Abstract
To rapidly evaluate the safety and efficacy of COVID-19 vaccine candidates, prioritizing vaccine trial sites in areas with high expected disease incidence can speed endpoint accrual and shorten trial duration. Mathematical and statistical forecast models can inform the process of site selection, integrating available data sources and facilitating comparisons across locations. We recommend the use of ensemble forecast modeling – combining projections from independent modeling groups – to guide investigators identifying suitable sites for COVID-19 vaccine efficacy trials. We describe an appropriate structure for this process, including minimum requirements, suggested output, and a user-friendly tool for displaying results. Importantly, we advise that this process be repeated regularly throughout the trial, to inform decisions about enrolling new participants at existing sites with waning incidence versus adding entirely new sites. These types of data-driven models can support the implementation of flexible efficacy trials tailored to the outbreak setting.
Keywords: Efficacy trial, Trial planning, Forecast model, Ensemble modeling
1. Introduction
The COVID-19 pandemic is a public health emergency, and there is an urgent need for effective vaccines to limit morbidity and mortality. Efforts are underway to accelerate all steps in the vaccine development pathway [1]. Large randomized field trials are crucial for determining the safety and efficacy of candidates to inform regulatory decisions [2]. In these trials, many thousands of eligible and consenting participants across multiple sites are enrolled and individually randomized to vaccine or control. These trials are event driven, where an expected primary endpoint is laboratory-confirmed symptomatic disease [3], with infection regardless of symptoms as a valuable secondary endpoint [4]. Selecting vaccine trial sites where disease incidence is highest during the study period can accelerate the accrual of endpoints.
Mathematical and statistical models are recognized as valuable tools for planning infectious disease clinical trials [5]. They can be used to optimize design features such as cluster size or to examine the validity of the trial’s statistical analysis [6]. The use of spatially explicit forecast models to select vaccine trial sites was first explored during the 2015–2016 Zika epidemic [7]. These forecast models synthesize available data to make projections about which sites might have the highest future disease incidence.
An important value of models is that they standardize projections across locations. Trends in raw reported numbers of cases depend heavily on the sensitivity of the underlying surveillance system. Case definitions and access to care and testing may vary over time and space. Models that integrate many data sources, such as reported cases, test positivity, hospitalizations and deaths, can facilitate more meaningful comparisons across locations. Forecasts provide estimates along with the uncertainty associated with those estimates to make best use of the available information.
Models can incorporate many features to capture the complex dynamics of infectious diseases. Incidence is expected to vary widely over time and between locations, as a function of control measures in place, patterns of introduction, seasonality, and other sources of variability. Mathematical models naturally account for prior circulation of the virus and any buildup of population-level immunity. Areas that have already experienced substantial outbreaks may be less suitable for inclusion, and this would be reflected in projections. Models can explicitly capture correlation due to movement between nearby sites or between sites and a common hub [8]. Models can also reflect relevant population-level features associated with expected incidence, such as density, race/ethnicity, age distribution, and educational status.
We recommend the use of ensemble modeling, whereby multiple modeling groups prepare independent projections and these are combined to guide decision-making. Individual models can be agent-based, compartmental, or statistical, can use different assumptions and data sources, but are all tasked with the same question of which sites are likely to have the highest disease incidence over a moderate time horizon. Ensemble modeling has been shown to be more robust for complex systems than specialist models and better able to capture the complete range of possible outcomes [9]. The strength of ensemble modeling has been shown for diseases like influenza [10], dengue [11] and Ebola [12]. Ensemble modeling for COVID-19, like the COVID-19 Forecast Hub, is similarly more robust [13].
In addition to using forecast modeling for initial site selection, we propose that modeling be repeated at regular intervals throughout the trial. In the context of outbreaks, trials should be flexible to allow new sites to be added in response to evolving epidemiology [14]. Some sites will have lower than projected incidence during the trial period. For example, local policies or voluntary changes in behavior could effectively reduce transmission, meaning that the site is no longer a “hotspot.” The modeling results can guide investigators deciding whether to continue to enroll new participants from existing sites, or to enroll new sites in emerging hotspots.
In this paper, we describe a simplified framework for the use of ensemble modeling to guide the selection and continued evaluation of sites for a vaccine efficacy trial, with a focus on the COVID-19 pandemic.
2. Ensemble modeling for trial planning
2.1. Individual model structure
Individual modeling teams are welcome to contribute to the consensus model. We assume that models would already be built for general public health planning, so they would not be constructed only for this effort, though they may need to be modified. Investigators can leverage existing groups or form new groups of modelers. Participating modeling teams would be provided with a list of all candidate sites being explored. This list of sites may be based on previous engagement between the trial investigators and potential research partners. For a multi-country trial, this may include several sites per country from multiple countries.
Participating models must meet a minimum set of requirements. Suggested guidelines are the ability to: (i) capture all geographic areas in the candidate list of sites, (ii) disaggregate to at least the first administrative level (e.g. state, province), though finer levels may be preferred for certain planning activities, (iii) project the COVID-19 symptomatic cumulative incidence, i.e. the number of new symptomatic infections of any severity divided by the total population size during a pre-specified period (three months suggested), and (iv) produce a minimum of 1000 simulated epidemics. Models must also be screened for internal consistency and basic plausibility when compared to historical trends.
For each site, each model must generate a probabilistic predictive distribution for quantities of interest, such as the symptomatic cumulative incidence. These are bins of 1% width centered around whole numbers [0.5, 1.5%), [1.5, 2.5%), [2.5, 3.5%) and so on. The bin that includes 0% is narrower [0, 0.5%).
2.2. Model aggregation
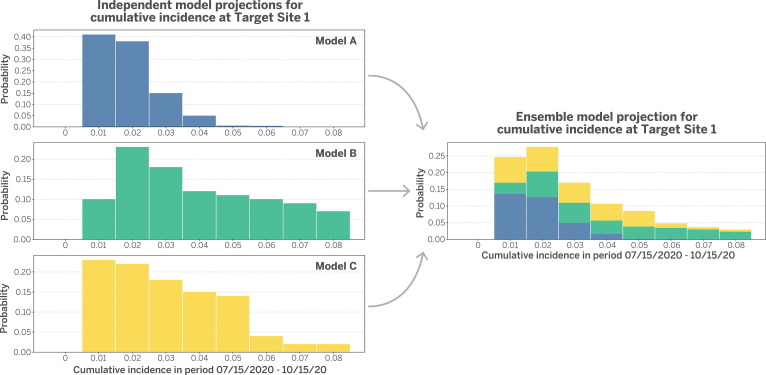
For each site, probabilistic predictive distributions are aggregated across models using stacking. Fig. 1 describes a hypothetical model stacking procedure for a target site, per Ray and Reich [9)] For simplicity and transparency, each model is assigned an equal weight, which is one over the number of models as done by the COVID-19 Forecast Hub [13]. If a participating team has developed more than one model, they must specify which model is primary and will contribute to the aggregate. More complex weighting schemes exist that preferentially weight models that performed best in previous rounds after an appropriate burn-in period [9].
Fig. 1.
Hypothetical model stacking procedure for a target trial site. The procedure integrates projections from three independent models. For each model, cumulative incidence during a target period is projected and then summarized in bins of 1% width (left). Models are equally weighted and then stacked in an ensemble model projection (right). This process is repeated for each site. Figure modeled after Ray and Reich (2018).
For each site, we can use the combined predictive distribution to produce summary statistics. Suggested summary statistics are: (i) median incidence value, (ii) 25th percentile incidence value, (iii) 75th percentile incidence value, and (iv) probability incidence value is [0, 0.5%) (probability of a very small or no outbreak).
To present this information in a way that is easy for trial investigators to explore, we recommend reporting stacked projections, summary statistics, and basic information about the sites in an interactive tool, like the R Shiny platform [15]. This allows the end user to sort the table or select rows for closer examination. In this way, they could select a subset of “best rows” and view these together to approximate the formation of a trial. Fig. 2 is a sample screenshot from such a program (code provided in Supplemental Materials).
Fig. 2.
Screenshot from an interactive tool to display ensemble model projections and associated summary statistics. The rows are sortable and can be selected or deselected to form a hypothetical trial. Additional columns can be added to describe site features that are useful to investigators.
By generating a range of possible outcomes, models can capture the stochasticity of future transmission, including scenarios where incidence is much lower or much higher than the median projection. Where incidence is highly variable with the potential to be very low, it may be preferable to include a larger number of sites to guard against the chance of accruing no efficacy data.
2.3. Site selection
The goal of ensemble modeling is to provide a simple and informative resource rather than a definitive recommendation. Investigators will simultaneously consider many operational, political, and scientific factors. To provide context, we describe several other key considerations.
To ensure a high-quality trial, sites should have adequate capacity for testing, safety monitoring, active surveillance, and high participant retention. Nonetheless, sites with projected high incidence but poorer capacity should not be excluded if there is a potential role for mobile trial teams, as was used in the Ebola ring vaccination trial in Guinea [16]. Approval for the trial may be at the national or sub-national level, with flexibility to identify the particular target population when investigators are ready to start enrollment.
For multi-country trials, investigators must weigh including multiple sites per country against including more countries. On one hand, given the complexity of country-specific procedures for approving clinical research, it may be easier to include multiple sites per country from fewer countries overall. On the other hand, the global community must ensure equitable access to potentially effective vaccines. Broad representation also increases generalizability of the trial results, as it can best capture the effectiveness of vaccine candidates in diverse settings. These include variations in population age profile, race/ethnicities, climate, background presence of non-pharmaceutical interventions, and co-circulation of other coronaviruses.
Including many different geographic locations makes trials more robust to changes in the epidemic. While China was once the center of the COVID-19 epidemic, several treatment trials initiated there were underpowered due to waning transmission [17]. As other countries adopt more effective control strategies, incidence would likely decline, but it is less likely to wane in all areas, and new sites can also be added. Experience with Zika in the Americas provides a useful counter-example, though, where trials were not possible because incidence dramatically declined everywhere [18]. If that were to occur, the ensemble modeling process would be useful for assessing trial feasibility.
2.4. Model evaluation
Finally, the ensemble modeling process should be evaluated by comparing model projections to subsequently observed data. An evaluation procedure could be conducted prior to each new round of modeling, before investigators want to make decisions about adding new trial sites. This process could assess how well model-projected rankings corresponded to observed rankings of hardest hit sites. Where there is a lot of uncertainty in which sites will have highest incidence, as reflected in low correlation, investigators may feel more comfortable making future decisions based on logistical or political considerations rather than purely on model rankings.
This evaluation procedure could also be conducted formally after the trial ends to compare model-projected and observed cumulative incidence and observed incidence during the target time periods. These types of reports are very useful for understanding the role of modeling as a tool for real-time decision-making in outbreaks [13], [19].
3. Discussion
We describe an ensemble modeling procedure to inform site selection for a vaccine efficacy trial planned during an ongoing epidemic. By prioritizing sites with highest projected disease incidence, investigators can accelerate the pace of endpoint accrual. Mathematical and statistical models synthesize the best available evidence to guide this planning. We focus on COVID-19 as a motivating example, but the general principles apply to other emerging infectious diseases.
We present a highly simplified procedure to reduce the burden on modeling groups to prepare results and potentially enable more groups to participate. For example, models could, but not be required to, explicitly account for the impact of vaccination on transmission dynamics. The assumption is that population vaccine coverage will be relatively low even in large trials, and that the rank ordering of sites is similar in less complex models. This is a recommended minimum structure, but other relevant practical questions will likely emerge that can be explored as add-ons. For example, the modeling results can be used to answer questions about expected duration of the trial as a function of enrollment rates and expected incidence. Nonetheless, it is important to remember that projections can be very uncertain, particularly as they depend upon rapidly changing policies and human behavior. Thus, we focus on simple output for the purposes of prioritization, acknowledging that other important questions may be difficult to answer precisely.
In addition to identifying geographic locations, models could also be used to explore targeted enrollment in sub-populations defined by age, occupation, or other covariates. Models could also guide the design of post-licensure observational studies for continued evaluation of vaccine effectiveness.
4. Conclusions
It is a top priority to rapidly evaluate the safety and efficacy of candidate COVID-19 vaccines. Data-driven models can help to optimize site selection and contribute to accelerating trials in a setting where every day counts.
CRediT authorship contribution statement
Natalie E. Dean: Conceptualization, Writing - original draft, Writing - review & editing, Funding acquisition. Ana Pastore y Piontti: Conceptualization, Visualization, Writing - review & editing. Zachary J. Madewell: Software, Visualization, Writing - review & editing. Derek A.T Cummings: Writing - review & editing. Matthew D.T. Hitchings: Writing - review & editing. Keya Joshi: Writing - review & editing. Rebecca Kahn: Writing - review & editing. Alessandro Vespignani: Conceptualization, Writing - review & editing. M. Elizabeth Halloran: Conceptualization, Writing - review & editing. Ira M. Longini: Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Funding: This work was supported by the National Institutes of Health R01-AI139761 (NED, MEH, IML, AV, APP, ZJM). R37-AI032042 (MEH).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2020.09.031.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lurie N., Saville M., Hatchett R., et al. Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 2.Dean N.E., Gsell P.-S., Brookmeyer R., et al. Design of vaccine efficacy trials during public health emergencies. Sci. Transl. Med. 2019;11(499):eaat0360. doi: 10.1126/scitranslmed.aat0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. An international randomised trial of candidate vaccines against COVID-19. Geneva, Switzerland: 2020.(https://www.who.int/blueprint/priority-diseases/key-action/Outline_CoreProtocol_vaccine_trial_09042020.pdf)
- 4.Lipsitch M, Kahn R, Mina MJ. Antibody testing will enhance the power and accuracy of COVID-19-prevention trials. Nat. Med. 2020;26:818–819. doi: 10.1038/s41591-020-0887-3. [DOI] [PubMed] [Google Scholar]
- 5.Halloran M.E., Auranen K., Baird S., et al. Simulations for designing and interpreting intervention trials in infectious diseases. BMC Med. 2017;15(1):223. doi: 10.1186/s12916-017-0985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellan S.E., Pulliam J.R.C., Pearson C.A.B., et al. Statistical power and validity of Ebola vaccine trials in Sierra Leone: A simulation study of trial design and analysis. Lancet Infect. Dis. 2015;15(6):703–710. doi: 10.1016/S1473-3099(15)70139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ZIKAVAT Collaboration, Asher J, Barker C, et al. Preliminary results of models to predict areas in the Americas with increased likelihood of Zika virus transmission in 2017. bioRxiv. 2017;https://doi.org/10.1101/187591.
- 8.Zhang Q., Sun K., Chinazzi M., et al. Spread of Zika virus in the Americas. Proc. Natl. Acad. Sci. 2017;114(22):E4334–E4343. doi: 10.1073/pnas.1620161114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ray E.L., Reich N.G. Prediction of infectious disease epidemics via weighted density ensembles. PLoS Comput. Biol. 2018;14(2):1–23. doi: 10.1371/journal.pcbi.1005910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reich NG, McGowan CJ, Yamana TK, et al. Accuracy of real-time multi-model ensemble forecasts for seasonal influenza in the U.S. PLoS Comput. Biol. 2019;15(11):1–19. doi: 10.1371/journal.pcbi.1007486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson M.A., Apfeldorf K.M., Dobson S., et al. An open challenge to advance probabilistic forecasting for dengue epidemics. Proc. Natl. Acad. Sci. U. S. A. 2019;116(48):24268–24274. doi: 10.1073/pnas.1909865116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viboud C., Sun K., Gaffey R., et al. The RAPIDD Ebola forecasting challenge: Synthesis and lessons learnt. Epidemics. April 2017;2018(22):13–21. doi: 10.1016/j.epidem.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray E.L., Wattanachit N., Niemi J., et al. Ensemble Forecasts of Coronavirus Disease 2019 (COVID-19) in the U. S. medRxiv. 2020 [Google Scholar]
- 14.Dean N.E., Gsell P.-S., Brookmeyer R., et al. Creating a Framework for Conducting Randomized Clinical Trials during Disease Outbreaks. N. Engl. J. Med. 2020;382:1366–1369. doi: 10.1056/NEJMsb1905390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang W, Cheng J, Allaire J, et al. Shiny: web application framework for R. R package version. 2017 [Google Scholar]
- 16.Gsell P.S., Camacho A., Kucharski A.J., et al. Ring vaccination with rVSV-ZEBOV under expanded access in response to an outbreak of Ebola virus disease in Guinea, 2016: an operational and vaccine safety report. Lancet Infect. Dis. 2017;17(12):1276–1284. doi: 10.1016/S1473-3099(17)30541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao B., Wang Y., Wen D., et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vannice K.S., Cassetti M.C., Eisinger R.W., et al. Demonstrating vaccine effectiveness during a waning epidemic: A WHO / NIH meeting report on approaches to development and licensure of Zika vaccine candidates. Vaccine. 2019;37(6):863–868. doi: 10.1016/j.vaccine.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chretien J.P., Riley S., George D.B. Mathematical modeling of the West Africa Ebola epidemic. Elife. 2015;4(DECEMBER2015):1–15. doi: 10.7554/eLife.09186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.