Abstract
The capsid protein (CA) of HIV-1 plays essential roles in multiple steps of the viral replication cycle by assembling into functional capsid core, controlling the kinetics of uncoating and nuclear entry, and interacting with various host factors. Targeting CA represents an attractive yet underexplored antiviral approach. Of all known CA-targeting small molecule chemotypes, the peptidomimetic PF74 is particularly interesting because it binds to the same pocket used by a few important host factors, resulting in highly desirable antiviral phenotypes. However, further development of PF74 entails understanding its pharmacophore and mitigating its poor metabolic stability. We report herein the design, synthesis, and evaluation of a large number of PF74 analogs aiming to provide a comprehensive chemical profiling of PF74 and advance the understanding on its detailed binding mechanism and pharmacophore. The analogs, containing structural variations mainly in the aniline domain and / or the indole domain, were assayed for their effect on stability of CA hexamers, antiviral activity, and cytotoxicity. Selected analogs were also tested for metabolic stability in liver microsomes, alone or in the presence of a CYP3A inhibitor. Collectively, our studies identified important pharmacophore elements and revealed additional binding features of PF74, which could aid in future design of improved ligands to better probe the molecular basis of CA-host factor interactions, design strategies to disrupt them, and ultimately identify viable CA-targeting antiviral leads.
Keywords: HIV-1, capsid-targeting antivirals, PF74
Graphical Abstract
1. Introduction
As part of the virally-encoded Gag polyprotein [1,2], human immunodeficiency virus type 1 (HIV-1) capsid protein (CA) plays critical roles in viral assembly and multiple post-entry events during viral replication [3]. CA is a key component of the Gag polyprotein, and CA-CA interactions drive the assembly of Gag [4] into the mature viral particle. During viral maturation [5,6], ~1500 CA monomers assemble into the cone-shaped viral capsid core, composed primarily of CA hexamers and exactly 12 asymmetrically-distributed CA pentamers. Maintaining tightly regulated capsid core stability is critical to the replication cycle of HIV-1: the capsid core provides a protected environment to allow efficient initiation and elongation of viral DNA during reverse transcription, and shields viral DNA from cytosolic DNA sensing [7]. In addition, CA interacts with various host factors [8,9], such as TRIM5α [10,11], cleavage and polyadenylation specific factor 6 (CPSF6) [12,13], nucleoporins 153 [14–16] and 358 [17–19] (NUP153, NUP358), MxB [20,21], and Cyclophilin A (CypA) [22–24], to enable various post-entry events [25], including reverse transcription, cytoplasmic trafficking, uncoating, nuclear entry, site of integration, and evasion of innate immunity. Therefore, CA-targeting compounds that perturb the stability of the viral capsid core and interfere with important CA-host interactions, may confer both early and late stage antiviral phenotypes.
Structurally, CA is highly helical (Figure 1, A), containing 7 helices in the N-terminal domain (CANTD) and 4 helices in the C-terminal domain (CACTD) [26–28]. Extensive contacts between CANTD and both the CANTD and CACTD of an adjacent CA monomer provide the molecular basis of hexameric capsid assembly, as well as various virus-host interactions. Although a few small molecule binding sites have been reported [29], the pocket (Figure 1, A and B) formed around H3 and H4 of the CANTD (cyan) and H8 and H9 of CACTD of an adjacent monomer (green) is of particular significance. This pocket accommodates the binding of PF74 [28,30,31], a potent and well-characterized [32,33] peptidomimetic CA-targeting compound (Figure 1, B and C), as well as a smaller CA-targeting chemotype represented by BI-2 (Figure 1, E). Importantly, this pocket is also used by host factors Nup153 and CPSF6 [30,31]. Nup153 is a nucleoporin important for the nuclear transport of viral pre-integration complexes (PICs), whereas CPSF6 transports HIV-1 PICs to transcriptionally active chromatin [34]. Consistent with the binding versatility of this pocket, PF74 displayed a concentration-dependent bimodal mechanism [32,33] for its antiviral activity: it competes against host factors CPSF6 and NUP153 to affect nuclear entry at lower concentrations; and blocks uncoating and reverse transcription at higher concentrations, presumably by altering inter-hexamer interactions [28].
Figure 1.
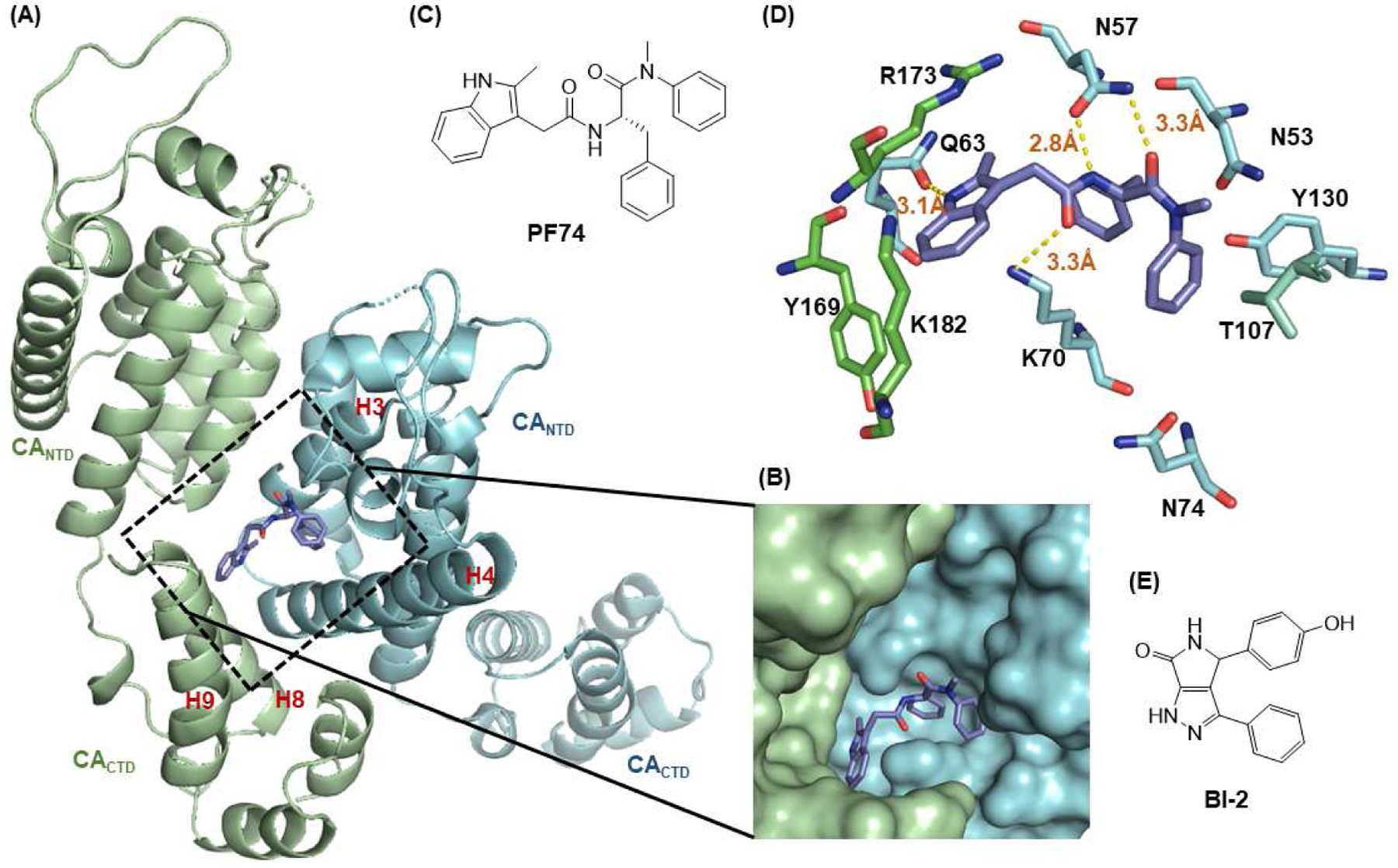
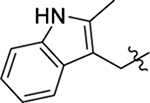
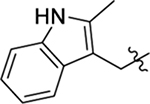
Structure of HIV-1 CA and an important ligand binding pocket. (A) The highly helical structure of CA dimer (PDB: 4FXZ [28]). The pocket formed around H3 and H4 of the CANTD (cyan), and H8 and H9 of the adjacent CACTD (green) accommodates the binding of multiple ligands, including host factors Nup153 and CPSF6, and small molecules PF74 and BI-2; (B) close-up view of the pocket bound by PF74; (C) the chemical structure of PF74; (D) important residues providing key interactions with PF74; (E) the chemical structure of BI-2.
The well-defined binding mode of PF74 entails a network of molecular interactions between PF74 and the binding pocket (Figure 1, D) [28,30,31]. Most of the binding interactions are located in the CANTD featuring a few key H-bonds: two between the terminal amide of N57 and the PF74 backbone carbonyl and NH of the phenylalanine moiety; one between K70 and the other PF74 backbone carbonyl; and one between Q63 and the PF74 indole ring NH. In addition, the aniline moiety of PF74 appears to make contact with N53 (via the N-methyl group), and A105, T107, and Y130 (via the phenyl ring), and the phenylalanine moiety of PF74 has hydrophobic interactions with residues M66 and L69. Interestingly, the indole moiety of PF74 is in close proximity to three residues in the adjacent CANTD: Y169, R173 and K182 [28]. However, these molecular details have not been extensively explored for the design of improved PF74 analogs. The Zhan and Liu group recently reported a new PF74-based chemotype where the indole moiety was replaced by the simpler and synthetically more accessible 1,2,3-triazole ring, though substantially decreased potency (by >10-fold) was observed [35,36]. We report herein a comprehensive chemical profiling on PF74 via chemical synthesis of a large number of PF74 analogs, biological assays and molecular modeling. Sites for structural diversification are shown in Figure 2. Although some analogs are known from two previous patent applications [37,38], the comprehensive SAR required to inform details of ligand binding and formulate the pharmacophore remains unknown.
Figure 2.
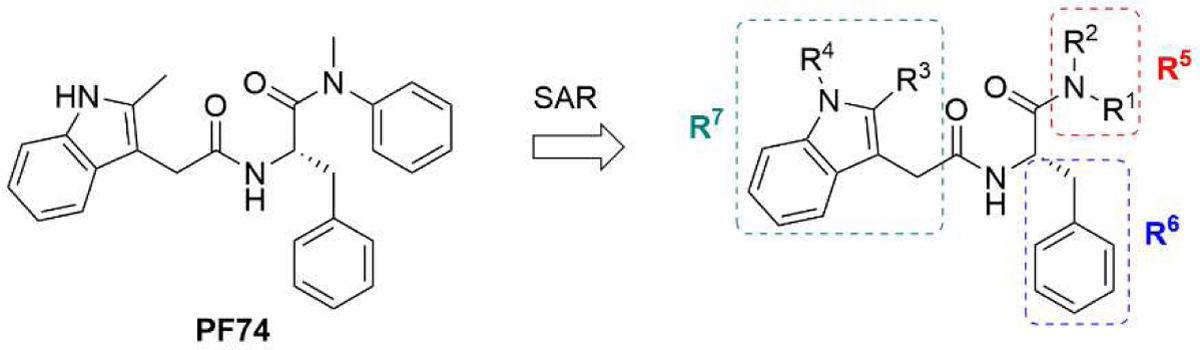
The plan for PF74 chemical profiling. R1-R7 indicates sites for structural modifications.
2. Results and discussion
2.1. Chemistry
The general synthetic strategy for analogs 1–46 is delineated in Scheme 1. Briefly, indoleacetic acid derivatives (a) were reacted with methyl L-phenylalanine hydrochloride in the presence of HATU and DIPEA in DMF to afford intermediates (b). Upon hydrolysis, b were converted to carboxylic acid intermediates c which were treated with various amines to produce PF74 analogs 1–46. The synthesis of analogs 47–72 is outlined in Scheme 2. Commercially available (tert-butoxycarbonyl)-L-phenylalanine (d) was treated with various amines using HATU or T3P as the coupling agent in the presence of DIPEA to afford carboxamide intermediates e. Removal of the Boc protecting group of e yielded f which were reacted with indoleacetic acid derivatives to give PF74 analogs 47–72. The synthesis of analogs 73–78 was accomplished using a route similar to the synthesis of 1–72 and the details are outlined in the supplemental information.
Scheme 1.
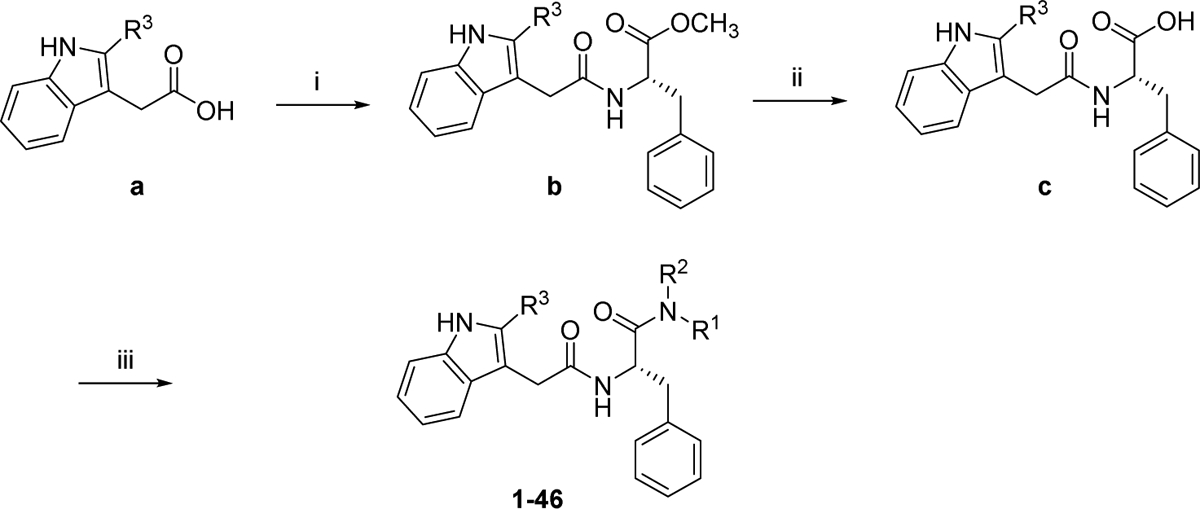
Synthetic strategy for analogs 1–46. Reagents and conditions: (i) methyl L-phenylalaninate hydrochloride, HATU, DIPEA, DMF, rt, 12 h; (ii) LiOH, MeOH/H2O, rt, overnight; (iii) amine, HATU (or T3P), DIPEA, DMF, rt, 12 h.
Scheme 2.
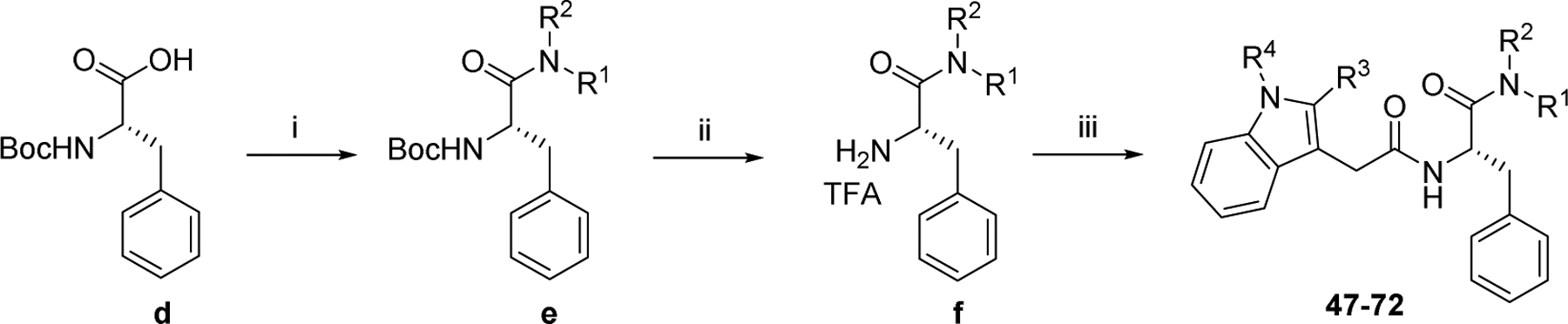
Synthetic strategy for analogs 47–72. Reagents and conditions: (i) amine, HATU (or T3P), DIPEA, DMF, rt, 12 h; (ii) TFA; DCM, rt, 4–6 h; (iii) indoleacetic acid analogue, HATU, DIPEA, DMF, rt, 12 h.
2.2. Biological assays and SAR
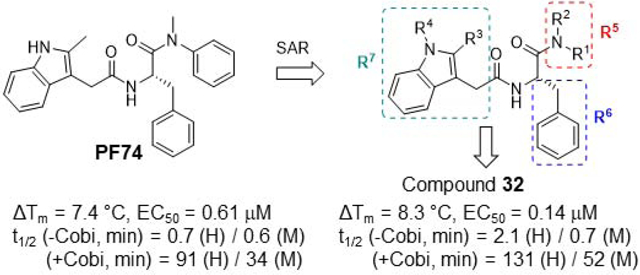
The chemical profiling was based on the structure-activity relationships (SARs) of two main structural domains of PF74: the aniline domain (the right end) and the indole domain (the left end). The activity was assessed first with a biophysical protein stability assay, the thermal shift assay, where the effect of the compound was measured by the change in protein melting point compared to DMSO control (ΔTm). A positive value in ΔTm indicates a stabilizing effect on the protein. All compounds were then screened at 20 μM against HIV-1 in a cell-based assay to determine antiviral activity. Compounds demonstrating inhibition were then further screened at 2 μM against HIV-1. Promising compounds were further evaluated for antiviral EC50 valules. All compounds were tested for cytotoxicity either by screening at 100 or 50 μM, or determination of CC50 values. For some compounds, % inhibition at two concentrations (2 μM and 20 μM) was reported instead of the EC50. In general, a good agreement between the stability assay and the antiviral assay was observed. The parent compound PF74 was resynthesized and tested in these assays (1, ΔTm = 7.4 °C, EC50 = 0.61 μM, CC50 = 76 μM). Many compounds were also tested for plasma and liver microsomal stability. Molecular modeling was performed for a few selected compounds to help understand the SAR.
2.2.1. SAR of R1, R2 and R3
This part of the SAR concerns mainly the aniline domain of PF74 (R1 and R2) and the methyl effect (R3) on the indole ring, with the synthesis of 46 analogs (Table 1). Most prominently, R2 was highly sensitive to SAR as all unsubstituted analogs (R2 = H, 11–21, 38–45) were essentially inactive in both the CA stability and antiviral assays, whereas all closely-related analogs with a small alkyl group (R2 = methyl: 2–7, 23–37; or R2 = ethyl: 22, 46) remained active in both assays. This SAR observation strongly suggests that the molecular interactions between the small alkyl group (R2 = methyl or ethyl) and the N53 and T107 residues (Figure 1, D) are essential for ligand binding. By contrast, the methyl group at the 2 position of the indole ring (R3) appears to be largely expendable. Removing this methyl group generally led to reduced CA stability but improved antiviral activity (Table 1: 23 vs 1, 24 vs 2, 26 vs 3). Therefore, the methyl group did not produce discernible benefits to the overall activity profile, and without the methyl group analog synthesis could be simplified. As for R1, the SAR showed a strong bias favoring a phenyl ring over heterocyclic rings. A prominent SAR observation regarding R1 was that bioisosteric replacement of the phenyl ring with a pyrazole (8) or pyridine (9, 36) resulted in complete loss of anti-HIV-1 activity (EC50 > 20 μM) and no effect on CA stability (ΔTm = 0 °C). Although another heterocycle, a pyridazine, in the aniline domain produced a moderate effect on CA hexamer stability and anti-HIV-1 activity (10: ΔTm = 3.2 °C, EC50 = 3.7 μM), this level of activity still reflects a significant decrease when compared with analogs that have a phenyl ring. These observations inform a clear preference of R1 being a phenyl ring over a heterocyclic ring. Remarkably, para-substitution with a chlorine on the otherwise inactive pyridine analog (36) generated an analog (37) with a moderate effect on CA hexamer stability (ΔTm = 4.9 °C) and potent antiviral activity (EC50 = 0.26 μM). A similar halogen effect was observed with a para-bromine substituent, which drastically enhanced both the CA hexamer stability and anti-HIV-1 activity (32: ΔTm = 8.3 °C, EC50 = 0.14 μM vs 23: ΔTm = 6.1 °C, EC50 = 0.46 μM). Importantly, analogs with a meta-chlorine (31), meta-bromine (33) and para- (28) or meta-fluorine (29) all exhibited substantially reduced potencies, strongly suggesting that there could be a halogen bond interaction (see docking studies). Finally, electron-donating groups at the para position of the phenyl ring, such as OMe (24) and Me (26) also led to largely improved antiviral activity (24, 26 vs 23), though significant improvement in CA hexamer stability was not observed. For these electron-donating groups, substitution at the meta position was similarly undesirable (25 vs 24, 27 vs 26).
Table 1.
Anti-HIV-1 activity, cytotoxicity, and CA hexamer stability profiles of 1–46 (R1, R2 and R3).
 | ||||||
|---|---|---|---|---|---|---|
| Compd | R1 | R2 | R3 | EC50a(μM) or % inhibition at 2 / 20 μM | CC50b (μM) | TSA ΔTm (°C)c |
| PF74 (1) |  |
Me | Me | 0.61 ± 0.2 | 76 ± 9 | 7.4 |
| 2 |  |
Me | Me | 0.63 ± 0.1 | > 100 | 8.4 |
| 3 | 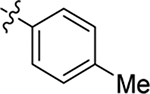 |
Me | Me | 0.31 ± 0.1 | 72 ± 3 | 9.1 |
| 4 | 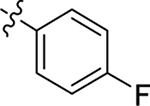 |
Me | Me | 0.67 ± 0.03 | > 50 | 7.0 |
| 5 | 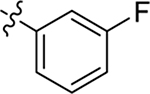 |
Me | Me | 0.69 ± 0.01 | > 50 | 6.2 |
| 6 | 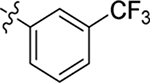 |
Me | Me | 0.58 ± 0.2 | 18 ± 1 | 5.4 |
| 7 | 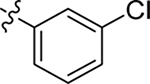 |
Me | Me | 1.2 ± 0.7 | < 50 | 7.6 |
| 8 | 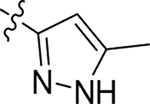 |
Me | Me | > 20 | < 100 | 0 |
| 9 | 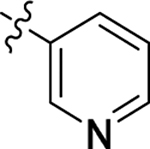 |
Me | Me | > 20 | > 100 | 0 |
| 10 | 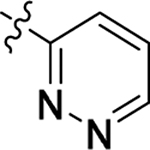 |
Me | Me | 3.7 ± 0.4 | > 100 | 3.2 |
| 11 | 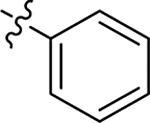 |
H | Me | > 20 | 40 ± 0.4 | 0.7 |
| 12 | 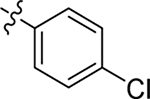 |
H | Me | > 20 | > 100 | 0 |
| 13 | 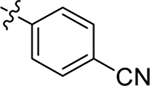 |
H | Me | > 20 | > 100 | −0.6 |
| 14 | 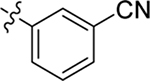 |
H | Me | > 20 | > 100 | −0.5 |
| 15 | 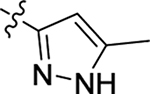 |
H | Me | > 20 | > 100 | 0 |
| 16 | 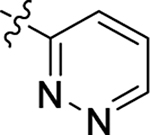 |
H | Me | > 20 | > 100 | 0 |
| 17 | 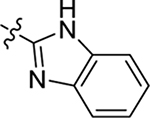 |
H | Me | 11 ± 0.8 | 15 ± 0.4 | −0.6 |
| 18 | 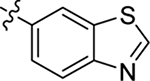 |
H | Me | 10 ± 0.7 | 49 ± 1 | 1.4 |
| 19 | 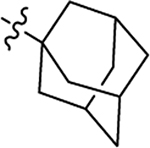 |
H | Me | > 20 | > 100 | −0.8 |
| 20 | 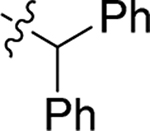 |
H | Me | > 20 | > 100 | 0 |
| 21 | 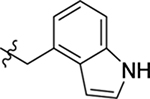 |
H | Me | > 20 | 36 ± 4 | 0 |
| 22 |  |
Et | Me | 0.25 ± 0.03 | 82 ± 10 | 8.5 |
| 23 | 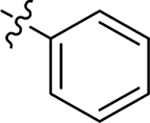 |
Me | H | 0.46 ± 0.1 | > 100 | 6.1 |
| 24 | 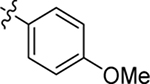 |
Me | H | 0.23 ± 0.02 | > 100 | 6.2 |
| 25 | 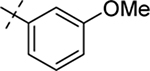 |
Me | H | 41/97 | > 50 | 1.3 |
| 26 | 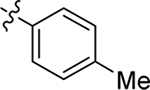 |
Me | H | 0.28 ± 0.04 | 66 ± 6 | 6.6 |
| 27 | 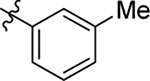 |
Me | H | 37/98 | > 50 | 1.6 |
| 28 | 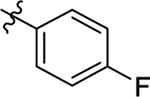 |
Me | H | 1.8 ± 0.1 | > 100 | 3.3 |
| 29 | 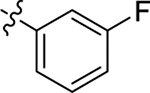 |
Me | H | 39/97 | ~ 50 | 2.5 |
| 30 | 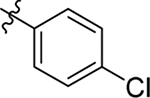 |
Me | H | 0.45 ± 0.05 | < 50 | 4.6 |
| 31 | 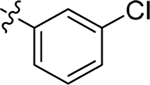 |
Me | H | 38/99.5 | < 50 | 4.9 |
| 32 | 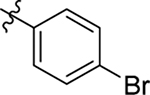 |
Me | H | 0.14 ± 0.03 | 45 ± 7 | 8.3 |
| 33 | 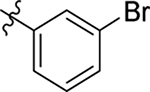 |
Me | H | 2.0 ± 0.1 | <50 | 3.5 |
| 34 | 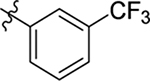 |
Me | H | 16/96 | < 50 | 2.8 |
| 35 | 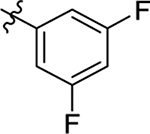 |
Me | H | 2.5 ± 0.2 | < 50 | 2.3 |
| 36 | 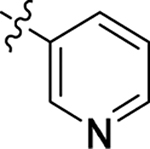 |
Me | H | > 20 | > 100 | −0.8 |
| 37 | 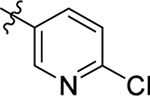 |
Me | H | 0.26 ± 0.02 | 57 ± 0.3 | 4.9 |
| 38 |  |
H | H | > 20 | 96 ± 6 | 0 |
| 39 | 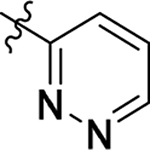 |
H | H | > 20 | > 100 | 0.8 |
| 40 | 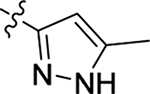 |
H | H | > 20 | > 100 | 0 |
| 41 | 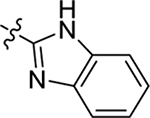 |
H | H | > 20 | 44 ± 0.5 | −0.6 |
| 42 | 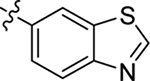 |
H | H | > 20 | 68 ± 9 | 0 |
| 43 | 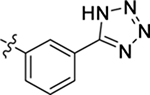 |
H | H | > 20 | > 100 | −0.7 |
| 44 | 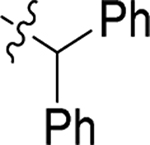 |
H | H | > 20 | > 100 | −1.6 |
| 45 | 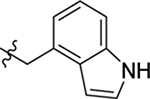 |
H | H | > 20 | < 100 | 0.5 |
| 46 |  |
Et | H | 0.44 ± 0.001 | 70 ± 2 | 6.4 |
Concentration of compound inhibiting HIV-1 replication by 50%, expressed as the mean ± standard deviation from at least two independent experiments.
Concentration of compound causing 50% cell death, expressed as the mean ± standard deviation from at least two independent experiments.
TSA: thermal shift assay. ΔTm: change of CA hexamer melting point in presence of compound compared to DMSO control.
2.2.2. SAR of R4
Having established the strong preference for a phenyl ring (R1) in the aniline domain, the requirement of a small alkyl group (R2) on the aniline N, and the tolerance of removing the methyl group (R3) on the indole ring, we turned our attention to the N-1 position (R4) of the indole domain for further SAR. All compounds synthesized herein include an unsubstituted or monosubstituted phenyl ring as R1, and a methyl or ethyl group as R2 to maximize the potency. First, we added an ethyl group to the N-1 position of the indole ring to probe the tolerance of modifying the H-bonding capability of the indole NH. Of the six analogs synthesized (47–52), five demonstrated drastically improved CA hexamer stability (ΔTm = 10.7–11.9 °C vs 7.4 °C for PF74), as well as enhanced anti-HIV-1 activity (EC50 = 0.22–0.33 μM vs 0.61 μM for PF74). The only exception, compound 52 (ΔTm = 9.2 °C, EC50 = 0.49 μM) was also better than PF74. As for the mono-substituent on the phenyl ring, no significant difference was observed between electron-donating and -withdrawing groups (48 vs 49, 51). Next, we synthesized analogs with a bigger R4 (isopropyl) group. These analogs (54–59) showed similar HIV-1 inhibition but a decreased effect on CA hexamer stability when compared with the ethyl series (47–52). These SAR trends suggest binding changes as compared to the PF74 binding mode (Figure 1, D) where the indole NH forms an important H-bond with Q63. The improved CA hexamer stability and antiviral activity may reflect favorable interactions formed by the ethyl or isopropyl group with their surrounding residues (see docking studies). Since R3 SAR tolerates the deletion of the methyl group, we also synthesized a series of compounds (61–72) without the methyl (R3 = H) and with either an ethyl (61–66) or an isopropyl group (67–72) at R4. These compounds exhibited a similar activity profile to their R3 methyl congeners. Significantly, the totality of the SAR in Table 2 confirmed a strong preference for para-substitution over meta-substitution on the R1 phenyl ring for both the CA hexamer stability and antiviral activity (Table 2: 51 vs 52, 58 vs 59, 63 vs 64, 65 vs 66, 70 vs 71). Another notable SAR was that replacing the R2 methyl group with an ethyl conferred decreased anti-HIV-1 activity (Table 2: 53 vs 47, 60 vs 54, 72 vs 67). Our overall SAR identified the methyl group as the optimal R2 substituent. Finally, it should be noted that all analogs with either ethyl or isopropyl in the N-1 position (R4) were more cytotoxic than the N-1 unsubstituted compounds, suggesting that the benefit of alkylating the indole NH could be limited.
Table 2.
Anti-HIV-1 activity, cytotoxicity, and CA hexamer stability profiles of 47–72 (R4).
 | |||||||
|---|---|---|---|---|---|---|---|
| Compd | R1 | R2 | R3 | R4 | EC50 (μM) or % inhibition at 2 / 20 μM | CC50 (μM) | TSA μTm (°C) |
| PF74 (1) |  |
Me | Me | H | 0.61 ± 0.2 | 76 ± 9 | 7.4 |
| 47 |  |
Me | Me | Et | 0.28 ± 0.1 | 21 ± 1 | 10.8 |
| 48 |  |
Me | Me | Et | 0.23 ± 0.05 | 18 ± 0.1 | 11.0 |
| 49 | 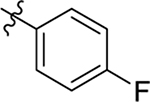 |
Me | Me | Et | 0.22 ± 0.1 | 19 ± 3 | 10.7 |
| 50 | 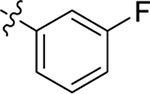 |
Me | Me | Et | 0.33 ± 0.1 | 20 ± 2 | 10.8 |
| 51 | 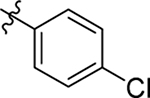 |
Me | Me | Et | 0.24 ± 0.09 | 27 ± 11 | 11.9 |
| 52 | 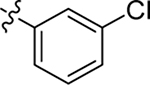 |
Me | Me | Et | 0.49 ± 0.2 | 16 ± 6 | 9.2 |
| 53 |  |
Et | Me | Et | 0.56 ± 0.2 | 20 ± 2 | 10.0 |
| 54 |  |
Me | Me | iPr | 0.28 ± 0.02 | 16 ± 1 | 7.8 |
| 55 | 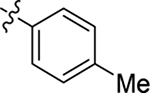 |
Me | Me | iPr | 0.63 ± 0.3 | 10 ± 2 | 8.4 |
| 56 | 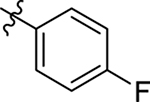 |
Me | Me | iPr | 0.32 ± 0.02 | 20 ± 0.8 | 7.5 |
| 57 | 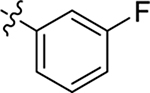 |
Me | Me | iPr | 0.29 ± 0.02 | 17 ± 2 | 7.0 |
| 58 | 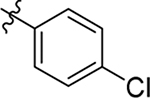 |
Me | Me | iPr | 0.15 ± 0.01 | 14 ± 0.6 | 8.5 |
| 59 | 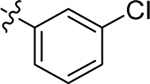 |
Me | Me | iPr | 0.36 ± 0.05 | 14 ± 2 | 5.5 |
| 60 | 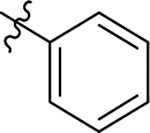 |
Et | Me | iPr | 39/99.7 | 23 ± 0.2 | 7.0 |
| 61 | 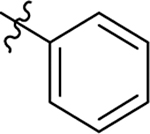 |
Me | H | Et | 0.31 ± 0.02 | 23 ± 0.2 | 6.7 |
| 62 | 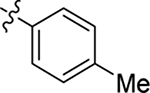 |
Me | H | Et | 0.34 ± 0.1 | 14 ± 1 | 8.6 |
| 63 | 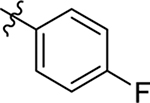 |
Me | H | Et | 0.27 ± 0.01 | 18 ± 0.9 | 7.1 |
| 64 | 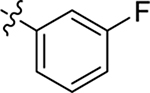 |
Me | H | Et | 0.84 ± 0.06 | 31 ± 5 | 5.9 |
| 65 | 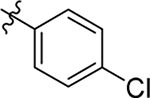 |
Me | H | Et | 0.25 ± 0.01 | 16 ± 2 | 8.6 |
| 66 | 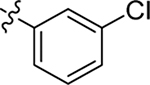 |
Me | H | Et | 2.0 ± 0.2 | 20 ± 0.3 | 5.8 |
| 67 |  |
Me | H | iPr | 0.33 ± 0.01 | 21 ± 4 | 6.0 |
| 68 | 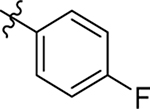 |
Me | H | iPr | 0.37 ± 0.02 | 18 ± 4 | 6.5 |
| 69 | 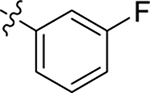 |
Me | H | iPr | 0.32 ± 0.04 | 12 ± 2 | 5.8 |
| 70 | 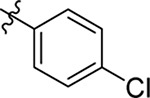 |
Me | H | iPr | 0.32 ± 0.02 | 10 ± 4 | 7.1 |
| 71 | 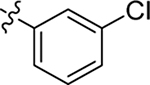 |
Me | H | iPr | 1.3 ± 0.3 | 13 ± 2 | 4.5 |
| 72 | 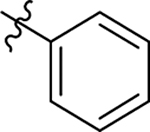 |
Et | H | iPr | 0.87 ± 0.05 | 15 ± 0.6 | 5.7 |
Concentration of compound inhibiting HIV-1 replication by 50%, expressed as the mean ± standard deviation from at least two independent experiments.
Concentration of compound causing 50% cell death, expressed as the mean ± standard deviation from at least two independent experiments.
TSA: thermal shift assay. ΔTm: change of CA hexamer melting point in presence of compound compared to DMSO control.
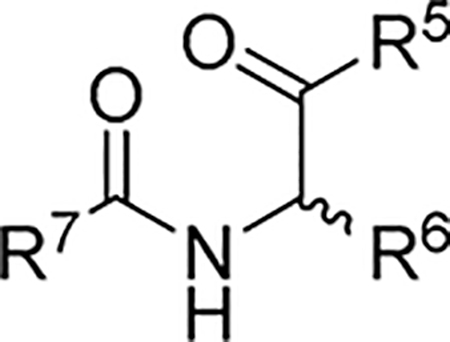
2.2.3. SAR of R5, R6 and R7
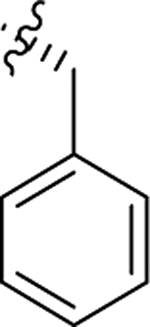
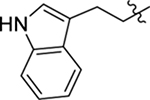
As part of our extended SAR studies, we synthesized and tested analogs with skeletal changes in aniline (R5), the side chain of phenylalanine (R6), and indole (R7) domains (Table 3). Significantly, replacing aniline with a phenylpyrrolidine-like structure led to a complete loss of activity (73 and 74), and removing aniline also had little to no effect on CA hexamer stability and abrogated antiviral activity (75). These observations confirmed that the aniline domain was crucial to maintaining CA hexamer stability and anti-HIV-1 activity. Analog 76 which has an extended indole ring linker (one more methylene) showed substantially reduced CA hexamer stability and antiviral activity when compared with 23. R6 SAR was not extensively pursued, though replacement of phenylalanine with a D-phenylglycine (77) led to a weak effect on CA hexamer stability (ΔTm = 1.5 °C) and a complete loss of antiviral activity (EC50 > 20 μM). Addition of a meta-bromine on the phenylalanine ring (78) also conferred significantly reduced CA hexamer stability and antiviral activity compared to PF74 (78 vs PF74).
Table 3.
Anti-HIV-1 activity, cytotoxicity, and CA hexamer stability profiles of 73–78 (R5, R6, and R7).
 | ||||||
|---|---|---|---|---|---|---|
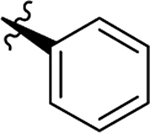
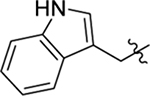
| Compd | R5 | R6 | R7 | EC50a (μM) or % inhibition at 2 / 20 μM | CC50b (μM) | TSA ΔTm (°C)c |
| 23 |  |
 |
 |
0.46 ± 0.1 | > 100 | 6.1 |
| 73 | 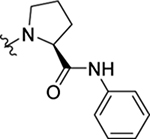 |
 |
 |
> 20 | > 50 | 0.6 |
| 74 | 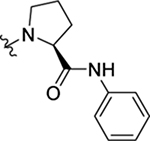 |
 |
 |
> 20 | > 50 | 0 |
| 75 | OH |  |
 |
> 20 | > 50 | −0.6 |
| 76 | 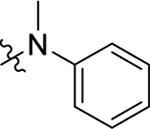 |
 |
 |
0/98 | > 50 | 2.3 |
| 77 | 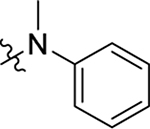 |
 |
 |
>20 | > 50 | 1.5 |
| 78 | 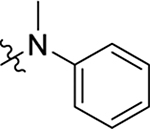 |
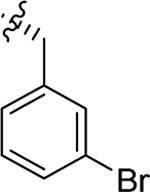 |
 |
0/94 | < 50 | 3.4 |
| PF74 (1) | 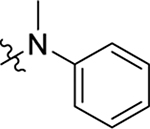 |
 |
 |
0.61 ± 0.2 | 76 ± 9 | 7.4 |
Concentration of compound inhibiting HIV-1 replication by 50%, expressed as the mean ± standard deviation from at least two independent experiments.
Concentration of compound causing 50% cell death, expressed as the mean ± standard deviation from at least two independent experiments.
TSA: thermal shift assay. ΔTm: change of CA hexamer melting point in presence of compound compared to DMSO control.
2.3. Stability Studies in Plasma and Liver Microsomes
A major barrier to further developing PF74 or its analogs is the prohibitively low metabolic stability. To gauge the medicinal chemistry value of PF74 analogs we tested their plasma stability in human and mouse plasma, and conducted metabolic stability assays in human liver microsomes (HLMs) and mouse liver microsomes (MLMs). All compounds showed excellent plasma stability without appreciable degradation after 6 hours (Table 4). Results from the microsomal stability assay indicated that, like PF74, all analogs suffer from metabolic instability with very short half-life (t1/2). Although the addition of an electron-withdrawing group, such as F (28, 63) or Br (32), to the phenyl ring increased the microsomal metabolic half-life, all analogs remained highly susceptible to NADPH-dependent metabolism. Peptidomimetics are known to be good substrates for CYP3A, the major liver metabolizing enzyme [39]. We hypothesized that a CYP3A inhibitor could prevent PF74 analogs from quick phase I metabolism to allow a therapeutically meaningful half-life. Toward this end, pre-incubation of two analogs, PF74 and its bromine congener 32, with CYP3A inhibitor Cobicistat [40] (Cobi) in the liver microsomes drastically improved their microsomal metabolic stability, as the half-life of PF74 increased by 130-fold (HLM) and 57-fold (MLM); and the half-life of 32 increased by 62-fold (HLM) and 74-fold (MLM) (Table 4). These results strongly indicate that CYP3A may indeed play an important role in the metabolism of PF74 analogs, and that the use of a CYP3A inhibitor in combination therapy settings could be a potential strategy to enhance their antiviral efficacy.
Table 4.
Summary of in vitro stability in plasma and liver microsomes.
| Compound Number | Plasma Stability | Microsomal Stability | ||
|---|---|---|---|---|
| t1/2 (h) | Phase I, t1/2 (min) | |||
| Human | Mouse | HLMa (+Cobi) | MLMb (+Cobi) | |
| PF74 (1)c | >6 | >6 | 0.7 (91) | 0.6 (34) |
| 2 | >6 | >6 | 1 | 0.7 |
| 3 | >6 | >6 | 0.9 | 0.5 |
| 4 | >6 | >6 | 1 | 0.5 |
| 22 | >6 | >6 | 0.9 | 0.5 |
| 24 | >6 | >6 | 1.6 | 0.6 |
| 26 | >6 | >6 | 1.5 | 0.6 |
| 28 | >6 | >6 | 1.3 | 0.5 |
| 32c | >6 | >6 | 2.1 (131) | 0.7 (52) |
| 35 | >6 | >6 | 0.9 | 0.6 |
| 37 | >6 | >6 | 2.1 | 0.7 |
| 47 | >6 | >6 | 1 | 0.5 |
| 49 | >6 | >6 | 1 | 0.5 |
| 54 | >6 | >6 | 0.6 | 0.5 |
| 56 | >6 | >6 | 0.6 | 0.5 |
| 61 | >6 | >6 | 0.9 | 0.5 |
| 63 | >6 | >6 | 1.3 | 0.5 |
| 67 | >6 | >6 | 0.9 | 0.5 |
| 68 | >6 | >6 | 0.9 | 0.5 |
| Verapamil | ND | ND | 15.1 | 4.2 |
HLM: human liver microsome
MLM: mouse liver microsome
Microsomal stability measured in the absence and presence of CYP3A inhibitor Cobi.
2.4. Molecular modeling
To help understand some of the afore-mentioned SAR, we performed molecular modeling with selected compounds based on the co-crystal structure of native full length HIV-1 capsid protein bound to PF74 (PDB code: 4XFZ) [28]. First, compound 23, a 3-H indole analog of PF74, interacted with the native form of the CA-hexamer in a similar fashion to that of PF74 (Figure 3A). Observed key binding interactions include i) extensive H-bonding between the terminal amide of N53 and carbonyl and NH of the phenylalanine backbone in PF74, between K70 and the other carbonyl in PF74, and between Q63 and the indole NH of PF74; ii) cation-π interaction between protonated K70 and aromatic moieties, such as, benzyl, aniline, and both aromatic rings of the indole moiety. Given the proximity to the aniline moiety, potential increased interactions with two key residues, N74 of the CANTD and Q179 of the adjacent CACTD, could play important roles in increasing the potency of these compounds. A substitution on the aniline ring could elongate its reach to interact with other surrounding residues. As ascertained by superposed docking results (Figure 3B), presence of a halogen atom at the para-position was found to rotate the aniline moiety such that it achieves a maximal proximity with N74 and Q179 for potential halogen-bonding. Specifically, halogen substituents (Cl or Br) of compounds 32 (olive), 37 (magenta), and 58 (azure) are oriented such that they could interact with the carbonyl of N74 through halogen-bonding (Figure 3B, boxed). Similar halogen-bonding may also be possible with the carbonyl of Q179. This halogen-bonding could account for increased potency of these compounds over their non-halogen congeners (32 vs 23, 37 vs 36, 58 vs 54). By contrast, introduction of a halogen atom at the meta-position of aniline moiety may not be desirable, as exemplified with 3-bromo aniline analog 33 in which the 3-Br led to an orientation such that halogen-bonding may not be possible (arrows, Figure 3C) and the cation-π interaction may be lost, resulting in a less favorable docking score than its 4-bromo analog 32 (Glide score: −5.7 for 33 vs −7.8 for 32). These docking results were consistent with the observed activity profiles of 32 and 33. To understand the unexpected increase in potency observed for the N-isopropyl substituted indole analogs over their NH congeners, docking experiments were performed with analog 58. As depicted in Figure 3D, the N-isopropyl group of the indole ring was found to fit nicely into the cavity created around residues Q63, M66, Q67, Y169, R173, K182. The extra space occupied in this cavity and the resulting molecular interactions may compensate for the loss of the key H-bond predicted for the free N-H with Q63. Docking scores and ligand interaction diagrams (LIDs) for additional analogs are described in the supplemental information.
Figure 3.
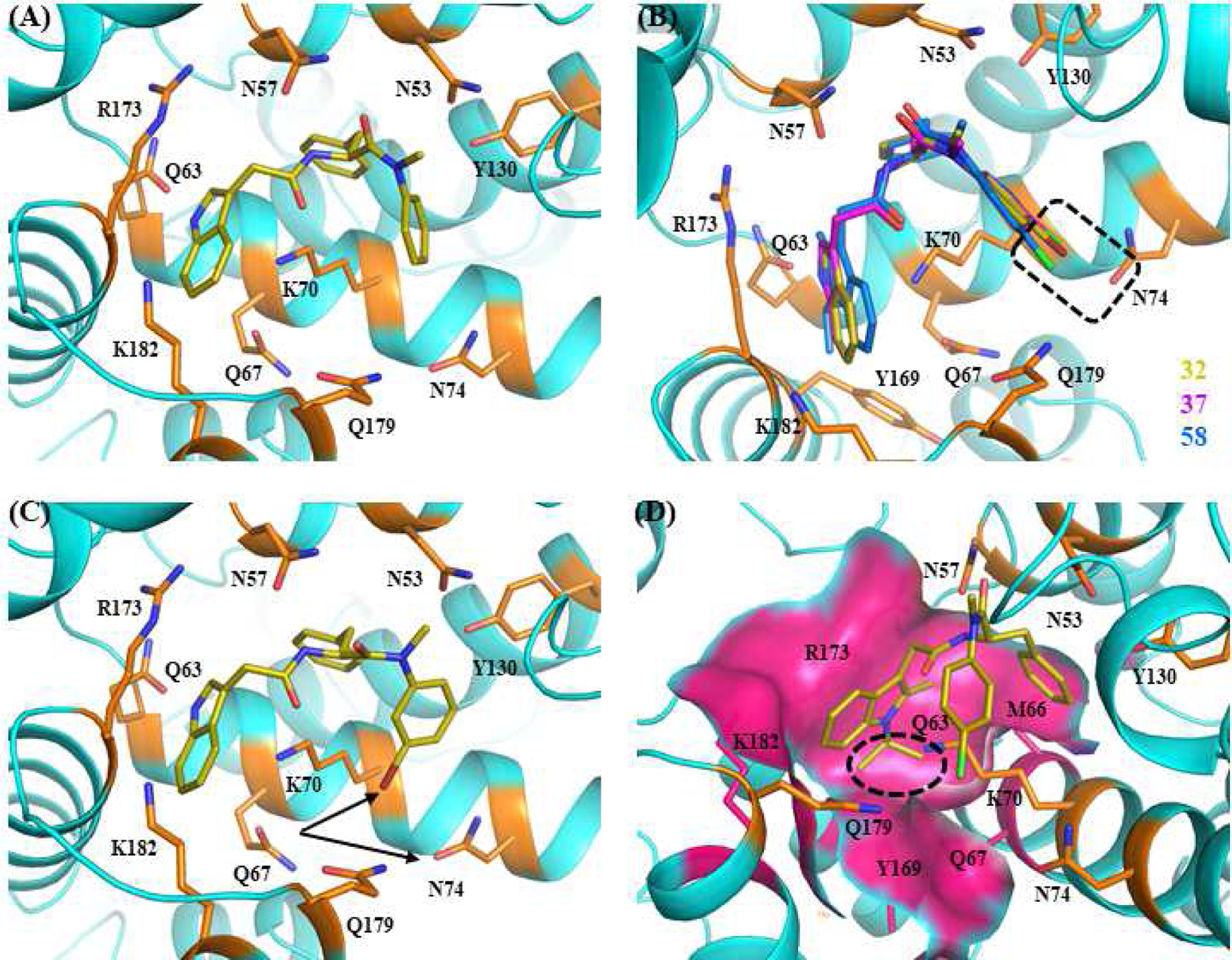
Docking poses of selected compounds based on native full length HIV-1 capsid protein bound to PF74 (PDB code: 4XFZ). (A) Predicted binding mode of 23. Glide score (kcal/mol): −6.3. (B) Superimposition of predicted binding mode of 32, 37, and 58. Glide scores (kcal/mol): 32(−7.8), 37(−7.8), 58(−6.0)). (C) Predicted binding mode of 33. Glide score (kcal/mol): −5.7. (D) Predicted fitting of the N-isopropyl group (circled) on the indole moiety of 58 within the cavity formed around residues Q63, M66, Q67, Y169, R173, K182. The nitrogen, oxygen, bromine, and chlorine atoms are colored blue, red, green, and cherry red, respectively.
3. Conclusions
PF74, a peptidomimetic and HIV-1 CA-targeting antiviral, binds to a pocket between viral CANTD and the adjacent CACTD, the same pocket where a few host factors important for viral replication also bind. Although structurally well-characterized and potent in cell culture, PF74 suffers from prohibitively low metabolic stability, and its complete SAR informing the optimal pharmacophore remains unknown. In this study, we performed a comprehensive chemical profiling of PF74 concerning structural modifications at seven different sites. CA hexamer stability and antiviral assays on these analogs revealed a few key pharmacophore elements, including a benzene ring, a para-halogen substitution and a small alkyl N-substitution at the aniline domain, and the tolerance of the C3 and NH of the indole domain for modifications. Combining these preferred pharmacophore elements yielded analogs with largely improved CA hexamer stability and anti-HIV-1 activity when compared with PF74. Molecular modeling identified an important halogen bond between the aniline domain and N74 or Q179, and a cavity that could accommodate the N-alkyl group of the indole ring. We also demonstrated that analogs of PF74 largely lacked metabolic stability, and that their metabolic stability can be drastically improved in the presence of Cobi, suggesting that combination therapy with a CYP3A inhibitor could salvage the therapeutic values of PF74 and its analogs.
4. Experimental section
4.1. Chemistry
General Procedures.
All commercial chemicals were used as supplied unless otherwise indicated. Flash chromatography was performed on a Teledyne Combiflash RF-200 with RediSep columns (silica) and indicated mobile phase. 1H and 13C NMR spectra were recorded on a Varian 600 MHz or Bruker 400 spectrometer. Mass data were acquired using an Agilent 6230 TOF LC/MS spectrometer. All NMR and mass spectrometers are located in the shared instrument rooms at the Center for Drug Design, University of Minnesota.
4.1.1. General Procedure for Synthesis of 1–46
To a solution of indoleacetic acid derivative c (1 equiv.) in DMF (3 mL) were add HATU or T3P (2 equiv.) and DIPEA (3 equiv.), and the mixture was stirred at room temperature for 20 min before an amine (1.2 equiv.) was added. The mixture was stirred at room temperature overnight. Upon completion of the reaction, H2O was added and the reaction mixture was extracted with EtOAc (3×20 mL). The organic phases were combined and washed with brine, dried over anhydrous MgSO4, filtered and concentrated. The product was purified by combi-flash on silica gel using EtOAc in hexane (yield 45–89%).
4.1.2. (S)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-N,3-diphenylpropanamide (1).
1H NMR (600 MHz, DMSO-d6) δ 10.68 (s, 1H), 8.27 (d, J = 7.0 Hz, 1H), 7.41 – 7.25 (m, 6H), 7.18 (d, J = 7.6 Hz, 1H), 7.12 – 6.85 (m, 4H), 6.84 – 6.79 (m, 3H), 4.41 (s, 1H), 3.51 – 3.31 (m, 2H), 3.15 (s, 3H), 2.82 (d, J = 11.5 Hz, 1H), 2.67 (d, J = 11.5 Hz, 1H), 2.23 (s, 3H); 13C NMR (151 MHz, DMSO) δ 171.5, 171.0, 143.3, 138.0, 135.4, 133.3, 130.0, 129.2, 128.8, 128.4, 128.2, 128.0, 126.7, 120.3, 118.4, 118.4, 110.5, 105.3, 52.2, 37.6, 37.4, 31.3, 11.7; HRMS (ESI) m/z calcd for C27H26N3O2 [M − H]− 424.2031, found 424.2029.
4.1.3. (S)-N-(4-methoxyphenyl)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (2).
1H NMR (600 MHz, DMSO-d6) δ 10.65 (s, 1H), 8.19 (d, J = 7.6 Hz, 1H), 7.24 (d, J = 7.7 Hz, 1H), 7.19 – 7.12 (m, 1H), 7.19 – 7.03 (m, 4H), 6.90 – 6.87 (m, 3H), 6.80 – 6.81 (m, 3H), 4.34 – 4.35 (m, 1H), 3.71 (s, 3H), 3.46 – 3.31 (m, 2H), 3.06 (s, 3H), 2.79 (dd, J = 13.1, 3.7 Hz, 1H), 2.65 – 2.61 (m, 1H), 2.20 (s, 3H); 13C NMR (150 MHz, DMSO-d6) δ 171.8, 170.9, 158.9, 138.1, 135.9, 135.4, 133.3, 129.3, 129.1, 128.8, 128.5, 126.7, 120.2, 118.4, 118.4, 115.0, 110.4, 105.3, 55.8, 52.00, 37.7, 37.5, 31.3, 11.7; HRMS (ESI) m/z calcd for C28H30N3O3 [M + H]+ 456.2282, found 456.2280.
4.1.4. (S)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenyl-N-(p-tolyl)propanamide (3).
1H NMR (600 MHz, DMSO-d6) δ 10.65 (s, 1H), 8.21 (d, J = 6.2 Hz, 1H), 7.24 (d, J = 7.5 Hz, 1H), 7.16 – 7.10 (m, 3H), 7.10 – 7.04 (m, 5H), 6.91 – 6.89 (m, 1H), 6.82 – 6.78 (m, 3H), 4.38 – 4.35 (m, 1H), 3.42 – 3.31 (m, 2H), 3.08 (s, 3H), 2.80 – 2.78 (m, 1H), 2.27 – 2.61 (m, 1H), 2.27 (s, 3H), 2.20 (s, 3H); 13C NMR (150 MHz, DMSO-d6) δ 171.6, 170.9, 140.7, 138.0, 137.6, 135.4, 133.3, 130.4, 129.3, 128.8, 128.4, 127.7, 126.7, 120.3, 118.4, 118.4, 110.5, 105.3, 52.0, 37.6, 31.3, 21.0, 11.7; HRMS (ESI) m/z calcd for C28H30N3O2 [M + H]+ 440.2333, found 440.2334.
4.1.5. (S)-N-(4-fluorophenyl)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (4).
1H NMR (400 MHz, DMSO-d6) δ 10.67 (s, 1H), 8.25 (d, J = 7.6 Hz, 1H), 7.31 – 7.29 (m, 1H), 7.22 – 7.15 (m, 7H), 6.94 – 6.92 (m, 2H), 6.87 – 6.85 (m, 3H), 4.34 – 4.31 (m, 1H), 3.44 – 3.33 (m, 2H), 3.11 (s, 3H), 2.87 – 2.83 (m, 1H), 2.71 – 2.68 (m, 1H), 2.24 (s, 3H); HRMS (ESI) m/z calcd for C27H27FN3O2 [M + H]+ 444.2082, found 444.2080.
4.1.6. (S)-N-(3-fluorophenyl)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (5).
1H NMR (600 MHz, CD3OD) δ 7.35 − 7.32 (m, 2H), 7.25 (d, J = 8.0 Hz, 1H), 7.19 − 7.08 (m, 5H), 7.03 (t, J = 7.5 Hz, 1H), 6.95 (t, J = 7.4 Hz, 1H), 6.83 (s, 1H), 6.78 (d, J = 7.1 Hz, 2H), 6.65 (s, 1H), 4.61 (t, J = 7.2 Hz, 1H), 3.59 − 3.52 (m, 2H), 3.14 (s, 3H), 2.85 (dd, J = 12.8, 7.7 Hz, 1H), 2.64 (dd, J = 13.2, 7.5 Hz, 1H), 2.30 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 174.2, 173.0, 164.2 (d, JCF = 247.6 Hz), 145.3, 137.6, 137.1, 134.8, 132.2 (d, JCF = 8.8 Hz), 130.1, 129.6, 129.5, 128.0, 124.7, 121.7, 119.9, 118.5, 116.3 (d, JCF = 20.5 Hz), 115.9 (d, JCF = 22.6 Hz), 111.4, 104.7, 53.3, 39.2, 37.9, 32.3, 11.4; HRMS (ESI) m/z calcd for C27H25FN3O2[M − H]− 442.1936, found 442.1933.
4.1.7. (S)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenyl-N-(3(trifluoromethyl)phenyl)propanamide (6).
1H NMR (600 MHz, CD3OD) δ 7.63 (d, J = 7.7 Hz, 1H), 7.57 (s, 1H), 7.49 (t, J = 7.8 Hz, 1H), 7.34 (d, J = 7.8 Hz, 1H), 7.25 (d, J = 8.1 Hz, 2H), 7.19 − 7.12 (m, 4H), 7.03 (t, J = 7.5 Hz, 1H), 6.96 (t, J = 7.4 Hz, 1H), 6.78 (d, J = 7.2 Hz, 2H), 4.55 − 4.51 (m, 1H), 3.60 − 3.54 (m, 2H), 3.15 (s, 3H), 2.86 (dd, J = 13.3, 7.6 Hz, 1H), 2.66 (dd, J = 13.1, 7.2 Hz, 1H), 2.31 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 174.21, 173.03, 144.59, 137.56, 137.11, 134.75, 132.99 (q, J = 33.2 Hz), 132.76, 131.80, 130.09, 129.68, 129.57, 128.03, 126.03, 125.44, 124.9 (q, J = 271.1 Hz), 121.72, 119.92, 118.51, 111.42, 104.76, 53.29, 39.32, 37.98, 32.29, 11.38; HRMS (ESI) m/z calcd for C28H25F3N3O2 [M − H]− 492.1904, found 492.1908.
4.1.8. (S)-N-(3-chlorophenyl)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (7).
1H NMR (600 MHz, CD3OD) δ 7.34 (d, J = 7.7 Hz, 2H), 7.29 (t, J = 7.9 Hz, 1H), 7.25 (d, J = 8.0 Hz, 1H), 7.21 − 7.15 (m, 4H), 7.03 (t, J = 7.5 Hz, 1H), 6.97 − 6.95 (m, 2H), 6.80 (d, J = 7.1 Hz, 2H), 4.58 (t, J = 7.3 Hz, 1H), 3.60 − 3.53 (m, 2H), 3.12 (s, 3H), 2.85 (dd, J = 13.1, 7.8 Hz, 1H), 2.66 (dd, J = 13.1, 7.2 Hz, 1H), 2.32 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 174.2, 173.0, 145.1, 137.6, 137.1, 136.0, 134.8, 132.0, 130.2, 129.7, 129.6 129.5, 128.8, 128.0, 127.3, 121.7, 119.9, 118.5, 111.4, 104.8, 53.3, 39.4, 37.9, 32.3, 11.4; HRMS (ESI) m/z calcd for C27H25ClN3O2 [M − H]− 458.1640, found 458.1644.
4.1.9. (S)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-N-(5-methyl-1H-pyrazol-3-yl)-3-phenylpropanamide (8).
1H NMR (400 MHz, DMSO-d6) δ 10.71 (s, 1H), 8.35 – 8.11 (m, 1H), 7.31 (d, J = 7.9 Hz, 1H), 7.28 – 7.14 (m, 6H), 6.95 (t, J = 7.0 Hz, 1H), 6.85 (t, J = 7.1 Hz, 1H), 5.54 – 5.50 (m, 1H), 3.59 – 3.42 (m, 2H), 3.33 (s, 3H), 3.22 (dd, J = 13.3, 2.6 Hz, 1H), 2.82 – 2.79 (m, 1H), 2.41 (s, 3H), 2.23 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 171.4, 171.0, 158.7, 144.5, 138.6, 135.4, 133.4, 129.5, 128.8, 128.7, 128.6, 126.8, 120.4, 118.5, 118.4, 110.6, 105.1, 102.2, 54.4, 37.0, 31.5, 29.6, 14.9, 11.7; HRMS (ESI) m/z calcd for C25H28N5O2 [M + H]+ 430.2238, found 430.2240.
4.1.10. (S)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenyl-N-(pyridin-3-yl)propanamide (9).
1H NMR (400 MHz, DMSO-d6) δ 10.72 (s, 1H), 8.56 (t, J = 10.2 Hz, 1H), 8.42 – 8.40 (m, 2H), 8.08 (d, J = 8.2 Hz, 1H), 7.33 (d, J = 7.8 Hz, 1H), 7.25 – 7.13 (m, 6H), 7.07 (d, J = 5.8 Hz, 2H), 6.98 – 6.94 (m, 1H), 6.86 (t, J = 7.4 Hz, 1H), 4.58 – 4.54 (m, 1H), 4.26 (t, J = 6.1 Hz, 2H), 3.47 (s, 3H), 3.03 – 2.98 (m, 1H), 2.92 – 2.78 (m, 1H), 2.25 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 171.9, 171.2, 149.8, 148.7, 138.1, 135.5, 133.4, 129.7, 128.9, 128.6, 126.8, 122.4, 120.4, 118.6, 118.4, 110.6, 105.2, 54.6, 41.5, 38.1, 31.7, 11.8; HRMS (ESI) m/z calcd for C26H27N4O2 [M + H]+ 427.2129, found 427.2126.
4.1.11. (S)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenyl-N-(pyridazin-3-yl)propanamide (10).
1H NMR (400 MHz, DMSO-d6) δ 10.68 (s, 1H), 9.15 (t, J = 2.9 Hz, 1H), 8.40 (d, J = 7.8 Hz, 1H), 7.76 (t, J = 6.0 Hz, 2H), 7.26 (d, J = 7.8 Hz, 1H), 7.19 – 7.10 (m, 6H), 6.93 (t, J = 7.1 Hz, 1H), 6.83 (t, J = 7.1 Hz, 1H), 4.66 – 4.54 (m, 1H), 3.41 – 3.33 (m, 5H), 3.18 – 3.11 (m, 1H), 2.86 – 2.80 (m, 1H), 2.23 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 172.9, 171.1, 159.5, 150.8, 138.0, 135.4, 133.4, 129.5, 129.3, 128.8, 128.6, 126.9, 125.2, 120.3, 118.5, 118.4, 110.5, 105.1, 52.8, 37.4, 35.6, 31.2, 11.7; HRMS (ESI) m/z calcd for C25H26N5O2 [M + H]+ 428.2081, found 428.2080.
4.1.12. (S)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-N,3-diphenylpropanamide (11).
1H NMR (600 MHz, CD3OD) δ 7.39 (d, J = 7.9 Hz, 2H), 7.33 (d, J = 7.8 Hz, 1H), 7.26 (t, J = 8.2 Hz, 3H), 7.12 (d, J = 6.7 Hz, 3H), 7.08 – 7.03 (m, 2H), 6.99 (d, J = 6.6 Hz, 2H), 6.95 (t, J = 7.4 Hz, 1H), 4.74 (dd, J = 13.4, 6.0 Hz, 1H), 3.60 (q, J = 16.7 Hz, 2H), 3.05 (dd, J = 13.7, 5.9 Hz, 1H), 2.91 (dd, J = 13.7, 8.0 Hz, 1H), 2.27 (s, 3H); HRMS (ESI) m/z calcd for C26H24N3O2 [M − H]− 410.1874, found 410.1877.
4.1.13. (S)-N-(4-chlorophenyl)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (12).
1H NMR (400 MHz, DMSO-d6) δ 10.71 (s, 1H), 10.21 (s, 1H), 8.19 (d, J = 8.1 Hz, 1H), 7.58 (d, J = 8.8 Hz, 2H), 7.46 – 7.25 (m, 4H), 7.28 – 7.11 (m, 5H), 6.95 (t, J = 7.5 Hz, 1H), 6.85 (t, J = 7.1 Hz, 1H), 4.64 – 4.61 (m, 1H), 3.48 – 3.45 (m, 2H), 3.03 (dd, J = 13.7, 4.9 Hz, 1H), 2.96 – 2.80 (m, 1H), 2.24 (s, 3H); HRMS (ESI) m/z calcd for C26H25ClN3O2 [M + H]+ 446.1630, found 446.1635.
4.1.14. (S)-N-(4-cyanophenyl)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (13).
1H NMR (600 MHz, DMSO) δ 10.68 (s, 1H), 10.51 (s, 1H), 8.25 (d, J = 7.9 Hz, 1H), 7.73 – 7.71 (m, 4H), 7.29 – 7.13 (m, 6H), 6.91 (t, J = 7.4 Hz, 1H), 6.81 (t, J = 7.3 Hz, 1H), 4.63 (dd, J = 13.4, 8.7 Hz, 1H), 3.46 – 3.41 (m, 2H), 3.01 (dd, J = 13.6, 4.7 Hz, 1H), 2.88 (dd, J = 13.6, 9.7 Hz, 1H), 2.21 (s, 3H); 13C NMR (151 MHz, DMSO) δ 171.6, 171.4, 143.4, 137.7, 135.4, 133.7, 133.4, 129.6, 128.8, 128.5, 126.8, 120.3, 119.7, 119.4, 118.5, 118.3, 110.6, 105.6, 105.1, 55.5, 37.9, 31.5, 11.7; HRMS (ESI) m/z calcd for C27H23N4O2 [M − H]− 435.1826, found 435.1836.
4.1.15. (S)-N-(3-cyanophenyl)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (14).
1H NMR (600 MHz, DMSO) δ 10.72 (s, 1H), 10.43 (s, 1H), 8.26 (d, J = 7.8 Hz, 1H), 8.04 (s, 1H), 7.76 (d, J = 3.4 Hz, 1H), 7.52 (m, 2H), 7.32 (d, J = 7.7 Hz, 1H), 7.28 – 7.13 (m, 6H), 6.95 (t, J = 7.4 Hz, 1H), 6.85 (t, J = 7.3 Hz, 1H), 4.64 (dd, J = 13.2, 8.3 Hz, 1H), 3.48 (q, J = 15.3 Hz, 2H), 3.04 (dd, J = 13.4, 4.6 Hz, 1H), 2.91 (dd, J = 13.4, 4.6 Hz, 1H), 2.25 (s, 3H); 13C NMR (151 MHz, DMSO) δ 171.4, 171.3, 140.0, 137.8, 135.4, 133.4, 130.7, 129.6, 128.8, 128.5, 127.3, 126.8, 124.3, 122.3, 120.3, 119.1, 118.5, 118.3, 112.0, 110.6, 105.1, 55.4, 37.9, 31.5, 11.7; HRMS (ESI) m/z calcd for C27H23N4O2 [M − H]− 435.1826, found 435.1837.
4.1.16. (S)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-N-(5-methyl-1H-pyrazol-3-yl)-3-phenylpropanamide (15).
1H NMR (400 MHz, DMSO-d6) δ 10.70 (s, 1H), 7.95 (d, J = 7.8 Hz, 1H), 7.32 (d, J = 7.6 Hz, 1H), 7.26 (d, J = 7.8 Hz, 1H), 7.24 – 7.12 (m, 8H), 6.95 (t, J = 6.9 Hz, 1H), 6.84 (t, J = 7.0 Hz, 1H), 4,67 – 4.61 (m, 1H), 3.51 – 3.47 (m, 2H), 2.99 (dd, J = 13.8, 4.7 Hz, 1H), 2.83 (dd, J = 13.6, 9.5 Hz, 1H), 2.24 (s, 3H), 2.19 (s, 3H); HRMS (ESI) m/z calcd for C24H26N5O2 [M + H]+ 416.2081, found 416.2085.
4.1.17. (S)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenyl-N-(pyridazin-3-yl)propanamide (16).
1H NMR (400 MHz, DMSO-d6) δ 11.33 (s, 1H), 10.70 (s, 1H), 8.98 (s, 1H), 8.39 – 8.20 (m, 1H), 8.14 (d, J = 7.9 Hz, 1H), 7.69 (dd, J = 9.0, 4.7 Hz, 1H), 7.31 – 7.19 (m, 6H), 6.95 (t, J = 7.5 Hz, 1H), 6.84 (t, J = 7.4 Hz, 1H), 4.84 – 4.78 (m, 1H), 3.47 (q, J = 15.3 Hz, 2H), 3.11 (dd, J = 13.6, 4.2 Hz, 1H), 2.91 (dd, J = 13.5, 10.2 Hz, 1H), 2.23 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 172.7, 171.5, 155.9, 149.0, 137.9, 135.4, 133.4, 129.8, 129.0, 128.9, 128.5, 126.8, 120.3, 118.7, 118.6, 118.3, 110.6, 105.1, 55.4, 37.6, 31.5, 11.7; HRMS (ESI) m/z calcd for C24H24N5O2 [M + H]+ 414.1925, found 414.1928.
4.1.18. (S)-N-(1H-benzo[d]imidazol-2-yl)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (17).
1H NMR (400 MHz, DMSO-d6) δ 12.01 (s, 1H), 11.78 (s, 1H), 10.73 (s, 1H), 8.12 – 8.00 (m, 1H), 7.51 – 7.39 (m, 2H), 7.34 – 7.15 (m, 7H), 7.09 – 7.15 (m, 2H), 6.95 (t, J = 7.5 Hz, 1H), 6.86 (t, J = 7.4 Hz, 1H), 4.73 – 4.69 (m, 1H), 3.49 (q, J = 15.4 Hz, 2H), 3.10 (dd, J = 13.6, 4.3 Hz, 1H), 2.93 (dd, J = 13.6, 9.8 Hz, 1H), 2.23 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 171.4, 137.7, 135.5, 133.5, 129.7, 128.9, 128.5, 126.9, 121.7, 120.4, 118.6, 118.3, 112.0, 110.6, 105.0, 55.1, 37.6, 31.5, 11.8; HRMS (ESI) m/z calcd for C27H26N5O2 [M + H]+ 452.2081, found 452.2079.
4.1.19. (S)-N-(benzo[d]thiazol-6-yl)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide(18).
1H NMR (400 MHz, DMSO-d6) δ 10.72 (s, 1H), 10.34 (s, 1H), 9.27 (s, 1H), 8.47 (s, 1H), 8.19 (d, J = 8.1 Hz, 1H), 8.01 (d, J = 8.8 Hz, 1H), 7.57 (d, J = 8.8, 1H), 7.34 (d, J = 7.8 Hz, 1H), 7.27 – 7.18 (m, 6H), 6.97 – 6.94 (m, 1H), 6.86 (t, J = 7.4 Hz, 1H), 4.71 – 4.68 (m, 1H), 3.54 – 3.45 (m, 2H), 3.07 (dd, J = 13.6, 4.9 Hz, 1H), 2.96 – 2.90 (m, 1H), 2.25 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 171.3, 170.9, 155.2, 149.7, 137.9, 136.9, 135.5, 134.7, 133.5, 129.7, 128.9, 128.5, 126.8, 123.4, 120.4, 119.4, 118.6, 118.4, 112.4, 110.6, 105.2, 55.3, 38.2, 31.6, 11.8; HRMS (ESI) m/z calcd for C27H25N4O2S [M + H]+ 469.1693, found 469.1691.
4.1.20. (2S)-N-((1s,3R)-adamantan-1-yl)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (19).
1H NMR (600 MHz, DMSO-d6) δ 10.71 (s, 1H), 7.66 (d, J = 8.1 Hz, 1H), 7.29 (d, J = 7.6 Hz, 1H), 7.24 (s, 1H), 7.18 – 7.08 (m, 4H), 6.93 (t, J = 7.3 Hz, 1H), 6.84 (t, J = 7.2 Hz, 1H), 4.42 – 4.40 (m, 1H), 3.40 (s, 2H), 2.84 (dd, J = 13.3, 4.6 Hz, 1H), 2.74 – 2.70 (m, 1H), 2.21 (s, 2H), 1.93 (s, 3H), 1.78 (s, 5H), 1.55 (s, 5H); 13C NMR (150 MHz, DMSO-d6) δ 170.7, 170.4, 138.1, 135.5, 133.4, 129.7, 128.8, 128.3, 126.5, 120.4, 118.6, 118.3, 110.6, 105.1, 54.4, 51.1, 41.2, 38.5, 36.4, 31.8, 29.2, 11.7; HRMS (ESI) m/z calcd for C30H34N3O2 [M − H]− 468.2657, found 468.2662.
4.1.21. (S)-N-benzhydryl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (20).
1H NMR (400 MHz, DMSO-d6) δ 10.72 (s, 1H), 8.87 (d, J = 8.3 Hz, 1H), 7.97 (d, J = 8.2 Hz, 1H), 7.52 – 7.05 (m, 16H), 7.01 – 6.85 (m, 1H), 6.89 – 6.79 (m, 1H), 6.07 (d, J = 8.2 Hz, 1H), 4.70 – 4.67 (m, 1H), 3.48 – 3.40 (m, 2H), 2.98 – 2.93 (m, 1H), 2.84 – 2.81 (m, 1H), 2.22 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 171.0, 170.9, 142.7, 138.0, 135.5, 133.4, 129.7, 128.9, 128.8, 128.4, 127.9, 127.5, 127.5, 127.3, 126.7, 120.4, 118.6, 118.3, 110.6, 105.1, 56.4, 54.3, 38.4, 31.7, 11.8; HRMS (ESI) m/z calcd for C33H32N3O2 [M + H]+ 502.2489, found 502.2490.
4.1.22. (S)-N-((1H-indol-4-yl)methyl)-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-3-phenylpropanamide (21).
1H NMR (400 MHz, DMSO-d6) δ 11.10 (s, 1H), 10.71 (s, 1H), 8.41 (t, J = 5.6 Hz, 1H), 7.93 (d, J = 8.4 Hz, 1H), 7.32 – 7.25 (m, 3H), 7.22 – 7.15 (m, 6H), 7.02 – 6.93 (m, 2H), 6.86 (t, J = 7.4 Hz, 1H), 6.78 (d, J = 7.1 Hz, 1H), 6.47 (s, 1H), 4.59 – 4.49 (m, 3H), 3.44 (d, J = 2.1 Hz, 2H), 2.97 (dd, J = 13.5, 4.8 Hz, 1H), 2.81 (dd, J = 13.5, 9.3 Hz, 1H), 2.22 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 171.4, 170.9, 138.2, 136.2, 135.5, 133.4, 130.3, 129.7, 128.9, 128.4, 126.6, 125.4, 121.2, 120.3, 118.6, 118.3, 117.7, 110.8, 110.6, 105.2, 99.9, 54.5, 41.1, 38.4, 31.7, 11.8; HRMS (ESI) m/z calcd for C29H29N4O2 [M + H]+ 465.2285, found 465.2281.
4.1.23. (S)-N-ethyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-N,3-diphenylpropanamide (22).
1H NMR (600 MHz, DMSO-d6) δ 10.67 (s, 1H), 8.22 (d, J = 7.8 Hz, 1H), 7.41 – 7.37 (m, 3H), 7.27 (d, J = 7.8 Hz, 1H), 7.18 – 7.10 (m, 6H), 6.92 (t, J = 7.5 Hz, 1H), 6.82 (t, J = 7.4 Hz, 1H), 6.82 – 6.76 (m, 2H), 4.29 (td, J = 8.8, 5.0 Hz, 1H), 3.72 (ddd, J = 12.5, 8.6, 6.3 Hz, 1H), 3.51 (ddd, J = 12.5, 9.3, 6.9 Hz, 1H), 3.43 (dd, J = 15.4, 1.8 Hz, 1H), 3.37 (dd, J = 15.4, 1.8 Hz, 1H), 2.82 (dd, J = 13.4, 4.5 Hz, 1H), 2.65 (dd, J = 13.4, 9.7 Hz, 1H), 2.22 (s, 3H), 0.97 (td, J = 7.2, 2.4 Hz, 3H); 13C NMR (100 MHz, DMSO-d6) δ 170.5 (one quaternary C merging), 141.0, 137.6, 134.9, 132.8, 129.4, 128.8, 128.5, 128.4, 128.0, 127.9, 126.3, 119.8, 118.0, 117.9, 110.0, 104.9, 51.9, 43.7, 37.1, 30.9, 12.7, 11.3; HRMS (ESI) m/z calcd for C28H30N3O2 [M + H]+ 440.2333, found 440.2330.
4.1.24. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (23).
1H NMR (600 MHz, CD3OD) δ 7.42 (d, J = 7.9 Hz, 1H), 7.37 – 7.31 (m, 4H), 7.18 – 7.05 (m, 5H), 7.03 – 6.94 (m, 2H), 6.75 (d, J = 7.3 Hz, 2H), 4.66 (t, J = 7.2 Hz, 1H), 3.61 (dd, J = 22.4, 15.8 Hz, 2H), 3.16 (s, 3H), 2.87 (dd, J = 13.3, 6.6 Hz, 1H), 2.64 (dd, J = 13.3, 6.6 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 172.7, 171.7, 142.5, 136.7, 136.5, 129.4, 128.7, 128.0, 127.9, 127.2, 127.0, 126.4, 123.5, 121.1, 118.6, 118.0, 110.9, 107.7, 51.9, 37.7, 36.6, 32.2; HRMS (ESI) m/z calcd for C26H24N3O2 [M − H]− 410.1874, found 410.1878.
4.1.25. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (24).
1H NMR (600 MHz, DMSO-d6) δ 10.76 (s, 1H), 8.27 (d, J = 7.8 Hz, 1H), 7.32 – 7.26 (m, 2H), 7.12 – 7.00 (m, 7H), 6.91 – 6.79 (m, 4H), 4.40 – 4.37 (m, 1H), 3.72 (s, 3H), 3.47 – 3.40 (m, 2H), 3.08 (s, 3H), 2.83 – 2.80 (m, 1H), 2.65 – 2.60 (m, 1H); 13C NMR (150 MHz, DMSO-d6) δ 171.7, 170.8, 158.9, 138.0, 136.4, 135.9, 129.3, 129.1, 128.5, 127.6, 126.8, 124.1, 121.2, 119.1, 118.6, 115.0, 111.6, 109.2, 55.8, 51.9, 37.7, 37.7, 32.5; HRMS (ESI) m/z calcd for C27H28N3O3 [M + H]+ 442.2125, found 442.2123.
4.1.26. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(3-methoxyphenyl)-N-methyl-3-phenylpropanamide (25).
1H NMR (600 MHz, CDCl3) δ 8.34 (s, 1H), 7.46 (d, J = 7.9 Hz, 1H), 7.37 (d, J = 8.2 Hz, 1H), 7.25 − 7.21 (m, 2H), 7.15 − 7.06 (m, 5H), 7.01 (s, 1H), 6.87 (d, J = 8.3 Hz, 1H), 6.71 − 6.70 (m, 2H), 6.51 (s, 1H), 6.25 (d, J = 8.2 Hz, 1H), 4.92 − 4.88 (m, 1H), 3.77 (s, 3H), 3.69 − 3.63 (m, 2H), 3.17 (s, 3H), 2.76 (dd, J = 13.4, 7.0 Hz, 1H), 2.58 (dd, J = 13.4, 7.0 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 171.4, 170.7, 160.5, 143.6, 136.4, 136.1, 130.4, 129.2, 128.3, 127.1, 126.7, 123.7, 122.4, 119.9, 119.3, 118.8, 114.1, 112.9, 111.3, 108.8, 55.5, 51.1, 38.9, 37.6, 33.4; HRMS (ESI) m/z calcd for C27H26N3O3 [M − H]− 440.1980, found 440.1989.
4.1.27. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(p-tolyl)propanamide (26).
1H NMR (600 MHz, DMSO-d6) δ 10.77 (s, 1H), 8.27 (s, 1H), 7.33 – 7.27 (m, 2H), 7.16 – 7.10 (m, 5H), 7.04 – 6.98 (m, 3H), 6.87 (t, J = 7.5 Hz, 1H), 6.81 – 6.79 (m, 2H), 4.43 – 4.41 (m, 1H), 3.44 – 3.39 (m, 2H), 3.07 (s, 3H), 2.81 – 2.79 (m 1H), 2.71 – 2.56 (m, 1H), 2.28 (s, 3H); 13C NMR (150 MHz, DMSO-d6) δ 171.5, 170.8, 140.7, 137.9, 137.6, 136.4, 130.3, 129.2, 128.4, 127.6, 127.6, 126.7, 124.1, 121.2, 119.1, 118.6, 111.5, 109.1, 51.9, 37.7, 37.5, 32.5, 21.0; HRMS (ESI) m/z calcd for C27H28N3O2 [M + H]+ 426.2176, found 426.2176.
4.1.28. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(m-tolyl)propanamide (27).
1H NMR (600 MHz, CD3OD) δ 10.37 (s, 1H), 7.71 (d, J = 7.6 Hz, 1H), 7.42 (d, J = 7.9 Hz, 1H), 7.33 (d, J = 8.1 Hz, 1H), 7.20 − 7.07 (m, 8H), 6.98 (t, J = 7.4 Hz, 1H), 6.78 − 6.76 (m, 3H), 4.65 (q, J = 7.5 Hz, 1H), 3.63 − 3.57 (m, 2H), 3.11 (s, 3H), 2.86 (dd, J = 13.3, 7.2 Hz, 1H), 2.64 (dd, J = 13.2, 7.5 Hz, 1H), 2.23 (s, 3H); 13C NMR (100 MHz, CD3OD) δ 174.1, 173.1, 143.8, 141.1, 138.1, 137.9, 130.6, 130.2, 130.0, 129.4, 129.0, 128.5, 127.8, 125.4, 125.0, 122.6, 120.0, 119.4, 112.3, 109.2, 53.3, 39.4, 38.0, 33.7, 21.2; HRMS (ESI) m/z calcd for C27H26N3O2 [M − H]− 424.2031, found 424.2035.
4.1.29. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (28).
1H NMR (400 MHz, DMSO-d6) δ 10.77 (s, 1H), 8.30 (d, J = 7.8 Hz, 1H), 7.38 (d, J = 7.9 Hz, 1H), 7.31 (d, J = 8.1 Hz, 1H), 7.20 – 7.18 (m, 6H), 7.08 – 7.02 (m, 2H), 6.93 – 6.87 (m, 3H), 4.39 – 4.34 (m, 1H), 3.45 – 3.43 (m, 2H), 3.11 (s, 3H), 2.90 – 2.85 (m 1H), 2.68 – 2.74 (m, 1H); HRMS (ESI) m/z calcd for C26H25FN3O2 [M + H]+ 430.1925, found 430.1927.
4.1.30. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (29).
1H NMR (600 MHz, CD3OD) δ 10.37 (s, 1H), 7.82 (d, J = 7.6 Hz, 1H), 7.44 (d, J = 7.9 Hz, 1H), 7.34 − 7.28 (m, 2H), 7.19 − 7.13 (m, 3H), 7.10 − 7.05 (m, 3H), 6.98 (t, J = 7.5 Hz, 1H), 6.82 − 6.77 (m, 3H), 6.59 (s, 1H), 4.62 (dd, J = 14.8, 7.4 Hz, 1H), 3.64 − 3.58 (m, 2H), 3.12 (s, 3H), 2.88 (dd, J = 13.1, 7.8 Hz, 1H), 2.68 (dd, J = 13.2, 7.1 Hz, 1H); 13C NMR (100 MHz, CD3OD) δ 174.2, 173.1, 164.2 (d, JCF = 247.4 Hz), 145.4 (d, JCF = 9.8 Hz), 138.2, 137.8, 132.1 (d, JCF = 9.2 Hz), 130.2, 129.5, 128.5, 128.0, 125.0, 124.7, 122.6, 120.0, 119.4, 116.2 (d, JCF = 21.2 Hz), 115.9 (d, JCF = 22.9 Hz), 112.3, 109.2, 53.4, 39.3, 37.9, 33.6; HRMS (ESI) m/z calcd for C26H23FN3O2 [M − H]− 428.1780, found 428.1781.
4.1.31. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(4-chlorophenyl)-N-methyl-3-phenylpropanamide (30).
1H NMR (600 MHz, DMSO-d6) δ 10.77 (s, 1H), 8.37 (d, J = 7.5 Hz, 1H), 7.39 − 7.37 (m, 2H), 7.34 (d, J = 7.8 Hz, 1H), 7.28 (d, J = 8.1 Hz, 1H), 7.14 − 7.11 (m, 5H), 7.05 − 6.99 (m, 2H), 6.89 − 6.84 (m, 3H), 4.36 − 4.33 (m, 1H), 3.48 – 3.41 (m, 2H), 3.07 (s, 3H), 2.82 (dd, J = 13.3, 5.0 Hz, 1H), 2.66 (dd, J = 12.9, 9.5 Hz, 1H); 13C NMR (150 MHz, DMSO-d6) δ 171.4, 170.9, 142.1, 137.8, 136.4, 132.7, 129.8, 129.3, 128.6, 127.6, 126.9, 124.2, 121.3, 119.1, 118.7, 111.6, 110.0, 109.1, 52.0, 37.7, 37.5, 32.5; HRMS (ESI) m/z calcd for C26H23ClN3O2 [M − H]− 444.1484, found 444.1488.
4.1.32. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(3-chlorophenyl)-N-methyl-3-phenylpropanamide(31).
1H NMR (600 MHz, CD3OD) δ 7.46 (d, J = 7.9 Hz, 1H), 7.35 − 7.32 (m, 2H), 7.27 (t, J = 8.0 Hz, 1H), 7.22 − 7.16 (m, 4H), 7.11 − 7.09 (m, 2H), 7.00 (t, J = 7.5 Hz, 1H), 6.90 − 6.84 (m, 3H), 4.59 (t, J = 7.4 Hz, 1H), 3.66 − 3.60 (m, 2H), 3.11 (s, 3H), 2.89 (dd, J = 13.1, 8.1 Hz, 1H), 2.70 (dd, J = 13.1, 7.0 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 174.2, 173.0, 145.1, 138.1, 137.7, 135.9, 131.9, 130.2, 129.6, 129.5, 128.7, 128.5, 128.0, 127.2, 125.0, 122.6, 120.0, 119.4, 112.3, 109.2, 53.4, 39.4, 37.9, 33.6; HRMS (ESI) m/z calcd for C26H23ClN3O2 [M − H]− 444.1484, found 444.1498.
4.1.33. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(4-bromophenyl)-N-methyl-3-phenylpropanamide (32).
1H NMR (600 MHz, DMSO-d6) δ 10.77 (s, 1H), 8.37 (d, J = 7.0 Hz, 1H), 7.51 (d, J = 8.5 Hz, 2H), 7.34 (d, J = 7.4 Hz, 1H), 7.27 (d, J = 8.0 Hz, 1H), 7.14 – 7.11 (m, 3H), 7.01 – 7.04 (m, 3H), 6.93 – 6.77 (m, 3H), 4.35 – 4.32 (m, 1H), 3.40 (dt, J = 55.7, 17.8 Hz, 2H), 3.07 (s, 3H), 2.91 – 2.74 (m, 1H), 2.71 – 2.57 (m, 1H); HRMS (ESI) m/z calcd for C26H25BrN3O2 [M + H]+ 490.1125, found 490.1130.
4.1.34. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(3-bromophenyl)-N-methyl-3-phenylpropanamide (33).
1H NMR (600 MHz, DMSO-d6) δ 10.78 (s, 1H), 8.40 (d, J = 7.3 Hz, 1H), 7.52 (d, J = 7.2 Hz, 1H), 7.38 (d, J = 7.5 Hz, 1H), 7.32 − 7.26 (m, 3H), 7.20 − 7.16 (s, 4H), 7.07 (s, 1H), 7.01 (t, J = 7.5 Hz, 1H), 6.90 − 6.87 (m, 3H), 4.35 − 4.34 (m, 1H), 3.50 − 3.42 (m, 2H), 3.09 (s, 3H), 2.87 − 2.84 (m, 1H), 2.70 − 2.67 (m, 1H); 13C NMR (150 MHz, DMSO-d6) δ 171.4, 171.0, 144.8, 137.8, 136.4, 131.6, 131.1, 130.7, 129.3, 128.6, 127.6, 127.2, 126.9, 124.2, 122.1, 121.3, 119.1, 118.7, 111.6, 109.1, 52.2, 37.8, 37.5, 32.5; HRMS (ESI) m/z calcd for C26H23BrN3O2 [M − H]− 488.0979, found 488.0982.
4.1.35. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(3-(trifluoromethyl)phenyl)propanamide (34).
1H NMR (600 MHz, CDCl3) δ 8.76 (s, 1H), 7.53 (d, J = 7.8 Hz, 1H), 7.49 (d, J = 7.9 Hz, 1H), 7.42 (t, J = 7.8 Hz, 1H), 7.34 (d, J = 8.1 Hz, 1H), 7.22 − 7.11 (m, 7H), 6.91 (s, 1H), 6.78 − 6.77 (m, 2H), 6.34 (d, J = 8.0 Hz, 1H), 4.74 − 4.71 (m, 1H), 3.69 (s, 2H), 3.14 (s, 3H), 2.75 (dd, J = 12.9, 8.8 Hz, 1H), 2.64 (dd, J = 13.0, 6.1 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 171.5, 171.1, 143.0, 136.5, 135.7, 132.0 (q, JCF = 33.0 Hz), 131.2, 130.3, 129.1, 128.5, 127.1, 127.0, 124.9 (q JCF = 3.5 Hz), 123.9, 123.9, 123.4 (q, JCF = 272.7 Hz), 122.4, 119.8, 118.6, 111.5, 108.3, 51.3, 39.4, 37.6, 33.3; HRMS (ESI) m/z calcd for C27H23F3N3O2 [M − H]− 478.1748, found 478.1750.
4.1.36. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(3,5-difluorophenyl)-N-methyl-3-phenylpropanamide (35).
1H NMR (600 MHz, CDCl3) δ 8.36 (s, 1H), 7.51 (d, J = 7.9 Hz, 1H), 7.38 (d, J = 8.2 Hz, 1H), 7.24 − 7.13 (m, 6H), 7.06 (s, 1H), 6.83 − 6.82 (m, 2H), 6.75 − 6.73 (m, 1H), 6.26 − 6.23 (m, 2H), 4.75 − 4.74 (m, 1H), 3.71 (s, 2H), 3.09 (s, 3H), 2.76 − 2.73 (m, 1H), 2.67 − 2.64 (m, 1H); 13C NMR (150 MHz, CDCl3) δ 171.2, 170.9, 162.9 (d, JCF = 250.6 Hz), 162.8 (d, JCF = 250.5 Hz),136.4, 135.7, 129.3, 128.5, 127.1, 127.0, 123.7, 122.6, 120.0, 118.7 111.4, 111.0 (d, JCF = 23.1 Hz), 108.6, 103.8, 51.4, 39.3, 37.4, 33.3; HRMS (ESI) m/z calcd for C26H22F2N3O2 [M − H]− 446.1686, found 446.1690.
4.1.37. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(pyridin-3-yl)propanamide (36).
1H NMR (600 MHz, DMSO-d6) δ 10.79 (s, 1H), 8.57 (s, 1H), 8.36 (s, 1H), 8.22 (d, J = 8.0 Hz, 1H), 7.37 (d, J = 7.9 Hz, 1H), 7.30 (d, J = 8.0 Hz, 1H), 7.19 – 7.17 (m, 5H), 7.05 – 7.05 (m, 4H), 6.89 (t, J = 7.4 Hz, 1H), 4.56 – 4.54 (m, 1H), 4.25 – 4.22 (m, 2H), 3.50 (s, 3H), 3.00 – 2.97 (m, 1H), 2.85 – 2.82 (m, 1H); 13C NMR (150 MHz, DMSO-d6) δ 171.9, 171.3, 149.7, 148.7, 138.0, 136.5, 129.6, 128.6, 127.6, 126.8, 124.2, 122.3, 121.3, 119.1, 118.7, 111.7, 109.1, 54.7, 41.5, 38.0, 32.8; HRMS (ESI) m/z calcd for C25H25N4O2 [M + H]+ 413.1972, found 413.1972.
4.1.38. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(6-chloropyridin-3-yl)-N-methyl-3-phenylpropanamide (37).
1H NMR (600 MHz, CDCl3) δ 8.44 (s, 1H), 7.50 (d, J = 7.7 Hz, 1H), 7.38 (d, J = 8.2 Hz, 1H), 7.24 − 7.13 (m, 8H), 7.06 (s, 1H), 6.84 (d, J = 7.3 Hz, 2H), 6.25 (s, 1H), 4.60 − 4.56 (m, 1H), 3.70 (s, 2H), 3.10 (s, 3H), 2.75 (dd, J = 12.6, 9.6 Hz, 1H), 2.64 (dd, J = 12.9, 5.7 Hz, 1H); 13C NMR (150 MHz, CDCl3) δ 171.5, 171.1, 150.5, 148.3, 138.1, 138.1, 136.4, 135.5, 129.2, 128.7, 127.3, 127.0, 124.8, 123.7, 122.6, 120.0, 118.6, 111.4, 108.5, 51.3, 39.2, 37.8, 33.2; HRMS (ESI) m/z calcd for C25H22ClN4O2 [M − H]− 445.1437, found 445.1442.
4.1.39. (S)-2-(2-(1H-indol-3-yl)acetamido)-N,3-diphenylpropanamide (38).
1H NMR (600 MHz, DMSO) δ 10.82 (s, 1H), 10.10 (s, 1H), 8.32 (d, J = 8.1 Hz, 1H), 7.58 – 7.41 (m, 4H), 7.33 – 7.22 (m, 8H), 7.08 – 6.91 (m, 2H), 4.71 (dd, J = 9.1, 5.0 Hz, 1H), 3.59 – 3.51 (m, 2H), 3.05 (dd, J = 13.6, 4.8 Hz, 1H), 2.91 (dd, J = 13.4, 9.6 Hz, 1H); 13C NMR (151 MHz, DMSO) δ 171.1, 170.6, 139.1, 138.0, 136.3, 129.6, 129.1, 128.5, 127.6, 126.8, 124.0, 123.8, 121.3, 119.7, 119.1, 118.7, 111.6, 109.1, 55.1, 38.2, 32.7; HRMS (ESI) (−) m/z calcd for C25H22N3O2 [M − H]− 396.1718, found 396.1716.
4.1.40. (S)-2-(2-(1H-indol-3-yl)acetamido)-3-phenyl-N-(pyridazin-3-yl)propanamide (39).
1H NMR (400 MHz, DMSO-d6) δ 11.37 (s, 1H), 10.80 (s, 1H), 8.98 (s, 1H), 8.35 – 8.23 (m, 2H), 7.69 (dd, J = 9.0, 4.7 Hz, 1H), 7.36 – 7.29 (m, 4H), 7.28 – 7.19 (m, 3H), 7.11 – 6.96 (m, 2H), 6.89 (t, J = 7.4 Hz, 1H), 4.87 – 4.83 (m, 1H), 3.61 – 3.42 (m, 2H), 3.12 (dd, J = 13.6, 4.2 Hz, 1H), 2.90 (dd, J = 13.5, 10.3 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 172.7, 171.4, 155.9, 149.0, 137.9, 136.5, 129.8, 129.0, 128.5, 127.6, 126.9, 124.2, 121.3, 119.1, 118.7, 111.6, 109.1, 55.4, 37.7, 32.7; HRMS (ESI) m/z calcd for C23H22N5O2 [M + H]+ 400.1768, found 400.1771.
4.1.41. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(5-methyl-1H-pyrazol-3-yl)-3-phenylpropanamide (40).
1H NMR (400 MHz, DMSO-d6) δ 10.81 (s, 1H), 8.24 (d, J = 8.3 Hz, 1H), 7.43 – 7.30 (m, 2H), 7.28 – 7.13 (m, 5H), 7.13 – 7.00 (m, 2H), 6.92 (t, J = 7.4 Hz, 1H), 5.64 – 5.58 (m, 1H), 3.58 – 3.51 (m, 2H), 3.17 (dd, J = 13.7, 3.0 Hz, 1H), 2.91 – 2.79 (m, 1H), 2.40 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 171.3, 171.1, 157.7, 144.3, 138.3, 136.5, 129.5, 128.6, 127.6, 126.9, 124.2, 121.4, 119.2, 118.7, 111.7, 109.1, 103.2, 53.6, 37.1, 32.7, 14.8; HRMS (ESI) m/z calcd for C23H24N5O2 [M + H]+ 402.1925, found 402.1925.
4.1.42. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(1H-benzo[d]imidazol-2-yl)-3-phenylpropanamide (41).
1H NMR (400 MHz, DMSO-d6) δ 12.03 (s, 1H), 11.79 (s, 1H), 10.81 (s, 1H), 8.29 (s, 1H), 7.51 – 7.35 (m, 3H), 7.32 (d, J = 8.0 Hz, 3H), 7.28 – 7.18 (m, 3H), 7.16 – 7.01 (m, 4H), 6.91 (t, J = 7.5 Hz, 1H), 4.84 – 4.72 (m, 1H), 3.64 – 3.49 (m, 2H), 3.13 (dd, J = 13.6, 4.4 Hz, 1H), 2.93 (dd, J = 13.6, 9.9 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 171.4, 137.8, 136.5, 129.8, 128.6, 127.6, 126.9, 124.2, 121.4, 119.1, 118.8, 111.7, 109.0, 55.2, 37.7, 32.7; HRMS (ESI) m/z calcd for C26H24N5O2 [M + H]+ 438.1925, found 438.1926.
4.1.43. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(benzo[d]thiazol-6-yl)-3-phenylpropanamide (42).
1H NMR (400 MHz, DMSO-d6) δ 10.82 (s, 1H), 10.37 (s, 1H), 9.27 (s, 1H), 8.50 (d, J = 6.2 Hz, 1H), 8.34 (d, J = 8.8 Hz, 1H), 8.02 (d, J = 8.8 Hz, 1H), 7.59 – 7.56 (m, 1H), 7.42 (d, J = 7.9 Hz, 1H), 7.33 – 7.21 (m, 6H), 7.10 – 7.03 (m, 2H), 6.92 (t, J = 7.4 Hz, 1H), 4.77 – 4.72 (m, 1H), 3.60 – 3.54 (m, 2H), 3.11 – 3.06 (m, 1H), 2.97 – 2.91 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 171.2, 170.9, 155.2, 149.7, 137.9, 136.9, 136.5, 134.7, 129.7, 128.5, 127.7, 126.8, 124.2, 123.4, 121.3, 119.4, 119.2, 118.7, 112.4, 111.7, 109.2, 55.4, 38.2, 32.8; HRMS (ESI) m/z calcd for C26H23N4O2S [M + H]+ 455.1536, found 455.1538.
4.1.44. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-(3-(1H-tetrazol-5-yl)phenyl)-3-phenylpropanamide (43).
1H NMR (400 MHz, DMSO-d6) δ 10.80 (s, 1H), 10.35 (s, 1H), 8.37 (s, 1H), 8.31 (d, J = 8.0 Hz, 1H), 7.72 – 7.70 (m, 2H), 7.52 (t, J = 8.8 Hz, 1H), 7.41 (d, J = 7.9 Hz, 1H), 7.34 – 7.20 (m, 6H), 7.13 – 6.99 (m, 2H), 6.91 (t, J = 7.1 Hz, 1H), 4.82 – 4.62 (m, 1H), 3.67 – 3.48 (m, 2H), 3.08 (dd, J = 13.7, 5.1 Hz, 1H), 2.99 – 2.86 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 171.3, 171.1, 140.1, 137.9, 136.5, 130.3, 129.7, 128.6, 127.7, 126.9, 124.2, 122.3, 122.2, 121.3, 119.2, 118.7, 118.1, 111.6, 109.1, 63.6, 55.4, 38.1, 32.7; HRMS (ESI) m/z calcd for C26H24N7O2 [M + H]+ 466.1986, found 466.1991.
4.1.45. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-benzhydryl-3-phenylpropanamide (44).
1H NMR (400 MHz, DMSO-d6) δ 10.81 (s, 1H), 8.89 (d, J = 8.4 Hz, 1H), 8.13 (d, J = 8.3 Hz, 1H), 7.39 (d, J = 7.9 Hz, 1H), 7.33 – 7.25 (m, 5H), 7.25 – 7.18 (m, 9H), 7.13 (d, J = 7.1 Hz, 2H), 7.07 – 7.03 (m, 2H), 6.91 (t, J = 7.4 Hz, 1H), 6.08 (d, J = 8.4 Hz, 1H), 4.76 – 4.71 (m, 1H), 3.56 – 3.45 (m, 2H), 2.97 (dd, J = 13.5, 5.3 Hz, 1H), 2.90 – 2.77 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 171.0, 170.9, 142.7, 142.6, 138.1, 136.5, 129.7, 128.8, 128.5, 127.9, 127.7, 127.6, 127.5, 127.3, 126.7, 124.2, 121.3, 119.2, 118.7, 111.7, 109.2, 56.4, 54.4, 38.7, 32.8; HRMS (ESI) m/z calcd for C32H30N3O2 [M + H]+ 488.2333, found 488.2337.
4.1.46. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-((1H-indol-4-yl)methyl)-3-phenylpropanamide (45).
1H NMR (400 MHz, DMSO-d6) δ 11.11 (s, 1H), 10.81 (s, 1H), 8.46 (t, J = 5.6 Hz, 1H), 8.13 (d, J = 8.5 Hz, 1H), 7.40 – 7.28 (m, 4H), 7.24 – 7.15 (m, 5H), 7.07 – 6.98 (m, 3H), 6.91 (t, J = 7.4 Hz, 1H), 6.78 (d, J = 7.1 Hz, 1H), 6.48 (s, 1H), 4.65 – 4.56 (m, 1H), 4.53 (d, J = 5.5 Hz, 2H), 3.51 (s, 2H), 3.00 (dd, J = 13.5, 4.8 Hz, 1H), 2.82 (dd, J = 13.5, 9.5 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 171.5, 170.9, 138.3, 136.5, 136.2, 130.3, 129.7, 128.4, 127.7, 126.6, 126.6, 125.4, 124.2, 121.3, 121.2, 119.1, 118.7, 117.7, 111.6, 110.8, 109.2, 99.9, 54.6, 41.1, 38.5, 32.9; HRMS (ESI) m/z calcd for C28H27N4O2 [M + H]+ 451.2129, found 451.2132.
4.1.47. (S)-2-(2-(1H-indol-3-yl)acetamido)-N-ethyl-N,3-diphenylpropanamide (46).
1H NMR (600 MHz, DMSO-d6) δ 10.76 (s, 1H), 8.28 (d, J = 8.0 Hz, 1H), 7.36 – 7.34 (m, 3H), 7.32 – 7.30 (m, 2H), 7.11 – 7.08 (m, 4H), 7.04 – 7.00 (m, 2H), 6.85 – 6.78 (m, 3H), 4.38 – 4.22 (m, 1H), 3.68 – 3.64 (m, 1H), 3.52 – 3.41 (m, 3H), 2.83 – 2.80 (m, 1H), 2.65 – 2.61 (m, 1H), 0.95 (t, J = 7.0 Hz, 3H); 13C NMR (150 MHz, DMSO-d6) δ 170.9, 170.8, 162.7, 141.5, 137.9, 136.4, 129.9, 129.3, 128.9, 128.5, 128.4, 127.6, 126.8, 124.1, 121.2, 119.1, 118.6, 111.6, 109.2, 52.3, 44.2, 37.7, 36.2, 13.1; HRMS (ESI) m/z calcd for C27H28N3O2 [M + H]+ 426.2176, found 426.2181.
4.1.48. General Procedure for Synthesis of 47–72
To a solution of indoleacetic acid derivative (1 equiv.) in DMF (3 mL) were added HATU (2 equiv.) and DIPEA (3 equiv.), and the mixture was stirred at room temperature for 20 min before compound f (1.2 equiv.) was added. The mixture was stirred at room temperature overnight. Upon completion of the reaction, H2O was added and the reaction mixture was extracted with EtOAc (3×20 mL). The organic phases were combined and washed with brine, dried over anhydrous MgSO4, filtered and concentrated. The product was purified by combi-flash on silica gel using EtOAc in hexane (yield 55–86%).
4.1.49. (S)-2-(2-(1-ethyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (47).
1H NMR (600 MHz, CD3OD) δ 7.44 (d, J = 7.6 Hz, 1H), 7.35 − 7.29 (m, 5H), 7.14 − 6.96 (m, 7H), 6.69 (d, J = 7.4 Hz, 2H), 4.66 – 4.62 (m, 1H), 4.15 (q, J = 7.2 Hz, 2H), 3.60 − 3.53 (m, 2H), 3.16 (s, 3H), 2.83 (dd, J = 13.4, 6.2 Hz, 1H), 2.59 (dd, J = 13.4, 8.2 Hz, 1H), 2.28 (s, 3H), 1.27 (t, J = 7.2 Hz, 3H); 13C NMR (150 MHz, CD3OD) δ 174.0, 173.1, 143.9, 137.8, 137.0, 135.3, 130.9, 130.1, 129.4, 129.1, 128.6, 127.8, 121.8, 120.1, 118.8, 109.8, 105.1, 53.2, 39.0, 38.7, 38.1, 32.5, 15.5, 10.0; HRMS (ESI) m/z calcd for C29H30N3O2 [M − H]− 452.2344, found 452.2340.
4.1.50. (S)-2-(2-(1-ethyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(p-tolyl)propanamide (48).
1H NMR (600 MHz, CD3OD) δ 7.37 (d, J = 7.8 Hz, 0.6H), 7.33 (d, J = 7.9 Hz, 1H), 7.28 (d, J = 8.2 Hz, 1H), 7.14 − 7.05 (m, 7H), 6.97 (t, J = 7.4 Hz, 1H), 6.86 (s, 1H), 6.69 (d, J = 7.8 Hz, 2H), 4.64 (q, J = 7.9 Hz, 1H), 4.13 (q, J = 7.2 Hz, 2H), 3.58 − 3.51 (m, 2H), 3.11 (s, 3H), 2.81 (dd, J = 13.4, 6.3 Hz, 1H), 2.57 (dd, J = 13.4, 8.1 Hz, 1H), 2.31 (s, 3H), 2.26 (s, 3H), 1.26 (t, J = 7.2 Hz, 3H); 13C NMR (100 MHz, CD3OD) δ 173.9, 173.2, 141.3, 139.6, 137.8, 137.0, 135.3, 131.4, 130.1, 129.4, 129.1, 128.3, 127.8, 121.9, 120.1, 118.9, 109.8, 105.1, 53.1, 39.0, 38.7, 38.1, 32.6, 21.1, 15.5, 10.0; HRMS (ESI) m/z calcd for C30H32N3O2 [M − H]− 466.2500, found 466.2502.
4.1.51. (S)-2-(2-(1-ethyl-2-methyl-1H-indol-3-yl)acetamido)-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (49).
1H NMR (600 MHz, CD3OD) δ 7.55 (d, J = 7.5 Hz, 1H), 7.36 (d, J = 7.8 Hz, 1H), 7.31 (d, J = 8.2 Hz, 1H), 7.18 − 7.08 (m, 4H), 7.04 − 6.94 (m, 5H), 6.78 (d, J = 7.3 Hz, 2H), 4.57 (q, J = 7.4 Hz, 1H), 4.17 (q, J = 7.2 Hz, 2H), 3.60 − 3.54 (m, 2H), 3.13 (s, 3H), 2.86 (dd, J = 13.4, 7.0 Hz, 1H), 2.64 (dd, J = 13.4, 7.7 Hz, 1H), 2.30 (s, 3H), 1.29 (t, J = 7.2 Hz, 3H); 13C NMR (150 MHz, CD3OD) δ 174.0, 173.2, 163.4 (d, JCF = 247.0 Hz), 140.0 (d, JCF = 3.1 Hz), 130.7 (d, JCF = 8.7 Hz), 130.2, 129.5, 129.1, 127.9, 121.8, 120.1, 118.9, 117.5, 117.4, 109.8, 105.2, 53.1, 39.1, 38.7, 38.1, 32.5, 15.5, 10.0; HRMS (ESI) m/z calcd for C29H29FN3O2 [M − H]− 470.2249, found 470.2245.
4.1.52. (S)-2-(2-(1-ethyl-2-methyl-1H-indol-3-yl)acetamido)-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (50).
1H NMR (600 MHz, CD3OD) δ 7.53 (d, J = 6.9 Hz, 1H), 7.37 − 7.30 (m, 3H), 7.18 − 7.16 (m, 1H), 7.13 − 7.08 (m, 4H), 6.99 (t, J = 7.4 Hz, 1H), 6.84 − 6.77 (m, 3H), 6.65 (s, 1H), 4.63 − 4.60 (m, 1H), 4.17 (q, J = 7.2 Hz, 2H), 3.62 − 3.55 (m, 2H), 3.14 (s, 3H), 2.86 (dd, J = 13.2, 7.1 Hz, 1H), 2.65 (dd, J = 13.2, 7.5 Hz, 1H), 2.31 (s, 3H), 1.29 (t, J = 7.2 Hz, 3H); 13C NMR (150 MHz, CD3OD) δ 174.1, 173.0, 164.2 (d, JCF = 247.6 Hz), 145.4 (d, JCF = 9.9 Hz), 137.7, 137.2, 135.3, 132.2 (d, JCF = 9.2 Hz), 130.2, 129.5, 129.1, 127.9, 124.7, 121.8, 120.1, 118.8, 116.3 (d, JCF = 21.3 Hz), 115.9 (d, JCF = 22.9 Hz), 53.3, 39.2, 38.7, 37.9, 32.5, 15.5, 10.1; HRMS (ESI) m/z calcd for C29H29FN3O2 [M − H]− 470.2249, found 470.2248.
4.1.53. (S)-N-(4-chlorophenyl)-2-(2-(1-ethyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (51).
1H NMR (600 MHz, CD3OD) δ 7.37 (d, J = 7.8 Hz, 1H), 7.32 − 7.28 (m, 3H), 7.19 − 7.09 (m, 5H), 6.99 (t, J = 7.4 Hz, 1H), 6.89 (s, 1H), 6.79 (d, J = 7.2 Hz, 2H), 4.58 (t, J = 7.3 Hz, 1H), 4.17 (q, J = 7.2 Hz, 2H), 3.61 − 3.55 (m, 2H), 3.12 (s, 3H), 2.86 (dd, J = 13.3, 7.3 Hz, 1H), 2.65 (dd, J = 13.3, 7.4 Hz, 1H), 2.31 (s, 3H), 1.29 (t, J = 7.2 Hz, 3H); 13C NMR (150 MHz, CD3OD) δ 174.0, 173.1, 142.5, 137.7, 137.1, 135.3, 135.0, 130.8, 130.3, 130.2, 129.5, 129.2, 127.9, 121.8, 120.1, 118.9, 109.8, 105.2, 53.2, 39.2, 38.7, 38.0, 32.5, 15.5, 10.1; HRMS (ESI) m/z calcd for C29H29ClN3O2 [M − H]− 486.1954, found 486.1950.
4.1.54. (S)-N-(3-chlorophenyl)-2-(2-(1-ethyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (52).
1H NMR (600 MHz, CD3OD) δ 7.55 (d, J = 6.6 Hz, 1H), 7.37 (d, J = 7.8 Hz, 1H), 7.32 (s, 1H), 7.26 (s, 1H), 7.19 (s, 1H), 7.12 (s, 1H), 7.09 (t, J = 7.5 Hz, 1H), 6.99 (s, 1H), 6.94 (s, 1H), 6.78 (d, J = 7.2 Hz, 2H), 4.57 (dd, J = 14.4, 7.2 Hz, 1H), 4.16 (q, J = 7.2 Hz, 2H), 3.61 (s, 1H), 3.55 (s, 1H), 3.10 (s, 2H), 2.85 (dd, J = 13.2, 7.6 Hz, 1H), 2.65 (dd, J = 13.2, 7.2 Hz, 1H), 2.31 (s, 2H), 1.28 (t, J = 7.2 Hz, 3H); 13C NMR (100 MHz, CD3OD) δ 174.1, 173.0, 145.1, 137.6, 137.1, 136.0, 135.3, 131.9, 130.2, 129.6, 129.5, 129.2, 128.7, 128.0, 127.3, 121.9, 120.1, 118.9, 109.8, 105.2, 53.3, 39.3, 38.7, 37.9, 32.5, 15.5, 10.1; HRMS (ESI) m/z calcd for C29H29ClN3O2 [M − H]− 486.1954, found 486.1950.
4.1.55. (S)-N-ethyl-2-(2-(1-ethyl-2-methyl-1H-indol-3-yl)acetamido)-N,3-diphenylpropanamide (53).
1H NMR (600 MHz, CDCl3) δ 7.40 − 7.28 (m, 5H), 7.19 − 7.04 (m, 6H), 6.88 (s, 1H), 6.66 (d, J = 7.4 Hz, 2H), 6.16 (d, J = 8.2 Hz, 1H), 4.67 – 4.63 (m, 1H), 4.13 (q, J = 7.2 Hz, 2H), 3.76 − 3.70 (m, 1H), 3.63 − 3.50 (m, 3H), 2.72 (dd, J = 13.3, 7.0 Hz, 1H), 2.51 (dd, J = 13.3, 7.1 Hz, 1H), 2.29 (s, 3H), 1.33 (t, J = 7.2 Hz, 3H), 1.03 (t, J = 7.2 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 170.8, 170.6, 140.9, 136.2, 135.7, 134.1, 129.6, 129.3, 128.5, 128.2, 127.7, 126.6, 121.1, 119.4, 118.0, 108.8, 104.0, 51.2, 44.5, 38.7, 38.0, 32.5, 15.3, 12.8, 10.0; HRMS (ESI) m/z calcd for C30H32N3O2 [M − H]− 466.2500, found 466.2502.
4.1.56. (S)-2-(2-(1-isopropyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (54).
1H NMR (600 MHz, CD3OD) δ 7.47 (d, J = 8.3 Hz, 1H), 7.34 − 7.31 (m, 4H), 7.12 (t, J = 7.3 Hz, 1H), 7.07 − 7.03 (m, 5H), 6.94 (t, J = 7.4 Hz, 1H), 6.69 (d, J = 7.4 Hz, 2H), 4.73 − 4.69 (m, 1H), 4.64 – 4.62 (m, 1H), 3.58 − 3.51 (m, 2H), 3.15 (s, 3H), 2.83 (dd, J = 13.5, 6.2 Hz, 1H), 2.59 (dd, J = 13.4, 8.2 Hz, 1H), 2.29 (s, 3H), 1.58 (s, 3H), 1.57 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 174.0, 173.1, 143.9, 137.8, 136.1, 135.4, 130.8, 130.1, 129.8, 129.4, 128.6, 127.8, 121.5, 119.8, 119.0, 112.1, 105.3, 53.2, 48.4, 39.0, 38.1, 32.5, 21.8, 11.2; HRMS (ESI) m/z calcd for C30H32N3O2 [M − H]− 466.2500, found 466.2501.
4.1.57. (S)-2-(2-(1-isopropyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(p-tolyl)propanamide (55).
1H NMR (600 MHz, CDCl3) δ 7.48 (d, J = 8.3 Hz, 1H), 7.39 (d, J = 7.8 Hz, 1H), 7.14 − 7.03 (m, 7H), 6.80 − 6.66 (m, 4H), 6.17 (d, J = 8.2 Hz, 1H), 4.80 − 4.76 (m, 1H), 4.69 − 4.64 (m, 1H), 3.62 – 3.54 (m, 2H), 3.14 (s, 3H), 2.71 (dd, J = 13.4, 6.9 Hz, 1H), 2.52 (dd, J = 13.4, 7.0 Hz, 1H), 2.36 (s, 3H), 2.30 (s, 3H), 1.63 (s, 3H), 1.61 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 171.3, 170.8, 140.0, 138.0, 136.2, 134.8, 134.3, 130.3, 129.2, 128.4, 128.2, 127.1, 126.6, 120.7, 119.1, 118.1, 111.1, 104.2, 50.9, 47.2, 38.7, 37.6, 32.5, 21.7, 21.1, 11.2; HRMS (ESI) m/z calcd for C31H34N3O2 [M − H]− 480.2656, found 486.2659.
4.1.58. (S)-N-(4-fluorophenyl)-2-(2-(1-isopropyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (56).
1H NMR (600 MHz, CD3OD) δ 7.57 (d, J = 7.5 Hz, 0.4H), 7.49 (d, J = 8.3 Hz, 1H), 7.35 (d, J = 7.8 Hz, 1H), 7.18 − 7.12 (m, 3H), 7.07 − 7.02 (m, 4H), 6.97 − 6.95 (m, 2H), 6.80 (d, J = 7.1 Hz, 2H), 4.76 − 4.72 (m, 1H), 4.57 (t, J = 7.2 Hz, 1H), 3.60 − 3.54 (m, 2H), 3.13 (s, 8H), 2.87 (dd, J = 13.4, 7.0 Hz, 1H), 2.65 (dd, J = 13.4, 7.7 Hz, 1H), 2.32 (s, 3H), 1.60 (s, 3H), 1.59 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 174.1, 173.2, 163.4 (d, JCF = 247.0 Hz), 137.8, 136.1, 135.4, 130.7 (d, JCF = 8.6 Hz), 130.2, 129.9, 129.5, 127.9, 121.5, 119.8, 119.0, 117.5, 117.4, 112.1, 105.4, 53.2, 48.4, 39.1, 38.1, 32.5, 21.8, 11.2; HRMS (ESI) m/z calcd for C30H31FN3O2 [M − H]− 484.2406, found 484.2402.
4.1.59. (S)-N-(3-fluorophenyl)-2-(2-(1-isopropyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (57).
1H NMR (600 MHz, CD3OD) δ 7.54 (d, J = 6.7 Hz, 0.8H), 7.47 (d, J = 8.3 Hz, 1H), 7.36 − 7.30 (m, 2H), 7.17 − 7.03 (m, 5H), 6.95 (t, J = 7.4 Hz, 1H), 6.83 − 6.77 (m, 3H), 6.65 (s, 1H), 4.74 − 4.70 (m, 1H), 4.63 − 4.59 (m, 1H), 3.60 − 3.54 (m, 2H), 3.13 (s, 4H), 2.85 (dd, J = 13.1, 7.1 Hz, 1H), 2.65 (dd, J = 13.0, 7.5 Hz, 1H), 2.31 (s, 4H), 1.59 (s, 3H), 1.58 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 174.1, 173.0, 164.2 (d, JCF = 247.5 Hz), 145.4 (d, JCF = 9.6 Hz), 137.7, 136.1, 135.5, 132.2 (d, JCF = 9.2 Hz), 130.2, 129.8, 129.5, 127.9, 124.7, 121.5, 119.8, 119.0, 116.2 (d, JCF = 21.0 Hz), 115.9 (d, JCF = 22.4 Hz), 112.1, 105.3, 53.3, 48.4, 39.2, 37.9, 32.5, 21.8, 11.2; HRMS (ESI) m/z calcd for C30H31FN3O2 [M − H]− 484.2406, found 484.2402.
4.1.60. (S)-N-(4-chlorophenyl)-2-(2-(1-isopropyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (58).
1H NMR (600 MHz, CD3OD) δ 7.60 (d, J = 7.5 Hz, 0.5H), 7.48 (d, J = 8.3 Hz, 1H), 7.35 (d, J = 7.8 Hz, 1H), 7.27 (d, J = 8.5 Hz, 2H), 7.17 − 7.11 (m, 3H), 7.04 (t, J = 7.6 Hz, 1H), 6.95 (t, J = 7.4 Hz, 1H), 6.87 − 6.79 (m, 4H), 4.75 − 4.70 (m, 1H), 4.59 − 4.56 (m, 1H), 3.59 − 3.53 (m, 2H), 3.11 (s, 3H), 2.86 (dd, J = 13.3, 7.2 Hz, 1H), 2.65 (dd, J = 13.2, 7.5 Hz, 1H), 2.31 (s, 3H), 1.59 (s, 3H), 1.58 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 174.1, 173.1, 142.5, 137.8, 136.1, 135.4, 135.0, 130.8, 130.3, 130.2, 129.9, 129.5, 127.9, 121.5, 119.8, 119.0, 112.1, 105.4, 53.2, 48.4, 39.2, 38.0, 32.5, 21.8, 11.2; HRMS (ESI) m/z calcd for C30H31ClN3O2 [M − H]− 500.2110, found 500.2109.
4.1.61. (S)-N-(3-chlorophenyl)-2-(2-(1-isopropyl-2-methyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (59).
1H NMR (600 MHz, CD3OD) δ 7.57 (d, J = 6.3 Hz, 0.4H), 7.47 (d, J = 8.3 Hz, 1H), 7.37 (d, J = 7.8 Hz, 1H), 7.32 − 7.26 (m, 2H), 7.19 − 7.12 (m, 3H), 7.04 (t, J = 7.6 Hz, 1H), 6.97 − 6.94 (m, 2H), 6.80 − 6.79 (m, 3H), 4.75 − 4.70 (m, 1H), 4.58 − 4.55 (m, 1H), 3.60 − 3.54 (m, 2H), 3.11 (s, 3H), 2.85 (dd, J = 13.2, 7.5 Hz, 1H), 2.66 (dd, J = 13.1, 7.2 Hz, 1H), 2.32 (s, 3H), 1.59 (s, 3H), 1.58 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 174.1, 173.0, 145.1, 137.6, 136.1, 136.0, 135.5, 131.9, 130.2, 129.9, 129.6, 129.5, 128.7, 128.0, 127.3, 121.5, 119.8, 119.0, 112.1, 105.3, 53.3, 48.4, 39.3, 38.0, 32.5, 21.8, 11.2; HRMS (ESI) m/z calcd for C30H31ClN3O2 [M − H]− 500.2110, found 500.2112.
4.1.62. (S)-N-ethyl-2-(2-(1-isopropyl-2-methyl-1H-indol-3-yl)acetamido)-N,3-diphenylpropanamide (60).
1H NMR (600 MHz, CDCl3) δ 7.48 (d, J = 8.2 Hz, 1H), 7.40 − 7.34 (m, 4H), 7.15 − 7.03 (m, 6H), 6.88 (s, 1H), 6.66 (d, J = 7.4 Hz, 2H), 6.16 (d, J = 8.0 Hz, 1H), 4.69 − 4.63 (m, 2H), 3.76 − 3.70 (m, 1H), 3.62 − 3.51 (m, 3H), 2.73 (dd, J = 13.3, 6.9 Hz, 1H), 2.51 (dd, J = 13.3, 7.0 Hz, 1H), 2.30 (s, 3H), 1.63 (s, 3H), 1.62 (s, 3H), 1.03 (t, J = 7.1 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 170.8, 170.6, 140.9, 136.2, 134.8, 134.3, 129.6, 129.3, 128.5, 128.4, 128.2, 126.6, 120.7, 119.1, 118.1, 111.1, 104.2, 53.4, 51.2, 47.2, 44.5, 38.7, 32.5, 21.7, 12.8, 11.2; HRMS (ESI) m/z calcd for C31H34N3O2 [M − H]− 480.2656, found 486.2657.
4.1.63. (S)-2-(2-(1-ethyl-1H-indol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (61).
1H NMR (600 MHz, CD3OD) δ 7.41 (d, J = 7.9 Hz, 1H), 7.36 − 7.34 (m, 4H), 7.16 − 7.08 (m, 4H), 7.03 (s, 1H), 7.01 − 6.99 (m, 3H), 6.75 (d, J = 7.3 Hz, 2H), 4.67 – 4.64 (m, 1H), 4.15 (q, J = 7.3 Hz, 2H), 3.62 – 3.56 (m, 2H), 3.17 (s, 3H), 2.88 (dd, J = 13.4, 6.5 Hz, 1H), 2.65 (dd, J = 13.4, 8.1 Hz, 1H), 1.40 (t, J = 7.2 Hz, 3H); 13C NMR (150 MHz, CD3OD) δ 174.0, 173.2, 143.9, 138.0, 137.5, 130.9, 130.1, 129.4, 129.4, 129.2, 128.6, 127.8, 127.6, 122.6, 120.0, 119.9, 110.4, 108.7, 53.4, 41.7, 39.1, 38.1, 33.5, 15.8; HRMS (ESI) m/z calcd for C28H28N3O2 [M − H]− 438.2187, found 428.2190.
4.1.64. (S)-2-(2-(1-ethyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenyl-N-(p-tolyl)propanamide (62).
1H NMR (600 MHz, CD3OD) δ 7.76 (d, J = 7.7 Hz, 1H), 7.41 (d, J = 7.9 Hz, 1H), 7.32 (d, J = 8.3 Hz, 1H), 7.14 − 7.07 (m, 7H), 7.00 − 6.98 (m, 2H), 6.83 (s, 2H), 6.75 (d, J = 7.5 Hz, 2H), 4.68 − 4.64 (m, 1H), 4.11 (q, J = 7.3 Hz, 2H), 3.60 − 3.54 (m, 2H), 3.11 (s, 3H), 2.86 (dd, J = 13.4, 6.5 Hz, 1H), 2.63 (dd, J = 13.4, 8.0 Hz, 1H), 2.31 (s, 3H), 1.36 (t, J = 7.3 Hz, 3H); 13C NMR (100 MHz, CD3OD) δ 173.9, 173.3, 141.3, 139.5, 138.0, 137.5, 131.3, 130.2, 129.4, 129.2, 128.3, 127.8, 127.6, 122.6, 120.0, 119.9, 110.4, 108.7, 53.2, 41.6, 39.1, 38.1, 33.5, 21.1, 15.8; HRMS (ESI) m/z calcd for C29H30N3O2 [M − H]− 452.2343, found 452.2340.
4.1.65. (S)-2-(2-(1-ethyl-1H-indol-3-yl)acetamido)-N-(4-fluorophenyl)-N-methyl-3-phenylpropanamide (63).
1H NMR (600 MHz, CD3OD) δ 7.44 (d, J = 7.9 Hz, 1H), 7.34 (d, J = 8.3 Hz, 1H), 7.19 − 7.13 (m, 5H), 7.04 (s, 1H), 7.02 – 6.99 (m, 4H), 6.83 (d, J = 7.1 Hz, 2H), 4.58 (t, J = 7.4 Hz, 1H), 4.14 (d, J = 7.2 Hz, 2H), 3.62 − 3.56 (m, 2H), 3.12 (s, 3H), 2.90 (dd, J = 13.3, 7.3 Hz, 1H), 2.69 (dd, J = 13.3, 7.6 Hz, 1H), 1.39 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, CD3OD) δ 174.0, 173.3, 163.4 (d, J = 247.0 Hz), 140.0 (d, J = 3.1 Hz), 137.9, 137.5, 130.7 (d, J = 8.7 Hz), 130.2, 129.5, 129.2, 127.9, 127.6, 122.6, 120.0, 119.9, 117.4 (d, J = 23.0 Hz), 110.4, 108.8, 53.2, 41.7, 39.2, 38.1, 33.4, 15.8; HRMS (ESI) m/z calcd for C28H27FN3O2 [M − H]− 456.2093, found 456.2093.
4.1.66. (S)-2-(2-(1-ethyl-1H-indol-3-yl)acetamido)-N-(3-fluorophenyl)-N-methyl-3-phenylpropanamide (64).
1H NMR (600 MHz, CD3OD) δ 7.90 (d, J = 6.8 Hz, 1H), 7.45 (d, J = 7.9 Hz, 1H), 7.35 − 7.29 (m, 2H), 7.19 − 7.06 (m, 6H), 7.01 (t, J = 7.5 Hz, 1H), 6.83 − 6.80 (m, 3H), 6.62 (s, 1H), 4.65 (s, 1H), 4.61 (s, 1H), 4.15 (q, J = 7.3 Hz, 1H), 3.64 (s, 1H), 3.58 (s, 1H), 3.13 (s, 1H), 2.90 (dd, J = 13.2, 7.5 Hz, 1H), 2.70 (dd, J = 13.2, 7.4 Hz, 1H), 1.39 (t, J = 7.3 Hz, 1H); 13C NMR (150 MHz, CD3OD) δ 174.1, 173.1, 164.2 (d, J = 247.5 Hz), 145.4 (d, J = 9.6 Hz), 137.8, 137.5, 132.1 (d, J = 9.1 Hz), 130.2, 129.5, 129.2, 128.0, 127.6, 124.7, 122.6, 120.0, 119.9, 116.2 (d, J = 21.2 Hz), 115.9 (d, J = 22.7 Hz), 110.4, 108.7, 53.4, 41.7, 39.3, 37.9, 33.4, 15.8; HRMS (ESI) m/z calcd for C28H27FN3O2 [M − H]− 456.2093, found 456.2094.
4.1.67. (S)-N-(4-chlorophenyl)-2-(2-(1-ethyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (65).
1H NMR (600 MHz, CD3OD) δ 7.45 (d, J = 7.9 Hz, 1H), 7.36 (d, J = 8.3 Hz, 1H), 7.27 (d, J = 8.7 Hz, 2H), 7.19 − 7.14 (m, 4H), 7.06 (s, 1H), 7.01 (t, J = 7.5 Hz, 1H), 6.85 − 6.84 (m, 4H), 4.59 (t, J = 7.4 Hz, 1H), 4.16 (q, J = 7.2 Hz, 2H), 3.60 (s, 2H), 3.12 (s, 3H), 2.91 (dd, J = 13.3, 7.5 Hz, 1H), 2.70 (dd, J = 13.3, 7.4 Hz, 1H), 1.40 (t, J = 7.2 Hz, 3H); 13C NMR (150 MHz, CD3OD) δ 174.0, 173.1, 142.6, 137.9, 137.5, 135.0, 130.8, 130.2, 129.5, 129.2, 128.0, 127.6, 122.6, 120.0, 119.9, 110.4, 108.8, 53.3, 41.7, 39.2, 38.0, 33.4, 15.9; HRMS (ESI) m/z calcd for C28H27ClN3O2 [M − H]− 472.1797, found 472.1800.
4.1.68. (S)-N-(3-chlorophenyl)-2-(2-(1-ethyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (66).
1H NMR (600 MHz, CD3OD) δ 7.46 (d, J = 7.9 Hz, 1H), 7.35 − 7.31 (m, 2H), 7.26 (t, J = 7.9 Hz, 1H), 7.20 − 7.13 (m, 5H), 7.07 (s, 1H), 7.01 (t, J = 7.5 Hz, 1H), 6.91 − 6.83 (m, 3H), 4.58 (t, J = 7.4 Hz, 1H), 4.15 (q, J = 7.3 Hz, 2H), 3.63 − 3.58 (m, 2H), 3.11 (s, 3H), 2.89 (dd, J = 13.2, 7.9 Hz, 1H), 2.70 (dd, J = 13.2, 7.1 Hz, 1H), 1.40 (t, J = 7.3 Hz, 3H); 13C NMR (100 MHz, CD3OD) δ 174.1, 173.1, 145.1, 137.8, 137.6, 135.9, 131.9, 130.2, 129.6, 129.5, 129.2, 128.8, 128.0, 127.6, 127.3, 122.6, 120.0, 119.9, 110.4, 108.8, 53.4, 41.7, 39.4, 37.9, 33.4, 15.8; HRMS (ESI) m/z calcd for C28H27ClN3O2 [M − H]− 472.1797, found 472.1799.
4.1.69. (S)-2-(2-(1-isopropyl-1H-indol-3-yl)acetamido)-N-methyl-N,3-diphenylpropanamide (67).
1H NMR (600 MHz, CD3OD) δ 7.32 − 7.28 (m, 2H), 7.25 − 7.23 (m, 3H), 7.10 (s, 1H), 7.06 − 6.98 (m, 4H), 6.91 − 6.89 (m, 3H), 6.65 (d, J = 7.3 Hz, 2H), 4.62 − 4.54 (m, 2H), 3.53 − 3.47 (m, 2H), 3.07 (s, 3H), 2.79 (dd, J = 13.4, 6.5 Hz, 1H), 2.56 (dd, J = 13.4, 8.1 Hz, 1H), 1.40 − 1.38 (m, 6H); 13C NMR (150 MHz, CD3OD) δ 174.1, 173.2, 143.9, 137.92, 137.4, 130.9, 130.1, 129.4, 129.4, 129.1, 128.6, 127.8, 124.2, 122.5, 120.1, 119.9, 110.6, 108.9, 53.4, 48.2, 39.1, 38.1, 33.6, 23.0; HRMS (ESI) m/z calcd for C29H30N3O2 [M − H]− 452.2344, found 452.2346.
4.1.70. (S)-N-(4-fluorophenyl)-2-(2-(1-isopropyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (68).
1H NMR (600 MHz, CD3OD) δ 7.44 (d, J = 7.9 Hz, 1H), 7.38 (d, J = 8.3 Hz, 1H), 7.21 (s, 1H), 7.17 − 7.13 (m, 5H), 7.02 − 6.99 (m, 4H), 6.83 (d, J = 7.0 Hz, 2H), 4.71 − 4.67 (m, 1H), 4.58 (t, J = 7.4 Hz, 1H), 3.63 − 3.58 (m, 2H), 3.12 (s, 3H), 2.91 (dd, J = 13.3, 7.3 Hz, 1H), 2.69 (dd, J = 13.3, 7.5 Hz, 1H), 1.50 − 1.48 (m, 6H); 13C NMR (150 MHz, CD3OD) δ 174.0, 173.3, 163.4 (d, J = 246.9 Hz), 140.0 (d, J = 3.1 Hz), 137.9, 137.4, 130.7 (d, J = 8.4 Hz), 130.2, 129.5, 129.1, 127.9, 124.2, 122.5, 120.1, 119.9, 117.5, 117.3, 110.6, 109.0, 53.3, 48.2, 39.2, 38.1, 33.5, 23.00, 22.98; HRMS (ESI) m/z calcd for C29H29FN3O2 [M − H]− 470.2249, found 470.2245.
4.1.71. (S)-N-(3-fluorophenyl)-2-(2-(1-isopropyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (69).
1H NMR (600 MHz, CD3OD) δ 7.44 (d, J = 7.9 Hz, 1H), 7.37 (d, J = 8.3 Hz, 1H), 7.31 − 7.27 (m, 1H), 7.21 (s, 1H), 7.18 − 7.05 (m, 6H), 7.00 (t, J = 7.5 Hz, 1H), 6.82 − 6.78 (m, 3H), 6.61 (s, 1H), 4.70 − 4.65 (m, 1H), 4.61 (t, J = 7.3 Hz, 1H), 3.63 – 3.58 (m, 2H), 3.12 (s, 3H), 2.89 (dd, J = 13.2, 7.5 Hz, 1H), 2.69 (dd, J = 13.2, 7.4 Hz, 1H), 1.48 − 1.47 (m, 6H); 13C NMR (150 MHz, CD3OD) δ 174.1, 173.1, 164.2 (d, J = 247.5 Hz), 145.4 (d, J = 9.5 Hz), 137.8, 137.4, 132.1 (d, J = 9.2 Hz), 130.2, 129.5, 129.1, 128.0, 124.7, 124.2, 122.5, 120.1, 119.9, 116.2 (d, J = 21.1 Hz), 115.9 (d, J = 22.6 Hz), 110.6, 108.9, 53.4, 48.2, 39.3, 37.9, 33.5, 23.0; HRMS (ESI) m/z calcd for C29H29FN3O2 [M − H]− 470.2249, found 470.2249.
4.1.72. (S)-N-(4-chlorophenyl)-2-(2-(1-isopropyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (70).
1H NMR (600 MHz, CD3OD) δ 7.45 (d, J = 7.9 Hz, 1H), 7.39 (d, J = 8.3 Hz, 1H), 7.26 (d, J = 8.7 Hz, 2H), 7.22 (s, 1H), 7.18 − 7.13 (m, 4H), 7.01 (t, J = 7.5 Hz, 1H), 6.85 − 6.83 (m, 4H), 4.72 − 4.67 (m, 1H), 4.58 (t, J = 7.3 Hz, 1H), 3.61 (s, 2H), 3.12 (s, 3H), 2.90 (dd, J = 13.3, 7.6 Hz, 1H), 2.70 (dd, J = 13.3, 7.3 Hz, 1H), 1.50 − 1.48 (m, 6H); 13C NMR (150 MHz, CD3OD) δ 174.1, 173.1, 142.5, 137.9, 137.4, 135.0, 130.8, 130.2, 129.5, 129.1, 128.0, 124.2, 122.5, 120.1, 119.9, 110.6, 109.0, 53.3, 48.2, 39.2, 38.0, 33.5, 23.01, 22.99; HRMS (ESI) m/z calcd for C29H29ClN3O2 [M − H]− 486.1954, found 486.1950.
4.1.73. (S)-N-(3-chlorophenyl)-2-(2-(1-isopropyl-1H-indol-3-yl)acetamido)-N-methyl-3-phenylpropanamide (71).
1H NMR (600 MHz, CD3OD) δ 7.90 (d, J = 6.7 Hz, 0.7H), 7.45 (d, J = 7.9 Hz, 1H), 7.37 (d, J = 8.3 Hz, 1H), 7.31 (d, J = 8.0 Hz, 1H), 7.26 − 7.12 (m, 8H), 7.00 (t, J = 7.4 Hz, 1H), 6.83 (d, J = 7.2 Hz, 2H), 4.70 − 4.66 (m, 1H), 4.59 − 4.56 (m, 1H), 3.64 − 3.59 (m, 2H), 3.10 (s, 3H), 2.89 (dd, J = 13.1, 7.9 Hz, 1H), 2.69 (dd, J = 13.1, 7.0 Hz, 1H), 1.49 − 1.47 (m, 6H); 13C NMR (100 MHz, CD3OD) δ 174.1, 173.1, 145.1, 137.7, 137.4, 135.9, 131.9, 130.2, 129.6, 129.4, 129.1, 128.7, 128.0, 127.2, 124.2, 122.5, 120.1, 119.9, 110.6, 108.9, 53.4, 48.2, 39.4, 37.9, 33.6, 23.0; HRMS (ESI) m/z calcd for C29H29ClN3O2 [M − H]− 486.1954, found 486.1950.
4.1.74. (S)-N-ethyl-2-(2-(1-isopropyl-1H-indol-3-yl)acetamido)-N,3-diphenylpropanamide (72).
1H NMR (600 MHz, CDCl3) δ 7.44 (d, J = 7.9 Hz, 1H), 7.37 − 7.33 (m, 4H), 7.26 (s, 1H), 7.22 (t, J = 7.6 Hz, 1H), 7.15 − 7.06 (m, 5H), 6.71 (d, J = 7.3 Hz, 2H), 6.22 (d, J = 8.2 Hz, 1H), 4.70 − 4.63 (m, 2H), 3.77 − 3.71 (m, 1H), 3.65 (q, J = 16.9 Hz, 2H), 3.56 − 3.50 (m, 1H), 2.76 (dd, J = 13.3, 7.2 Hz, 1H), 2.56 (dd, J = 13.3, 6.9 Hz, 1H), 1.54 – 1.50 (m, 6H), 1.03 (t, J = 7.2 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 170.7, 170.7, 140.9, 136.2, 136.0, 129.6, 129.31, 128.5, 128.2, 128.2, 127.7, 126.6, 123.1, 121.7, 119.4, 119.0, 109.6, 107.5, 51.3, 47.1, 44.5, 38.8, 33.5, 22.8, 22.8, 12.8; HRMS (ESI) m/z calcd for C30H32N3O2 [M − H]− 466.2500, found 466.2503.
4.1.75. Synthesis of 73–78 is described in supplemental information.
4.1.76. 1-((2-(2-methyl-1H-indol-3-yl)acetyl)-L-phenylalanyl)-N-phenylpyrrolidine-2-carboxamide (73).
1H NMR (600 MHz, CD3OD) δ 7.53 (d, J = 8.0 Hz, 2H), 7.31 − 7.23 (m, 4H), 7.12 − 7.07 (m, 4H), 7.05 − 7.01 (m, 3H), 6.94 (t, J = 7.2 Hz, 1H), 4.85 − 4.83 (m, 1H), 4.53 − 4.51 (m, 1H), 3.78 − 3.75 (m, 1H), 3.57 − 3.51 (m, 3H), 3.08 (dd, J = 14.0, 5.3 Hz, 1H), 2.82 (dd, J = 14.0, 8.3 Hz, 1H), 2.24 (s, 3H), 2.18 − 2.13 (m, 1H), 2.06 − 1.99 (m, 2H), 1.92 − 1.89 (m, 1H); 13C NMR (150 MHz, CD3OD) δ 174.4, 172.3, 172.2, 139.7, 137.7, 137.1, 134.8, 130.5, 129.8, 129.7, 129.4, 127.7, 125.3, 121.7, 121.3, 119.9, 118.5, 111.4, 104.7, 62.3, 53.9, 38.0, 32.4, 30.4, 26.0, 11.3; HRMS (ESI) m/z calcd for C31H31N4O3 [M − H]− 507.2402, found 507.2403.
4.1.77. 1-((2-(1H-indol-3-yl)acetyl)-L-phenylalanyl)-N-phenylpyrrolidine-2-carboxamide (74).
1H NMR (600 MHz, CD3OD) δ 7.54 (d, J = 8.5 Hz, 2H), 7.41 (d, J = 7.9 Hz, 1H), 7.34 − 7.29 (m, 3H), 7.13 − 7.08 (m, 7H), 7.03 (s, 1H), 6.98 (t, J = 7.4 Hz, 1H), 4.88 – 4.86 (m, 1H), 4.53 – 4.51 (m, 1H), 3.78 − 3.74 (m, 1H), 3.64 − 3.58 (m, 2H), 3.54 − 3.50 (m, 1H), 3.10 (dd, J = 13.9, 5.4 Hz, 1H), 2.85 (dd, J = 13.9, 8.5 Hz, 1H), 2.16 − 2.12 (m, 1H), 2.05 − 1.99 (m, 2H), 1.91 − 1.86 (m, 1H); 13C NMR (150 MHz, CD3OD) δ 174.5, 172.3, 172.3, 139.7, 138.1, 137.9, 130.4, 129.8, 129.4, 128.5, 127.7, 125.3, 125.0, 122.6, 121.3, 120.0, 119.4, 112.3, 109.1, 62.3, 54.0, 38.1, 33.6, 30.3, 26.0; HRMS (ESI) m/z calcd for C30H29N4O3 [M − H]− 493.2245, found 493.2245.
4.1.78. (S)-2-(3-(1H-indol-3-yl)propanamido)-N-methyl-N,3-diphenylpropanamide (76).
1H NMR (600 MHz, CD3OD) δ 7.50 (d, J = 7.9 Hz, 1H), 7.33 − 7.28 (m, 5H), 7.17 − 7.14 (m, 3H), 7.05 (t, J = 7.5 Hz, 1H), 6.97 − 6.94 (m, 2H), 6.86 − 6.81 (m, 3H), 4.60 (t, J = 7.4 Hz, 1H), 3.14 (s, 3H), 2.98 − 2.94 (m, 2H), 2.87 − 2.83 (m, 1H), 2.61 − 2.49 (m, 3H); 13C NMR (150 MHz, CD3OD) δ 175.3, 173.4, 145.0, 138.2, 138.1, 130.7, 130.2, 129.4, 129.2, 128.6, 128.6, 127.8, 123.0, 122.3, 119.5, 119.3, 114.9, 112.2, 53.2, 39.3, 38.1, 37.7, 22.4; HRMS (ESI) m/z calcd for C27H26N3O2 [M − H]− 424.2031, found 424.2033.
4.1.79. (R)-2-(2-(1H-indol-3-yl)acetamido)-N-methyl-N,2-diphenylacetamide (77).
1H NMR (600 MHz, CD3OD) δ 7.44 (d, J = 7.9 Hz, 1H), 7.34 − 7.29 (m, 5H), 7.22 (t, J = 7.3 Hz, 1H), 7.18 − 7.15 (m, 3H), 7.07 (t, J = 7.5 Hz, 1H), 6.99 − 6.95 (m, 2H), 6.89 (d, J = 7.4 Hz, 2H), 5.48 (s, 1H), 3.69 − 3.63 (m, 2H), 3.18 (s, 3H); 13C NMR (100 MHz, CD3OD) δ 174.0, 171.8, 143.7, 138.1, 137.8, 130.8, 129.6, 129.5, 129.3, 129.1, 129.0, 128.5, 125.0, 122.6, 120.0, 119.5, 112.3, 109.2, 56.4, 38.3, 33.5; HRMS (ESI) m/z calcd for C25H22N3O2 [M − H]− 396.1718, found 396.1720.
4.1.80. (S)-3-(3-bromophenyl)-N-methyl-2-(2-(2-methyl-1H-indol-3-yl)acetamido)-N-phenylpropanamide (78).
1H NMR (600 MHz, CD3OD) δ 7.41 − 7.36 (m, 3H), 7.32 − 7.29 (m, 2H), 7.24 (d, J = 8.0 Hz, 1H), 7.03 − 6.99 (m, 4H), 6.96 − 6.94 (m, 2H), 6.68 (d, J = 7.5 Hz, 1H), 4.67 − 4.64 (m, 1H), 3.58 − 3.52 (m, 2H), 3.17 (s, 3H), 2.83 (dd, J = 13.5, 6.4 Hz, 1H), 2.61 (dd, J = 13.4, 8.2 Hz, 1H), 2.31 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 174.3, 172.8, 143.8, 140.6, 137.1, 134.7, 133.1, 131.2, 131.0, 130.9, 129.7, 129.5, 128.9, 128.5, 123.3, 121.7, 119.9, 118.5, 111.4, 104.7, 53.1, 38.6, 38.0, 32.3, 11.4; HRMS (ESI) m/z calcd for C27H25BrN3O2 [M − H]− 502.1136, found 502.1140.
4.2. Thermal Shift Assays (TSAs) to screen compounds for HIV-1 CA hexamer stability
Compounds were screened for their effect on CA stability using purified covalently-crosslinked hexameric CAA14C/E45C/W184A/M185A (CA121). CA121 cloned in a pET11a expression plasmid was kindly provided by Dr. Owen Pornillos (University of Virginia, Charlottesville, VA). Protein was expressed in E. coli BL21(DE3)RIL and purified as reported previously [27]. The TSA has been previously described [41–43]. Briefly, the TSA was conducted on the PikoReal Real-Time PCR System (Thermo Fisher Scientific) or the QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific). Each reaction contained 10 μL of 15 μM CA121 in 50 mM sodium phosphate buffer (pH 8.0), 10 μL of 2x Sypro Orange Protein Gel Stain (Life Technologies) in 50 mM sodium phosphate buffer (pH 8.0) and 0.2 μL of DMSO (control) or compound. Compounds were tested at a final concentration of 20 μM. The plate was heated from 25 to 95 °C with a heating rate of 0.2 °C/10 sec. The fluorescence intensity was measured with an Ex range of 475–500 nm and Em range of 520–590 nm. The differences in the melting temperature (ΔTm) of CA hexamer in DMSO (T0) verses in the presence of compound (Tm) were calculated using the following formula: ΔTm=Tm−T0.
4.3. Virus Production
The wild-type laboratory HIV-1 strain, HIV-1NL4–3 [44], was produced using a pNL4–3 vector that was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. HIV-1NL4–3 was generated by transfecting HEK 293FT cells in a T75 flask with 10 μg of the pNL4–3 vector and FuGENE®HD Transfection Reagent (Promega). Supernatant was harvested 48–72 hours post-transfection and transferred to MT2 cells for viral propagation. Virus was harvested when syncytia formation was observed, which took 3–5 days. The viral supernatant was then concentrated using 8% w/v PEG 8,000 overnight at 4 °C, followed by centrifugation for 40 min at 3,500 rpm. The resulting viral-containing pellet was concentrated 10-fold by resuspension in DMEM without FBS and stored at −80 °C.
4.4. Anti-HIV-1 and Cytotoxicity assays
Anti-HIV-1 activity of PF74 and PF74-related analogs were examined in TZM-GFP cells. The potency of HIV-1 inhibition by a compound was based on its inhibitory effect on viral LTR-activated GFP expression compared with that of compound-free (DMSO) controls. Briefly, TZM-GFP cells were plated at density of 1 × 104 cells per well in a 96-well plate. 24 h later, media was replaced with increasing concentrations of compound. 24 h post treatment, cells were exposed to an HIV-1 strain (MOI = 1). After incubation for 48 h, anti-HIV-1 activity was assessed by counting the number of GFP positive cells on a Cytation™ 5 Imaging Reader (BioTek) and 50% effective concentration (EC50) values were determined.
Cytotoxicity of each compound was also determined in TZM-GFP cells. Cells were plated at a density of 1× 104 cells per well in a 96-well plate and were continuously exposed to increasing concentrations of a compound for 72 hours. The number of viable cells in each well was determined using a Cell Proliferation Kit II (XTT), and 50% cytotoxicity concentration (CC50) values were determined. All the cell-based assays were conducted in duplicate and in at least two independent experiments.
For the EC50 and CC50 dose responses, values were plotted in GraphPad Prism 5 and analyzed with the log (inhibitor) vs. normalized response - variable slope equation. Final values were calculated in each independent assay and the average values were determined. Statistical analysis (calculation of standard deviation) was performed by using Microsoft Excel.
4.5. ADME assays
Microsomal stability assay.
The in vitro microsomal stability assay was conducted in duplicate in mouse and human liver microsomal systems, which were supplemented with nicotinamide adenine dinucleotide phosphate (NADPH) as a cofactor. Briefly, a compound (1 μM final concentration) was pre-incubated, in the absence or presence of 0.5 μM Cobicistat (CYP 3A inhibitor, purchased from medchemexpress.com and verified with LCMS), with the reaction mixture containing liver microsomal protein (0.5 mg/mL final concentration) and MgCl2 (1 mM final concentration) in 0.1 M potassium phosphate buffer (pH 7.4) at 37 °C for 15 minutes. The reaction was initiated by addition of 1 mM NADPH, followed by incubation at 37 °C. A negative control was performed in parallel in the absence of NADPH to measure any chemical instability or non-NADPH dependent enzymatic degradation for each compound. At various time points (0, 5, 15, 30, and 60 min), 1 volume of reaction aliquot was taken and quenched with 3 volumes of acetonitrile containing an appropriate internal standard and 0.1% formic acid. The samples were then vortexed and centrifuged at 15,000 rpm for 5 min at 4 °C. The supernatants were collected and analyzed by LC-MS/MS to determine the in vitro metabolic half-life (t1/2).
Plasma stability assay.
The plasma stability assay was performed in duplicate by incubating each selected compound (1 μM final concentration) in normal mouse and human plasma diluted to 80% with 0.1 M potassium phosphate buffer (pH 7.4) at 37 °C. At 0 and 3 h, aliquots of the plasma mixture were taken and quenched with 3 volumes of acetonitrile containing an appropriate internal standard and 0.1% of formic acid. The quenched samples were filtered through the 0.2 μm Agilent Captiva filtration plates and analyzed by LC-MS/MS.
4.6. Molecular modeling
Molecular modeling was performed using the Schrödinger small molecule drug discovery suite 2019–1 [45]. The crystal structure of native full length HIV-1 capsid protein in complex with PF74 as reported by Sarafianos et al. was retrieved from the protein data bank (PDB code: 4XFZ). The above structure was analyzed using Maestro [46] (Schrödinger Inc. [47]) and subjected to a docking protocol that involves several steps including preparing the protein of interest, grid generation, ligand preparation, and docking. The crystal structure was refined using the protein preparation wizard [46] (Schrödinger Inc. [47]) in which missing hydrogen atoms, side chains, and loops were added using prime and minimized using the OPLS 3e force field [48] to optimize the hydrogen bonding network and converge the heavy atoms to an rmsd of 0.3 Å. In the processed crystal structure, the two chains (A & B) were separated and analyzed separately. The receptor grid generation tool in Maestro (Schrödinger Inc.) [47] was used to define an active site around the native ligand PF74 to cover all the residues within 12 Å. All the compounds were drawn using Maestro and subjected to Lig Prep [49] to generate conformers, possible protonation at pH of 7±2 that serves as an input for docking process. All the dockings were performed using Glide XP [50] (Glide [51], version 6.9) with the van der Waals radii of nonpolar atoms for each of the ligands scaled by a factor of 0.8. The solutions were further refined by post docking and minimization under implicit solvent to account for protein flexibility. The residue numbers of HIV-1 capsid protein used in the discussion and the figures were based on the native full length HIV-1 capsid protein.
Supplementary Material
Highlights.
Comprehensive SAR on HIV-1 CA-targeting antiviral PF74.
Multiple sites of structural modification explored with 78 analogs synthesized.
Identified a key halogen bond along with a few other pharmacophore elements.
Metabolic stability drastically improved using a CYP3A inhibitor
Molecular modeling corroborated observed SAR.
Acknowledgments
This research was supported by the National Institutes of Health (R01AI120860 to SGS and ZW). We thank the Minnesota Supercomputing Institute for molecular modeling resources.
Abbreviations
- HIV
human immunodeficiency virus
- CA
capsid protein
- CPSF6
cleavage and polyadenylation specific factor 6
- CypA
Cyclophilin A
- CANTD
CA N-terminal domain
- CACTD
CA C-terminal domain
- PICs
preintegration complexes
- SAR
structure-activity-relationship
- HATU
1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate
- T3P
2,4,6-Tripropyl-1,3,5,2,4,6-trioxatriphosphorinane-2,4,6-trioxide
- DIPEA
N,N-Diisopropylethylamine
- TSA
thermal shift assay
- HLM
human liver microsome
- MLM
mouse liver microsome
- LIDs
ligand interaction diagrams
- EC50
50% effective concentration
- CC50
50% cytotoxicity concentration
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data related to this article can be found at
Declaration of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Freed EO HIV-1 Gag Proteins: Diverse Functions in the Virus Life Cycle. Virology 1998, 251, 1–15. [DOI] [PubMed] [Google Scholar]
- [2].Bell NM; Lever AML HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol. 2013, 21, 136–144. [DOI] [PubMed] [Google Scholar]
- [3].Campbell EM; Hope TJ HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol 2015, 13, 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Scarlata S; Carter C Role of HIV-1 Gag domains in viral assembly. BBA. Biomembranes 2003, 1614, 62–72. [DOI] [PubMed] [Google Scholar]
- [5].Freed EO HIV-1 assembly, release and maturation. Nat. Rev. Microbiol 2015, 13, 484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sundquist WI; Kräusslich H-G HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012, 2, a006924–a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Le Sage V; Mouland AJ; Valiente-Echeverría F Roles of HIV-1 capsid in viral replication and immune evasion. Virus Res. 2014, 193, 116–129. [DOI] [PubMed] [Google Scholar]
- [8].Hilditch L; Towers GJ A model for cofactor use during HIV-1 reverse transcription and nuclear entry. Curr. Opin. Virol 2014, 4, 32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ambrose Z; Aiken C HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology 2014, 454–455, 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Stremlau M; Owens CM; Perron MJ; Kiessling M; Autissier P; Sodroski J The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [DOI] [PubMed] [Google Scholar]
- [11].Sayah DM; Sokolskaja E; Berthoux L; Luban J Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 2004, 430, 569–573. [DOI] [PubMed] [Google Scholar]
- [12].Saito A; Henning MS; Serrao E; Dubose BN; Teng S; Huang J; Li X; Saito N; Roy SP; Siddiqui MA; Ahn J; Tsuji M; Hatziioannou T; Engelman AN; Yamashita M Capsid-CPSF6 Interaction is Dispensable for HIV-1 Replication in Primary Cells but is Selected during Virus Passage in Vivo. J. Virol 2016, 90, 6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bejarano DA; Peng K; Laketa V; Börner K; Jost KL; Lucic B; Glass B; Lusic M; Müller B; Kräusslich H-G HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. eLife 2019, 8, e41800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Woodward CL; Prakobwanakit S; Mosessian S; Chow SA Integrase Interacts with Nucleoporin NUP153 To Mediate the Nuclear Import of Human Immunodeficiency Virus Type 1. J. Virol 2009, 83, 6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Matreyek KA; Yücel SS; Li X; Engelman A Nucleoporin NUP153 Phenylalanine-Glycine Motifs Engage a Common Binding Pocket within the HIV-1 Capsid Protein to Mediate Lentiviral Infectivity. PLOS Pathog. 2013, 9, e1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Buffone C; Martinez-Lopez A; Fricke T; Opp S; Severgnini M; Cifola I; Petiti L; Frabetti S; Skorupka K; Zadrozny KK; Ganser-Pornillos BK; Pornillos O; Di Nunzio F; Diaz-Griffero F Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells. J. Virol 2018, 92, e00648–00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Meehan AM; Saenz DT; Guevera R; Morrison JH; Peretz M; Fadel HJ; Hamada M; van Deursen J; Poeschla EM A Cyclophilin Homology Domain-Independent Role for Nup358 in HIV-1 Infection. PLOS Pathog. 2014, 10, e1003969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dharan A; Talley S; Tripathi A; Mamede JI; Majetschak M; Hope TJ; Campbell EM KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection. PLOS Pathog. 2016, 12, e1005700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mamede JI; Damond F; Bernardo A. d.; Matheron S; Descamps D; Battini J-L; Sitbon M; Courgnaud V Cyclophilins and nucleoporins are required for infection mediated by capsids from circulating HIV-2 primary isolates. Sci. Rep-UK 2017, 7, 45214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fricke T; White TE; Schulte B; de Souza Aranha Vieira DA; Dharan A; Campbell EM; Brandariz-Nuñez A; Diaz-Griffero F MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 2014, 11, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xu B; Pan Q; Liang C Role of MxB in Alpha Interferon-Mediated Inhibition of HIV-1 Infection. J. Virol 2018, 92, e00422–00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim K; Dauphin A; Komurlu S; McCauley SM; Yurkovetskiy L; Carbone C; Diehl WE; Strambio-De-Castillia C; Campbell EM; Luban J Cyclophilin A protects HIV-1 from restriction by human TRIM5α. Nat. Microbiol 2019, 4, 2044–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Franke EK; Yuan HEH; Luban J Specific incorporation of cyclophilin A into HIV-1 virions. Nature 1994, 372, 359–362. [DOI] [PubMed] [Google Scholar]
- [24].Thali M; Bukovsky A; Kondo E; Rosenwlrth B; Walsh CT; Sodroski J; Göttlinger HG Functional association of cyclophilin A with HIV-1 virions. Nature 1994, 372, 363–365. [DOI] [PubMed] [Google Scholar]
- [25].Novikova M; Zhang Y; Freed EO; Peng K Multiple Roles of HIV-1 Capsid during the Virus Replication Cycle. Virol. Sin 2019, 34, 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao G; Perilla JR; Yufenyuy EL; Meng X; Chen B; Ning J; Ahn J; Gronenborn AM; Schulten K; Aiken C; Zhang P Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497, 643–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pornillos O; Ganser-Pornillos BK; Kelly BN; Hua Y; Whitby FG; Stout CD; Sundquist WI; Hill CP; Yeager M X-ray structures of the hexameric building block of the HIV capsid. Cell 2009, 137, 1282–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gres AT; Kirby KA; KewalRamani VN; Tanner JJ; Pornillos O; Sarafianos SG X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 2015, 349, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Goudreau N; Lemke CT; Faucher AM; Grand-Maitre C; Goulet S; Lacoste JE; Rancourt J; Malenfant E; Mercier JF; Titolo S; Mason SW Novel inhibitor binding site discovery on HIV-1 capsid N-terminal domain by NMR and X-ray crystallography. ACS Chem. Biol 2013, 8, 1074–1082. [DOI] [PubMed] [Google Scholar]
- [30].Price AJ; Jacques DA; McEwan WA; Fletcher AJ; Essig S; Chin JW; Halambage UD; Aiken C; James LC Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly. PLOS Pathog. 2014, 10, e1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bhattacharya A; Alam SL; Fricke T; Zadrozny K; Sedzicki J; Taylor AB; Demeler B; Pornillos O; Ganser-Pornillos BK; Diaz-Griffero F; Ivanov DN; Yeager M Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. USA 2014, 111, 18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shi J; Zhou J; Shah VB; Aiken C; Whitby K Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J. Virol 2011, 85, 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Saito A; Ferhadian D; Sowd GA; Serrao E; Shi J; Halambage UD; Teng S; Soto J; Siddiqui MA; Engelman AN; Aiken C; Yamashita M Roles of Capsid-Interacting Host Factors in Multimodal Inhibition of HIV-1 by PF74. J. Virol 2016, 90, 5808–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sowd GA; Serrao E; Wang H; Wang W; Fadel HJ; Poeschla EM; Engelman AN A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2016, 113, E1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wu G; Zalloum WA; Meuser ME; Jing L; Kang D; Chen CH; Tian Y; Zhang F; Cocklin S; Lee KH; Liu X; Zhan P Discovery of phenylalanine derivatives as potent HIV-1 capsid inhibitors from click chemistry-based compound library. Eur. J. Med. Chem 2018, 158, 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sun L; Huang T; Dick A; Meuser ME; Zalloum WA; Chen CH; Ding X; Gao P; Cocklin S; Lee KH; Zhan P; Liu X Design, synthesis and structure-activity relationships of 4-phenyl-1H-1,2,3-triazole phenylalanine derivatives as novel HIV-1 capsid inhibitors with promising antiviral activities. Eur. J. Med. Chem 2020, 190, 112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bondy SS; Chou C-H; Link JO; TSE WC, Antiviral agents, WO 2015/130966 A1, September 3, 2015. [Google Scholar]
- [38].Chen P; Zhou D; Pryde D; Bell A; Li T; He Z; Cai Z-W, Novel antiviral compounds, WO 2012/065062 A1, May 18, 2012. [Google Scholar]
- [39].Wacher VJ; Silverman JA; Zhang Y; Benet LZ Role of P-Glycoprotein and Cytochrome P450 3A in Limiting Oral Absorption of Peptides and Peptidomimetics. J. Pharm. Sci-US 1998, 87, 1322–1330. [DOI] [PubMed] [Google Scholar]
- [40].Xu L; Liu H; Murray BP; Callebaut C; Lee MS; Hong A; Strickley RG; Tsai LK; Stray KM; Wang Y; Rhodes GR; Desai MC Cobicistat (GS-9350): A Potent and Selective Inhibitor of Human CYP3A as a Novel Pharmacoenhancer. ACS Med. Chem. Lett 2010, 1, 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lo M-C; Aulabaugh A; Jin G; Cowling R; Bard J; Malamas M; Ellestad G Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem 2004, 332, 153–159. [DOI] [PubMed] [Google Scholar]
- [42].Miyazaki Y; Doi N; Koma T; Adachi A; Nomaguchi M Novel In Vitro Screening System Based on Differential Scanning Fluorimetry to Search for Small Molecules against the Disassembly or Assembly of HIV-1 Capsid Protein. Front. Microbiol 2017, 8, 1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pantoliano MW; Petrella EC; Kwasnoski JD; Lobanov VS; Myslik J; Graf E; Carver T; Asel E; Springer BA; Lane P; Salemme FR High-Density Miniaturized Thermal Shift Assays as a General Strategy for Drug Discovery. J. Biomol. Screen 2001, 6, 429–440. [DOI] [PubMed] [Google Scholar]
- [44].Adachi A; Gendelman HE; Koenig S; Folks T; Willey R; Rabson A; Martin MA Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol 1986, 59, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Schrödinger Small-Molecule Drug Discovery Suite 2015–4, Schrödinger, Llc, New York, Ny, 2015., 2015. [Google Scholar]
- [46].Madhavi Sastry G; Adzhigirey M; Day T; Annabhimoju R; Sherman W Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aid. Mol. Des 2013, 27, 221–234. [DOI] [PubMed] [Google Scholar]
- [47].Schrödinger Schrödinger Release 2015–4: Schrödinger Suite 2015–4 (a)Protein Preparation Wizard; Epik, Schrödinger, Llc, New York, Ny, 2015; Impact, Schrödinger, Llc, New York, Ny, 2015; Prime, Schrödinger, Llc, New York, Ny, 2015., 2015. [Google Scholar]
- [48].Jorgensen WL; Maxwell DS; Tirado-Rives J Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc 1996, 118, 11225–11236. [Google Scholar]
- [49].Schrödinger Schrödinger Release 2015–4: Ligprep, Schrödinger, Llc, New York, Ny, 2015., 2015. [Google Scholar]
- [50].Friesner RA; Murphy RB; Repasky MP; Frye LL; Greenwood JR; Halgren TA; Sanschagrin PC; Mainz DT Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein–Ligand Complexes. J. Med. Chem 2006, 49, 6177–6196. [DOI] [PubMed] [Google Scholar]
- [51].Schrödinger Release 2015–4: Glide Version 6.9, Schrödinger, Llc, New York, Ny, 2015., 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


