10.1002/ajhb.21045. [DOI] [PubMed] [Google Scholar]
- Murray P.J. Amino acid auxotrophy as a system of immunological control nodes. Nat. Immunol. 2016;17:132–139. doi: 10.1038/ni.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.J., Murray A.B. Anorexia of infection as a mechanism of host defense. Am. J. Clin. Nutr. 1979;32:593–596. doi: 10.1093/ajcn/32.3.593. [DOI] [PubMed] [Google Scholar]
- Murray C., Griffin É.W., O’Loughlin E., Lyons A., Sherwin E., Ahmed S., Stevenson N.J., Harkin A., Cunningham C. Interdependent and independent roles of type I interferons and IL-6 in innate immune, neuroinflammatory and sickness behaviour responses to systemic poly I:C. Brain Behav. Immun. 2015;48:274–286. doi: 10.1016/j.bbi.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M., Xiao Y., Zhou X., Chang J.-H., Chang M., Cheng X., Blonska M., Lin X., Sun S.-C. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norata G.D.D., Caligiuri G., Chavakis T., Matarese G., Netea M.G.G., Nicoletti A., O’neill L.A.J.J., Marelli-Berg F.M.M., O’Neill L.A.J., Marelli-Berg F.M.M. Elsevier Inc.; 2015. The Cellular and Molecular Basis of Translational Immunometabolism. [DOI] [PubMed] [Google Scholar]
- O’Neill L.A.J., Artyomov M.N. Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 2019;19:273–281. doi: 10.1038/s41577-019-0128-5. [DOI] [PubMed] [Google Scholar]
- Okin D., Medzhitov R. The Effect of Sustained Inflammation on Hepatic Mevalonate Pathway Results in Hyperglycemia. Cell. 2016;165:343–356. doi: 10.1016/j.cell.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Villagómez D., Van Kaer L. Intestinal Intraepithelial Lymphocytes: Sentinels of the Mucosal Barrier. Trends Immunol. 2018;39:264–275. doi: 10.1016/j.it.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabha D., Baker K.D. Drosophila gains traction as a repurposed tool to investigate metabolism. Trends Endocrinol. Metab. 2014;25:518–527. doi: 10.1016/j.tem.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Pålsson-McDermott E.M., O’Neill L.A.J. Targeting immunometabolism as an anti-inflammatory strategy. Cell Res. 2020;30:300–314. doi: 10.1038/s41422-020-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott E.M., Curtis A.M., Goel G., Lauterbach M.A.R., Sheedy F.J., Gleeson L.E., van den Bosch M.W.M., Quinn S.R., Domingo-Fernandez R., Johnston D.G., et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel A., Teixeira A., Haddad E., Wood E.G., Steinman R.M., Longhi M.P. Direct type I IFN but not MDA5/TLR3 activation of dendritic cells is required for maturation and metabolic shift to glycolysis after poly IC stimulation. PLoS Biol. 2014;12:e1001759. doi: 10.1371/journal.pbio.1001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek V., Tian H., Winograd N., Benkovic S.J. Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science. 2020;368:283–290. doi: 10.1126/science.aaz6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., Beghini F., Manghi P., Tett A., Ghensi P., et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell. 2019;176:649–662.e20. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H.J., Patel B.M. TNF-α and cancer cachexia: Molecular insights and clinical implications. Life Sci. 2017;170:56–63. doi: 10.1016/j.lfs.2016.11.033. [DOI] [PubMed] [Google Scholar]
- Patel S., Alvarez-Guaita A., Melvin A., Rimmington D., Dattilo A., Miedzybrodzka E.L., Cimino I., Maurin A.C., Roberts G.P., Meek C.L., et al. GDF15 Provides an Endocrine Signal of Nutritional Stress in Mice and Humans. Cell Metab. 2019;29:707–718.e8. doi: 10.1016/j.cmet.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.J., Everts B. Dendritic cell metabolism. Nat. Rev. Immunol. 2015;15:18–29. doi: 10.1038/nri3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Pearce E.J. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Walsh M.C., Cejas P.J., Harms G.M., Shen H., Wang L.-S., Jones R.G., Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.J., Camporez J.G., Kursawe R., Titchenell P.M., Zhang D., Perry C.J., Jurczak M.J., Abudukadier A., Han M.S., Zhang X.-M.M., et al. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell. 2015;160:745–758. doi: 10.1016/j.cell.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A.T., Doedens A.L., Palazon A., Tyrakis P.A., Cheung K.P., Johnson R.S., Goldrath A.W. Constitutive Glycolytic Metabolism Supports CD8+ T Cell Effector Memory Differentiation during Viral Infection. Immunity. 2016;45:1024–1037. doi: 10.1016/j.immuni.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G.C., Malachowski W.P., DuHadaway J.B., Muller A.J. Discovery of IDO1 Inhibitors: From Bench to Bedside. Cancer Res. 2017;77:6795–6811. doi: 10.1158/0008-5472.CAN-17-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest C., Tontonoz P. Inter-organ cross-talk in metabolic syndrome. Nat. Metab. 2019;1:1177–1188. doi: 10.1038/s42255-019-0145-5. [DOI] [PubMed] [Google Scholar]
- Protzer U., Maini M.K., Knolle P.A. Nature Publishing Group; 2012. Living in the liver: Hepatic infections. [DOI] [PubMed] [Google Scholar]
- Pryor R., Norvaisas P., Marinos G., Best L., Thingholm L.B., Quintaneiro L.M., De Haes W., Esser D., Waschina S., Lujan C., et al. Host-Microbe-Drug-Nutrient Screen Identifies Bacterial Effectors of Metformin Therapy. Cell. 2019;178:1299–1312.e29. doi: 10.1016/j.cell.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Villa M., Sanin D.E., Buck M.D., O’Sullivan D., Ching R., Matsushita M., Grzes K.M., Winkler F., Chang C.-H., et al. Acetate Promotes T Cell Effector Function during Glucose Restriction. Cell Rep. 2019;27:2063–2074.e5. doi: 10.1016/j.celrep.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn R.A., Melnik A.V., Vrbanac A., Fu T., Patras K.A., Christy M.P., Bodai Z., Belda-Ferre P., Tripathi A., Chung L.K., et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature. 2020;579:123–129. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racanelli V., Rehermann B. The liver as an immunological organ. Hepatology. 2006;43(2, Suppl 1):S54–S62. doi: 10.1002/hep.21060. [DOI] [PubMed] [Google Scholar]
- Raez L.E., Papadopoulos K., Ricart A.D., Chiorean E.G., Dipaola R.S., Stein M.N., Rocha Lima C.M., Schlesselman J.J., Tolba K., Langmuir V.K., et al. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2013;71:523–530. doi: 10.1007/s00280-012-2045-1. [DOI] [PubMed] [Google Scholar]
- Rao S., Schieber A.M.P., O’Connor C.P., Leblanc M., Michel D., Ayres J.S. Pathogen-Mediated Inhibition of Anorexia Promotes Host Survival and Transmission. Cell. 2017;168:503–516.e12. doi: 10.1016/j.cell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappez L., Stadler M., Triana S., Phapale P., Heikenwalder M., Alexandrov T. Spatial single-cell profiling of intracellular metabolomes in situ. bioRxiv. 2019 doi: 10.1101/510222. [DOI] [Google Scholar]
- Raud B., Roy D.G., Divakaruni A.S., Tarasenko T.N., Franke R., Ma E.H., Samborska B., Hsieh W.Y., Wong A.H., Stüve P., et al. Etomoxir Actions on Regulatory and Memory T Cells Are Independent of Cpt1a-Mediated Fatty Acid Oxidation. Cell Metab. 2018;28:504–515.e7. doi: 10.1016/j.cmet.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann B., Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- Rieckmann J.C., Geiger R., Hornburg D., Wolf T., Kveler K., Jarrossay D., Sallusto F., Shen-Orr S.S., Lanzavecchia A., Mann M., Meissner F. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 2017;18:583–593. doi: 10.1038/ni.3693. [DOI] [PubMed] [Google Scholar]
- Robinson M.W., Harmon C., O’Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell. Mol. Immunol. 2016;13:267–276. doi: 10.1038/cmi.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F., Macosko E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronda C., Chen S.P., Cabral V., Yaung S.J., Wang H.H. Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods. 2019;16:167–170. doi: 10.1038/s41592-018-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E.D., Spiegelman B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart S.P., Vassallo B.G., Angeletti D., Hutchinson D.S., Morgan A.P., Takeda K., Hickman H.D., McCulloch J.A., Badger J.H., Ajami N.J., et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171:1015–1028.e13. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart S.P., Herz J., Vassallo B.G., Hunter A., Wall M.K., Badger J.H., McCulloch J.A., Anastasakis D.G., Sarshad A.A., Leonardi I., et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 2019;365:80. doi: 10.1126/science.aaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D., Costea P.I., Godneva A., Kalka I.N., Bar N., et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- Roy D.G., Chen J., Mamane V., Ma E.H., Muhire B.M., Sheldon R.D., Shorstova T., Koning R., Johnson R.M., Esaulova E., et al. Methionine Metabolism Shapes T Helper Cell Responses through Regulation of Epigenetic Reprogramming. Cell Metab. 2020;31:250–266.e9. doi: 10.1016/j.cmet.2020.01.006. [DOI] [PubMed] [Google Scholar]
- Russell D.G., Huang L., VanderVen B.C. Immunometabolism at the interface between macrophages and pathogens. Nat. Rev. Immunol. 2019;19:291–304. doi: 10.1038/s41577-019-0124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos I., Colaço H.G., Neves-Costa A., Seixas E., Velho T.R., Pedroso D., Barros A., Martins R., Carvalho N., Payen D., et al. CXCL5-mediated recruitment of neutrophils into the peritoneal cavity of <em>Gdf15</em>-deficient mice protects against abdominal sepsis. bioRxiv. 2019 doi: 10.1101/613901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., Sabatini D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheja L., Heeren J. The endocrine function of adipose tissues in health and cardiometabolic disease. Nat. Rev. Endocrinol. 2019;15:507–524. doi: 10.1038/s41574-019-0230-6. [DOI] [PubMed] [Google Scholar]
- Schieber A.M.P., Ayres J.S. Thermoregulation as a disease tolerance defense strategy. Pathog. Dis. 2016;74:1–15. doi: 10.1093/femspd/ftw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper H.S., Prakken B., Kalkhoven E., Boes M. Adipose tissue-resident immune cells: key players in immunometabolism. Trends Endocrinol. Metab. 2012;23:407–415. doi: 10.1016/j.tem.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdelci S., Rendeiro A.F., Rathert P., You W., Lin J.G., Ringler A., Hofstätter G., Moll H.P., Gürtl B., Farlik M., et al. MTHFD1 interaction with BRD4 links folate metabolism to transcriptional regulation. Nat. Genet. 2019;51:990–998. doi: 10.1038/s41588-019-0413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Chapman N.M., Wen J., Guy C., Long L., Dhungana Y., Rankin S., Pelletier S., Vogel P., Wang H., et al. Amino Acids License Kinase mTORC1 Activity and Treg Cell Function via Small G Proteins Rag and Rheb. Immunity. 2019;51:1012–1027.e7. doi: 10.1016/j.immuni.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair L.V., Rolf J., Emslie E., Shi Y.B., Taylor P.M., Cantrell D.A. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallanzani R.G., Zemmour D., Xiao T., Jayewickreme T., Li C., Bryce P.J., Benoist C., Mathis D. Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman J.R. Measuring energy metabolism in the mouse - theoretical, practical, and analytical considerations. Front. Physiol. 2013;4:34. doi: 10.3389/fphys.2013.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 2016;21:75–80. doi: 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl P.L., Salmén F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., Giacomello S., Asp M., Westholm J.O., Huss M., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- Stark R., Grzelak M., Hadfield J. RNA sequencing: the teenage years. Nat. Rev. Genet. 2019;20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- Stein M., Lin H., Jeyamohan C., Dvorzhinski D., Gounder M., Bray K., Eddy S., Goodin S., White E., Dipaola R.S. Targeting tumor metabolism with 2-deoxyglucose in patients with castrate-resistant prostate cancer and advanced malignancies. Prostate. 2010;70:1388–1394. doi: 10.1002/pros.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein E.M., DiNardo C.D., Fathi A.T., Pollyea D.A., Stone R.M., Altman J.K., Roboz G.J., Patel M.R., Collins R., Flinn I.W., et al. Molecular remission and response patterns in patients with mutant-IDH2 acute myeloid leukemia treated with enasidenib. Blood. 2019;133:676–687. doi: 10.1182/blood-2018-08-869008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub R.H., Cutolo M., Buttgereit F., Pongratz G. Energy regulation and neuroendocrine-immune control in chronic inflammatory diseases. J. Intern. Med. 2010;267:543–560. doi: 10.1111/j.1365-2796.2010.02218.x. [DOI] [PubMed] [Google Scholar]
- Stubbington M.J.T., Rozenblatt-Rosen O., Regev A., Teichmann S.A. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358:58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Li T., Song X., Huang L., Zang Q., Xu J., Bi N., Jiao G., Hao Y., Chen Y., et al. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. USA. 2019;116:52–57. doi: 10.1073/pnas.1808950116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C., Rao T.N., Winnay J.N., Garcia-Martin R., Grinspoon S.K., et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefts E., Gannon M., Wasserman D.H. The liver. Curr. Biol. 2017;27:R1147–R1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A., Debelius J., Brenner D.A., Karin M., Loomba R., Schnabl B., Knight R. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A., Gollwitzer E.S., Pattaroni C., Lopez-Mejia I.C., Riva E., Pernot J., Ubags N., Fajas L., Nicod L.P., Marsland B.J. Dietary Fiber Confers Protection against Flu by Shaping Ly6c- Patrolling Monocyte Hematopoiesis and CD8+ T Cell Metabolism. Immunity. 2018;48:992–1005.e8. doi: 10.1016/j.immuni.2018.04.022. [DOI] [PubMed] [Google Scholar]
- Tsai V.W.W., Lin S., Brown D.A., Salis A., Breit S.N. Anorexia-cachexia and obesity treatment may be two sides of the same coin: role of the TGF-b superfamily cytokine MIC-1/GDF15. Int. J. Obes. 2016;40:193–197. doi: 10.1038/ijo.2015.242. [DOI] [PubMed] [Google Scholar]
- Tsai V.W.W., Husaini Y., Sainsbury A., Brown D.A., Breit S.N. The MIC-1/GDF15-GFRAL Pathway in Energy Homeostasis: Implications for Obesity, Cachexia, and Other Associated Diseases. Cell Metab. 2018;28:353–368. doi: 10.1016/j.cmet.2018.07.018. [DOI] [PubMed] [Google Scholar]
- Tschöp M.H., Speakman J.R., Arch J.R.S., Auwerx J., Brüning J.C., Chan L., Eckel R.H., Farese R.V., Jr., Galgani J.E., Hambly C., et al. A guide to analysis of mouse energy metabolism. Nat. Methods. 2011;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura Y., Fuhrer T., Li H., Lawson M.A., Zimmermann M., Yilmaz B., Zindel J., Ronchi F., Sorribas M., Hapfelmeier S., et al. Antibodies Set Boundaries Limiting Microbial Metabolite Penetration and the Resultant Mammalian Host Response. Immunity. 2018;49:545–559.e5. doi: 10.1016/j.immuni.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde L.-A., Guo X.J., Barbaric L., Smith A.M., Oguin T.H., 3rd, Thomas P.G., Murray P.J. Stress Kinase GCN2 Controls the Proliferative Fitness and Trafficking of Cytotoxic T Cells Independent of Environmental Amino Acid Sensing. Cell Rep. 2016;17:2247–2258. doi: 10.1016/j.celrep.2016.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berghe G. The role of the liver in metabolic homeostasis: implications for inborn errors of metabolism. J. Inherit. Metab. Dis. 1991;14:407–420. doi: 10.1007/BF01797914. [DOI] [PubMed] [Google Scholar]
- Van den Eynde B.J., van Baren N., Baurain J.-F. Is There a Clinical Future for IDO1 Inhibitors After the Failure of Epacadostat in Melanoma? Annual Review of Cancer Biology. 2020;4:241–256. [Google Scholar]
- Vaquero J.J., Kinahan P. Positron Emission Tomography: Current Challenges and Opportunities for Technological Advances in Clinical and Preclinical Imaging Systems. Annu. Rev. Biomed. Eng. 2015;17:385–414. doi: 10.1146/annurev-bioeng-071114-040723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickovic S., Eraslan G., Salmén F., Klughammer J., Stenbeck L., Schapiro D., Äijö T., Bonneau R., Bergenstråhle L., Navarro J.F., et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconti A., Le Roy C.I., Rosa F., Rossi N., Martin T.C., Mohney R.P., Li W., de Rinaldis E., Bell J.T., Venter J.C., et al. Interplay between the human gut microbiome and host metabolism. Nat. Commun. 2019;10:4505. doi: 10.1038/s41467-019-12476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Huen S.C., Luan H.H., Yu S., Zhang C., Gallezot J.D., Booth C.J., Medzhitov R. Opposing Effects of Fasting Metabolism on Tissue Tolerance in Bacterial and Viral Inflammation. Cell. 2016;166:1512–1525.e12. doi: 10.1016/j.cell.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Luan H.H., Medzhitov R. An evolutionary perspective on immunometabolism. Science. 2019;363:80. doi: 10.1126/science.aar3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the Origin of Cancer Cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Warburg O., Wind F., Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O., Gawehn K., Geissler A.W. Stoffwechsel der weißen Blutzellen. Zeitschrift Fur Naturforsch. B. 1958;13 doi: 10.1515/znb-1958-0806. [DOI] [PubMed] [Google Scholar]
- Weber D.D., Aminazdeh-Gohari S., Kofler B. Ketogenic diet in cancer therapy. Aging (Albany N.Y.) 2018;10:164–165. doi: 10.18632/aging.101382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger B.D., Gobet C., Yeung J., Martin E., Jimenez S., Betrisey B., Foata F., Berger B., Balvay A., Foussier A., et al. The Mouse Microbiome Is Required for Sex-Specific Diurnal Rhythms of Gene Expression and Metabolism. Cell Metab. 2019;29:362–382.e8. doi: 10.1016/j.cmet.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S.P., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick A.N., Drury D.R., Nakada H.I., Wolfe J.B. Localization of the primary metabolic block produced by 2-deoxyglucose. J. Biol. Chem. 1957;224:963–969. [PubMed] [Google Scholar]
- Wieman H.L., Wofford J.A., Rathmell J.C. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol. Biol. Cell. 2007;18:1437–1446. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Sanin D.E.E., Everts B., Chen Q., Qiu J., Buck M.D.D., Patterson A., Smith A.M.M., Chang C.-H.H., Liu Z., et al. Type 1 Interferons Induce Changes in Core Metabolism that Are Critical for Immune Function. Immunity. 2016;44:1325–1336. doi: 10.1016/j.immuni.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymore Brand M., Wannemuehler M.J., Phillips G.J., Proctor A., Overstreet A.-M., Jergens A.E., Orcutt R.P., Fox J.G. The Altered Schaedler Flora: Continued Applications of a Defined Murine Microbial Community. ILAR J. 2015;56:169–178. doi: 10.1093/ilar/ilv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Grijalva A., Skowronski A., van Eijk M., Serlie M.J., Ferrante A.W., Jr. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K.E., Bittinger M.A., Su S.M., Fantin V.R. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- Yen K., Travins J., Wang F., David M.D., Artin E., Straley K., Padyana A., Gross S., DeLaBarre B., Tobin E., et al. AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations. Cancer Discov. 2017;7:478–493. doi: 10.1158/2159-8290.CD-16-1034. [DOI] [PubMed] [Google Scholar]
- Yin Y., Choi S.-C., Xu Z., Perry D.J., Seay H., Croker B.P., Sobel E.S., Brusko T.M., Morel L. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 2015;7:274ra18. doi: 10.1126/scitranslmed.aaa0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York A.G., Williams K.J., Argus J.P., Zhou Q.D., Brar G., Vergnes L., Gray E.E., Zhen A., Wu N.C., Yamada D.H., et al. Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling. Cell. 2015;163:1716–1729. doi: 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Jin W., Wu R., Li J., Park S.-A., Tu E., Zanvit P., Xu J., Liu O., Cain A., Chen W. High Glucose Intake Exacerbates Autoimmunity through Reactive-Oxygen-Species-Mediated TGF-β Cytokine Activation. Immunity. 2019;51:671–681.e5. doi: 10.1016/j.immuni.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhang S., Liu Y., He X., Qu M., Xu G., Wang H., Huang M., Pan J., Liu Z., et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 2020;6:22. doi: 10.1038/s41421-020-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Xu M.J., Gao B. Hepatocytes: a key cell type for innate immunity. Cell. Mol. Immunol. 2016;13:301–315. doi: 10.1038/cmi.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A., Lee D., Shim H. Metabolic positron emission tomography imaging in cancer detection and therapy response. Semin. Oncol. 2011;38:55–69. doi: 10.1053/j.seminoncol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570:462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]