Abstract
Fibro/Adipogenic Progenitors (FAPs) are muscle-interstitial progenitors mediating pro-myogenic signals that are critical for muscle homeostasis and regeneration. In myopathies, the autocrine/paracrine constraints controlling FAP adipogenesis are released causing fat infiltrates. Here, by combining pharmacological screening, high-dimensional mass cytometry and in silico network modeling with the integration of single-cell/bulk RNA sequencing data, we highlighted the canonical WNT/GSK/β-catenin signaling as a crucial pathway modulating FAP adipogenesis triggered by insulin signaling. Consistently, pharmacological blockade of GSK3, by the LY2090314 inhibitor, stabilizes β-catenin and represses PPARγ expression abrogating FAP adipogenesis ex vivo while limiting fatty degeneration in vivo. Furthermore, GSK3 inhibition improves the FAP pro-myogenic role by efficiently stimulating, via follistatin secretion, muscle satellite cell (MuSC) differentiation into mature myotubes. Combining, publicly available single-cell RNAseq datasets, we characterize FAPs as the main source of WNT ligands inferring their potential in mediating autocrine/paracrine responses in the muscle niche. Lastly, we identify WNT5a, whose expression is impaired in dystrophic FAPs, as a crucial WNT ligand able to restrain the detrimental adipogenic differentiation drift of these cells through the positive modulation of the β-catenin signaling.
Subject terms: Cell biology, Molecular biology
Introduction
A limited injury of the adult skeletal muscle triggers a self-repair program orchestrated by a regenerative process involving multiple cell types that cooperate to restore and preserve muscle homeostasis [1–17]. However, the regeneration machinery dithers in aging individuals [18, 19] and fails in patients affected by muscle-related disorders such as myopathies [20]. Fibro/adipogenic progenitors (FAPs) are interstitial progenitors of mesenchymal origin [21] that contribute to muscle homeostasis [17] and regeneration by transiently supporting the activation and differentiation of muscle satellite cells (MuSCs) [7, 13, 16, 17, 22]. Aside from their supporting activity in muscle regeneration, FAPs have the potential to differentiate into adipocytes and myofibroblasts that, in the diseased tissue, fill the muscle interstitium [23, 24]. Embryonic pathways, such as Hedgehog and Notch, have been recently implicated in controlling FAP differentiation and the dysregulation of their signaling cascade, in dystrophic models, is suggested to account for the deviation from FAP physiological role [25, 26]. Wingless-related integration site (WNT) signaling also plays a pivotal role in adult muscle regeneration by influencing MuSC stemness, self-renewal, proliferation and differentiation [27–30]. The potential role played by WNT proteins in modulating FAP activation and/or differentiation has not been investigated yet. Here we show that the Glycogen Synthase Kinase 3 (GSK3), a crucial hub of the WNT signaling, controls FAP adipogenesis. Specifically, GSK3 blockade fully abrogates FAP adipogenesis ex vivo while limiting the intramuscular fat infiltrations that accompany muscle damage upon glycerol injection in vivo. By using single-cell mass cytometry, we observed that CTNNB1 (β-catenin) downregulation marks FAPs undergoing adipogenesis. We also observed that FAPs express high levels of WNT5a and that this expression is impaired in FAPs isolated from dystrophic mice, suggesting that an altered autocrine circuit may contribute to the adipogenic drift of FAPs in dystrophic muscles. In summary, our study uncovers the WNT/GSK3/β-catenin axis as a new and previously unexplored pathway contributing to control FAP adipogenesis and muscle fatty degeneration. Modulating the WNT pathway, either by targeting GSK3 or by restoring autocrine WNT5a signaling in FAPs, is a promising strategy to counteract intramuscular fat infiltrations in myopathies.
Materials and methods
Mouse models
C57BL/6J (RRID:IMSR JAX:000664) and C57BL/10ScSn-Dmdmdx/J mice (RRID:IMSR_JAX:001801) (hereafter referred to as wt and mdx, respectively) were purchased from the Jackson Laboratories. In our study, sexes were equally balanced between genotypes. Young (45-day-old), adult (3.5 month) wt and young (45-day-old), old (18-month-old) mdx mice were employed in this work. Mice were bred and maintained according to the standard facility procedures. All experimental studies were conducted according to the rules of good animal experimentation I.A.C.U.C. n̊ 432 of March 12 2006 and under ethical approval released on 11-12-2012 from Italian Ministry of Health, protocol #20/01-D.
Murine primary cells
FAPs and MuSCs were isolated from the hind limbs of male and female wt and mdx mice. Cell culture procedure were performed as described previously [31]. Briefly, freshly sorted FAPs were resuspended in FAP-GM consisting of DMEM GlutaMAX® (25 mM Glc) supplemented with 20% FBS, 10 mM Hepes, 1 mM sodium pyruvate, and 100 U/ml P/S. Freshly purified MuSCs were resuspended in growth medium (MuSCs-GM) consisting of DMEM GlutaMAX® (25 mM Glc) supplemented with 20% FBS, 10% donor horse serum, 2% chicken embryo extract, 10 mM Hepes, 1 mM sodium pyruvate, and 100 U/ml P/S.
Isolation of murine primary cells from skeletal muscles using MACS technology
FAPs and MuSCs from wt, mdx and CTX-injured mice were isolated through magnetic-activated cell sorting (MACS, Miltenyi) according to [31]. Mice were sacrificed through cervical dislocation and the hind limbs were washed with 70% ethanol. The skin was removed and the limbs were ablated and placed in ice-cold Hank’s Balanced Salt Solution (HBSS, Gibco) supplemented with 100 U/ml penicillin/streptomycin (P/S, Roche) and 0.2% bovine serum albumin (BSA, AppliChem). After mechanical dissociation under sterile hood, the tissue was washed with HBSS and centrifuged at 700 × g for 10 min at 4 °C. The minced tissue was weighted and resuspended in enzymatic mix, in proportion (4 ml/g), consisting of 2 mg/ml collagenase A, 2.4 U/ml dispase II, and 10 µg/ml DNase I (Roche) in Dulbecco’s Phosphate Buffered Saline (DPBS, BioWest) w/calcium and magnesium. Tissue preparations were incubated in water bath at 40 °C (not in immersion), under gentle shaking for 1 h (vortexing after 30 min). After digestion, the suspension was diluted with two volumes of HBSS and filtered through 100 µm cell strainers (Corning) and then centrifuged at 700 × g for 10 min at 4 °C. Pellets were resuspended in 10 ml of HBSS and filtered through 70 µm cell strainers. After separation, red blood cells were lysed in RBC Lysis Buffer (Santa Cruz) for 150 s in ice and then diluted in HBSS and filtered through 40 µm cell strainers. Note that cell strainers were activated and washed with 5 ml HBSS. For MACS, pellets were resuspended in 500 μl of magnetic beads buffer consisting of 0.5% BSA, 2 mM EDTA in 1× PBS. Cell suspensions were filtered through a 30 µm pre-separation filter (Miltenyi) and the mononuclear cells were counted and centrifuged at 700 × g for 10 min at 4 °C. The cell suspension was selected with magnetic bead-conjugated antibodies according to manufacturer’s instructions. Lin− cells were selected for CD31 and CD45. Anti-α7-integrin antibody was used to separate satellite cells while FAPs were obtained with positive selection with anti-Sca1 (non-HSC) antibody.
FAP culture, differentiation, and treatment
FAPs were cultured in growth medium (GM) and plated at the cell density of 1.5 × 104 cell/cm2. For in vitro expansion, 200,000 cells were plated in 100 mm diameter dishes in Cytogrow (Resnova, catalog TGM-9001-B) and cultured for 4 days (P0). Cells were detached (P1) and used for specific experiments.
Adipogenic differentiation was carried out incubating cells in adipocyte differentiation medium (ADM) consisting of GM supplemented with 1 µg/ml insulin, 0.5 mM 3-isobutyl-1-methylxanthine, and 1 µM Dexamethasone for 3 days. Adipocyte maturation was achieved with additional incubation (2 days) in adipocyte maintenance medium (AMM) consisting of GM supplemented with 1 µg/ml insulin.
For fibrogenic differentiation, cells were plated at the cell density of 0.75 × 104 cell/cm2 and, after a period of attachment (2 days), were incubated with 5 ng/ml TGF-beta 1 (PeproTech, catalog 100-21) for 3 days in DMEM supplemented with 2% horse serum.
For pharmacological and cytokine treatments, LY2090314 (Selleckchem, catalog S7063), MG132 (Selleckchem, catalog S2619), and PNU-74654 (Selleckchem, catalog S8429) were reconstituted in DMSO according to manufacturer’s instructions. Recombinant mouse Wnt10b (R&D System, catalog 2110-WN) and Recombinant human/mouse WNT5a (R&D System, catalog 645-WN-010) were reconstituted in sterile PBS 1× according to manufacturer’s recommendations. Culture media and concentrations are specified in “Results” and “Figure legends”.
MuSC culture, differentiation, and treatment
To reduce fibroblasts contamination, freshly sorted wt and mdx MuSCs were pre-plated for 2 h in pre-warmed MuSCs-GM. Pre-plated MuSCs were seeded in MuSC-GM at the cell density of 1.5 × 104 cells/cm2 in Matrigel-coated 96-well plates. After 3 days, MuSC-GM was fully refreshed and cells cultured in the presence of LY2090314 or vehicle for 3 additional days, allowing spontaneous myotube formation.
Kinase inhibitor screening
The Published Kinase Inhibitor Set (PKIS) was obtained from Structural Genomics Consortium and detailed information regarding chemical structures, kinase inhibition potency and kinase specificity can be found at ChEMBL (http://chembl.blogspot.co.uk/2013/05/pkis-data-in-chembl.html). To screen the PKIS, mdx FAPs were seeded on 384-well plate at the density of 1500 cells/well. Twenty-four hours after seeding cells were treated with the PKIS compounds at 1 and 0.1 μM for 6 additional days. DMSO and Trichostatin A (TSA) have been used as negative and positive controls, respectively. Compounds were transferred from a 10 mM DMSO stock solution to assay plates by acoustic droplet ejection (ATS-100, EDC biosystems, USA).
Cardiotoxin and glycerol muscle injury experiments
For muscle injury experiments, mice were first anesthetized with an intramuscular injection of saline solution containing ketamine (5 mg/ml) and xylazine (1 mg/ml). For cardiotoxin muscle injury, 20 µl of 10 µM cardiotoxin solution was administered into tibialis anterior (TA), quadriceps, and gastrocnemius muscles. Three days after injury FAPs were purified from limb muscles. For glycerol muscle injury 50 µl of 50% v/v of hypertonic solution of glycerol was administered intramuscularly into TA, quadriceps, and gastrocnemius muscles. The contralateral limb was equally injected with saline solution as control. Mice were randomly allocated in each experimental group without any type of blinding.
In vivo administration of LY2090314
LY2090314 (Selleckchem, catalog S7063) was dissolved in DMSO at 100 mM at the concentration of 0.0512 mg/μl. Three days after glycerol injury, 25 mg/Kg of LY2090314 were resuspended in corn oil (5% LY2090314/95% corn oil) and administered interperitoneally for 3 consecutive days. Control mice were treated with vehicle (5% DMSO/95% corn oil). Mice were randomly allocated in each experimental group without any type of blinding.
Flow cytometry
CD45−/CD31−/a7-integrin− and Sca1+ FAPs were routinely assessed for the expression of platelet-derived growth factor receptor-a (PDGFRa) used as a distinctive FAP marker. Immunophenotype characterization was performed by incubating 4.0 × 105 cells/ml for 30 min with antibodies against CD140a (PDGFRa-APC, eBioscience), diluted 1:50 in Flow Cytometry (FC) buffer (1% BSA in PBS 1×). Live/death cells were discriminated using propidium iodide (viability dye). For each sample 10,000 events were acquired using BD FACSCALIBUR (BD Biosciences).
Single-cell mass cytometry
For single-cell profiling via CyTOF2, amplified FAPs, from three independent mice, were maintained in FAP-GM or induced to differentiate into adipocytes by incubating cells with ADM. After 72 h, cells were detached using trypsin and 2.0 × 106 cells, for each condition, were employed for downstream procedures. Dead cells were stained incubating with Cell-ID Cisplatin-194Pt (Fluidigm) at final concentration of 1 μM. The samples from the three independent replicates were barcoded incubating with the appropriate combination of palladium isotopes from the Cell-ID™ 20-Plex Pd Barcoding Kit (Fluidigm, Catalog #201060) at the final concentration of 5 μM. The staining was quenched by adding Maxpar cell staining buffer (Fluidigm). Labeling with lanthanides-conjugated antibodies against surface and intracellular antigens was performed in a single tube according to the manufacturer’s instructions. The full list of antibodies is provided as Supplementary Table S2. After cell labelling, samples were incubated for 1 h at RT with Cell-ID™ Intercalator-Ir (Fluidigm) diluted at the final concentration of 125 nM in Maxpar Fix and Perm Buffer (Fluidigm). Cells were resuspended at the final concentration of 2.5 × 105 cells per ml and filtered with a 30 μm cell strainer cap into 5 ml polystyrene tubes. Single-cell data were acquired using mass cytometry platform CyTOF2. The instrument was tuned and mass calibrated according to manufacturer’s instructions. Samples were acquired at a rate of <400 events/s. Data were then converted to .fcs file and normalized using the manufacturer’s settings.
Analysis of single-cell mass cytometry data
After data acquisition, the normalized .fsc file with single-cell data were debarcoded with the Debarcoder Software (Fluidigm). The debarcoded.fcs files were then uploaded to the Cytobank server (Cytobank, Inc.) for data processing. Cells were initially manually gated from debris basing on the DNA content identified through Iridium (Ir) intercalator. Doublets were then excluded according to the event length parameter and single live cells were finally manually gated out by using Cisplatin (Pt) intercalator signal. t-distributed stochastic neighbor embedding (t-SNE) analysis was performed on gated live single cells launching the viSNE implementation on Cytobank.org [32] on the populations of interest identified through the following surface markers chosen for clustering cells: SCA1, CD140a, CD140b, CD146, CD90.2, CD34. At least 20,000 live single cells were analyzed for each sample.
Analysis of single-cell RNAseq datasets
Droplet-based single-cell 3′-end transcriptomics data of limb muscle mononuclear cells were retrieved from Tabula Muris Consortium [33] and from the recently published work of Giordani et al. [34]. In both experiments, mice had comparable ages (ranging from 10 to 15 weeks) allowing dataset integration. Data were processed using Seurat R package [35, 36], merged, and the expression values transformed to logarithmic scale. Features (2000 genes per dataset) exhibiting high cell-to-cell variation have been considered for downstream analyses and a linear transformation (z-score) has been applied. Principal component analysis indicated the minimum number of useful dimensions for the downstream analyses. The first nine components have been considered. For cell cluster identification, spatial resolution was set at 0.1 and the t-SNE algorithm applied to visualize cell clusters. t-SNE maps have been computed with Rtsne package [37]. For each cell cluster, cell ontology classes were identified on the basis of the biomarker information collected in the Myo-REG database [38] coupled with the biomarkers provided by Giordani et al. [34].
Analysis of bulk RNAseq datasets
For bulk RNAseq analysis, Marinkovic et al. [26] and Malecova et al. [39] datasets have been used and independently analyzed before any comparison between them. Data have been processed from htseq counts using DESeq2 package [40]. For Marinkovic dataset, three replicates from the two conditions (mdx and wild type) have been used. For Malecova dataset, the two available replicates for the two conditions (mdx and wild type) have been used. For both datasets, Ensemble gene identifiers have been mapped on M. musculus gene symbols. Statistical significance threshold for differentially expressed genes was set with an adjusted p value < 0.05 (false discovery rate correction for multiple comparisons).
Histological analysis
TA muscles were dissected and embedded in optimal cutting temperature medium and snap-frozen in liquid nitrogen for 10 s. Embedded muscles were stored at −80 °C prior to cryo-sectioning. Muscles were transversally cryo-sectioned (10 μm thickness slices) using a Leica cryostat. Sections were collected sequentially on different Superfrost glass slides (Thermo Fisher Scientific), thus adjacent sections were separated at least 100 μm from each other. Cryosections were performed for the entire length of the muscles. For the hematoxylin and eosin stain (H&E), sections were fixed in 2% paraformaldehyde (PFA) for 10 min at RT. After washing twice with 1× PBS, sections were incubated in hematoxylin solution for 20 min. The slides were washed for 10 min with gently running tap water and then rinsed in dd-H2O. Eosin staining was carried out for 1 h. Sections were ethanol-dehydrated (one wash in 90% followed by three washes in 100%), clarified with the Histo-Clear solution, and covered with coverslips using Eukitt mounting medium.
Immunoblot analysis
Cells were harvested removing culture medium and washing with PBS 1×. Cells were lysed in ice-cold Radio Immuno-Precipitation Assay lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 1% Nonidet P-40, 1 mM EGTA, 5 mM MgCl2, 0.1% SDS) supplemented with 1 mM PMSF, 1 mM orthovanadate, 1 mM NaF, protease inhibitor mixture 1×, inhibitor phosphatase mixture II 1×, and inhibitor phosphatase mixture III 1×. The insoluble material was separated at 15,500 × g for 30 min at 4 °C and total protein concentration was assayed on supernatants using Bradford reagent. Protein denaturation was performed with NuPAGE® LDS Sample Buffer supplemented with NuPAGE® Sample Reducing Agent, by heating at 95 °C for 10 min. SDS-PAGE and transfer were performed on 4–15% Bio-Rad Mini PROTEAN® TGX™/CRITERION® and Trans-Blot® Turbo™ mini or midi nitrocellulose membranes using a Trans-Blot® Turbo™ transfer System (Bio-Rad), respectively. Nonspecific binding sites were blocked using 5% nonfat dried milk in TBS-0.1% Tween-20 (TBS-T) for 1 h at RT under shaking. Primary antibodies were diluted according to manufacturer’s instruction and incubated overnight at 4 °C under shaking. HRP-conjugated secondary antibodies were diluted 1:3000 in 5% nonfat dried milk in TBS-T and incubated for 1 h at RT under shaking. Immunolabeling was detected with Clarity™ Western ECL Blotting Substrates (Bio-Rad) using the Las-3000 imaging system (Fujifilm). Densitometric quantitation of bands was performed with Fiji (Image J, NIH). The full list and catalog numbers of primary and secondary antibodies is provided in Supplementary Table S6.
Real-time PCR
Total RNA extraction was performed with TRIzol® reagent according to manufacturer’s instruction with minimal adaptation. Precipitation was carried out adding 10 μg of glycogen to the aqueous phase and incubating samples at −20 °C, overnight. RNA concentration was assessed using NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific). Samples were diluted using nuclease-free water to 25–100 ng/μl, according to the need and reverse-transcribed into cDNA with PrimeScript™ RT Reagent Kit (Takara) on 500–1000 ng. cDNA was diluted to 25 ng/μl with Easy-Dilution Reagent provided by the RT Reagent Kit. Real-Time PCR was performed on 50 ng of cDNA (2 μl) using SYBR Premix Ex Taq (Tli RNaseH Plus) (Takara) to a final volume of 20 μl. Relative expression was estimated by 2−ΔCt as described while different runs were compared by the 2−Δ ΔCt method [41].
Viability assay
Freshly isolated FAPs were seeded at the cell density of 1.5 × 104 cells/cm2 in 96-well plates. After 2 days FAPs were incubated in FAP-GM with increasing doses of LY2090314 (ranging from 1 to 100 nM) for 48 h. Cell viability was evaluated with CellTiter-Glo Luminescent Viability Assay (Promega) according to manufacturer’s instructions. Briefly, CellTiter-Glo reagent was added at a ratio of 1:1 to cell culture medium and mixed for 2 min on an orbital shaker. Samples were transferred to a black-walled 96-well plate and luminescence was recorded through Victor Multilabel plate reader (PerkinElmer). Background consisted of FAP-GM incubated in the absence of FAPs and processed as described above. Data are presented as relative luminescence units.
Oil Red O (ORO) staining for lipid droplets
ORO stock solution (3.5 mg/ml in 2-propanol) were diluted 3:2 (ORO:ddH2O) and filtered twice through 0.22 μm syringe filter. Culture media were removed and cells were fixed with 2% PFA solution in PBS 1× for 20 min at room temperature (RT). After fixation, cells were washed twice with PBS 1× and permeabilized with 0.5% Triton X-100 for 5 min. After washing twice with PBS 1×, cells were incubated with ORO-diluted solution for 10 min at RT, in dark and under gentle shaking. Cells were washed 4 times with PBS 1× (5 min each wash) and counterstained with Hoechst 33342 (2 μg/ml) for 5 min at RT. After washing, sample were maintained in PBS 1× supplemented with 0.002% sodium azide and stored at 4 °C until acquisition via fluorescence microscopy.
Immunofluorescence
Cells and sections were fixed in 2% PFA for 10 min at RT. For immunocytochemistry, fixed cells were washed twice in PBS 1× and permeabilized with 0.5% Triton X-100 in PBS 1× for 5 min. Blocking of unspecific binding sites consisted in 10% FBS, 0.1% Triton X-100 in PBS 1× for 1 h. Primary antibodies were diluted in blocking solution and incubated overnight at 4 °C. After washing four times with 0.1% Triton X-100 in PBS 1×, host-specific secondary antibodies were diluted in blocking solution, and incubated for 30 min at RT. Primary and secondary antibodies were diluted according to the manufacturer’s recommendations. Finally, cells were washed four times with 0.1% Triton X-100 in PBS 1× and counterstained with Hoechst solution (2 μg/ml Hoechst 33342, 0.1% Triton X-100 in 1× PBS) for 5 min at RT.
For Perilipin, PPARγ, PDGFRa and Laminin muscle section immunofluorescence, fixed sections were washed twice in PBS 1× and permeabilized with 0.3% Triton X-100 in PBS 1× for 30 min at RT. Blocking was performed using 10% Goat Serum, 1% Glycine, 0.1% Triton X-100 in 1× PBS for 1 h at RT. Anti-Perilipin, PPARγ, PDGFRa and Laminin primary antibodies were diluted 1:400 in blocking solution and incubated overnight at 4 °C. Slices were washed four times with 0.1% Triton X-100 in 1× PBS and incubated for 30′ at RT with host-specific secondary antibodies and/or with Alexa Fluor™ 488 Phalloidin. Nuclei were revealed by incubating samples for 10 min with Hoechst solution. Sections were washed twice in PBS 1× and mounted with Aqua Poly/Mount (Polysciences) mounting medium. The full list and catalog numbers of primary and secondary antibodies is provided in Supplementary Table S6.
Boolean network analysis
The Boolean networks describing Wnt and Insulin signaling pathways have been merged and automatically retrieved from Myo-REG [38]. This produced a network (N; E) in which the nodes N are the biological entities and the edges E are the activations (E+) and inhibitions (E−). Activation states of each node are represented by a discrete variable (0 for non-active nodes, 1 for active nodes).
To each node nk ∈ N a Boolean function Fk is associated, specifying how the value of that node can change depending on the incoming interactions (activation state of source nodes). Boolean Functions are reported in Supplementary Table S4. The synchronous or asynchronous updating schemes of the functions F1, …, Fn, at each discrete time step makes the network evolve from one state to another. When a successor state leads the network to a state has been already reached before, the network reaches an attractor, which can be a single state (steady state) or a cycle between two or more states.
The R package BoolNet has been applied to assess the network robustness, to analyze the network dynamics, and to generate the attractors associated to specific cell phenotypes [42]. Robustness analysis has been performed by comparing the percentage of identical attractors of the naïve network against 100 randomly generated networks. Perturbations (bitflip) ranging from 1 to 100 has been applied in each network-to-network comparison. The code is available in GitHub at the following link (https://github.com/Alessandro-Palma/wnt).
Network analysis
Differentially expressed Wnt genes between mdx and wild-type FAPs were extracted from two recently published datasets of bulk RNAseq FAP preparations [26, 39]. In both experiments, FAPs were purified from rodents of comparable ages (young wild-type and mdx mice) allowing dataset integration. Wnt-related genes have been retrieved by UniProt, using the keyword “Wnt signaling pathway”. MEST and Sox6 have been added to the list of Wnt pathway-related genes. The signaling network has been created by using the casual interactions collected in SIGNOR [43]. FAP Wnt signaling pathway was retrieved from Myo-REG to complete the signaling network. The resulting table of interactions has been transformed into a graph with Cytoscape [44]. Node sizes have been mapped on the adjusted p value from the two datasets, while their color has been mapped according to the log2 fold change in the comparisons between mdx and wild-type FAPs.
Image acquisition
Images of immunolabeled cells and section were acquired using the automated florescence microscope DMI6000B (Leica). A total of nine images per well were captured using the “matrix screener” mode. Images were nonoverlapping and homogenously distributed across the well surface. Alternatively, for high-resolution images of sections and β-catenin dots, micrographs were captured using the confocal microscope Olympus IX-81 at 20× and 60× magnitude. H&E sections were captured in brightfield microscopy using Zeiss Lab A1 AX10 microscope at the 20× magnification. Micrograph stitching to obtain a whole section image was performed using the “Stitching” plug-in in Fiji.
Image analysis
Intramuscular fat infiltrates were evaluated by measuring the extent of Perilipin-positive area using Fiji. The following setting were used: split channels, subtract background, threshold (50, 255), measure area. β-catenin immunoreactivity from confocal microscopy images was assessed using the same modalities.
Micrographs from the kinase inhibitor screening were analyzed in unbiased fashion by evaluating the FAP adipogenesis and fibrogenesis. Adipogenic and fibrogenic index were evaluated in unbiased fashion by measuring the extent of ORO-positive area and αSMA-positive area (in pixels) for each micrograph. Results are reported in Table S1. Fibrogenic differentiation of FAPs was estimated in unbiased fashion using CellProfiler [45] through a dedicated pipeline that recognizes, for each captured field, the αSMA-positive area (expressed in pixels). The modules of the pipeline were routinely adjusted in agreement with the image magnifications. The average αSMA-positive area was normalized over the average number of cells per field.
Adipogenic differentiation was estimated in unbiased fashion using CellProfiler through a dedicated pipeline that recognizes, for each captured field, the ORO-positive area (expressed in pixels). The modules of the pipeline were routinely adjusted in agreement with the image magnifications. The average ORO-positive area was normalized over the average number of cells per field. Alternatively, adipogenic differentiation of FAPs was estimated by counting the number of FAP-derived adipocytes or PPARγ-positive FAPs over the total cells in each field. Positive objects were manually scored using Fiji by two independent collaborators.
Statistical analysis
Data are represented as mean ± SEM of at least three independent experimental samples (n = 3) unless otherwise mentioned. In addition, in cell studies, for each biological triplicate we performed at least technical duplicates to control experimental errors. Statistical significance between two groups was estimated using the unpaired Student’s t test assuming a two-tailed distribution. Comparisons between three or more groups were performed using one-way or two-way ANOVA without correction for multiple comparisons and assuming a two-tailed distribution. The statistical significance is defined as p value where *p < 0.05; **p < 0.01; ***p < 0.001. All statistical analyses were performed using Prism 7 (GraphPad).
Results
GSK3 inhibitors impair FAP adipogenesis ex vivo
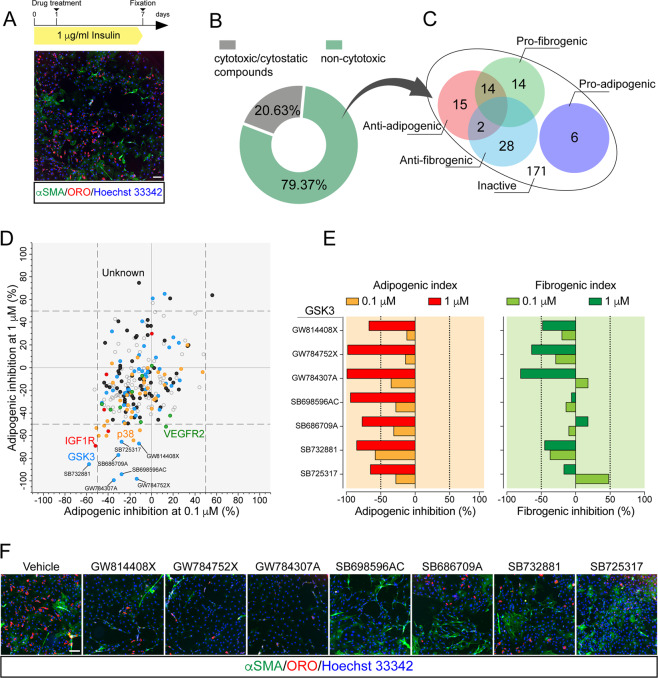
To identify crucial hubs affecting FAP adipogenesis, we applied a fluorescence microscopy-based screening [46] of a small molecule library targeting the mouse kinome. FAPs were purified as CD45−/CD31−/ITGA7−/SCA1+ from the hind limbs of mdx mice and cultured in pro-adipogenic condition (1 μg/ml insulin) for 7 days [31, 47] (Fig. 1a). Freshly isolated (P0) SCA1+ FAPs co-express CD140α (Platelet-Derived Growth Factor Receptor α [PDGFRα]) (Fig. S1a, c), as reported by others [7, 16, 17, 48]. The expression of PDGFRα remained unchanged upon 4 days of in vitro expansion (P1), confirming the purity and stability of FAP preparations (Fig. S1b–d). Kinase inhibitors (KIs) from the PKIS library, were added 24 h after plating at two concentrations (0.1 and 1 μM) (Fig. 1a). FAP adipogenesis was assessed by ORO staining. Treated samples were also immunostained with anti-α-Smooth Muscle Actin (α-SMA) antibody to reveal FAP-derived myofibroblasts (Fig. 1a). Compounds were classified (see “Material and methods” for details) as noncytotoxic (79.37%) or cytotoxic/cytostatic (20.63%) at these concentrations (Fig. 1b and Table S1). Among the noncytotoxic molecules, we focused on those affecting FAP differentiation, either positively or negatively (Table S1). Specifically, the drugs were defined as active only when the adipogenic/fibrogenic index was higher (induction) or lower (inhibition) than that observed in the negative control (DMSO). We arbitrarily chose an inhibition or induction by more than 50% to define a drug as a hit. This strategy allowed the identification of 79 KIs that were able to interfere with FAP differentiation trajectories (Fig. 1c and Table S1). To single out compounds specifically interfering with adipogenesis, we filtered the hit list to remove drugs that, in addition to inhibiting FAP adipogenesis, had the side effect of promoting FAP fibrogenesis (14 compounds) (Fig. 1c and Table S1). Next, we focused on the 15 compounds that selectively impaired adipogenesis (Fig. 1d and Table S1). This list of dose-dependent anti-adipogenic drugs (Fig. 1d, e) was found to be enriched in molecules targeting the kinase GSK3 and, to a lesser extent, Mitogen-Activated Protein Kinase 14 (MAPK14, p38). Among the GSK3-targeting molecules, in addition to their anti-adipogenic effect (Fig. 1e), GW784307A and GW784752X also impaired the spontaneous differentiation of FAPs into myofibroblasts when tested at 1 μM (Fig. 1f). In our experimental setup adipogenesis is induced by insulin. Consistently, we observed that a couple of inhibitors targeting the Insulin-like Growth Factor 1 Receptor also inhibited adipogenesis (Fig. 1d and Table S1). Overall, the results summarized in Fig. 1 point to GSK3 as a kinase playing a role in modulating FAP differentiation.
Fig. 1. GSK inhibitors affect the differentiation potential of FAPs.
a Experimental plan to screen the PKI library. Representative vehicle-treated sample (10× magnification, scale bar: 100 μm) showing spontaneous differentiation of FAPs into adipocytes (red, ORO staining) and myofibroblasts (green, stained with anti-αSMA antibody). b Screening compounds were classified as safe or cytotoxic/cytostatic. Cytotoxic/cytostatic compounds were defined for their capability to reduce by more than 30% the number of Hoechst 33342-positive nuclei at the end of the assay. c Venn diagrams summarizing the number and class of hits. d Scatter plot representing the inhibition of adipogenesis of the safe compounds at the two tested concentrations (0.1 and 1 μM). Dashed vertical and horizontal lines indicate the cutoff used to define a “hit” (±50% of the effect vs. DMSO). KIs targeting the same kinase are labeled with the same color. e Bar plot representing the percentage of adipogenic (left) and myofibrogenic (right) inhibition for the active compounds targeting GSK3. Dashed vertical lines indicate the cutoff for hit selection. f Representative immunofluorescence images (10× magnification, scale bar: 100 μm) showing FAP adipogenesis and myofibrogenesis upon treatment with GSK3-targeting compounds. Nuclei (blue) were revealed using Hoechst 33342.
LY2090314 inhibits FAP adipogenesis ex vivo and limits fatty degeneration in vivo
Prompted by the screening results, we sought to investigate the role of GSK3 in modulating FAP adipogenesis. To avoid the redundancy of the library compounds and to increase the translational impact of our findings, we set out to use LY2090314 as a representative GSK3 inhibitor. LY2090314 is a highly selective GSK3 inhibitor, currently tested in phase I/II studies as an antineoplastic agent [49–51].
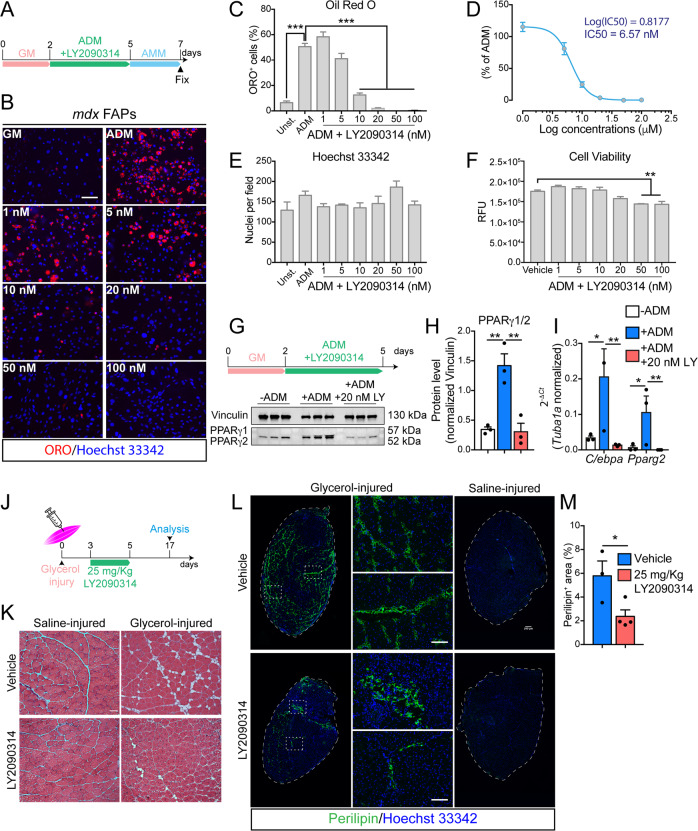
FAPs from mdx mice were induced to differentiate into adipocytes by incubating cells in ADM for 3 days followed by 2 additional days in AMM (Fig. 2a). Pro-adipogenic conditions promoted adipogenic differentiation of FAPs (Fig. 2a, b). Short-term treatment with LY2090314 (Fig. 2a) inhibited, in a dose-dependent manner, mdx FAP differentiation into adipocytes (Fig. 2b, c) with an IC50 of 6.57 nM (Fig. 2d) without affecting cell number (Fig. 2e). A slight decrease in cell viability can be appreciated at concentrations in the order of two-digit nanomolar (Fig. 2f). For further characterization, we chose 20 nM as the minimum noncytotoxic concentration of LY2090314 producing the maximum anti-adipogenic effect. FAPs from physiological or regenerating environments (as in the case of dystrophin deficiency or myotoxin injury) are characterized by a different activation state and response to external stimuli [26, 39] (Fig. S2a). We established that 20 nM LY2090314 was equally effective in inhibiting adipogenic differentiation of FAPs purified from wild-type (wt), mdx and cardiotoxin-injured mice suggesting that its inhibitory activity is independent from the FAP activation state (Fig. S2b). To address the molecular perturbation induced by LY2090314, downstream of GSK3 and to dissect the transcriptional circuit affecting adipogenic differentiation, we investigated peroxisome proliferator-activated receptor γ (PPARγ) expression [47] upon GSK3 inhibition by LY2090314. Three days of incubation in ADM promoted PPARγ1/2 expression (Fig. 2g–i). Interestingly, 20 nM LY2090314 fully abrogated PPARγ1/2 induction (Fig. 2g, h and Fig. S2c). Consistently, the positive transcriptional loop linking CCAAT/enhancer-binding protein alpha (C/ebpa) and Pparg expression was disrupted upon exposure to LY2090314 (Fig. 2i). In addition, short-time treatment (48 h) with LY2090314 did not affect the expression of fibrogenic genes upon transforming growth factor β (TGF-β) stimulation, as revealed by monitoring αSMA expression and small mother against decapentaplegic member 3 (SMAD3) phosphorylation (Fig. S2d, e). Collectively, these results confirm the anti-adipogenic effect of GSK3 inhibition and point to GSK3 as a crucial hub in the mechanism controlling the transcriptional activation of the adipogenic program.
Fig. 2. LY2090314 restricts FAP adipogenesis ex vivo while limiting fatty degeneration in vivo.
a Experimental plan for FAP differentiation. GM growth medium, ADM adipocyte differentiation medium, AMM adipocyte maintenance medium. b Representative ORO staining (20× magnification; scale bar: 100 μm) of mdx FAPs differentiated into adipocytes (red) in the presence of increasing doses (from 1 to 100 nM) of LY2090314 (n = 3). Nuclei (blue) were revealed using Hoechst 33342. Control samples were incubated with DMSO as vehicle. c Bar plot reporting the fraction of ORO-positive FAPs in each condition. The statistical significance was estimated by One-way ANOVA. d Plot reporting the non-linear regression analysis for the IC50 calculation of adipogenic inhibition at increasing doses of LY2090314 in FAPs undergoing adipogenesis. e Bar plot representing the average number of nuclei per field in each condition. The statistical significance was estimated by one-way ANOVA. f Viability assay of FAPs undergoing adipogenesis cultured with increasing doses of LY2090314 (RFU relative fluorescence unit). The statistical significance was estimated by one-way ANOVA. g Experimental plan describing the timing of assay. Western blot of PPARγ1/2 protein levels (n = 3). h Bar plot representing the normalized densitometric signal of PPARγ1/2. The statistical significance was estimated by one-way ANOVA. i Quantitative PCR (qPCR) of C/ebpa, and Pparg2 in FAPs undergoing adipogenesis in the presence of 20 nM LY2090314. The statistical significance was estimated by one-way ANOVA. j Experimental plan to induce fatty degeneration in mice hind limbs. k Representative hematoxylin and eosin staining (20× magnification; scale bar: 200 μm) of TAs injected either with glycerol or saline solution from LY2090314 (25 mg/Kg) and vehicle (DMSO)-treated mice (vehicle, n = 3; LY2090314, n = 4). l Representative immunofluorescence of intramuscular adipocytes (green) in TA sections (10× magnification, scale bar: 200 μm), stained with anti-perilipin 1 antibody. TA section reconstructions from mice treated with LY2090314 or vehicle in glycerol- and saline-injured limbs are reported. Insets (20× magnification, scale bar: 100 μm) showing higher magnification images. Nuclei (blue) were revealed using Hoechst 33342. m Bar plot reporting the average percentage of perilipin-positive area over the total sections. The statistical significance was estimated by Student’s t test. All data are represented as mean ± SEM and the statistical significance is defined as *p < 0.05; **p < 0.01; ***p < 0.001.
Ectopic adipose tissue is a hallmark of myopathies and/or incomplete regeneration [23, 24]. However, mdx muscles display intramuscular adiposity only at late stages. By contrast, intramuscular injection of a hypertonic solution of glycerol triggers degenerative changes that progressively lead to the accumulation of adipocytes in 7–10 days [16, 25, 52]. We used this model to assess whether the in vivo inhibition of GSK3 could limit intramuscular adipocyte infiltrations. To this end, we injured 3-month-old wt muscles (TA, gastrocnemius, and quadriceps) with 50% v/v of glycerol. LY2090314 (25 mg/Kg) [49] or vehicle (DMSO) were administered intraperitoneally for 3 consecutive days starting from the 3rd day after injury, avoiding any interferences with the early activation of the muscle stem cell niche (Fig. 2j). Glycerol-induced degeneration was already evident at 24 h after injection since the glycerol-injected mouse paw and limb reported a reduced mobility (Fig. S2f). After 17 days, saline-injured TA muscles showed no infiltrating adipocytes (Fig. 2k). In contrast, adipose areas were found between myofibers of glycerol-injected TAs (Fig. 2k). Likewise, antibody against perilipin-1 did not stain saline-injured TAs, while glycerol-injured muscles where markedly stained (Fig. 2l). Similarly, PPARγ expression in PDGFRα-positive FAPs was not identified in uninjured muscle sections while they become evident after glycerol injection (Fig. S2g). Short-term administration of LY2090314 limited glycerol-induced fatty degeneration (Fig. 2k), as revealed by the reduced extent and size of infiltrating Perilipin 1-positive adipocytes as well as by the reduced number of PPARγ-committed PDGFRα-positive FAPs (Fig. 2l, m and Fig. S2g). Regardless of LY2090314 administration, muscle fiber recovery was complete at 17 days from glycerol damage (Fig. S2h).
Canonical β-catenin signaling governs FAP adipogenesis
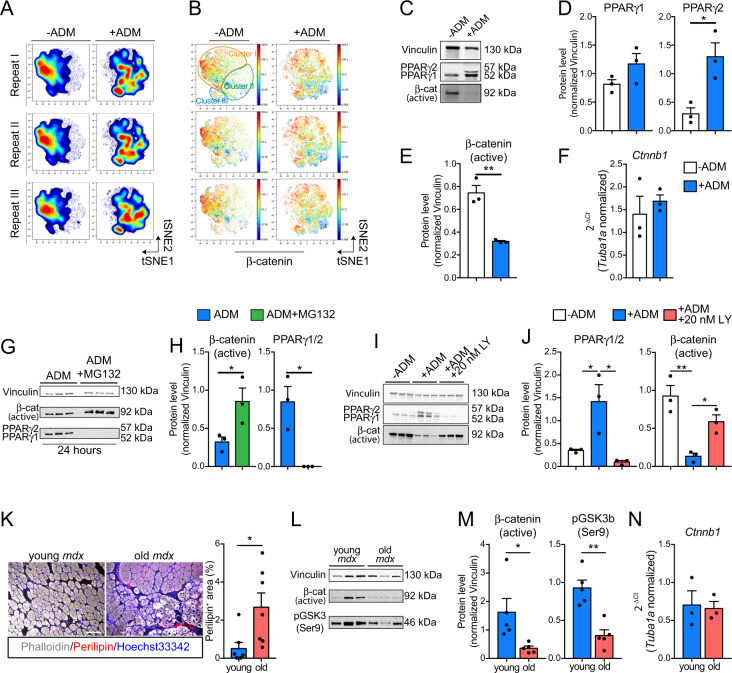
One of the GSK3 best characterized functions is the modulation of CTNNB1 (β-catenin) stability [53]. We applied high-dimensional single-cell mass cytometry to profile the dynamic changes of the FAP antigen repertoire and monitored β-catenin levels at single-cell resolution. FAPs were purified from three mdx mice, expanded in culture for 4 days (P1), and stimulated to differentiate into adipocytes by incubating cells with ADM (Fig. S3a). Three days after adipogenic induction, cells were harvested, barcoded with palladium isotopes, combined in a single tube, and stained with an antibody panel which includes 18 heavy metal-conjugated antibodies that recognize FAPs and other muscle-resident mononuclear cells (Fig. S3a, b and Table S2). FAPs were profiled in a single run in a CyTOF2 instrument. Signals were debarcoded and single-cell data analyzed by the Cytobank software suite [54] (Fig. S3a). Cells co-expressing SCA1 and PDGFRα neither express the leucocyte marker CD45 nor MuSCs antigens such as ITGA7 (α7-integrin) and CXCR4 (Fig. S3c). To trace the dynamic changes in the FAP antigen repertoire upon adipogenic induction, we applied the t-SNE algorithm [32] by specifying FAP antigens (PDGFRα, SCA1, CD34, PDGFRβ, CD90.2, and CD146) [7, 16, 26, 39] as clustering features (see “Materials and methods” for details). The resulting viSNE maps revealed, with high reproducibility, a dynamic rearrangement of the FAP immunophenotype during the adipogenic differentiation (Fig. 3a, Fig. S3d). FAP heterogeneity is appreciated at single-cell resolution (Fig. S3e), where surface antigen expression identifies FAP subpopulations (clusters) (Fig. S3f). Notably, cluster I highly expresses the FAP antigens PDGFRα, SCA1, CD34, PDGFRβ, and CD90.2 (Fig. S3e). Cluster II expresses lower levels of all these antigens, while Cluster III expresses higher levels of the adhesion marker CD146 (Fig. S3e). Adipogenic induction critically affected FAP antigen expressions. Notably, FAP antigens CD90.2, PDGFRβ, CD34, SCA1, and PDGFRα were significantly downregulated in cells undergoing adipogenesis as highlighted by their reduced signal intensity (Fig. S3g). A single exception is for CD146, whose expression is maintained during FAP adipogenesis (Fig. S3g). Intriguingly, changes in the antigen expression were coupled to a significant drift in cell abundance from one cluster to the others (Fig. S3h). β-catenin is highly expressed in FAPs cultivated in GM (Fig. 3b). By contrast, ADM exposure significantly reduced β-catenin expression (Fig. 3b and Fig. S3i), suggesting that β-catenin downregulation marks FAPs committed to adipogenesis. Consistently, PPARγ was upregulated during FAP adipogenesis, with a significant increase in isoform 2 (Fig. 3c, d). Remarkably, we demonstrated that the levels of the active form of β-catenin were downregulated upon ADM exposure, by using a specific antibody which recognizes the non-phosphorylated β-catenin [β-cat (active)] (Fig. 3c, e). On the other hand, β-catenin transcripts (Ctnnb1) did not show a corresponding decrease, suggesting that the β-catenin modulation associated to FAP adipogenesis is regulated by post-translational events (Fig. 3f). Consistently, the proteasome inhibitor MG132 prevented β-catenin degradation and PPARγ expression (Fig. 3g, h) thus demonstrating the pivotal role of β-catenin degradation in modulating the differentiation potentials of FAPs. Accordingly, GSK3 inhibition by LY2090314 promoted β-catenin stabilization while limiting PPARγ expression in FAPs undergoing adipogenesis (Fig. 3i, j). LY2090314 promoted β-catenin stabilization even shortly after treatment (24 h), as confirmed by western blot and immunofluorescence analysis, showing an increase of membrane and nuclear β-catenin staining area and intensity (Fig. S4a–c).
Fig. 3. β-catenin degradation marks FAPs undergoing adipogenesis.
a Contour maps obtained by projecting, onto a two-dimensional plane, single-cell multiparametric data using the viSNE algorithm. Data from three independent FAP preparations, in the presence or absence of ADM, are shown. b Dot plot representations of the viSNE maps showing β-catenin expression (color coded, blue = low, red = high) across the reported conditions for each repeat. c Representative western blot showing the protein levels of both PPARγ isoforms, non-phospho (active) β-catenin and Vinculin in FAPs unstimulated or induced to differentiate into adipocytes upon ADM incubation (n = 3). d Bar graphs representing the densitometric values of PPARγ1, PPARγ2 in both conditions. e Bar graph showing the protein levels of non-phospho (active) β-catenin in both conditions. Protein levels were normalized to Vinculin. The statistical significance was estimated by Student's t test. f Relative expression of Ctnn1b in FAPs unstimulated or induced to differentiate into adipocytes upon ADM exposure (n = 3). g Western blot showing the protein levels of PPARγ, non-phospho (active) β-catenin and Vinculin in FAPs undergoing adipogenesis in the absence or presence of 20 μM MG132 (n = 3). (h) Bar graphs representing the densitometric values of PPARγ and non-phospho (active) β-catenin in both conditions. Protein levels were normalized to Vinculin. The statistical significance was estimated by Student's t test. i Western blot showing the protein levels of PPARγ, non-phospho (active) β-catenin and Vinculin in FAPs unstimulated or incubated with ADM and ADM supplemented with 20 nM LY2090314 (n = 3). j Bar graphs representing the densitometric values of PPARγ and non-phospho (active) β-catenin in all conditions. Protein levels were normalized to Vinculin. The statistical significance was estimated by One-way ANOVA. k Representative immunofluorescence (left) (20× magnification; scale bar, 100 μm) of TAs of young (1.5-month-old, n = 6) and old (18-month-old, n = 7) mdx mice. Infiltrating adipocytes (arrow heads) were revealed using antibodies against perilipin-1 while fibers were stained using phalloidin-488. Bar plot (right) reporting the fraction of perilipin positive area in young and old mdx TAs. Statistical significance was estimated by Student's t test. l Immunoblot showing the protein level of non-phospho (active) β-catenin, pGSK3 (Ser9) and Vinculin (n = 5) in FAPs from young (1.5-month-old) and old (18-month-old) mdx mice. m Bar plots showing the densitometric analysis for non-phospho (active) β-catenin and pGSK3 (Ser9) normalized over Vinculin, respectively. n Quantitative PCR (qPCR) for Ctnnb1 from FAPs purified from young (1.5-month-old) and old (18-month-old) mdx mice (n = 3). Statistical significance was estimated by Student's t test. All data are represented as mean ± SEM and the statistical significance is defined as *p < 0.05; **p < 0.01; ***p < 0.001.
Intramuscular fat infiltrates are observable in old mdx mice (18-month-old) (Fig. 3k), when FAPs also produce lower amounts of pro-myogenic cytokines such as Igf1 and Il6 (Fig. S4d) and become less efficient in supporting the myogenic program of MuSCs. We observe that FAPs from old mdx mice, have an increased expression of fibrogenic (Ctgf) and adipogenic genes (C/ebpa and Pparg1) (Fig. S4d), while showing reduced levels of active β-catenin (i.e., non phosphorylated) and increased amount of active GSK3 (i.e., non phosphorylated on Ser9) (Fig. 3l, m). Notably, Ctnnb1 levels in old mdx FAPs were unchanged (Fig. 3n), suggesting that β-catenin protein levels are under the same post-translational controls observed in ex vivo mdx FAPs upon adipogenic induction. Collectively these data support the notion that the GSK3/β-catenin axis modulates FAP adipogenesis and LY2090314 inhibits FAP adipogenic conversion by preventing β-catenin degradation.
LY2090314 promotes MuSC self-renewal while boosting the ability of FAPs to promote MuSCs myogenesis in an ex vivo co-culture system
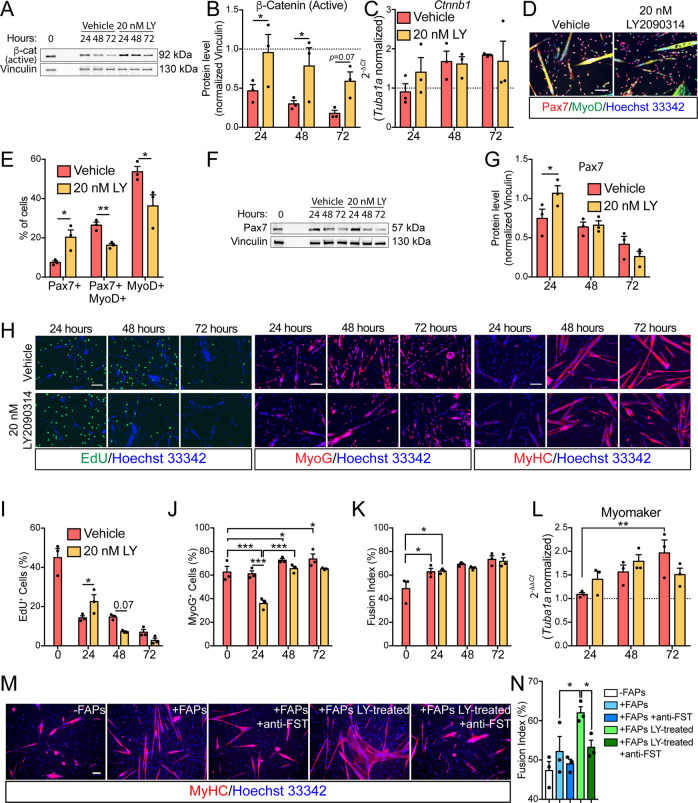
LY2090314 efficiently inhibits FAP conversion into adipocytes by modulating β-catenin stability. β-catenin also controls the differentiation fate of multiple cell types [55–58] including MuSCs [29]. We investigated MuSC self-renewal, proliferation and differentiation upon exposure to LY2090314. To this end MuSCs were purified from limb muscles of young mice and cultured ex vivo. First, we monitored the dynamic changes in β-catenin expression during spontaneous differentiation of MuSCs. Similarly to what observed in FAPs, β-catenin underwent a progressive downregulation in spontaneously differentiating MuSCs (Fig. 4a, b) while LY2090314 treatment prevented such decrease over time (Fig. 4a, b) without affecting β-catenin expression (Fig. 4c). This dynamic pattern suggested that β-catenin, upon LY2090314 exposure, is subjected to the same post-translational regulation observed in FAPs undergoing adipogenesis. After 24 h of drug treatment, GSK3 blockade by LY2090314 increased the fraction of renewing Pax7+ MuSCs (Fig. 4d, e) at the expenses of the proliferating ones, also expressing MyoD (Fig. 4d, e). Such increment in Pax7 expression is transient and limited to the first 24 h after LY2090314 treatment (Fig. 4f, g). Consistently, fluorescent microscopy analyses revealed that the increased MuSC self-renewal parallels with a significant reduction of MyoD-expressing cells (Fig. 4d, e), higher retention of EdU labelling (Fig. 4h, i), as well as blunted expression of myogenin (MyoG) in the first 24 h (Fig. 4h, j). Conversely, terminal differentiation and myogenic potential remained unaffected by LY2090314 as revealed by fusion index analysis and Myomaker expression (Fig. 4h, k, l).
Fig. 4. GSK3 inhibition promotes MuSC self-renewal and boosts the promyogenic ability of FAPs.
a Representative immunoblot showing the protein level of non-phospho (active) β-catenin and vinculin (n = 3) in MuSCs undergoing spontaneous myogenic differentiation in the presence of vehicle or 20 nM LY2090314. b Bar plot showing the densitometric analysis for non-phospho (active) β-catenin normalized over vinculin. c Quantitative PCR (qPCR) for Ctnnb1 in MuSCs undergoing spontaneous myogenic differentiation in the presence of vehicle or 20 nM LY2090314 (n = 3). The statistical significance was estimated by One-way ANOVA. d Representative Pax7/MyoD immunostaining (20× magnification; scale bar, 100 μm) in MuSCs after 24 h of culturing in the presence of vehicle or 20 nM LY2090314 (n = 3). e Bar plot showing the ratio of Pax7-, Pax7/MyoD-, and MyoD-positive cells. The statistical significance was estimated by two-way ANOVA. f Representative immunoblot showing the protein level of Pax7 in MuSCs undergoing spontaneous myogenic differentiation (n = 3). g Bar plot showing the densitometric values of Pax7 over vinculin. The statistical significance was estimated by two-way ANOVA. h Representative EdU labelling (left panel, 20× magnification; scale bar, 100 μm) in MuSCs undergoing spontaneous myogenic differentiation in the presence of vehicle or 20 nM LY2090314 (n = 3). Representative immunostaining against MyoG (central panel, 20× magnification; scale bar, 100 μm) in MuSCs undergoing spontaneous myogenic differentiation in the presence of vehicle or 20 nM LY2090314 (n = 3). Representative immunostaining against MyHC (left panel, 20× magnification; scale bar, 100 μm) in MuSCs undergoing spontaneous myogenic differentiation in the presence of vehicle or 20 nM LY2090314 (n = 3). Nuclei (blue) were revealed using Hoechst 33342. i Bar plot reporting the ratio of EdU-positive MuSCs. The statistical significance was estimated by two-way ANOVA. j Bar plot reporting the ratio of MyoG-positive MuSCs. The statistical significance was estimated by two-way ANOVA. k Bar plot reporting the percentage of fusion index of MuSCs-derived myotubes. The statistical significance was estimated by two-way ANOVA. l Quantitative PCR (qPCR) for Myomaker in MuSCs undergoing spontaneous myogenic differentiation in the presence of vehicle or 20 nM LY2090314 (n = 3). The statistical significance was estimated by One-way ANOVA. m Representative MyHC immunostaining (red, 20× magnification; scale bar, 100 μm) in MuSCs undergoing spontaneous myogenic differentiation in the presence of FAPs treated/untreated with 20 nM LY2090314 in the presence/absence of neutralizing anti-Follistatin antibodies (n = 3). n Bar plot reporting the percentage of fusion index of MuSCs-derived myotubes in each condition. The statistical significance was estimated by two-way ANOVA. Nuclei (blue) were revealed using Hoechst 33342. All data are represented as mean ± SEM and the statistical significance is defined as *p < 0.05; **p < 0.01; ***p < 0.001.
FAPs-derived stimuli are crucial for MuSC differentiation upon injury. In this context, we asked whether LY2090314 can alter the pro-myogenic role of FAPs on MuSCs in an ex vivo co-culture system. Two days exposure of FAPs with 20 nM LY2090314 better promoted MuSC differentiation into multinucleated myotubes than FAPs treated with the vehicle alone (Fig. 4m, n). Among the multitude of pro-myogenic stimuli produced by FAPs and active on MuSCs [7, 9, 59–62], we focused on follistatin (FST) since we recently highlighted that β-catenin controls FST expression in FAPs [31]. Consistently, neutralizing FST-antibodies fully abrogated the promyogenic effect of LY2090314-treated FAPs on MuSCs (Fig. 4m, n). Collectively, our data uncover that GSK3 blockade in FAPs improved FAP promyogenic capacities in a FST dependent manner.
FAPs are the main source of WNT ligands
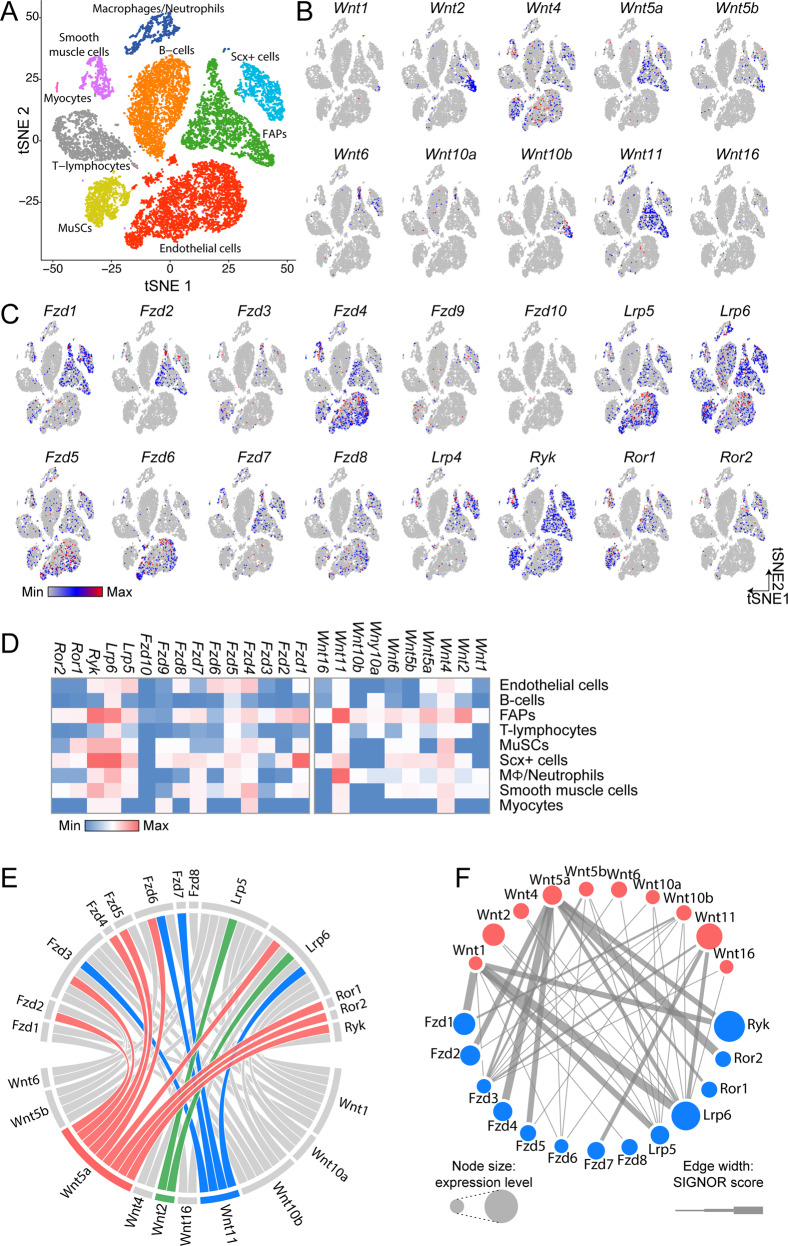
The GSK/β-catenin axis is controlled by the WNT pathway. To investigate the Wnt-mediated cross talk between different muscle cell types, we re-analyzed the single-cell transcriptomes of muscle mononuclear cells recently profiled by Giordani et al. and by the Tabula Muris consortium [33, 34]. The two single-cell RNAseq (scRNAseq) datasets were merged and reprocessed using the R package Seurat [35]. Principal component analysis identified separated cell clusters and showed good concordance between the datasets (Fig. S5a, b). Cell population clusters were generated using the t-SNE algorithm [37]. In the resulting t-SNE map, cell population clusters were conserved across datasets demonstrating high reproducibility between different experiments (Fig. S5b, c). Cell ontology identities were assigned on the basis of the expression of markers annotated in Myo-REG [38] and by Giordani et al. [34] (Fig. S5d–m) allowing the mapping of nine cell populations onto the t-SNE map (endothelial cells, B cells, FAPs, T-lymphocytes, MuSCs, Scx+ cells, macrophages/neutrophils, smooth muscle cells, and myocytes) (Fig. 5a). Each population is characterized by a different transcriptional signature (Fig. S5n). Next, we interrogated this integrated single-cell dataset to explore the expression of WNT ligands and receptors in the different cell populations (Fig. 5a–d). Intriguingly, a large majority of events in the FAP cluster expresses the canonical Wnt family member 2 (Wnt2) and noncanonical WNT members, including Wnt11 and Wnt5a (Fig. 5b). Notably, as reported by others, the MuSC cluster expresses Wnt4 [63] while the cluster of macrophages/neutrophils expresses high levels of Wnt11 [64] (Fig. 5d). Conversely, WNT receptors, including Frizzled (Fzds), low-density lipoprotein receptor-related proteins (LRPs) and noncanonical receptors such as Ryk, Ror1 and Ror2, albeit expressed by FAPs, were found to be also expressed, to a variable extent and with a different profile, in most muscle mononuclear cell populations (Fig. 5c, d). Signaling relations for all WNT ligands and receptors were retrieved from the SIGNOR database [43], thus connecting all WNTs and their receptors in the dataset. Analysis of the specificity of WNT ligand interactions reveals a potential highly intricate cell cross talk (Fig. 5e). The fact that most of WNT ligands are produced by FAPs suggests that FAP-secreted WNTs have the potential to influence muscle residing cells in a paracrine manner. FAPs are not only the main source of WNT ligands but also express cognate receptors. This suggests the possibility that FAP-produced WNT ligands can act in an autocrine manner to modulate their own behavior (Fig. 5b–d, f and Fig. S6).
Fig. 5. FAPs are the main source of Wnt ligands in the muscle stem cell niche.
a t-SNE map of cell populations identified upon combining single-cell RNA sequencing experiments from Giordani et al. with those of Tabula Muris consortium. Data processing yielded nine clusters that were assigned to specific cell populations on the basis of their expression profiles. b t-SNE maps showing the expression of WNT ligands at single-cell resolution. c t-SNE maps showing the expression of canonical and noncanonical WNT receptors at single-cell resolution. d Heatmap representing the relative abundance of specific WNT ligands and receptor transcripts RNA in each cell population. e Chord diagram representing the relations between WNT ligands and their receptors as stored in the SIGNOR database. The relations for WNT11, WNT2, and WNT5a are highlighted in blue, green, and red, respectively. f Network describing the interaction between WNTs (red) with their receptors (blue) in FAPs. Node sizes are proportional to the transcript expression levels while edge thickness is related to the SIGNOR confidence score for the reported interaction.
WNT5a expression is downregulated in dystrophic FAPs
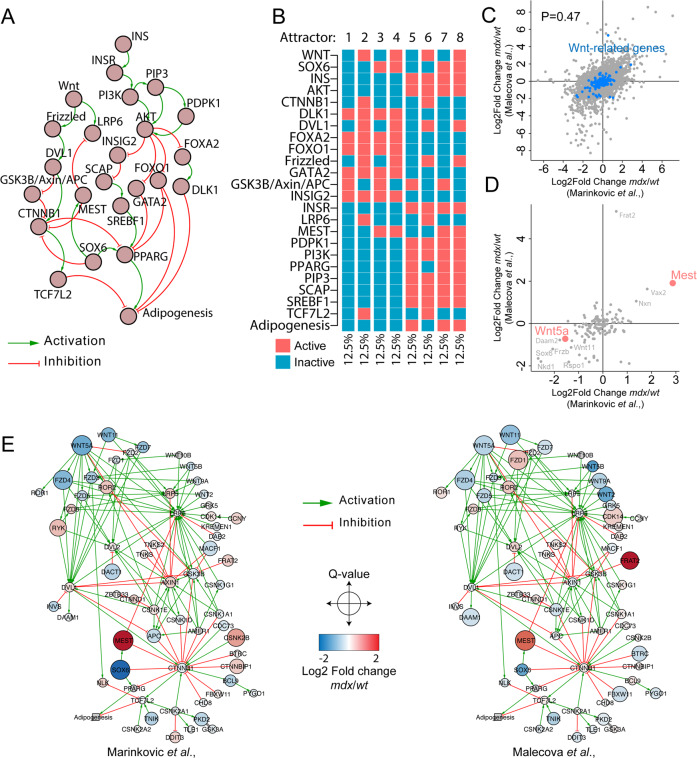
To understand the potential role of WNT ligands in controlling the adipogenic fate of FAPs, we exploited an in silico model that recapitulates the dynamics between insulin, which induces FAP adipogenesis [47], and WNT pathways. To construct a network describing the causal relations between the core components of insulin and WNT signaling pathways, we interrogated the Myo-REG database which collects manually-curated muscle cell-related intracellular interactions [38]. We retrieved a cross-regulatory network by merging the insulin (INS) and WNT pathways. The resulting network encompasses 24 nodes, including extracellular stimuli (WNT, INS), receptors (Frizzled, INSR), signal transducers, and transcription factors which accurately describe the INS-WNT cross talk (Fig. 6a and Table S3). Relationships between nodes are either positive (activations) or negative (inhibitions). A Boolean function was associated to each node (Table S4). The model was robust in comparison to 100 perturbations of 100 randomly generated networks (Fig. S7a). Characterization of the model attractors (equilibrium states) led to the identification of eight stable system states (attractor 1 to attractor 8), identified with equal frequencies (12.5%), representing known states predicting biological phenotypes (Fig. 6b). We characterized these attractors by clustering them according to the levels of the input signals (INS, WNT) and output phenotypes. The first four attractors (attractor 1 to attractor 4), are characterized by absence of INS (inactive). In these conditions, adipogenic differentiation is not observed (inactive PPARG) irrespective of the presence of the WNT stimulus (Fig. 6b). The remaining four attractors (attractor 5 to attractor 8), are characterized by active INS signaling, triggering a cascade that inactivates, via AKT, the PPARG inhibitors (GATA2, FOXO1, and DLK1), thus stimulating adipogenesis. Attractor six represents an exception in this respect. In the presence of a WNT ligand, the CTNNB1 destruction complex (GSK3b_AXIN_APC) is inactive (Fig. 6b). As a result, the active CTNNB1 node directly represses PPARγ, thus preventing adipogenesis (Fig. 6b). WNT-mediated inhibition of adipogenesis is not observed when transcription factor SOX6 (SOX6) and mesodermal specific transcript (MEST) are in an active state causing a negative modulation of CTNNB1 and LRP6, respectively (attractor 8) (Fig. 6b). Thus, the result of our model simulations supports a dominance of the negative modulation of adipogenesis, by the canonical β-catenin signaling, over the pro-adipogenic IGF/INS pathway, and confirms our observation that an active β-catenin is sufficient to impair adipogenesis even in the presence of insulin.
Fig. 6. Wnt5a expression is impaired in mdx FAPs.
a Network showing the casual signaling relations between signaling proteins in the Wnt and Insulin pathways. FAP-specific signaling information were retrieved from Myo-REG. b Heatmap showing the attractors in the simulation of the Wnt/Ins pathway. c Scatter plot showing the correlation between the log2 differences of the genes that were observed both in the Marinkovic et al. and Malecova et al. RNAseq datasets. d Scatter plot showing the correlation between the common genes between the Marinkovic et al. and Malecova et al. RNAseq datasets that are associate to Wnt signaling pathway according to UniProt Keywords. Genes in red are reported as significant in both datasets. e Networks reporting the signaling relations, retrieved from SIGNOR, of Wnt protein products Marinkovic et al. and Malecova et al. datasets. Nodes are color coded according to the Log2 fold change difference between mdx and wt FAPs.
Prompted by our findings, we sought to determine any possible alterations in the WNT signaling in FAPs from mdx mice. We interrogated two recently published RNAseq datasets generated by sequencing bulk RNA preparations of freshly isolated wt and mdx FAPs [26, 39]. Reassuringly, the Pearson correlation between the two datasets approaches 0.5, confirming result reproducibility across two independent experiments (Fig. 6c). We next identified 188 genes annotated as WNT-related according to the UniProt keyword “WNT signaling pathway” and added to this list SOX6 and MEST (Table S5). The RNA of 135 of these genes were identified in both experiments. Intriguingly, WNT5a and MEST were significantly down- and upregulated in mdx FAPs, respectively (Fig. 6d). To understand the signaling relationship among the protein products of the 135 Wnt-related genes, we mapped their expression on a causal network retrieved from SIGNOR [43]. The interrogation of the SIGNOR database showed that 66 of the 135 genes, that are common to the two datasets, were part of a highly connected causal network (Fig. S7b). To illustrate the cross talk between WNT signaling and the adipogenesis pathway we added the network controlling adipogenesis centered on PPARγ (Fig. S7c, d). For both datasets, we color coded the log2 fold change difference between mdx and wt FAPs (Fig. 6e). Overall, WNT-related networks define a global downregulation of WNT ligand and receptors while negative regulators of the signaling, such as MEST, are upregulated. Considering the role of such actors in the regulation of PPARγ, this evidence yields crucial insights into the altered cross talk between the WNT pathway and adipogenesis in FAPs from mdx mice.
WNT5a represses PPARγ expression and FAP adipogenesis through the activation of canonical β-catenin signaling
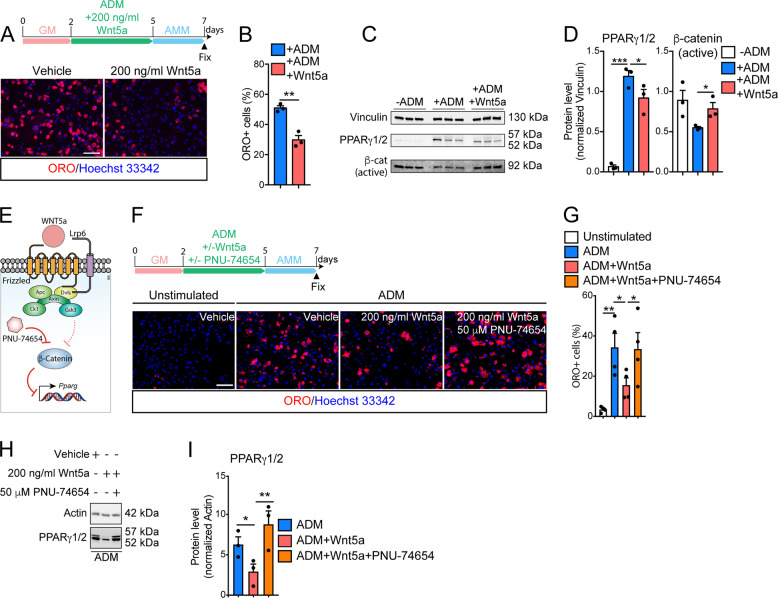
We focused our attention on WNT5a, which we found expressed by FAPs in their resting state (Fig. 5b, d) and after acute muscle damage [7]. WNT5a is significantly downregulated (Fig. 6d) in mdx mice in accordance with the higher propensity of these cells to engage the adipogenic program in dystrophic muscles [31, 62]. To verify our hypothesis, we treated purified mdx FAPs with 200 ng/ml of WNT5a (Fig. 7a). To reveal any possible WNT-related specific effect, we also included in our analysis WNT10b, which is also characterized as an adipogenesis inhibitor of pre-adipocyte cell lines [55, 65], but it was not significantly modulated in either dataset. Exogenous administration of WNT5a, during ADM, impaired FAP adipogenesis as revealed by the reduced fraction of ORO-positive cells (Fig. 7a, b). By contrast, increasing doses of WNT10b (ranging from 200 to 1000 ng/ml) did not alter the adipogenic program of FAPs (Fig. S7e), revealing a FAP-specific sensitivity for different WNT ligands.
Fig. 7. Wnt5a restrains FAP adipogenesis by activating the canonical β-catenin signaling.
a Representative ORO staining (20× magnification, scale bar:100 μm) of dystrophic FAPs differentiated in the presence of 200 ng/ml Wnt5a (n = 3). Control samples were incubated with vehicle. Nuclei (blue) were revealed with Hoechst 33342. b Bar plot reporting the fraction of FAP-derived adipocytes in both conditions described in a. The statistical significance was estimated by student t test. c Western blot showing the protein level of Vinculin, PPARγ1/2, and the active β-catenin in FAPs undergoing adipogenesis (n = 3). d Bar plots showing the densitometric values, normalized to Vinculin, of PPARγ1/2, and active β-catenin in each condition. e Schematic representation describing the WNT5a pathway in FAPs. f Representative ORO staining (20x magnification, scale bar:100 μm) of dystrophic FAPs differentiated in the presence of 200 ng/ml Wnt5a or in combination with 50 μm PNU-74654 (n = 3). g Bar plot reporting the fraction of FAP-derived adipocytes in both conditions. The statistical significance was estimated by one-way ANOVA. h Western blot showing the protein level of Actin and PPARγ1/2 (n = 3) upon treatment with 200 ng/ml Wnt5a or in combination with 50 μm PNU-74654 (n = 3). i Bar plots showing the densitometric values of PPARγ1/2, normalized to Actin, in each condition. The statistical significance was estimated by one-way ANOVA. All data are represented as mean ± SEM and the statistical significance is defined as *p < 0.05; **p < 0.01; ***p < 0.001.
WNT5a is s a noncanonical WNT that preferentially induces the activation of the JUN transcription factor via phosphorylation of Ser73 [66]. However, the phosphorylation status of Ser73 of JUN protein remained unaffected after WNT5a stimulation of differentiating FAPs (Fig. S7f). Thus, we considered the hypothesis that WNT5a affects FAP adipogenesis through the canonical β-catenin signaling. We first checked the phosphorylation status of the inhibitory residue (Ser9) of GSK3 as sign of the canonical β-catenin pathway activation. Exogenous administration of 200 ng/ml WNT5a significantly enhances the phosphorylation status of GSK3 inhibitory residue (Fig. S7g). Consistently, WNT5a treatment prevents β-catenin downregulation while limiting PPARγ expression in FAPs undergoing adipogenic differentiation (Fig. 7c, d). To demonstrate that WNT5a represses PPARγ in a β-catenin-depended manner (Fig. 7e), we used the highly selective β-catenin inhibitor, PNU-74654, in a combinatorial treatment with recombinant WNT5a during adipogenic differentiation of FAPs. β-catenin inhibition by PNU-74654 fully abrogates the anti-adipogenic effect of exogenous WNT5a (Fig. 7f–I). Thus, WNT5a, through the activation of canonical β-catenin signaling, restrains the ex vivo conversion of FAPs into adipocytes.
Overall our data describe for the first time a model whereby WNT5a may impact FAP adipogenesis through the modulation of the canonical GSK3/β-catenin axis and revealed that pharmacological manipulation of WNT signaling may help controlling ectopic fat deposition in dystrophic FAPs.
Discussion
FAPs are defined as SCA1- and PDGFRα-positive mesenchymal cells with the potential to differentiate into adipocytes and fibroblasts [7, 16]. In unperturbed muscles, FAPs contribute to muscle mass maintenance, MuSC pool preservation over time [17] as well as muscle regeneration [7, 9, 16, 61]. In a physiologically controlled regeneration, FAP activation at the site of damage is transient [7, 67] and FAP fibro/adipogenic potential is restrained by a variety of signals, including nitric oxide (NO), interleukin-4 (IL-4), Desert Hedgehog (DHH), tumor necrosis factor-α (TNFα), WNT1-inducible signaling pathway protein 1 (WISP1), and Neurogenic locus notch homolog protein (NOTCH) [6, 25, 26, 67–70]. Such different signals and stimuli possibly synergize to control FAP differentiation. In myopathies, the anti-adipogenic microenvironmental cues and FAP sensitivity to the niche constraints are attenuated and/or lost [26, 61, 62, 67, 71–74]. In this context, FAPs lose their beneficial role [9, 61, 71] and contribute to the deposition of intramuscular ectopic fat and fibrous tissue [23, 24, 67, 75]. We screened a collection of KIs with the aim of targeting kinases playing a role in the modulation of mdx FAP adipogenesis. The list of anti-adipogenic molecules was found to be enriched for compounds targeting the GSK3. GSK3 plays a central role in many processes and has been considered as a pharmacological target in a variety of physiopathological systems, including type 2 diabetes and cancer [76–78]. In this regard, small molecules that modulate its activity have been developed and have entered clinical trials. Among these, LY2090314 has been recently demonstrated to be effective in selectively blocking GSK3. LY2090314 restores insulin secretion in diabetic islets [76] and sensitizes cancer cells to apoptosis via GSK3 inhibition [49–51]. Consistently, we show that GSK3 inhibition by LY2090314 efficiently abrogates FAP adipogenesis at sub-micromolar concentrations and when administered in vivo limits intramuscular fat infiltrates in murine models of fatty degeneration. GSK3 plays a crucial role in regulating β-catenin intracellular concentrations. High-dimensional mass cytometry demonstrated that the FAP antigen repertoire is rearranged during FAP adipogenesis while β-catenin expression is downregulated as a consequence of its proteasomal degradation. Concomitantly, a robust upregulation of PPARγ is observed. Such divergent changes in protein levels are also found in old dystrophic FAPs when intramuscular fat infiltrates become apparent. Pharmacological treatment with LY2090314 disrupts this circuit, sustains β-catenin protein levels and impairs the adipogenic transcriptional cascade. Furthermore, we also investigated the possible impact of LY2090314 in MuSCs. Intriguingly, anti-adipogenic concentrations of LY2090314 were effective in sustaining PAX7 expression and promoting the self-renewal of ex vivo MuSCs. In addition, we also demonstrated that LY2090314 strengthens the paracrine directional interaction FAP-to-MuSC allowing optimal myotube formation of MuSCs, a process mainly mediated by the FAP-derived Follistatin. To characterize the relevance of the GSK3/β-catenin axis in controlling FAP adipogenesis we monitored the changes in the concentrations of soluble mediators that are enriched in the muscle stem cell niche, especially Wnt ligands, which are known to influence the activation of the GSK3/β-catenin axis. The Tabula Muris consortium recently reported the single-cell transcriptomes of 20 murine organs, including limb muscles [33]. More recently, Giordani et al. [34] reported a single-cell transcriptomic analysis of muscle mononuclear cell populations. The integration of the two datasets highlights FAPs as the main muscle cell population expressing Wnt ligands, especially Wnt2, WNT5a, and Wnt11, whose secretion possibly triggers autocrine and paracrine cellular responses. By combining published RNAseq data of bulk FAP RNA preparations, we highlight that Wnt signaling is impaired in dystrophic FAPs due to reduced expression of the majority of Wnt ligands and receptors. In particular, WNT5a, which is secreted by FAPs upon acute damage, is significantly downregulated. Moreover, MEST, a negative regulator of β-catenin and LRP6, is upregulated, thus contributing to a reduced sensitivity of FAPs to the canonical Wnt/β-catenin signaling. We recently applied MS-based proteomics to uncover intrinsic metabolic defects in dystrophic FAPs [26, 31]. Proteome profiling revealed that MEST protein levels are also upregulated in dystrophic FAPs, while β-catenin is downregulated [31]. We demonstrated that such circuit is also sensitive to nutritional interventions. Specifically, high-fat diet rewires the FAP proteome and restores oxidative metabolism in dystrophic FAPs. Such condition correlates with a reduction of MEST protein levels, while the β-catenin/follistatin axis is activated [31]. Altogether, these data indicate that the intracellular levels of β-catenin are crucial in controlling the balance between the beneficial and detrimental role of FAPs. Finally, we show that exogenous administration of WNT5a, but not Wnt10b, impairs ex vivo FAP adipogenesis revealing a Wnt-specific effect in FAPs. WNT5a is a noncanonical Wnt ligand reported to activate the planar cells polarity (PCP) Wnt pathway [66]. We demonstrated that, in FAPs, WNT5a affects adipogenesis by repressing PPARγ expression in a β-catenin-dependent manner as shown by the rescue of adipogenesis obtained by pharmacological inhibition of the β-catenin transcriptional complex. Moreover, our data are also supported by other reports showing that in different cellular and physiological contexts WNT5a can act through the canonical β-catenin-dependent signaling probably relying on the cell specific receptor context [79, 80]. In summary, this work uncovers a previously unappreciated role of WNT5a in modulating insulin-mediated FAP adipogenesis while identifying in the GSK3/β-catenin axis a new potential pharmacological target that can be modulated to counteract intramuscular fat infiltrates in dystrophic muscles.
Supplementary information
Acknowledgements
This work has been supported by the European Research Council (ERC) (grant N°. 322749) and by the Foundation AIRC for Research on Cancer (AIRC) (grant N°. 14135), and partly supported by the AIRC Investigator Grant (IG 2013) to G.C.; A Reggio was supported by Fondazione Umberto Veronesi. The PKIS library was supplied by GlaxoSmithKline, LLC, and the Structural Genomic Consortium under an open access Material Transfer and Trust Agreement: http://www.sgc-unc.org. This work was made possible at IRBM by the CNCCS s.c.a.r.l. initiative.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by P. Salomoni
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Alessio Reggio, Marco Rosina, Alessandro Palma
Contributor Information
Luisa Castagnoli, Email: castagnoli@uniroma2.it.
Gianni Cesareni, Email: cesareni@uniroma2.it.
Supplementary information
The online version of this article (10.1038/s41418-020-0551-y) contains supplementary material, which is available to authorized users.
References
- 1.Besson V, Smeriglio P, Wegener A, Relaix F, Nait Oumesmar B, Sassoon DA, et al. PW1 gene/paternally expressed gene 3 (PW1/Peg3) identifies multiple adult stem and progenitor cell populations. Proc Natl Acad Sci USA. 2011;108:11470–5. doi: 10.1073/pnas.1103873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christov C, Chrétien F, Abou-Khalil R, Bassez G, Vallet G, Authier F-J, et al. Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol Biol Cell. 2007;18:1397–409. doi: 10.1091/mbc.E06-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dellavalle A, Maroli G, Covarello D, Azzoni E, Innocenzi A, Perani L, et al. Pericytes resident in postnatal skeletal muscle differentiate into muscle fibres and generate satellite cells. Nat Commun. 2011;2:499. doi: 10.1038/ncomms1508. [DOI] [PubMed] [Google Scholar]
- 4.Du H, Shih C-H, Wosczyna MN, Mueller AA, Cho J, Aggarwal A, et al. Macrophage-released ADAMTS1 promotes muscle stem cell activation. Nat Commun. 2017;8:669. doi: 10.1038/s41467-017-00522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fry CS, Johnson DL, Ireland ML, Noehren B. ACL injury reduces satellite cell abundance and promotes fibrogenic cell expansion within skeletal muscle: ACL injury induces muscle maladaptations. J Orthop Res. 2017;35:1876–85. doi: 10.1002/jor.23502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heredia JE, Mukundan L, Chen FM, Mueller AA, Deo RC, Locksley RM, et al. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–88. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joe AWB, Yi L, Natarajan A, Le Grand F, So L, Wang J, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–63. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juban G, Saclier M, Yacoub-Youssef H, Kernou A, Arnold L, Boisson C, et al. AMPK activation regulates LTBP4-dependent TGF-β1 secretion by pro-inflammatory macrophages and controls fibrosis in duchenne muscular dystrophy. Cell Rep. 2018;25:2163–76.e6. doi: 10.1016/j.celrep.2018.10.077. [DOI] [PubMed] [Google Scholar]
- 9.Kuswanto W, Burzyn D, Panduro M, Wang KK, Jang YC, Wagers AJ, et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity. 2016;44:355–67. doi: 10.1016/j.immuni.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liadaki K, Casar JC, Wessen M, Luth ES, Jun S, Gussoni E, et al. β4 integrin marks interstitial myogenic progenitor cells in adult murine skeletal muscle. J Histochem Cytochem. 2012;60:31–44. doi: 10.1369/0022155411428991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell KJ, Pannérec A, Cadot B, Parlakian A, Besson V, Gomes ER, et al. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol. 2010;12:257–66. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- 12.Mounier R, Théret M, Arnold L, Cuvellier S, Bultot L, Göransson O, et al. AMPKα1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab. 2013;18:251–64. doi: 10.1016/j.cmet.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–37. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pannerec A, Formicola L, Besson V, Marazzi G, Sassoon DA. Defining skeletal muscle resident progenitors and their cell fate potentials. Development. 2013;140:2879–91. doi: 10.1242/dev.089326. [DOI] [PubMed] [Google Scholar]
- 15.Saclier M, Yacoub-Youssef H, Mackey AL, Arnold L, Ardjoune H, Magnan M, et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells. 2013;31:384–96. doi: 10.1002/stem.1288. [DOI] [PubMed] [Google Scholar]
- 16.Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–52. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 17.Wosczyna MN, Konishi CT, Perez Carbajal EE, Wang TT, Walsh RA, Gan Q, et al. Mesenchymal stromal cells are required for regeneration and homeostatic maintenance of skeletal muscle. Cell Rep. 2019;27:2029–35.e5. doi: 10.1016/j.celrep.2019.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almada AE, Wagers AJ. Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat Rev Mol Cell Biol. 2016;17:267–79. doi: 10.1038/nrm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blau HM, Cosgrove BD, Ho ATV. The central role of muscle stem cells in regenerative failure with aging. Nat Med. 2015;21:854–62. doi: 10.1038/nm.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emery AE. The muscular dystrophies. BMJ. 1998;317:991–5. doi: 10.1136/bmj.317.7164.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vallecillo-García P, Orgeur M, vom Hofe-Schneider S, Stumm J, Kappert V, Ibrahim DM, et al. Odd skipped-related 1 identifies a population of embryonic fibro-adipogenic progenitors regulating myogenesis during limb development. Nat Commun. 2017;8. 10.1038/s41467-017-01120-3. [DOI] [PMC free article] [PubMed]
- 22.Mathew SJ, Hansen JM, Merrell AJ, Murphy MM, Lawson JA, Hutcheson DA, et al. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138:371–84. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uezumi A, Ito T, Morikawa D, Shimizu N, Yoneda T, Segawa M, et al. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci. 2011;124:3654–64. doi: 10.1242/jcs.086629. [DOI] [PubMed] [Google Scholar]
- 24.Hogarth MW, Defour A, Lazarski C, Gallardo E, Diaz Manera J, Partridge TA, et al. Fibroadipogenic progenitors are responsible for muscle loss in limb girdle muscular dystrophy 2B. Nat Commun. 2019;10:2430. doi: 10.1038/s41467-019-10438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopinke D, Roberson EC, Reiter JF. Ciliary Hedgehog Signaling Restricts Injury-Induced Adipogenesis. Cell. 2017;170:340–51.e12. doi: 10.1016/j.cell.2017.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marinkovic M, Fuoco C, Sacco F, Cerquone Perpetuini A, Giuliani G, Micarelli E, et al. Fibro-adipogenic progenitors of dystrophic mice are insensitive to NOTCH regulation of adipogenesis. Life Sci Alliance. 2019;2. 10.26508/lsa.201900437. [DOI] [PMC free article] [PubMed]
- 27.Bentzinger CF, Wang YX, Dumont NA, Rudnicki MA. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 2013;14:1062–72. doi: 10.1038/embor.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Grand F, Jones AE, Seale V, Scimè A, Rudnicki MA. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–47. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones AE, Price FD, Le Grand F, Soleimani VD, Dick SA, Megeney LA, et al. Wnt/β-catenin controls follistatin signalling to regulate satellite cell myogenic potential. Skelet Muscle. 2015;5:14. doi: 10.1186/s13395-015-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–10. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 31.Reggio A, Rosina M, Krahmer N, Palma A, Petrilli LL, Maiolatesi G, et al. Metabolic reprogramming of fibro/adipogenic progenitors facilitates muscle regeneration. Life Sci Alliance. 2020;3. 10.26508/lsa.202000660. [DOI] [PMC free article] [PubMed]
- 32.Amir ED, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–52. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–72. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giordani L, He GJ, Negroni E, Sakai H, Law JYC, Siu MM, et al. High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol Cell. 2019;74:609–21.e6. doi: 10.1016/j.molcel.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 35.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–20. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive Integration of single-cell data. Cell. 2019;177:1888–902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Maaten L. Accelerating t-SNE using tree-based algorithms. J Mach Learn Res. 2014;15:3221–45. [Google Scholar]
- 38.Palma A, Perpetuini AC, Ferrentino F, Fuoco C, Gargioli C, Giuliani G, et al. Myo-REG: a portal for signaling interactions in muscle regeneration. Front Physiol. 2019;10. 10.3389/fphys.2019.01216. [DOI] [PMC free article] [PubMed]
- 39.Malecova B, Gatto S, Etxaniz U, Passafaro M, Cortez A, Nicoletti C, et al. Dynamics of cellular states of fibro-adipogenic progenitors during myogenesis and muscular dystrophy. Nat Commun. 2018;9. 10.1038/s41467-018-06068-6. [DOI] [PMC free article] [PubMed]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Müssel C, Hopfensitz M, Kestler HA. BoolNet-an R package for generation, reconstruction and analysis of Boolean networks. Bioinformatics. 2010;26:1378–80. doi: 10.1093/bioinformatics/btq124. [DOI] [PubMed] [Google Scholar]
- 43.Licata L, Lo Surdo P, Iannuccelli M, Palma A, Micarelli E, Perfetto L, et al. SIGNOR 2.0, the SIGnaling Network Open Resource 2.0: 2019 update. Nucleic Acids Res. 2020;48:D504–10. doi: 10.1093/nar/gkz949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerquone Perpetuini A, Giuliani G, Reggio A, Cerretani M, Santoriello M, Stefanelli R, et al. Janus effect of glucocorticoids on differentiation of muscle fibro/adipogenic progenitors. Sci Rep. 2020;10:5363. doi: 10.1038/s41598-020-62194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reggio A, Spada F, Rosina M, Massacci G, Zuccotti A, Fuoco C, et al. The immunosuppressant drug azathioprine restrains adipogenesis of muscle Fibro/Adipogenic Progenitors from dystrophic mice by affecting AKT signaling. Sci Rep. 2019;9:4360. doi: 10.1038/s41598-019-39538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wosczyna MN, Biswas AA, Cogswell CA, Goldhamer DJ. Multipotent progenitors resident in the skeletal muscle interstitium exhibit robust BMP-dependent osteogenic activity and mediate heterotopic ossification. J Bone Miner Res. 2012;27:1004–17. doi: 10.1002/jbmr.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atkinson JM, Rank KB, Zeng Y, Capen A, Yadav V, Manro JR, et al. Activating the Wnt/β-catenin pathway for the treatment of melanoma-application of LY2090314, a novel selective inhibitor of glycogen synthase kinase-3. PLoS ONE. 2015;10:e0125028. doi: 10.1371/journal.pone.0125028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizzieri DA, Cooley S, Odenike O, Moonan L, Chow KH, Jackson K, et al. An open-label phase 2 study of glycogen synthase kinase-3 inhibitor LY2090314 in patients with acute leukemia. Leuk Lymphoma. 2016;57:1800–6. doi: 10.3109/10428194.2015.1122781. [DOI] [PubMed] [Google Scholar]
- 51.Kunnimalaiyaan S, Schwartz VK, Jackson IA, Clark Gamblin T, Kunnimalaiyaan M. Antiproliferative and apoptotic effect of LY2090314, a GSK-3 inhibitor, in neuroblastoma in vitro. BMC Cancer. 2018;18:560. doi: 10.1186/s12885-018-4474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pisani DF, Bottema CDK, Butori C, Dani C, Dechesne CA. Mouse model of skeletal muscle adiposity: a glycerol treatment approach. Biochem Biophys Res Commun. 2010;396:767–73. doi: 10.1016/j.bbrc.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 53.Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010;Chapter 10:Unit10.17. [DOI] [PMC free article] [PubMed]
- 55.Cawthorn WP, Bree AJ, Yao Y, Du B, Hemati N, Martinez-Santibañez G, et al. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone. 2012;50:477–89. doi: 10.1016/j.bone.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–50. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 57.Prestwich TC, Macdougald OA. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol. 2007;19:612–7. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sui Y, Liu Z, Park S-H, Thatcher SE, Zhu B, Fernandez JP, et al. IKKβ is a β-catenin kinase that regulates mesenchymal stem cell differentiation. JCI Insight. 2018;3. 10.1172/jci.insight.96660. [DOI] [PMC free article] [PubMed]
- 59.Biferali B, Proietti D, Mozzetta C, Madaro L. Fibro-adipogenic progenitors cross-talk in skeletal muscle: the social network. Front Physiol. 2019;10:1074. doi: 10.3389/fphys.2019.01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang X, Yang M, Shi Y, Xie M, Zhu M, Zheng X, et al. Interleukin-15 facilitates muscle regeneration through modulation of fibro/adipogenic progenitors. Cell Commun Signal. 2018;16. 10.1186/s12964-018-0251-0. [DOI] [PMC free article] [PubMed]
- 61.Lukjanenko L, Karaz S, Stuelsatz P, Gurriaran-Rodriguez U, Michaud J, Dammone G, et al. Aging disrupts muscle stem cell function by impairing matricellular WISP1 secretion from fibro-adipogenic progenitors. Cell Stem Cell. 2019;24:433–46.e7. doi: 10.1016/j.stem.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mozzetta C, Consalvi S, Saccone V, Tierney M, Diamantini A, Mitchell KJ, et al. Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old Mdx mice. EMBO Mol Med. 2013;5:626–39. doi: 10.1002/emmm.201202096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–59. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 64.Stefater JA, Lewkowich I, Rao S, Mariggi G, Carpenter AC, Burr AR, et al. Regulation of angiogenesis by a non-canonical Wnt-Flt1 pathway in myeloid cells. Nature. 2011;474:511–5. doi: 10.1038/nature10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bennett CN, Hodge CL, MacDougald OA, Schwartz J. Role of Wnt10b and C/EBPalpha in spontaneous adipogenesis of 243 cells. Biochem Biophys Res Commun. 2003;302:12–6. doi: 10.1016/s0006-291x(03)00092-5. [DOI] [PubMed] [Google Scholar]
- 66.Zuriaga MA, Fuster JJ, Farb MG, MacLauchlan S, Bretón-Romero R, Karki S, et al. Activation of non-canonical WNT signaling in human visceral adipose tissue contributes to local and systemic inflammation. Sci Rep. 2017;7:17326. doi: 10.1038/s41598-017-17509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lemos DR, Babaeijandaghi F, Low M, Chang C-K, Lee ST, Fiore D, et al. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat Med. 2015;21:786–94. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 68.Cordani N, Pisa V, Pozzi L, Sciorati C, Clementi E. Nitric oxide controls fat deposition in dystrophic skeletal muscle by regulating fibro-adipogenic precursor differentiation. Stem Cells. 2014;32:874–85. doi: 10.1002/stem.1587. [DOI] [PubMed] [Google Scholar]
- 69.Dong Y, Silva KAS, Dong Y, Zhang L. Glucocorticoids increase adipocytes in muscle by affecting IL-4 regulated FAP activity. FASEB J. 2014;28:4123–32. doi: 10.1096/fj.14-254011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferrand N, Béreziat V, Moldes M, Zaoui M, Larsen AK, Sabbah M. WISP1/CCN4 inhibits adipocyte differentiation through repression of PPARγ activity. Sci Rep. 2017;7:1749. doi: 10.1038/s41598-017-01866-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saccone V, Consalvi S, Giordani L, Mozzetta C, Barozzi I, Sandona M, et al. HDAC-regulated myomiRs control BAF60 variant exchange and direct the functional phenotype of fibro-adipogenic progenitors in dystrophic muscles. Genes Dev. 2014;28:841–57. doi: 10.1101/gad.234468.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mueller AA, van Velthoven CT, Fukumoto KD, Cheung TH, Rando TA. Intronic polyadenylation of PDGFRα in resident stem cells attenuates muscle fibrosis. Nature. 2016;540:276–9. doi: 10.1038/nature20160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abramowitz MK, Paredes W, Zhang K, Brightwell CR, Newsom JN, Kwon H-J, et al. Skeletal muscle fibrosis is associated with decreased muscle inflammation and weakness in patients with chronic kidney disease. Am J Physiol Ren Physiol. 2018;315:F1658–69. doi: 10.1152/ajprenal.00314.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moratal C, Raffort J, Arrighi N, Rekima S, Schaub S, Dechesne CA, et al. IL-1β- and IL-4-polarized macrophages have opposite effects on adipogenesis of intramuscular fibro-adipogenic progenitors in humans. Sci Rep. 2018;8. 10.1038/s41598-018-35429-w. [DOI] [PMC free article] [PubMed]
- 75.Contreras O, Rebolledo DL, Oyarzún JE, Olguín HC, Brandan E. Connective tissue cells expressing fibro/adipogenic progenitor markers increase under chronic damage: relevance in fibroblast-myofibroblast differentiation and skeletal muscle fibrosis. Cell Tissue Res. 2016;364:647–60. doi: 10.1007/s00441-015-2343-0. [DOI] [PubMed] [Google Scholar]
- 76.Sacco F, Seelig A, Humphrey SJ, Krahmer N, Volta F, Reggio A, et al. Phosphoproteomics reveals the GSK3-PDX1 axis as a key pathogenic signaling node in diabetic islets. Cell Metab. 2019;29:1422–32.e3. doi: 10.1016/j.cmet.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 77.Cuadrado A, Kügler S, Lastres-Becker I. Pharmacological targeting of GSK-3 and NRF2 provides neuroprotection in a preclinical model of tauopathy. Redox Biol. 2018;14:522–34. doi: 10.1016/j.redox.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu X, Stenson M, Abeykoon J, Nowakowski K, Zhang L, Lawson J, et al. Targeting glycogen synthase kinase 3 for therapeutic benefit in lymphoma. Blood. 2019;134:363–73. doi: 10.1182/blood.2018874560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okamoto M, Udagawa N, Uehara S, Maeda K, Yamashita T, Nakamichi Y, et al. Noncanonical Wnt5a enhances Wnt/β-catenin signaling during osteoblastogenesis. Sci Rep. 2014;4:4493. doi: 10.1038/srep04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.