Abstract
HNSCC is an immunologically active tumor with high levels of immune cell infiltration, high mutational burden and a subset of patients who respond to immunotherapy. One of the primary sources of mutations in HNSCC is the cytidine deaminase APOBEC3, which is a known participant in innate immunity. Why particular HNSCCs have higher rates of APOBEC mutations and how these mutations relate to the immune microenvironment remains unknown.
Utilizing whole exome and RNA-Seq datasets from TCGA HNSCCs we annotated APOBEC mutations, immune cell populations, activating and end effectors of immunity and neoantigens in order to interrogate the relationship between APOBEC mutations and the immune landscape.
Immune cell populations and composite scores of immune activation were tightly associated with APOBEC mutational burden (p = 0.04–1.17e-5). HNSCC had the highest levels of IFNy across cancer types with high APOBEC mutational burden, with the highest IFNy scores in HPV mediated HNSCC. Tumor specific neoantigens were significantly correlated with APOBEC mutational burden while other sources of neoantigens were not (0.53 [0.24, 0.76] p = 8e-5). The presence of a germline APOBEC polymorphism was more prevalent in non-white, non-black patients and within this group, patients with the polymorphism had higher APOBEC mutational burden (p = 0.002).
APOBEC mutations are tightly linked to immune activation and infiltration in HNSCC. Multiple mechanisms may exist within HNSCC leading to APOBEC mutations including immune upregulation in response to neoantigens and viral infection, via induction of IFNy. These mechanisms may be additive and not mutually exclusive, which could explain higher levels of APOBEC mutations in HPV mediated HNSCC.
Keywords: Head and neck cancer, Immunotherapy, APOBEC, Mutational signatures, Immune microenvironment
Introduction
The primary known risk factors for Head and Neck Squamous Cell Carcinoma (HNSCC) are tobacco smoke exposure and human papilloma virus (HPV) infection. Utilizing computational approaches for mutational signature annotation, the majority of mutations in HNSCC can be ascribed to aging, carcinogen exposure and activity of the apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3 (APOBEC3) family of cytosine deaminases. APOBECs have diverse functions but importantly, are involved in innate immunity and base excision repair [1,2].
In addition to HNSCC, APOBEC3 mutations are particularly common in lung, breast, bladder and cervical cancer, but present to some degree in all cancers [2]. The presence, quantity and functional importance of APOBEC3 mutations in cancer has been well established [2–5]. Why APOBEC mutations are so prevalent in certain cancers remains unclear. Considering the well described role of the APOBEC family in viral restriction and innate immunity, one theory that is particularly relevant to HNSCC is upregulation of APOBEC3 as part of the innate immune response to viral infection, resulting in off-target host genomic mutations [6]. We, and others, have shown that APOBEC3 mutations are particularly prominent in HPV mediated HNSCC (HPVmHNSCC) and that APOBEC3 mutations appear to be responsible for driver mutations in PIK3CA, the most frequently mutated gene in HPVmHNSCC [4,7]. While APOBEC mutagenesis is most prevalent in HPVmHNSCC, non-HPVmHNSCC also contain APOBEC3 mutations, suggesting that additional mechanisms must drive APOBEC mutagenesis as well [7]. For example, an APOBEC germline deletion polymorphism has been described in breast cancer and is associated with an increased rate of APOBEC mutations due to expression of an APOBEC3A-3B chimera, suggesting inherited factors may play a role in APOBEC mutagenesis [8,9].
HNSCC is known to be an immunologically “hot” tumor with immune cell infiltration levels amongst the highest of all cancers [10]. Despite this, and a moderate-high mutational burden, only 15–20% of HNSCC patients demonstrate response to immunotherapy with PD-1/PD-L1 checkpoint blockade (ICB) [2,11]. While expression of markers such as PD-L1, infiltration of immune cell populations such as CD8 T cells, and an “inflamed” tumor phenotype, as evidenced by IFNγ signatures, have been shown to correlate with response to immunotherapy, there are currently no accurate biomarkers to predict response to ICB in HNSCC [12]. Perhaps not surprisingly, HPVmHNSCC possesses higher levels of immune infiltration overall, CD8 T cells and markers of immune activation, compared to non-HPVmHNSCC [10]. Nonetheless, while HPVmHNSCC show faster and higher response rates to nivolumab, overall survival is the same as non-HPVmHNSCC [13]. APOBEC mutations have recently been shown to correlate with PD-L1 expression and response to immunotherapy in lung cancer, supporting the concept that APOBEC mutations are tightly linked to the immune environment of a tumor [14,15]. Here, we examined the relationship between APOBEC mutagenesis, the tumor immune microenvironment (TIME) and markers of response to immunotherapy, in HNSCC.
Methods
Datasets
Somatic exome, germline, transcriptome, and paired clinical datasets were originally obtained from the TCGA data portal, for 276 HNSCC. These files now exist in the Genomic Data Commons (https://gdc.cancer.gov). Clinical data from patients in TCGA from the University of Pittsburgh were merged with TCGA to increase accuracy and when discrepancies existed, preference was given to the data from the Pittsburgh Head and Neck Tumor database. APOBEC germline polymorphism data was gathered from published lists in TCGA [9].
APOBEC enrichment scores
Per sample APOBEC enrichment scores were calculated according to the methods described by Roberts [2,3,16]. Briefly, this is the fraction of all C/G mutations (CGm) that were TCW/WCA in nature (TCWm), normalized by the fraction of all C/G sites in the local 50 bp context (CGc) that were TCW motifs (TCWc)
Immune scores
IFNγ signature was calculated according to Ayers et al. [17]. CYT score was calculated according to Rooney et al. [18]. Estimate Immune Score was calculated according to Yoshihara et al. [19]. Immune deconvolution data was generated from RNA-Seq reads as previously described using Cibersort and ssGSEA [20,21]. Briefly, Cibersort is a validated computational approach that utilizes a leukocyte gene signature matrix to estimate the relative proportions of each cell type of interest in bulk RNA-Seq datasets. ssGSEA calculates a score for a gene signature for each pair of sample and gene set by comparing the ranks of the genes in the signature with the ranks of all other genes in the transcriptome, based on the degree to which the genes are up- or down- regulated. This approach has been orthogonally validated using immunofluorescence staining in samples with high concordance [21]. Further, these approaches have demonstrated strong correlation between each other in HNSCC [10]. Tumor purity was calculated with ASCAT [22].
Tumor antigens
Mutation-derived predicted class I binding neoantigens (mutation-induced neoantigens) were calculated as described by Rooney et al. [18]. A list of endogenous retroviruses (ERV) was obtained from Meyer et al and filtered for ERVs known to be transcriptionally silent based on Rooney et al. [18,23]. Expression levels of 60 Cancer Testis (CT) antigens known to be transcriptionally silent in normal tissue was obtained from Rooney et al. [18]. Expression values were dichotomized to zero and non-zero and the association between APOBEC enrichment and CT antigen expression was assessed by Wilcoxon rank-sum test.
Immunohistochemistry
40 HNSCC samples from patients at the University of Pittsburgh whose tumors were contributed to TCGA underwent Immunohistochemical (IHC) staining for CD8 (Roche, catalog # 790–4460), CD45RO (Roche, catalog # 790–2930), CD19 (Sigma, catalog # 119M-15), and Foxp3 (Spring Bioscience, catalog # M3972). Associations with immune deconvolution markers and APOBEC enrichment were assessed by linear regression.
Statistical analysis
The relationship between APOBEC enrichment, clinical variables, immune scores, immunohistochemistry, neoantigens and germline polymorphism status was assessed by linear regression models, Spearman’s correlation coefficient or/and Wilcoxon rank-sum test in the cohort overall, HPVmHNSCC and non-HPVmHNSCC. As enrichment score is right-skewed it was log-transformed. Survival analysis was conducted using Cox regression model. Models were built using stepwise method with HPV status and stage. Association between two categorical variables were assessed by Fisher’s exact test. P-values were adjusted for multiple testing using Benjamini-Hochberg method when necessary.
Results
Immune cell populations and activating/end effectors of immunity are tightly linked to APOBEC mutations
HNSCC is often an immunologically inflamed/infiltrated cancer. We hypothesized that variability in APOBEC mutation rates within HNSCC could be driven by the TIME and thus, that immune cell populations known to participate in anti-tumor immunity should correlate with APOBEC mutagenesis. In order to investigate this possibility we utilized deconvolution of RNA-Seq datasets from 276 HNSCC in TCGA to estimate immune cell populations and generate immune scores.
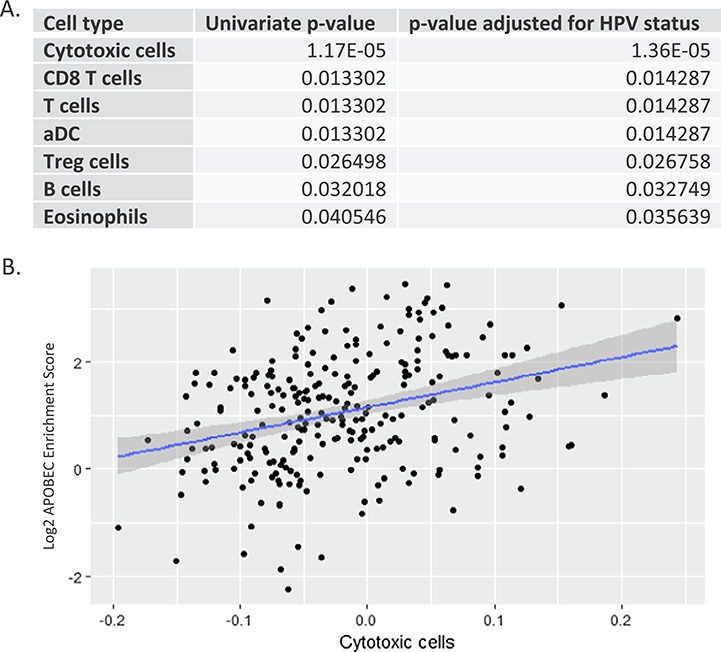
We first examined immune cell populations known to indicate inflamed tumors, finding that CD8 T cell populations were associated with APOBEC mutational burden (p = 0.013), even after adjusting for HPV status (p = 0.014) (Fig. 1A, Supplemental Fig. 1). Further, T cell populations overall, regulatory T cells (T-Regs), eosinophil, dendritic, cytotoxic cells and B cell populations were also associated with APOBEC mutations. (Fig. 1A, B, Supplemental Fig. 1A–F). The strongest correlation with APOBEC mutational burden was with cytotoxic cells in non-HPVmHNSCC (0.38 [0.24, 0.50], p = 2e-6, Spearman’s correlation). Contrary to this finding, cytotoxic cells did not positively correlate with the other known prominent mutational signatures in HNSCC, signature 1 (age) and signature 4 (smoking) (0 [−0.13, 0.13], p = 0.9 and −0.3 [−0.42, 0.19], p = 2e-6), respectively, Spearman’s correlation).
Fig. 1.
Immune cell populations are associated with APOBEC mutational burden. A. Univariate associations between immune cell populations and APOBEC mutations in descending order of p-values, demonstrating association of all populations and APOBEC mutations. B. Linear regression scatter plot demonstrating association of cytotoxic cell population with APOBEC mutational burden. Blue line represents linear regression fitting, Grey shadowing represents 95% confidence bands. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In order to examine if traditional immunohistochemical staining (IHC) of immune cell markers would also show an association with APOBEC mutations, IHC of CD8, CD45, CD19 and Foxp3 was performed on FFPE sections from tumors contributed to TCGA from the University of Pittsburgh, for which tissue blocks were available (n = 40). After controlling for multiple comparisons, APOBEC mutations were not statistically associated with CD8 T cell infiltration on IHC (p = 0.11), nor CD45, CD19 and Foxp3, despite their corresponding cell populations correlating with APOBEC mutations from the RNA-Seq data (Supplemental Fig. 2A). Because of this lack of association, we then compared immune cell populations by RNA-Seq and IHC, finding no statistical associations other than Foxp3 and T Regs (p = 6 e-6) (Supplemental Fig. 2B).
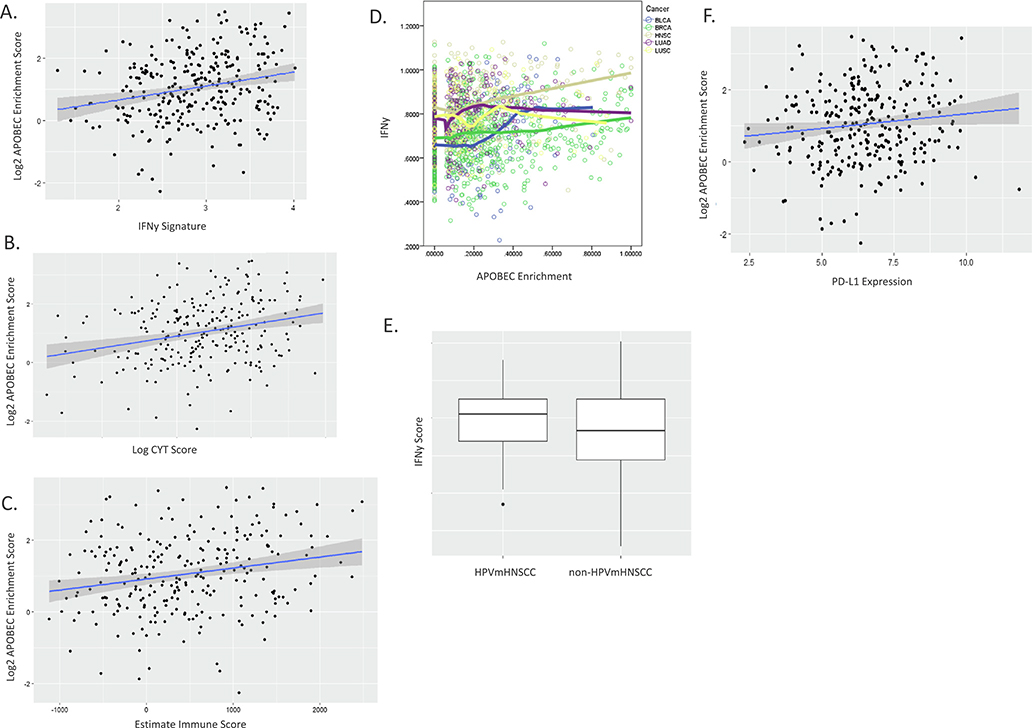
To explore in more detail the relationship between APOBEC mutations and the immune landscape, we examined the activating and effector molecules of immunity using an IFNγ score (composite score which has been shown to correlate with an inflamed tumor subtype and response to immunotherapy, CYT (a measure of the immune cytolytic activity using granzyme A (GZMA) and perforin (PRF1) which are known to be upregulated upon CD8+ T cell activation) and ESTIMATE immune score (measure of immune cell infiltration within a tumor), finding that each score was strongly positively associated with APOBEC mutations (p = 1e-4, p = 4e-5, p = 6e-4, respectively) (Fig. 2A–C). These associations remained significant after controlling for HPV status. APOBEC is known to be inducible by IFNγ [24]. Examining IFNγ across cancer types in TCGA known to have prominent APOBEC signatures, we found that HNSCC possesses the highest IFNγ expression of all the tumor types (Fig. 2D). Based on this observation, we examined the IFNγ score within HPVmHNSCC and non-HPVmHNSCC finding that HPVmHNSCC tumors have a trend towards elevated IFNγ scores (p = 0.09) (Fig. 2E). IFNγ is known to upregulate PD-L1 on tumors. Both IFNy score and PD-L1 have been shown to modestly predict response to immunotherapy. Therefore, we examined PD-L1 expression in relation to APOBEC mutations finding that they were associated univariately (p = 0.04) (Fig. 2F) and controlling for HPV status (p = 0.049).
Fig. 2.
Activating and end effector molecules and pathways of immunity are associated with APOBEC mutational burden. Linear regression scatter plots of APOBEC mutations vs IFNy score (A), CYT score (B) and Estimate Immune Score (C), demonstrating associations. D. IFNy expression levels are highest overall in HNSCC within the 5 highest APOBEC mutations burden cancers (BLCA: Bladder cancer, BRCA: Breast cancer, HNSC: Head and Neck cancer, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma). E. Box plot of IFNy score stratified by HPV showing higher level in HPVmHNSCC. Three horizontal lines represents first quartile, median, third quartile from bottom to top; the upper vertical line extends from the second quartile to the largest value no further than 1.5IQR (interquartile range) from the second quartile; the lower vertical line extends from the first quartile to the smallest value at most 1.5IQR from the first quartile; dots represent outliers that are more than 1.5 IQR away from first/second quartiles. F. Linear regression scatter plot demonstrating association between PL-L1 expression and APOBEC mutations.
Tumor specific neoantigen burden correlates with APOBEC mutations
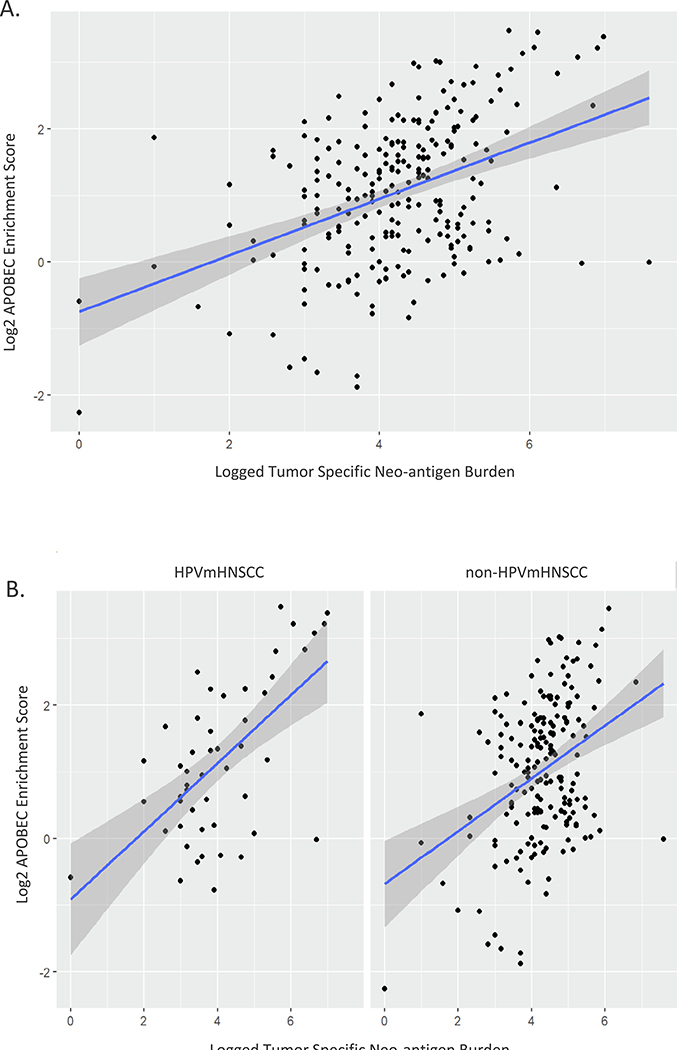
The host immune response to tumor cells has been shown to be at least partially driven by the presence of neoepitope-specific T cells [25]. Further, neoantigen burden, and type, have been shown to correlate with response to immunotherapy [26–28]. Knowing that APOBEC mutations appear to occur later in tumor evolution [29,30], we hypothesized that APOBEC activity may be driven by immune upregulation due to tumor associated antigens. In order to test this, we first generated mutation-derived predicted class I binding neoantigens (mutation-induced neoantigens). APOBEC mutation rate was strongly positively associated with mutation-induced neoantigens (p = 8e-12) (Fig. 3A). This association remained significant after controlling for HPV status (p = 5e-12) suggesting that mutation-induced neoantigens could be driving host immune response, and potentially APOBEC mutagenesis secondarily, independent of viral infection and viral-antigens (Fig. 3B). Interestingly, when HPVmHNSCC and non-HPVmHNSCC were analyzed separately, the correlation between HPVmHNSCC and APOBEC mutations was stronger than non-HPVmHNSCC (0.53 [0.24, 0.76] p = 8e-5 vs 0.29 [0.14, 0.42] p = 4e-5). In order to ensure that tumor purity was not driving the correlation between APOBEC mutational burden and neoantigen burden the association between APOBEC mutation rates and purity was examined for the entire cohort, HPVmHNSCC and non-HPVmHNSCC, revealing that none of the correlation coefficients were significant (−0.006 [−0.13, 0.13] p = 0.92, −0.007 [−0.30, 0.25] p = 0.96, 0.001 [−0.15, 0.15] p = 0.99, respectively). Further, the relationship between APOBEC mutations and mutation-induced neoantigens was examined using linear regression, controlling for HPV status and purity, revealing that mutation-induced neoantigens remained significant, while purity was not significant, in the model.
Fig. 3.
Tumor specific neo-antigens are strongly associated with APOBEC mutational burden. A. Linear regression scatter plot demonstrating strong association between TSAs and APOBEC mutations. B. Association persists after controlling for HPV status.
Numerous types of tumor associated antigens exist other than mutation-induced neoantigens, including cancer testis (CT) antigens. CT antigens are not expressed in healthy tissues, other than germ cells, but are expressed in some tumors. CT antigens have been shown to induce antigen-specific responses [31]. Using a list of 60 CT antigens that are transcriptionally silent in normal non-germline tissues, yet variably expressed in tumors, we examined the relationship between APOBEC mutations and CT gene expression. After controlling for multiple comparisons, there was no significant association.
Similar to CT antigens, Endogenous retroviruses (ERVs) are germline encoded and may be re-activated in cancer cells. Therefore, using 66 ERVs that have been shown to be expressed, we assessed if ERV antigens could be driving immune upregulation and APOBEC mutagenesis. While univariate linear regression revealed seven ERVs that associated with APOBEC mutations, after controlling for multiple comparisons, no association was significant (Supplemental Fig. 3).
APOBEC mutation rates and germline polymorphism status differ depending on race
Considering that APOBEC3 mutations were found to be tightly correlated with immune upregulation/infiltration in this dataset, and that immune cell infiltration correlates with survival in HNSCC [32,33], we examined the relationship between APOBEC3 mutations and clinical variables known to have prognostic significance in HNSCC (cohort characteristics summarized in Supplemental Table 1). There was no association between APOBEC3 mutation rate and grade, perineural invasion, or extracapsular extension univariately or controlling for HPV status (Supplemental Table 2, Supplemental Fig. 4A–C). We next examined APOBEC3 mutation rates, expression and survival. In order to control for potential confounders, a Cox regression model was built using stepwise strategies with enrichment score, stage, medical center and HPV status already in the model. No association was found between APOBEC3 mutations, APOBEC3A or 3B expression and OS, once all variables were considered.
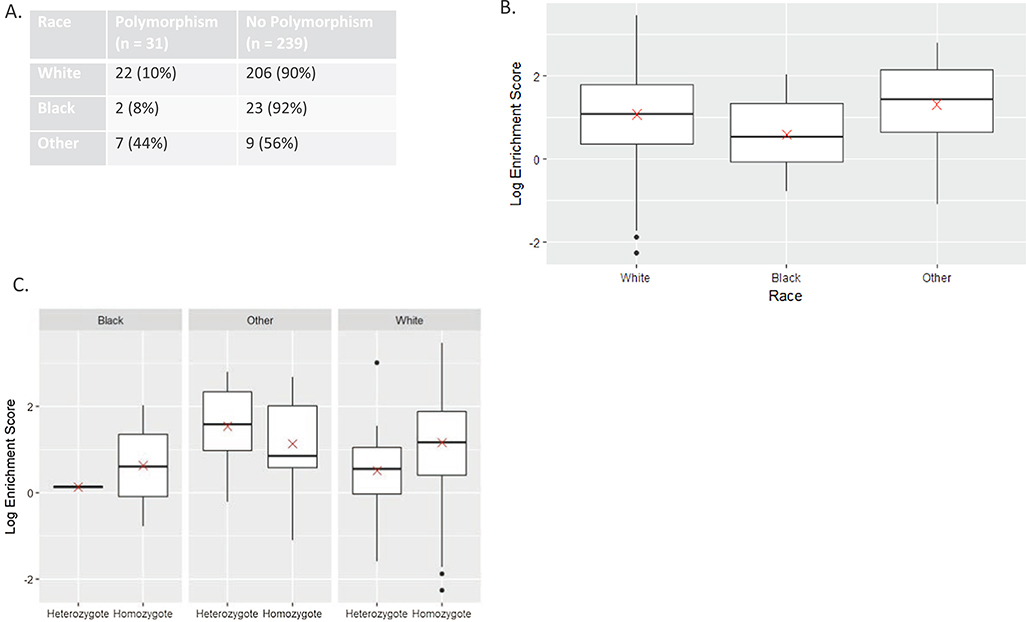
An APOBEC3 germline deletion polymorphism resulting in an APOBEC3A-B chimera has previously been reported to cause increased APOBEC mutation rates in breast cancer and oral cavity cancer in a Taiwanese population. In the Taiwanese population, presence of the polymorphism correlated with survival [34]. Therefore, we examined the relationship between the APOBEC3 germline polymorphism (3½70) and survival, finding no association (Supplemental Fig. 5A) (p = 0.53). Considering the racial differences between the Taiwanese cohort (predominately Asian) and TCGA (predominately White), we examined how race related to germline polymorphism status, finding that non-white, non-blacks (American Indians and Asians) had the highest rates of germline polymorphism, with nearly half of patients possessing this polymorphism (p = 0.002) (Fig. 4A). Because of this, we examined APOBEC mutation rates between race categories in TCGA, grouped as white, black and other, finding that APOBEC mutation rates trended towards being higher in the “other” race (p = 0.06) (Fig. 4B). Further, in the other race group, there was a trend towards those with the polymorphism having elevated APOBEC mutations rates compared to those without the polymorphism (Fig. 4C), albeit the sample size is small and thus there was not statistical significance (p = 0.50). Interestingly, this was different than the white and black race cohorts, which showed higher mutation rates in the patients without a polymorphism (p = 0.008) (Fig. 4C).
Fig. 4.
APOBEC mutation burden is affected by race and germline polymorphism status. A. Polymorphism status by race B. Box plot of APOBEC mutations burden by race demonstrating increase in “other” race. Red “x” represents mean C. Box plots of APOBEC mutational burden within each race by polymorphism status demonstrating a higher APOBEC mutations in “other” race in the presence of the polymorphism yet lower APOBEC mutations in the presence of the polymorphism in Black and White races. Heterozygote = polymorphism present, Homozygote = no polymorphism present. D. Box plot of APOBEC mutational burden in the cohort overall by polymorphism status demonstrating higher rates of mutations without the polymorphism.
Discussion
Currently, significant interest is focused on the field of immunooncology, spurred in part by the recent success of immune modulating therapeutics. HNSCC is known to be an immunologically active tumor, with high levels of tumor infiltrating T cells, high mutational burden and proven responsiveness to immunotherapy [10,11,35]. While mutational burden, neoantigen burden, PD-L1 expression, IFNy score, T cell infiltrates and an inflamed tumor phenotype have all been shown to correlate with response to immunotherapy, there are no reliable biomarkers for response to ICB in HNSCC and in fact, no validated biomarkers in HNSCC to predict tumor behavior, in general [28,36,37]. What is known is that HNSCC has an active immune landscape, numerous distinct mutational processes contributing to mutational burden and two primary etiologies. Taken together, HNSCC presents a unique opportunity among all cancers to study the interplay of the immune landscape and cancer development.
One established source of mutations in HNSCC is APOBEC3 mutagenesis. APOBEC mutations are of particular interest due to APOBECs role as both an endogenous mutator and as a participant in innate immunity as a viral restriction factor. Notably, HPVmHNSCCs possess the highest burden of APOBEC mutations, leading us, and others, to hypothesize that viral infection may drive APOBEC mutagenesis in HNSCC [7]. However, non-HPVmHNSCCs also possess variable levels of APOBEC mutations, suggesting that alternative pathways for triggering APOBEC3 mutations within HNSCC exist. Interestingly, Wang et al recently showed that APOBEC mutations corelate with response to immunotherapy in lung cancer while Boichard et al. reported that PD-L1 expression is related to APOBEC mutational burden across tumor types in TCGA [14,15]. Here, using computational approaches applied to whole exome and RNA-Seq datasets from TCGA HNSCCs, we examined the relationship between the TIME and APOBEC mutations.
Most notably, we found that the density of nearly all immune cell populations correlated with APOBEC mutational burden. The strongest correlation between immune cell subtypes and APOBEC mutations was found with cytotoxic cells. These associations were independent of HPV status. These findings support the concepts that: 1. the TIME is closely tied to APOBEC mutagenesis and 2. APOBEC mutational burden may be more related to the degree of tumor-specific immune activation, overall, as opposed to viral infection in particular. To further explore this concept, we examined activating and end effectors of immunity, including an IFNy score, CYT score and ESTIMATE Immune score. IFNy is known to be one of the major forces shaping the TIME and importantly, APOBEC is inducible by IFNy [24]. Here, IFNy scores correlated with APOBEC mutational burden. Although IFNy score was higher in HPVmHNSCC then in non-HPVmHNSCC overall, the correlation between APOBEC mutational burden and IFNy score was independent of HPV status. This again supports the concept that APOBEC mutations may be driven by the overall level of tumor-specific immune activation. It is possible that the additional immune activation from viral infection, in HPVmHNSCC, is additive, and could explain the elevated APOBEC mutational burden in HPVmHNSCC compared to non-HPVmHNSCC. Interestingly, we found that HNSCC have the highest levels of IFNy across all tumor types with high APOBEC mutational burden in TCGA. Similarly, PD-L1, which is known to relate to IFNy secretion, immune cell exhaustion, and response to immunotherapy, was also found to be tightly linked to APOBEC mutational burden. This further corresponds with the finding of elevated CYT score in high APOBEC mutation samples, as CYT is a marker of activated T cells.
Why certain tumors are more immunogenic than others remains an unanswered and complex question. Considerable emerging literature supports tumor antigens as a possible source driving immune activation [25]. Tumor associated antigens can arise from multiple sources including mutation-induced neoantigens, expression of CT antigens, ERVs and viral antigens from oncogenic viruses such as HPV. Here, we found that mutation-induced neoantigen burden was strongly positively correlated with APOBEC mutational burden. This was true independent of HPV status; however, the correlation was stronger in HPVmHNSCC, compared to non-HPVmHNSCC. This leads us to hypothesize that neoantigen burden, resulting in immune activation/ IFNy release, may be the primary driver of APOBEC mutations. This concept is in line with our finding of high levels of IFNy in HNSCC, and particularly, HPVmHNSCC. Other known sources of TSAs, such as CT antigens and ERVs were not found to be associated with APOBEC mutations. Contrary to this presented hypothesis is emerging evidence suggesting that higher neoantigen burden may actually represent a lack of immune surveillance, as opposed to triggering immune upregulation [38]. An alternative hypothesis is that APOBEC mutations actually cause immune upregulation by generating immunogenic neoantigens. However, work by McGranahan et al. suggest APOBEC mutations occur later in tumor evolution, which supports the concept that APOBEC mutations are an end result of immune activation, as opposed to causative [30]. It is important to note that APOBEC mRNA has been shown to be directly upregulated in response to HPV infection [39,40]. Similarly, the HPV protein E7 stabilizes APOBEC3A from high risk HPVs, prolonging its cellular half-life [41,42].
The APOBEC3 family includes 7 members. APOBEC3A and 3B are the most frequently studied in terms of mutations in cancer genomes. Using APOBEC-gene specific motifs, we and others, have shown that HNSCCs appear to possess both APOBEC3A and 3B mutations, but higher levels of APOBEC3A [7]. This is of interest as APOBEC3A appears to be specifically involved with anti-HPV restriction. This is in opposition to breast cancer, for example, where APOBEC3B appears to be the primary mutator [43]. Thus, not only are there likely multiple forces driving APOBEC mutagenesis in HNSCC, there are likely multiple different APOBEC3 family members active in a given cancer as well.
A germline deletion polymorphism has been described in breast cancer that causes expression of an APOBEC3A-3B chimera and increased APOBEC mutations. Subsequently, this polymorphism was identified in a population of oral cavity cancer patients from Taiwan. When we explored this polymorphism in HNSCC from TCGA we found low levels overall. However, the presence of the polymorphism was highly dependent on race, with non-white non-black patients possessing higher rates of the polymorphism. This is in line with published data on the rates of the polymorphism by continent of origin with Africans and Europeans having a low frequency (0.9% and 6%) and East Asians and Amerindians having higher rates (36.9% and 57.7%, respectively) [44]. In this cohort, non-white non-black patients trended towards having the highest rates of APOBEC mutations overall. Within this “other” cohort, made of Asians, Native Americans and Pacific Islanders, patients with the polymorphism again trended towards higher rates of APOBEC mutations compared those without the polymorphism. The cohort overall had the opposite finding, with higher rates of APOBEC mutations in the tumors from individuals without the polymophsim in white and black groups [9,34]. Thus, it appears there is a subset of HNSCCs whose APOBEC mutational burden is related to presence of a germline polymorphism, separate from the immune environment and viral infection; however, this population is a minority.
Overall, this data suggests that multiple mechanisms may exist within HNSCC that lead to APOBEC mutations. These mechanisms may be additive and not mutually exclusive, which would explain the higher levels of APOBEC mutations in HPVmHNSCC. If APOBEC mutational burden indeed reflects the additive effects of multiple sources of immune upregulation, the relationship of APOBEC mutations to ICB response should be examined alone, and in combination with existing known markers of response in HNSCC.
Supplementary Material
Acknowledgments
Funding
This work was funded in part by a Pilot grant from the American Head and Neck Society (Faden) NIH R01 CA206517-02 (Ferris), NIH P50 CA097190-13 (Ferris).
Footnotes
Declaration of Competing Interest
None.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.oraloncology.2019.07.020.
References
- [1].Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 2013;45:977–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Henderson S, Chakravarthy A, Su X, et al. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep 2014;7:1833–41. [DOI] [PubMed] [Google Scholar]
- [5].Hoopes J, Cortez L, Mertz T, et al. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep 2016;14:1273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Henderson S, Fenton T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol Med 2015;21:274–84. [DOI] [PubMed] [Google Scholar]
- [7].Faden DL, Thomas S, Cantalupo PG, et al. Multi-modality analysis supports APOBEC as a major source of mutations in head and neck squamous cell carcinoma. Oral Oncol 2017;74:8–14. [DOI] [PubMed] [Google Scholar]
- [8].Middlebrooks CD, Banday AR, Matsuda K, et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat Genet 2016;48:1330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nik-Zainal S, Wedge DC, Alexandrov LB, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet 2014;46:487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mandal R, Senbabaoglu Y, Desrichard A, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 2016;1 e89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ferris RL, Blumenschein G Jr., Fayette J, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim HR, Ha SJ, Hong MH, et al. PD-L1 expression on immune cells, but not on tumor cells, is a favorable prognostic factor for head and neck cancer patients. Sci Rep 2016;6:36956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ferris RL. AACR Abstract CT116; 2018.
- [14].Boichard A, Tsigelny IF, Kurzrock R. High expression of PD-1 ligands is associated with kataegis mutational signature and APOBEC3 alterations. Oncoimmunology 2017;6 e1284719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang S, Jia M, He Z, et al. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene 2018;37:3924–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Roberts SA, Lawrence MS, Klimczak LJ, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 2013;45:970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ayers M, Lunceford J, Nebozhyn M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Senbabaoglu Y, Gejman RS, Winer AG, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 2016;17:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Van Loo P, Nordgard SH, Lingjaerde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A 2010;107:16910–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mayer J, Blomberg J, Seal RL. A revised nomenclature for transcribed human endogenous retroviral loci. Mob DNA 2011;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stenglein MD, Burns MB, Li M, et al. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol 2010;17:222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fritsch EF, Hacohen N, Wu CJ. Personal neoantigen cancer vaccines: The momentum builds. Oncoimmunology 2014;3 e29311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gubin MM, Zhang X, Schuster H, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014;515:577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lauss M, Donia M, Harbst K, et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat Commun 2017;8:1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 2018;33:843–852.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Swanton C, McGranahan N, Starrett GJ, et al. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov 2015;5:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McGranahan N, Favero F, de Bruin EC, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med 2015;7:283ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gjerstorff MF, Andersen MH, Ditzel HJ. Oncogenic cancer/testis antigens: prime candidates for immunotherapy. Oncotarget 2015;6:15772–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gameiro SF, Ghasemi F, Barrett JW, et al. Treatment-naive HPV+ head and neck cancers display a T-cell-inflamed phenotype distinct from their HPV- counterparts that has implications for immunotherapy. Oncoimmunology 2018;7 e1498439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].de Ruiter EJ, Ooft ML, Devriese LA, et al. The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oncoimmunology 2017;6 e1356148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen TW, Lee CC, Liu H, et al. APOBEC3A is an oral cancer prognostic biomarker in Taiwanese carriers of an APOBEC deletion polymorphism. Nat Commun 2017;8:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kansy BA, Concha-Benavente F, Srivastava RM, et al. PD-1 status in CD8( + ) T cells associates with survival and anti-PD-1 therapeutic outcomes in head and neck cancer. Cancer Res 2017;77:6353–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Marty R, Kaabinejadian S, Rossell D, et al. MHC-I genotype restricts the oncogenic mutational landscape. Cell 2017;171:1272–1283.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vieira VC, Leonard B, White EA, et al. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. MBio 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mori S, Takeuchi T, Ishii Y, et al. Human papillomavirus 16 E6 upregulates APOBEC3B via the TEAD transcription factor. J Virol 2017;91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Warren CJ, Xu T, Guo K, et al. APOBEC3A functions as a restriction factor of human papillomavirus. J Virol 2015;89:688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Westrich JA, Warren CJ, Klausner MJ, et al. Human papillomavirus 16 E7 stabilizes APOBEC3A protein by inhibiting cullin 2-dependent protein degradation. J Virol 2018;92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Burns MB, Lackey L, Carpenter MA, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013;494:366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kidd JM, Newman TL, Tuzun E, et al. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet 2007;3 e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.