Highlights
-
•
Saliva is an eligible matrix for SARS-CoV-2 molecular detection and IgA measurement.
-
•
Saliva collection offers several advantages: safe, non-invasive and self-collection.
-
•
Positive molecular testing results were associated with disease duration.
-
•
The presence of salivary IgA was associated with pneumonia and CRP values.
Keywords: SARS-CoV-2, Naso-pharyngeal swab, Saliva, Salivary IgA
Abstract
Aim
This study aims to verify whether standardized saliva collection is suitable for SARS-CoV-2 molecular detection and IgA measurement.
Methods
43 COVID-19 inpatients and 326 screening subjects underwent naso-pharyngeal (NP)-swab and saliva collection (Salivette). Inpatients also underwent repeated blood collections to evaluate inflammation and organs involvement. In all patients and subjects, SARS-CoV-2 (gene E) rRT-PCR was undertaken in saliva and NP-swabs. Salivary IgA and serum IgA, IgG, IgM were measured on inpatients’ samples.
Results
NP-swabs and saliva were both SARS-CoV-2 positive in 7 (16%) or both negative in 35 (82%) out of 43 patients successfully included in the study. NP-swabs and saliva results did not perfectly match in one patient (saliva positive, NP-swab negative). Positive molecular results were significantly associated with disease duration (p = 0.0049). 326/326 screening subjects were SARS-CoV-2 negative on both NP-swabs and saliva. Among the 27 saliva samples tested for IgA, 18 were IgA positive. Salivary IgA positivity was associated with pneumonia (p = 0.002) and CRP values (p = 0.0183), not with other clinical and molecular data, or with serum immunoglubulins.
Conclusions
A standardized saliva collection can be adopted to detect SARS-CoV-2 infection in alternative to NP-swabs. Preliminary data on salivary IgA support the use of saliva also for patient monitoring.
1. Introduction
The rapid detection of SARS-CoV-2 is of utmost importance in identifying symptomatic and asymptomatic infected subjects, whose isolation has been proven effective in limiting viral spread [1], [2], [3]. For diagnosis of the disease, the World Health Organization (WHO) recommends naso-pharyngeal swab molecular testing in the presence of clinical and epidemiological factors [4]. Although it is sensitive and specific, molecular testing of naso-pharyngeal swabs has some limitations: sample collection requires expert personnel while the analytical process takes time and requires laboratories to implement molecular diagnostics [3], [5], [6]. Overall, these limitations, especially in a pandemic, might compromise the provision of numerous timely performed tests. In order to speed up analyses while enhancing health care workers’ safety, some authors have proposed swab self collection and/or naso-pharyngeal swabs pooling before analysis [7], [8], [9], [10]. Both proposals have drawbacks: 1. Patients’ self-collected swabs could generate false negative results due to sampling errors caused by the lack of specific training and the patient’s spontaneous resistance to correctly introducing the swab into his/her oral and/or nasal cavities; 2. Although pooling of up to 10 samples has been shown to produce reliable results, this approach could miss positive cases in the presence of a low viral load, i.e. the threshold cycle (Ct) of rRT-PCR is higher than 35 [8], [10]. A search should therefore be made for alternative strategies, in order to speed up SARS-CoV-2 identification without decreased test sensitivity.
Saliva testing has been proposed as a valid alternative to naso-pharyngeal swabs for detecting viral RNA sequences [5], [6], [11], [12], [13], [14], [15].
Saliva droplets, in fact, appear to be the main vehicle of viral spread, and the salivary glands are reportedly not only an infection target, but also a potential SARS-CoV-2 virus reservoir [16], [17]. In view of the potential infectivity of saliva, this biological fluid might be used for diagnostic purposes, having advantages over naso-pharyngeal swab, involving an easier collection procedure. The non-invasive nature of saliva collection would enhance patients’ compliance in repeated testing and the simplicity of the procedure facilitates self-collection without specific training being required. However, the collection mode should be carefully evaluated, as otherwise collection itself might incur a risk of disease spread [17]. Some Authors have suggested ‘cough up’ by clearing the throat for saliva sample collection [13], [14] and others, ‘passive drool’ [7], although saliva might also be collected by introducing different absorbent materials into the oral cavity, or by stimulation with citrate [12]. The saliva collection methods described, including spitting and cough, might generate harmful aerosol and droplets and might also result in patient-to-patient variability due to incorrect sample collection, ultimately decreasing test sensitivity. The search for a standardized saliva collection method that minimizes the risk of droplets or aerosol transmission is therefore recommended [12].
Another approach for SARS-CoV-2 diagnosis is the analysis of IgA, IgG and IgM class antibody by immunometric techniques in sera or by rapid immunochromatographic assays in whole blood [5], [6], [18]. Antibodies appear in blood one to two weeks after disease onset, their peak values starting to decline after three to four weeks [19]. Their identification in asymptomatic subjects might aid in the identification of healthy disease carriers. Antibodies could also be found in saliva, being blood-derived or directly produced by the mucosa-associated immune cells [12], [20]. Also, in this respect, saliva should offer advantages over serum/blood. In fact, although blood sampling is only minimally invasive, some patients, particularly children and their parents, might not be compliant. Since the majority of infected children are asymptomatic, they are a potential source of viral spread within their family, and in the community [21]. Screening children, therefore, appears to be a crucial step in limiting viral spread. To do this, repeated naso-pharyngeal swabs and/or blood testing might lower their compliance, but the direct search in saliva for viral RNA and antibodies might represent a non-invasive “one sample two tests” approach that enhances the success of screening programs, especially if new waves are expected in the near future, as has already occurred in countries that first experienced the pandemic [2]. Large-scale testing with contact-tracing and household quarantine are suggested as the most effective strategy to prevent a second pandemic wave and this strategy might benefit from easy to handle specimens alternative to naso-pharyngeal swabs, as saliva [22], [23].
The aim of the present study was to verify whether standardized and safe saliva collection is suitable for SARS-CoV-2 molecular detection and IgA class antibodies measurement.
2. Patients and methods
From 7 to 17 April 2020 we studied a total of 49 COVID-19 patients hospitalized at the University-Hospital of Padova (Italy) in two units: tropical and infectious diseases and semi-intensive care. Thirty-three were males (mean age 64 years, range 29–86 years) and 16 females (mean age 60 years, range 25–94 years), with no significant difference between the ages of the two groups (Student’s t test: 1.0369, p = 0.3051). Clinical data of the patients studied are reported in Table 1 .
Table 1.
Clinical data of the patients studied.
| Symptoms | Males (n = 33) | Females (n = 16) | Fisher's exact test |
|---|---|---|---|
| Fever >37.5 °C | 30 (91%) | 9 (56%) | p < 0.01 |
| Dyspnoea | 22 (67%) | 5 (31%) | p < 0.05 |
| Pneumonia | 29 (88%) | 10 (63%) | p = 0.06 |
| Gastrointestinal | 7 (21%) | 10 (63%) | p < 0.01 |
| Anosmia/Ageusia | 7 (21%) | 2 (13%) | p = 0.70 |
| Other | 18 (55%) | 9 (56%) | p = 1.00 |
Significant p values are reported in bold face.
On admission, all patients were SARS-CoV-2 positive at naso-pharyngeal swab. During hospitalization they underwent repeat naso-pharyngeal swab collection for SARS-CoV-2 molecular testing, performed as described by us elsewhere [24]. Blood samples were also obtained to evaluate cell blood count, coagulation parameters (Prothrombin Time-PT, Partial Thrombplastin Time-PTT) and biochemical markers evidencing any systemic inflammation (C-reactive protein-CRP and fibrinogen), renal (creatinine) and liver (aspartate aminotransferase-AST, alanine aminotransferase- ALT, Bilirubin, alkaline phosphatase-ALP and gamma-glutamyl transferase-GGT) function, as well as heart (troponin I and brain natriuretic peptide-BNP) and pancreatic (amylase) involvement, measured using standard laboratory methods.
Haematological and biochemical data at admission and study enrolment were retrieved from the Laboratory Information System (LIS) repository of the University-Hospital of Padova.
After patients had given their fully informed consent (Local Ethic Committee Nr. 27444), patients were enrolled in the study, and asked to collect a saliva sample immediately before a naso-pharyngeal swab was taken for analysis. Saliva was collected using Salivette® tubes containing a cotton swab without preparation (SARSTEDT AG & Co, Nümbrecht, Germany), patients being asked to remove the cotton swab from the Salivette® and chew it for about one minute to stimulate salivation. Finally, the patients had to return the swab with the absorbed saliva to the Salivette® and replace the stopper; 43/49 patients collected enough saliva and were successfully included for study purposes. The Salivette® were stored at 4 °C immediately after collection and centrifuged at 4000g for 5 min within 3 h from collection in order to ensure that saliva samples were clear. An aliquot of saliva (300 μL) was analysed for SARS-CoV-2 gene E by means of Real-Time reverse-transcription polymerase chain reaction (rRT-PCR), the remaining material being stored at −80 °C for no more than one month before testing for antibodies. In all cases, saliva SARS-CoV-2 molecular testing was performed in parallel with naso-pharyngeal swab analysis.
To verify whether threshold cycles (Ct) values resulting from rRT-PCR were correlated with decreasing SARS-CoV-2 viral load, Digital Droplet PCR (ddPCR) was performed on samples for which: (a) both saliva and naso-pharyngeal swab molecular testing was positive, and (b) sample volume was sufficient for performing the analysis.
All the procedures were undertaken following the manufacturer’s instructions for the QX200 AutoDG Droplet Digital PCR System using the One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad). The ddPCR reaction mixture contained 20× primers and probe mix (final concentrations of 900 and 250 nM, respectively), and 5 forμl RNA template in a final volume of 22 μL. Twenty microliters of each reaction mix was used to generate droplets with the AutoDG droplet generator (Bio-Rad). Droplet-partitioned samples were then transferred to a 96-well plate, sealed and cycled in a T100 Thermal Cycler (Bio-Rad) under the following cycling protocol: 50 °C for 1 h (reverse transcription), 95 °C for 10 min (DNA polymerase activation), followed by 45 cycles at 95 °C for 30 s (denaturation) and 60 °C for 1 min (annealing/extension) followed by enzyme deactivation at 98 °C for 10 min, and infinite 4-degree hold. The cycled plate was then transferred and read in the FAM and VIC channels using the QX200 Reader (Bio-Rad).
In a subset of 27 patients for whom saliva samples were available, salivary IgA were measured by means of an ELISA assay specific for IgA antibodies against S1 SARS-CoV-2 domain (Euroimmun Medizinische Laboradiagnostika, Luebeck, Germany) according to the manufacturer’s inserts after optimization for saliva. Seven saliva samples, two from healthy subjects and 5 from COVID-19 patients, were diluted 1:50, 1:100 (as recommended for serum) and 1:200. The absorbance at 450 nm was lower than the negative control in 4 serially diluted saliva samples. The 450 nm absorbance was measurable in three serially diluted clinical samples (sample#1: Abs450nm 1:50 = 0.608, Abs450nm 1:100 = 0.294, Abs450nm 1:200 = 0.165; sample#2: Abs450nm 1:50 = 0.265, Abs450nm 1:100 = 0.135, Abs450nm 1:200 = 0.081; sample#3: Abs450nm 1:50 = 0.204, Abs450nm 1:100 = 0.130, Abs450nm 1:200 = 0.063). For further saliva testing the 1:50 dilution was chosen. Salivary IgA were assayed using samples from 10 health care employers with negative naso-pharyngeal swabs results. The mean ± SD of the Abs450nm were 0.030 ± 0.02 and the mean ratio ± SD with respect to the calibrator was 0.05 ± 0.03. Patients’ salivary IgA were classified as positive when the ratio between saliva sample and the calibrator Abs450nm was higher than 0.300 [2*(Mean + 3SD of controls)].
Serum IgA, IgG and IgM at enrolment were measured by an ELISA assay based on whole-virus antigens (ENZY-WELL SARS-CoV-2, DIESSE Diagnostica Senese Spa, Monteriggioni, Siena, Italy) according to the manufacturer’s inserts.
From 22 to 30 April 2020, a total of 326 subjects were enrolled with the aim of screening the population and conducting a second, prospective phase of the study. Each subject underwent naso-pharyngeal swab and saliva sample collection by means of Salivette®. rRT-PCR was performed in parallel on naso-pharyngeal swab and saliva samples.
The statistical analysis of data was made with binary logistic regression, Spearman’s correlation, and Student’s t test for paired and unpaired data using STATA 13.1 statistical software (StataCorp, 4905 Lakeway Drive, USA).
3. Results
Naso-pharyngeal swabs collected at enrolment were positive for SARS-CoV-2 in 9/49 patients. Saliva was available for 43 patients, including 8/9 SARS-CoV-2 positive cases. Considering the 43 patients for whom saliva and naso-pharyngeal testing was performed in parallel, 7 cases shared positive and 35 shared negative results. For one patient, saliva was positive (Ct = 26) but the naso-pharyngeal swab was negative. Positive naso-pharyngeal swab or saliva results were correlated with the duration of symptoms, not with the duration of hospitalization, as shown in Table 2 .
Table 2.
Correlation between molecular testing results performed on naso-pharyngeal swab and saliva and disease duration.
| Naso-pharyngeal swab |
Saliva |
|||
|---|---|---|---|---|
| Disease duration | Positive (n = 9) | Negative (n = 40) | Positive (n = 8) | Negative (n = 35) |
| From symptoms onset Mean (95% CI) | 12 (9–15) days | 22 (18–25) days | 12 (9–16) days | 22 (18–25) days |
| Student’s t test | t = 2.95;p < 0.005 | t = 2.62;p < 0.05 | ||
| From hospitalization Mean (95% CI) | 14 (8–20) days | 17 (14–20) days | 14 (7–20) days | 16 (13–19) days |
| Student’s t test | t = 0.71; p = 0.48 | t = 0.68; p = 0.50 | ||
Significant p values are reported in bold face. CI: Confidence Interval.
The significant association between disease duration and naso-pharyngeal swab or saliva positive findings was confirmed at binary logistic regression analysis, other predictor variables, such as type of symptoms, age and gender not being significant (Table 3 ).
Table 3.
Logistic regression analyses considering positive molecular testing as the outcome variable, and age, gender, type and duration of symptoms as predictors.
| Naso-pharyngeal swab (n = 49) |
Saliva (n = 43) |
|||||
|---|---|---|---|---|---|---|
| Coefficient ± SE | p | 95% CI | Coefficient ± SE | p | 95% CI | |
| Age | 0.08 ± 0.07 | 0.26 | −0.06 to 0.23 | 0.09 ± 0.08 | 0.25 | −0.06 to 0.24 |
| Gender | −1.47 ± 1.52 | 0.34 | −4.46 to 1.53 | −0.28 ± 1.53 | 0.85 | −3.29 to 2.72 |
| Pneumonia | 1.97 ± 2.58 | 0.45 | −3.08 to 7.03 | 3.46 ± 2.98 | 0.25 | −2.38 to 9.30 |
| Fever | 0.45 ± 2.01 | 0.82 | −3.50 to 4.40 | 1.33 ± 2.14 | 0.53 | −2.86 to 5.52 |
| Gastrointestinal | 1.80 ± 1.45 | 0.22 | −1.05 to 4.65 | 2.49 ± 1.63 | 0.13 | −0.69 to 0.68 |
| Anosmia/Ageusia | 2.56 ± 2.05 | 0.21 | −1.46 to 6.57 | 3.34 ± 2.09 | 0.11 | −0.76 to 7.44 |
| Other symptoms | −1.85 ± 1.24 | 0.14 | −4.29 to 0.59 | −0.62 ± 1.29 | 0.63 | −3.15 to 1.92 |
| Duration of symptoms | −0.54 ± 0.23 | <0.05 | −0.99 to 0.09 | −0.61 ± 0.28 | <0.05 | −1.16 to −0.05 |
Significant p values are reported in bold face. SE: Standard Error; CI: Confidence Interval.
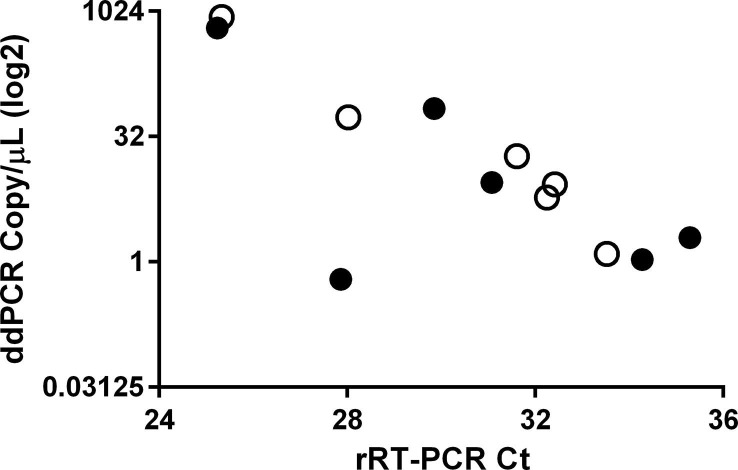
A significant correlation was found between the Ct values of the 7 double positive (naso-pharyngeal and saliva) samples (R-squared = 0.69, p < 0.05), being above 25 in both matrices, thus suggesting low viral loads. To further ascertain whether increasing Ct values were correlated with decreasing SARS-CoV-2 viral load, ddPCR was performed using the 6 samples for which both saliva and naso-pharyngeal swab materials were available. As shown in Fig. 1 , a significant inverse correlation was found for naso-pharyngeal swabs (Spearman’s r = −0.94, p < 0.05), but not saliva samples (Spearman’s r = −0.37, p = 0.50).
Fig. 1.
The correlation between threshold cycle (Ct) values obtained with rRT-PCR and SARS-CoV-2 copy number/μl (ddPCR). Correlation between threshold cycles (Ct) and the copy number/μl obtained with ddPCR (expressed in log2). Six SARS-CoV-2 positive naso-pharyngeal swabs (open circles) and corresponding saliva data (dots) are shown.
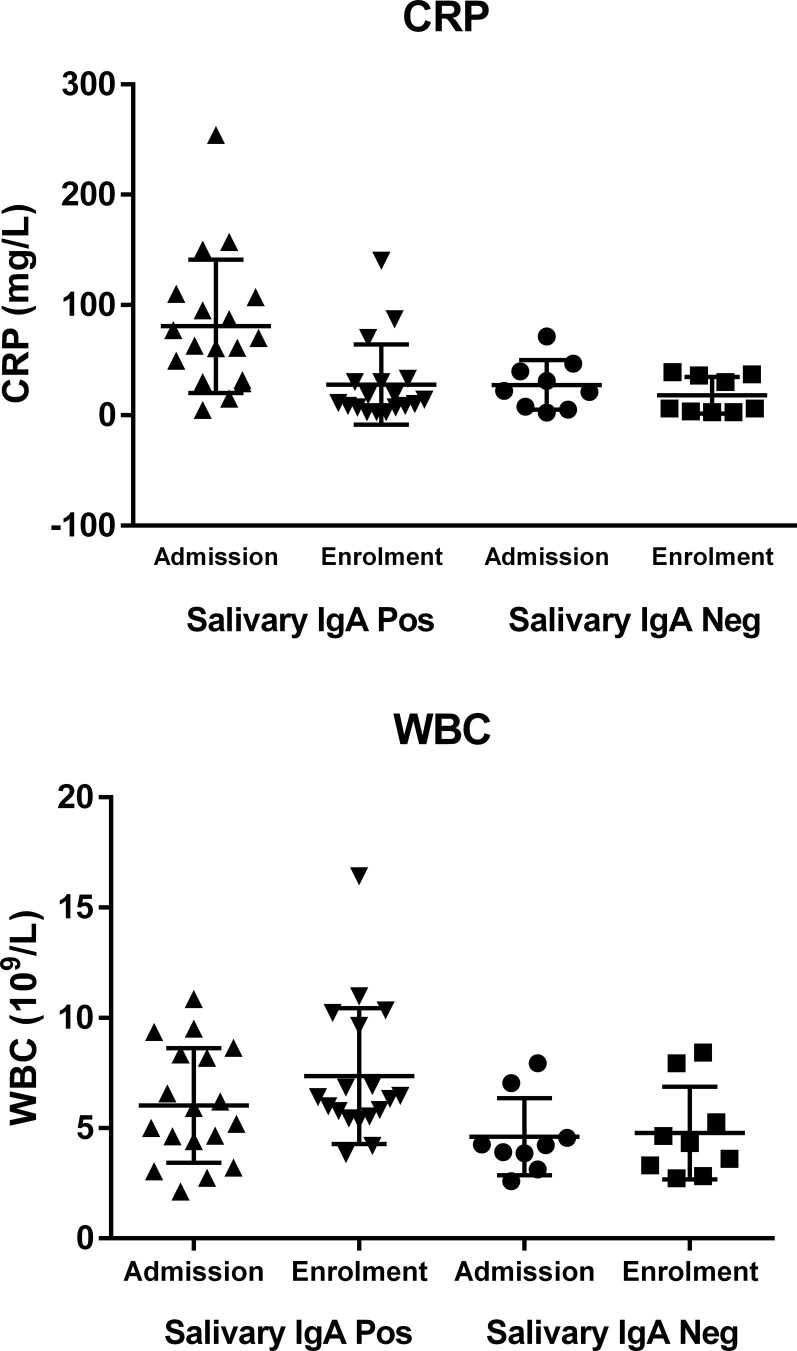
In a subset of 27 patients for whom saliva was available, salivary IgA were measured, positive results being found in 18 cases (67%). Salivary IgA results were not correlated with SARS-CoV-2 molecular naso-pharyngeal swab findings (Fisher's exact test: p = 0.65). Positive salivary IgA were significantly correlated with the presence of pneumonia (Fisher's exact test: p < 0.01), but not with the other clinical data (p:ns). Pneumonia was diagnosed in all the 18 patients with positive salivary IgA, and in 4/9 with negative salivary IgA findings. Of the 27 patients with salivary IgA data, serum antibody data were also available for 16 patients, 5 with negative and 11 with positive salivary IgA findings. Serum IgA and IgG were positive in all 16 patients, while serum IgM were positive in 9, and negative in 7, but not correlated with salivary IgA (Fisher's exact test: p = 0.60). In order to ascertain whether systemic inflammation and organ involvement might have any effect on salivary IgA, we evaluated haematological and coagulation parameters (PT, PTT) and biochemical data for systemic inflammation (CRP and fibrinogen), renal (creatinine) and liver (AST, ALT, Bilirubin, ALP and GGT) function, as well as heart (troponin I and BNP) and pancreatic (Amylase) involvement. Data were retrieved from cases in the LIS repository considering those at admission to the Hospital and those at enrolment (Supplementary Table 1). Positive salivary IgA were correlated with CRP values at admission (t = −2.52; p < 0.05) and with white blood cell count (WBC) at enrolment (t = −2.25; p < 0.05), as shown in Fig. 2 .
Fig. 2.
The correlation between salivary IgA and biochemical and haematological markers. The correlation between salivary IgA and biochemical and haematological parameters. The upper panel shows the correlation between salivary IgA positive or negative findings and CRP values obtained at admission and enrolment. The lower panel shows the correlation between salivary IgA and white blood cell count at admission and enrolment.
Findings at binary logistic regression analysis, performed considering salivary IgA as dependent, and CRP and WBC on admission and at enrolment as predictors, confirmed that CRP on admission is a significant variable (Table 4 ).
Table 4.
Logistic regression analyses considering salivary IgA as the outcome variable and age, gender, WBC and CRP at admission and enrolment as predictors.
| Salivary IgA (n = 27) |
|||
|---|---|---|---|
| Coefficient ± SE | p | 95% CI | |
| Age | −0.06 ± 0.06 | 0.34 | −0.18 to 0.06 |
| Gender | 0.59 ± 1.25 | 0.63 | −1.85 to 3.04 |
| WBC at admission | −1.17 ± 0.79 | 0.14 | −2.71 to 0.37 |
| WBC at enrolment | 1.73 ± 0.99 | 0.08 | −0.22 to 3.67 |
| CRP at admission | 0.09 ± 0.04 | <0.05 | 0.002 to 0.17 |
| CRP at enrolment | −0.02 ± 0.03 | 0.46 | −0.08 to 0.04 |
p values are reported in bold. SE: Standard Error; CI: Confidence Interval.
All the 326 subjects enrolled for screening had negative molecular test results for both naso-pharyngeal swab and saliva samples, thus confirming the 100% specificity of saliva molecular testing with respect to NP swab testing.
4. Discussion
On 21st February 2020, the first COVID-19 cases were identified in Italy, the rapid spread of infection leading to a war-like scenario. Other European countries soon experienced the same disaster, which continues to spread through other countries, such as the United States, Russia, India and Brazil [25], [26]. The rapid decision of national authorities to limit person-to-person contacts by a lock-down policy led to a progressive reduction in viral spread [27]. The present study was conducted at the University Hospital of Padova in early April 2020, when the positive rate of naso-pharyngeal swabs performed in our laboratory was 0.26% (13 positive tests out of 5,091 performed in the 10 day time-period of patient enrollment) against a 1.02% positivity rate (50/4,925) registered in the 10 previous days. The potential number of patients eligible for the study was therefore reduced at that time and most patients enrolled were in a recovery phase of the disease. This explains why the percentage of positive findings of naso-pharyngeal swabs in our inpatients series was low (18%), this result being in line with previous data in the literature showing negative or fluctuating SARS-CoV-2 positive findings in naso-pharyngeal swabs after two weeks from the onset of symptoms [14], [28], [29]. Moreover, the Ct values of positive swabs, the majority higher than 30, indicated that these patients had active infection with a low viral load, as expected when the disease persists for more than 15 days. The low viral load was confirmed in both saliva and naso-pharyngeal swabs by ddPCR, a reference procedure for nucleic acid quantification which should be taken into careful consideration in the near future in order to establish whether viral spread and disease severity are correlated with different viral loads [14], [30], [31]. In 6/49 patients, adequate saliva samples were unavailable for different reasons, including low compliance or low saliva production in patients with a severely compromised clinical status. Saliva rRT-PCR results were concordant with swab results not only in qualitative, but also in quantitative terms (i.e. Ct results). This finding further supports saliva as an alternative fluid to naso-pharyngeal swabs for SARS-CoV-2 diagnosis [11], [12], [13], [17]. The strength of the present study with respect to previous studies on saliva collection depended on our use of a standardized collection procedure that prevents droplet or aerosol release: saliva was collected for one minute, using a cotton swab without preparation to be chewed. The limitations in compliance that occurred among our patients were not related to the collection procedure per se, but to the extreme disease-related fatigue of some patients. In line with this, complete compliance was found among the 326 subjects enrolled for screening in the second and prospective phase of the study. The very low prevalence of infection registered during this second phase accounts for the lack of positive cases at both naso-pharyngeal swabs and saliva testing, which was confirmed to have 100% specificity.
The main routes of SARS-CoV-2 infection are the mouth and nose, salivary glands being infection site and reservoir [12], [16], [17]. While the virus persists a few days to a few weeks in the oral and nasal cavities, innate and adaptive immune responses take place. The paradigm of adaptive immunity, antibody production, is expected not only in blood but also in the infected mucosa and salivary glands [6]. In line with this, saliva may collect not only serum antibodies, but also locally produced IgA class antibodies. With this in mind, we measured in saliva IgA class antibodies elicited against the S1 domain of the spike protein, involved in ACE2 receptor binding [32], and compared findings with serum IgA, IgG and IgM data. Salivary IgA were positive in 67% of patients, unlike serum IgA and IgG, which were positive in all the cases studied. Different reasons might explain the differences in sensitivity between salivary and serum IgA: a) salivary IgA levels depends more than serum IgA on local mucosal immunity; b) in our study salivary and serum IgA recognize different antigens, the S1 domain of the spike protein and whole-virus antigens respectively. However, the sensitivity of our salivary IgA test results is higher with respect than that reported by Randad et al. [20], when the NAC S1 antigen was evaluated (8.7%). When the GeneScript S1 antigen was evaluated, the same authors found a high sensitivity (77%), but a low specificity (42%), whereas a specificity of 100% was achieved with our method. Our results are limited by the low number of saliva samples evaluated, especially for specificity (n = 9), due to restricted supplies during the pandemic. In agreement with Randad et al. [20], positive IgA salivary results were correlated with disease duration, and with the patient’s clinical status. Positive IgA class results were more frequent in patients with pneumonia, and they were correlated with the magnitude of systemic inflammatory response at disease onset. The lack of a direct association between salivary and serum immunoglobulins suggests that salivary IgA are produced by mucosal associated lymphocytes. It remains to be verified whether or not these immunoglobulins are protective against infection. The results of our study support saliva as a challenging alternative specimen type with respect to conventional naso-pharyngeal swabs and serum that might be helpful for large-scale screening to prevent and/or limit the second wave of SARS-CoV-2, which is more than expected especially if relaxation of lockdown measures become mandatory to enable economic activities and re-opening of schools [22], [23].
However the study has some limitations. First, the number of patients studied was limited, due to the success achieved in the outbreak management by our Region and evidenced by the relatively low prevalence of positive cases in the inpatient group. Second, results of ongoing studies aiming to provide evidence of protection from salivary IgA, are awaited.
5. Conclusions
In conclusion, our study supports that standardized and safe self-collection of saliva might be considered a valid alternative to naso-pharyngeal swab collection in detecting SARS-CoV-2 infection. We have also provided some preliminary data indicating that this fluid is of potential utility for monitoring local adaptive immunity by measuring IgA anti spike SARS-CoV-2 protein.
CRediT authorship contribution statement
Ada Aita: Conceptualization, Methodology, Data curation, Writing - original draft. Daniela Basso: Conceptualization, Methodology, Data curation, Writing - original draft. Anna Maria Cattelan: Resources, Investigation. Paola Fioretto: Resources, Investigation. Filippo Navaglia: Investigation. Francesco Barbaro: Resources, Investigation. Alice Stoppa: Investigation. Enrico Coccorullo: Investigation. Assunta Farella: Investigation. Aurora Socal: Investigation. Roberto Vettor: Writing - review & editing. Mario Plebani: Writing - review & editing.
Acknowledgments
The Authors thank Mrs. Monica Razetti, Mrs. Cinzia Centobene, Mrs. Tamara Avellone, Mrs. Daniela Rinaldi and Mrs. Maria Grazia Marzellan for their valuable technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2020.09.018.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Coronavirus disease 2019 (COVID-19) in the EU/EEA and the UK– ninth update, 2020, 1–50.
- 2.Leung K., Wu J.T., Liu D.i., Leung G.M. First-wave COVID-19 transmissibility and severity in China outside Hubei after control measures, and second-wave scenario planning: a modelling impact assessment. The Lancet. 2020;395(10233):1382–1393. doi: 10.1016/S0140-6736(20)30746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C.B.E.M. Reusken, E.K. Broberg, B. Haagmans, A. Meijer, V.M. Corman, A. Papa, et al., Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries, January 2020, Euro Surveill. 25 (6) (2020). http://doi.org/10.2807/1560-7917.ES.2020.25.6.2000082. [DOI] [PMC free article] [PubMed]
- 4.World Health Organization (WHO) Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. https://apps.who.int/iris/handle/10665/330676, 2020 (accessed 15 June 2020).
- 5.Loeffelholz M.J., Tang Y.-W. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg. Microbes Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): Current status, challenges, and countermeasures. Rev. Med. Virol. 2020;30(3) doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan P.S., Sailey C., Guest J.L., Guarner J., Kelley C., Siegler A.J. Detection of SARS-CoV-2 RNA and antibodies in diverse samples: Protocol to validate the sufficiency of provider-observed, home-collected blood, saliva, and oropharyngeal samples. JMIR Public Health Surveill. 2020;6(2) doi: 10.2196/19054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres I., Albert E., Navarro D. Pooling of nasopharyngeal swab specimens for SARS-CoV-2 detection by RT-PCR. J. Med. Virol. 2020:jmv.25971. doi: 10.1002/jmv.25971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Y.-P. Tu, R. Jennings, B. Hart, G.A. Cangelosi, R.C. Wood, K. Wehber, et al, Swabs collected by patients or health care workers for SARS-CoV-2 testing, N Engl. J. Med. (2020) NEJMc2016321. http://doi.org/10.1056/NEJMc2016321. [DOI] [PMC free article] [PubMed]
- 10.Wacharapluesadee S., Kaewpom T., Ampoot W., Ghai S., Khamhang W., Worachotsueptrakun K. Evaluating the efficiency of specimen pooling for PCR-based detection of COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.26005. jmv.26005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braz Silva P.H., Pallos D., Giannecchini S., To K.-K.-W. SARS-CoV-2: What Can Saliva Tell Us? Oral Dis. 2020 doi: 10.1111/odi.13365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J.J. Ceron, E. Lamy, S. Martinez-Subiela, P. Lopez-Jornet, F. Capela E Silva, P.D. Eckersall, A. Tvarijonaviciute, Use of Saliva for Diagnosis and Monitoring the SARS-CoV-2: A General Perspective, J. Clin. Med. 9 (5) (2020). http://doi.org/10.3390/jcm9051491. [DOI] [PMC free article] [PubMed]
- 13.To K.K.-W., Tsang O.T.-Y., Chik-Yan Yip C., Chan K.-H., Wu T.-C., Chan J.M.C. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. LancetInfect Dis. 2020 doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. Xu, Y. Li, F. Gan, Y. Du, Y. Yao, Salivary glands: Potential reservoirs for COVID-19 asymptomatic infection, J. Dent Res. (2020a) 22034520918518. http://doi.org/10.1177/0022034520918518. [DOI] [PubMed]
- 17.Xu R., Cui B., Duan X., Zhang P., Zhou X., Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int. J. Oral Sci. 2020;12(1):11. doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padoan A., Cosma C., Sciacovelli L., Faggian D., Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin. Chem. Lab Med. 2020 doi: 10.1515/cclm-2020-0443. [DOI] [PubMed] [Google Scholar]
- 20.P.R. Randad, N. Pisanic, K. Kruczynski, Y. C. Manabe, D. Thomas, A. Pekosz, et al., COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva, medRxiv. (2020). http://doi.org/10.1101/2020.05.24.20112300. [DOI] [PMC free article] [PubMed]
- 21.Zimmermann P., Curtis N. Coronavirus infections in children including COVID-19: an overview of the epidemiology, clinical features, diagnosis, treatment and prevention options in children. Pediatr. Infect. Dis. J. 2020;39(5):355–368. doi: 10.1097/INF.0000000000002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. Aleta, D. Martín-Corral, A. Pastore, Y. Piontti, M. Ajelli, M. Litvinova, M. Chinazzi, et al., Modelling the impact of testing, contact tracing and household quarantine on second waves of COVID-19, Nat. Hum. Behav. (2020). http://doi: 10.1038/s41562-020-0931-9. [DOI] [PMC free article] [PubMed]
- 23.Panovska-Griffiths J., Kerr C.C., Stuart R.M., Mistry D., Klein D.J., Viner R.M., Bonell C. Determining the optimal strategy for reopening schools, the impact of test and trace interventions, and the risk of occurrence of a second COVID-19 epidemic wave in the UK: a modelling study. Lancet Child Adolesc. Health. 2020;S2352–4642(20):30250–30259. doi: 10.1016/S2352-4642(20)30250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basso D., Aita A., Navaglia F., Franchin E., Fioretto P., Moz S. SARS-CoV-2 RNA identification in nasopharyngeal swabs: issues in pre-analytics. Clin. Chem. Lab Med. 2020 doi: 10.1515/cclm-2020-0749. [DOI] [PubMed] [Google Scholar]
- 25.Lai C.-C., Wang C.-Y., Wang Y.-H., Hsueh S.-C., Ko W.-C., Hsueh P.-R. Global epidemiology of coronavirus disease 2019 (COVID-19): disease incidence, daily cumulative index, mortality, and their association with country healthcare resources and economic status. Int. J. Antimicrob. Agents. 2020;55(4) doi: 10.1016/j.ijantimicag.2020.105946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization (2020). Coronavirus disease (COVID-19). Weekly epidemiological update 1. https://www.who.int/docs/default-source/coronaviruse/situation-reports/.
- 27.Remuzzi A., Remuzzi G. COVID-19 and Italy: what next? Lancet. 2020;395(10231):1225–1228. doi: 10.1016/S0140-6736(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J. (2020), Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg. Microbes Infect. 2020;2020(2):1–14. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joynt G.M., Wu W.K. Understanding COVID-19: what does viral RNA load really mean? Lancet Infect Dis. 2020;20(6):635–636. doi: 10.1016/S1473-3099(20)30237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20(6):697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.