Abstract
Water, collagen, and proteoglycans determine articular cartilage functionality. If altered, susceptibility to premature degeneration is increased. This study investigated the effects of enzymatic proteoglycan depletion on cartilage functionality as assessed by advanced Magnetic Resonance Imaging (MRI) techniques under standardized loading. Lateral femoral condylar cartilage-bone samples from patients undergoing knee replacement (n = 29) were serially imaged by Proton Density-weighted and T1, T1ρ, T2, and T2* mapping sequences on a clinical 3.0 T MRI scanner (Achieva, Philips). Using pressure-controlled indentation loading, samples were imaged unloaded and quasi-statically loaded to 15.1 N and 28.6 N, and both before and after exposure to low-concentrated (LT, 0.1 mg/mL, n = 10) or high-concentrated trypsin (HT, 1.0 mg/mL, n = 10). Controls were not treated (n = 9). Responses to loading were assessed for the entire sample and regionally, i.e. sub- and peri-pistonally, and zonally, i.e. upper and lower sample halves. Trypsin effects were quantified as relative changes (Δ), analysed using appropriate statistical tests, and referenced histologically. Histological proteoglycan depletion was reflected by significant sub-pistonal decreases in T1 (p = 0.003) and T2 (p = 0.008) after HT exposure. Loading-induced changes in T1ρ and T2* were not related. In conclusion, proteoglycan depletion alters cartilage functionality and may be assessed using serial T1 and T2 mapping under loading.
Subject terms: Medical research, Experimental models of disease, Biomaterials, Musculoskeletal system, Functional magnetic resonance imaging, Magnetic resonance imaging
Introduction
Cartilage degeneration is the hallmark change of osteoarthritis (OA), a potentially devastating and permanently debilitating condition that affects hundreds of millions of people globally, i.e. 3.3–3.6% of the world’s population. In the United States, radiographic evidence of OA is present in approximately 80% of the senior population, i.e. > 65 years of age, even though only 60% of these individuals have symptoms1. By and large, these findings have been confirmed for Germany, too2. Beyond the considerable impact on individual patients, OA has considerable societal and economic impact, which led to its declaration as a "priority disease" by the World Health Organisation3. Its medical and economic burden is projected to increase further as the principal risk factors, i.e. age and obesity, are projected to rise in incidence4.
Pathomechanistically, cartilage degeneration is the result of abnormal tissue remodelling secondary to predisposing tissue susceptibility, unfavourable biomechanical conditions, and persistent inflammation5. On a structural and compositional level, the tissue undergoes progressive changes of its key constituents, i.e. collagen and proteoglycans, that comprise the extracellular matrix (ECM). During early degeneration, proteoglycan content is lost, while the collagen network is increasingly disoriented and disrupted, which leads to increased water uptake, tissue swelling, and softening6. If progressive, the degenerative changes result in advanced tissue disintegration and, eventually, tissue loss.
In consideration of the deficits of clinical-standard morphological Magnetic Resonance Imaging (MRI) techniques in the detection of early, potentially reversible, cartilage degeneration7–9, quantitative MRI (qMRI) techniques and alternative biophysical methodologies such as solid-state nuclear magnetic resonance (NMR) have been studied in clinical and research contexts to assess the tissue beyond mere morphology10. While solid-state NMR spectroscopy provides atomic-level insight into the structure, dynamics, and functionality of cartilage and bone11–14, qMRI techniques spatially map the tissue’s distinct biophysical properties such as T2 or T1ρ relaxation characteristics in a pixel-wise manner, thereby providing markers of (ultra)structure and/or composition. Recently, qMRI techniques have been combined with mechanical loading to study the tissue in more functional contexts and beyond the unloaded configuration. For additional details, the reader is referred to related comprehensive review articles15,16. The rationale behind these approaches is to assess the tissue in more functional contexts by mapping its imaging features in both the unloaded and loaded configurations to determine changes as surrogate parameters of functionality. In a variety of in-vitro, in-situ, and in-vivo studies, physiological and pathological responses to loading could be discerned as promising indicators of the cartilage status in health and disease17–22. Hence, a solid body of scientific and clinical evidence indicates that aberrant response-to-loading patterns are associated with altered tissue integrity and functionality. Nonetheless, the interrelatedness of the tissue’s structural and compositional properties and resultant functionality remains to be fully understood as it determines the joint’s susceptibility to OA and OA progression.
Therefore, the objective of this study was to systematically investigate the role of proteoglycan depletion in load bearing of human articular cartilage. To this end, 20 human articular cartilage samples were exposed to trypsin in two different concentrations to simulate one of the earliest changes in cartilage degeneration and OA6 in a well-established pathomimetic model23. Nine samples served as controls. The samples’ response to loading as a marker of tissue functionality was assessed before and after incubation without (control study arm) or with trypsin (trypsin exposure study arms) using serial T1, T1ρ, T2, and T2* mapping under standardized indentation loading in three different configurations, i.e. unloaded, moderate loading, and strong loading. Based on this imaging-based basic research framework, we aimed to bring together compositional changes induced by controlled in-vitro degradation of cartilage and the imaging correlates of physiological and pathological functionality as assessed by the relative changes under loading of T1, T1ρ, T2, and T2*. Our hypotheses are i) that trypsin exposure alters cartilage composition and cartilage functionality as a function of concentration and ii) that T1, T1ρ, T2, and T2* are reflective of these alterations and thereby provided imaging features of physiological and pathological functionality.
Results
All 29 cartilage samples completed evaluation by qMRI and histology.
Histological evaluation of cartilage tissue adjacent to the cartilage sample revealed gross tissue integrity throughout as signified by mean Mankin sum scores of 1.9 ± 1.1 (range, 0–4), equalling Mankin Grade (MG) 0 in all tissues. Of note, most adjacent tissues had Mankin sum scores of 1 or 2, i.e. slight pre-existent signs of degeneration such as focal hypercellularity, surface fibrillation or Safranin-O de-staining. After incubation without trypsin exposure control samples did not demonstrate any structural or compositional changes on post-exposure evaluation, as indicated by intact tissue surface, regular cellularity, and no Safranin-O de-staining (Fig. 1). After exposure to low concentration of trypsin (LT), samples exhibited superficial Safranin-O de-staining indicative of slight proteoglycan depletion that was limited to the superficial tissue zone (Fig. 2). After exposure to high concentration of trypsin (HT), however, Safranin-O de-staining was progressive and involved the superficial and transitional tissue zones. No surface disintegration, cellular changes or extracellular (ECM) disruption were noted (Fig. 3).
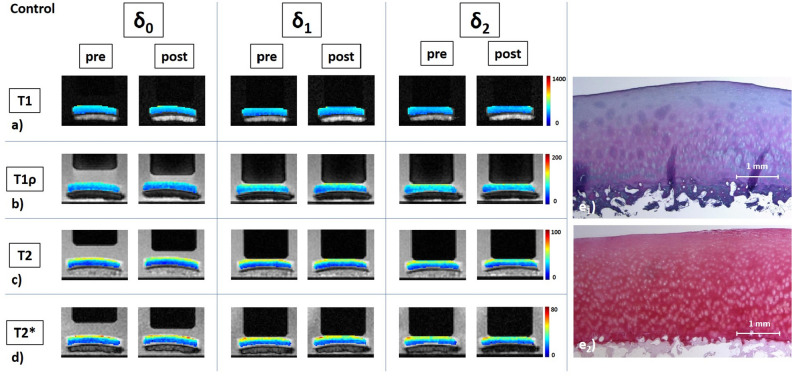
Figure 1.
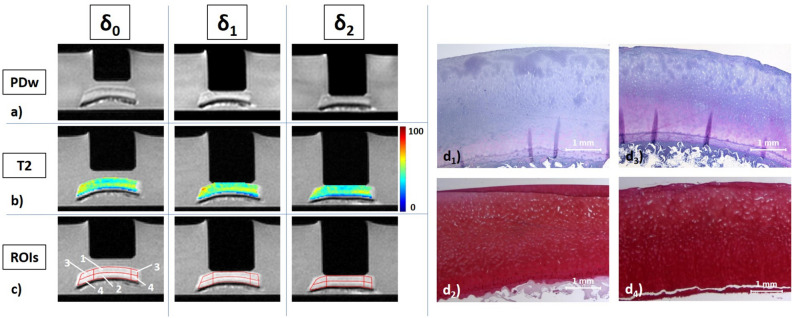
Details of a control cartilage sample and its response to loading as assessed by serial quantitative MR images as a function of loading intensity. Serially acquired T1 (a), T1ρ (b), T2 (c), and T2* (d) maps of a representative control sample before (pre) and after (post) subjection to incubation under standardized conditions (37 °C, 5% CO2, and 21% O2) without additional trypsin exposure. Loading intensity was controlled by set pressure levels and measurements were made in the unloaded configuration (δ0) and under loading to 15.1 N (δ1) and 28.6 N (δ2). Segmented color-coded relaxation maps were overlaid onto the corresponding morphological images. Unit of color codes on the right is [ms]. Of note, the loading piston’s diameter is 10 mm. Corresponding histological sections of the cartilage sample after Hematoxylin and eosin (e1) and Safranin O (e2) staining indicate structural and compositional integrity of the sample. More specifically, the tissue surface and tidemark were intact, while cellularity and Safranin-O staining intensity were regular.
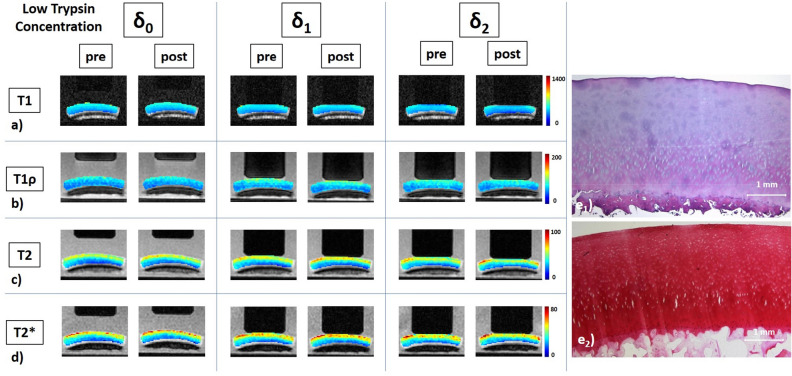
Figure 2.
Details of a low-concentration trypsin-treated cartilage sample, its response to loading, and corresponding histological sections. Serial quantitative T1 (a), T1ρ (b), T2 (c), and T2* (d) maps as well as corresponding histological sections (e) of a representative sample before and after exposure to low concentration of trypsin at 0.1 mg/mL for 2 h. Histological assessment revealed intact tissue surface and tidemark as well as regular cellularity (e1), but superficial Safranin-O de-staining indicative of proteoglycan depletion that was limited to the superficial tissue zone (e2). Otherwise, image details as in Fig. 1.
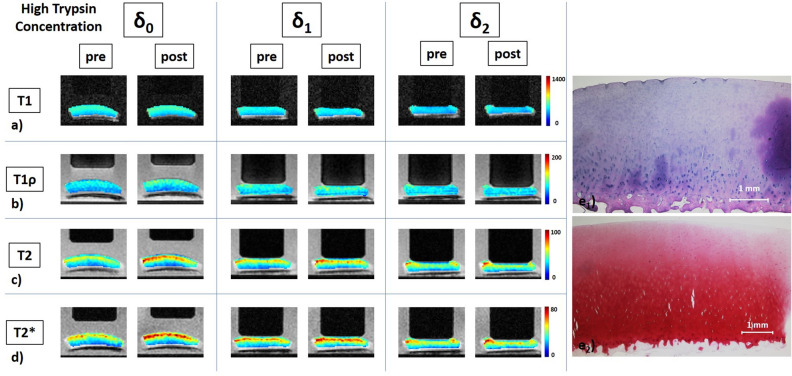
Figure 3.
Details of a high-concentration trypsin-treated cartilage sample, its response to loading, and corresponding histological sections. Serial quantitative T1 (a), T1ρ (b), T2 (c), and T2* (d) maps as well as corresponding histological sections (e) of a representative sample before and after exposure to high concentration of trypsin at 1.0 mg/mL for 2 h. Histological assessment revealed intact tissue surface and tidemark as well as regular cellularity (e1), but expansive Safranin-O de-staining indicative of advanced proteoglycan depletion of the superficial und transitional tissue zones (e2). Superficial artefacts secondary to histological preparation. Otherwise, image details as in Fig. 1.
In the following, δ0 refers to the absolute qMRI parameter values in the unloaded reference configuration, while δ1 and δ2 refer to the respective values under loading of 15.1 N (δ1) and 28.6 N (δ2). Consequently, relative changes in the values at δ1 or δ2 versus δ0 are referred to as Δ1 or Δ2.
In response to loading, T1, T1ρ, T2, and T2* underwent variable zonal and regional changes as a function of trypsin exposure and loading intensity. Table 1 outlines the relative changes of the unloaded reference configuration post-exposure (δ0post) as compared to the unloaded reference configuration pre-exposure (δ0pre). Table 2 outlines the relative changes of the different loading magnitudes, i.e. δ1 and δ2, as compared to the unloaded configuration δ0, both before and after trypsin exposure. Supplementary Table S3 details the absolute qMRI parameter values at δ0, δ1, and δ2.
Table 1.
Details of Δ0, i.e. relative changes of T1, T1ρ, T2, and T2* at δ0 during the pre- and post-exposure measurement series, as a function of trypsin exposure and region-of-interest.
| Δ0 | p value | |
|---|---|---|
| T1 | ||
| Low trypsin | ||
| Entire | 0.4 ± 2.9 | 0.846 |
| SPA | − 0.4 ± 3.2 | 0.625 |
| SPA upper | − 0.1 ± 1.8 | 0.922 |
| SPA lower | − 0.7 ± 6.5 | 0.625 |
| PPA | 1.4 ± 2.8 | 0.232 |
| PPA upper | 2.1 ± 2.3 | 0.027 |
| PPA lower | 0.7 ± 5.0 | 0.695 |
| High trypsin | ||
| Entire | 1.9 ± 3.5 | 0.131 |
| SPA | 1.9 ± 3.8 | 0.131 |
| SPA upper | 4.0 ± 3.3 | 0.004 |
| SPA lower | − 0.4 ± 6.1 | 0.846 |
| PPA | 2.1 ± 3.5 | 0.193 |
| PPA upper | 4.6 ± 3.2 | 0.004 |
| PPA lower | − 2.6 ± 5.2 | 0.275 |
| Controls | ||
| Entire | − 1.3 ± 1.3 | 0.027 |
| SPA | − 0.9 ± 1.6 | 0.084 |
| SPA upper | − 1.2 ± 2.0 | 0.105 |
| SPA lower | − 1.1 ± 2.5 | 0.275 |
| PPA | − 1.3 ± 2.2 | 0.131 |
| PPA upper | − 0.8 ± 2.3 | 0.275 |
| PPA lower | − 1.4 ± 3.7 | 0.275 |
| T1ρ | ||
| Low trypsin | ||
| Entire | 0.6 ± 7.9 | 0.922 |
| SPA | 0.4 ± 7.5 | 0.999 |
| SPA upper | 0.6 ± 8.6 | 0.999 |
| SPA lower | 0.2 ± 8.4 | 0.770 |
| PPA | − 0.7 ± 9.7 | 0.999 |
| PPA upper | 0.5 ± 11.4 | 0.922 |
| PPA lower | − 1.6 ± 9.9 | 0.999 |
| High trypsin | ||
| Entire | 3.3 ± 11.9 | 0.492 |
| SPA | 4.2 ± 13.4 | 0.322 |
| SPA upper | 5.6 ± 15.5 | 0.322 |
| SPA lower | 2.9 ± 12.6 | 0.557 |
| PPA | 1.4 ± 12.6 | 0.846 |
| PPA upper | 4.1 ± 12.3 | 0.557 |
| PPA lower | − 1.8 ± 14.5 | 0.770 |
| Controls | ||
| Entire | − 12.1 ± 8.1 | 0.002 |
| SPA | − 12.2 ± 7.7 | 0.002 |
| SPA upper | − 11.8 ± 9.2 | 0.002 |
| SPA lower | − 12.7 ± 9.4 | 0.004 |
| PPA | − 12.3 ± 9.3 | 0.004 |
| PPA upper | − 12.1 ± 8.8 | 0.004 |
| PPA lower | − 12.8 ± 11.2 | 0.004 |
| T2 | ||
| Low trypsin | ||
| Entire | 4.5 ± 9.7 | 0.232 |
| SPA | 4.3 ± 10.2 | 0.275 |
| SPA upper | 5.6 ± 11.1 | 0.193 |
| SPA lower | 2.5 ± 10.5 | 0.846 |
| PPA | 4.7 ± 9.5 | 0.105 |
| PPA upper | 6.0 ± 11.4 | 0.105 |
| PPA lower | 4.1 ± 10.6 | 0.432 |
| High trypsin | ||
| Entire | 9.5 ± 10.7 | 0.020 |
| SPA | 10.1 ± 10.2 | 0.014 |
| SPA upper | 10.9 ± 11.1 | 0.014 |
| SPA lower | 8.7 ± 12.1 | 0.049 |
| PPA | 9.5 ± 13.4 | 0.064 |
| PPA upper | 10.6 ± 13.4 | 0.049 |
| PPA lower | 8.5 ± 15.9 | 0.193 |
| Controls | ||
| Entire | − 12.4 ± 8.3 | 0.002 |
| SPA | − 12.5 ± 9.1 | 0.002 |
| SPA upper | − 11.2 ± 6.0 | 0.002 |
| SPA lower | − 14.6 ± 13.6 | 0.004 |
| PPA | − 12.1 ± 6.3 | 0.004 |
| PPA upper | − 9.9 ± 5.6 | 0.004 |
| PPA lower | − 16.4 ± 9.7 | 0.004 |
| T2* | ||
| Low trypsin | ||
| Entire | 5.8 ± 8.9 | 0.105 |
| SPA | 5.8 ± 8.8 | 0.105 |
| SPA upper | 6.5 ± 10.1 | 0.105 |
| SPA lower | 4.7 ± 12.2 | 0.375 |
| PPA | 5.3 ± 9.3 | 0.131 |
| PPA upper | 6.3 ± 10.9 | 0.160 |
| PPA lower | 4.9 ± 12.7 | 0.557 |
| High trypsin | ||
| Entire | 7.4 ± 8.5 | 0.014 |
| SPA | 8.5 ± 8.6 | 0.010 |
| SPA upper | 9.7 ± 9.3 | 0.020 |
| SPA lower | 8.5 ± 10.6 | 0.027 |
| PPA | 7.3 ± 12.9 | 0.275 |
| PPA upper | 7.5 ± 12.0 | 0.160 |
| PPA lower | 8.4 ± 19.6 | 0.432 |
| Controls | ||
| Entire | − 8.3 ± 9.1 | 0.014 |
| SPA | − 8.5 ± 10.7 | 0.037 |
| SPA upper | − 7.8 ± 7.5 | 0.010 |
| SPA lower | − 9.8 ± 16.1 | 0.105 |
| PPA | − 6.5 ± 9.4 | 0.049 |
| PPA upper | − 6.3 ± 9.2 | 0.049 |
| PPA lower | − 8.2 ± 15.6 | 0.049 |
Δ0 was calculated as Δ0 = ((δ0post/δ0pre) − 1) * 100 [%], where δ0post and δ0pre are the respective post- and pre-exposure absolute parameter values at δ0. Data are mean ± standard deviation [%]. Wilcoxon matched pairs test was used to assess differences between δ0pre and δ0post. Significant differences are indicated in bold type.
Table 2.
Details of Δ1 and Δ2, i.e. relative changes of T1, T1ρ, T2, and T2* at δ1 versus δ0 and at δ2 versus δ0 during the pre- and post-exposure measurement series, as a function of trypsin exposure, region-of-interest, and loading intensity.
| Pre exposure | Post exposure | p value | ||||
|---|---|---|---|---|---|---|
| Δ1pre | Δ2pre | Δ1post | Δ2post | Δ1pre vs. Δ1post | Δ2pre vs. Δ2post | |
| T1 | ||||||
| Low trypsin | ||||||
| Entire | − 3.7 ± 2.3 | − 7.4 ± 3.2 | − 3.4 ± 2.4 | − 7.1 ± 2.6 | 1.000 | 1.000 |
| SPA | − 6.0 ± 2.2 | − 10.2 ± 3.3 | − 5.0 ± 2.7 | − 10.4 ± 3.8 | 1.000 | 1.000 |
| SPA upper | − 8.6 ± 2.8 | − 15.5 ± 4.6 | − 8.0 ± 3.2 | − 15.4 ± 4.1 | 0.922 | 0.922 |
| SPA lower | − 2.6 ± 3.7 | − 3.5 ± 4.2 | − 0.9 ± 4.1 | − 3.8 ± 5.1 | 0.584 | 0.880 |
| PPA | − 0.6 ± 2.7 | − 3.5 ± 3.3 | − 1.0 ± 3.0 | − 3.1 ± 3.0 | 1.000 | 1.000 |
| PPA upper | − 0.9 ± 2.8 | − 4.7 ± 3.6 | − 1.8 ± 4 | − 4.3 ± 3.8 | 0.566 | 0.568 |
| PPA lower | 0.2 ± 4.7 | − 1.5 ± 5.0 | 0.4 ± 2.9 | − 0.5 ± 6.2 | 1.000 | 1.000 |
| High trypsin | ||||||
| Entire | − 5.0 ± 3.5 | − 10.4 ± 4.2 | − 6.4 ± 3.5 | − 11.5 ± 2.4 | 0.720 | 0.720 |
| SPA | − 6.3 ± 4.1 | − 12.6 ± 5.0 | − 9.0 ± 2.6 | − 15.2 ± 2.7 | 0.106 | 0.092 |
| SPA upper | − 9.6 ± 5.1 | − 17.2 ± 6.5 | − 15.1 ± 5.4 | − 23.0 ± 5.8 | 0.002 | 0.003 |
| SPA lower | − 2.0 ± 4.2 | − 6.7 ± 4.6 | − 0.8 ± 6.8 | − 4.6 ± 4.1 | 0.706 | 0.494 |
| PPA | − 2.7 ± 3.3 | − 6.7 ± 4.0 | − 2.4 ± 5.2 | − 6.2 ± 3.6 | 1.000 | 1.000 |
| PPA upper | − 4.0 ± 4.8 | − 8.4 ± 5.6 | − 4.8 ± 5.2 | − 9.0 ± 5.1 | 1.000 | 1.000 |
| PPA lower | − 1.7 ± 4.0 | − 4.3 ± 2.6 | 2.4 ± 8.8 | − 0.2 ± 6.1 | 0.172 | 0.152 |
| Controls | ||||||
| Entire | − 4.3 ± 2.5 | − 8.4 ± 2.1 | − 4.2 ± 2.2 | − 8.1 ± 3.1 | 1.000 | 1.000 |
| SPA | − 4.9 ± 2.4 | − 9.2 ± 2.3 | − 5.5 ± 3.1 | − 9.6 ± 3.2 | 0.650 | 0.745 |
| SPA upper | − 7.7 ± 4.5 | − 13.9 ± 4.1 | − 8.8 ± 4.0 | − 14.8 ± 3.3 | 0.594 | 0.594 |
| SPA lower | − 2.1 ± 2.8 | − 4.1 ± 3.6 | − 1.7 ± 3.7 | − 2.6 ± 5.0 | 0.908 | 0.908 |
| PPA | − 2.8 ± 2.9 | − 6.7 ± 4.4 | − 2.1 ± 1.8 | − 5.9 ± 4.0 | 0.768 | 0.768 |
| PPA upper | − 3.9 ± 3.1 | − 8.7 ± 5.0 | − 3.5 ± 2.7 | − 9.0 ± 5.1 | 1.000 | 1.000 |
| PPA lower | − 0.9 ± 3.6 | − 2.1 ± 4.7 | 0.2 ± 4.0 | − 1.2 ± 5.3 | 1.000 | 1.000 |
| T1ρ | ||||||
| Low trypsin | ||||||
| Entire | 13.3 ± 3.6 | 15.5 ± 6.2 | 12.2 ± 4.5 | 12.0 ± 7.4 | 0.367 | 0.140 |
| SPA | 15.5 ± 4.5 | 16.9 ± 7.9 | 13.8 ± 6.0 | 13.2 ± 7.2 | 1.000 | 1.000 |
| SPA upper | 21.3 ± 6.4 | 20.9 ± 7.9 | 18.8 ± 5.9 | 16.7 ± 8.3 | 0.324 | 0.324 |
| SPA lower | 9.4 ± 8.1 | 12.8 ± 11.3 | 8.7 ± 11.8 | 10.3 ± 12.5 | 0.765 | 0.082 |
| PPA | 9.1 ± 4.1 | 12.6 ± 4.6 | 9.9 ± 6.0 | 11.3 ± 10.0 | 1.000 | 1.000 |
| PPA upper | 12.6 ± 6 | 15.8 ± 5.2 | 13.6 ± 9.4 | 14.3 ± 11.4 | 1.000 | 1.000 |
| PPA lower | 5.4 ± 5.7 | 9.1 ± 6.1 | 5.5 ± 10.1 | 7.9 ± 13.1 | 1.000 | 1.000 |
| High trypsin | ||||||
| Entire | 13.5 ± 8.2 | 15.4 ± 9.4 | 12.6 ± 10.2 | 17.1 ± 26.2 | 1.000 | 1.000 |
| SPA | 16.1 ± 9.2 | 17.6 ± 10.7 | 14.4 ± 12.1 | 20.2 ± 32.9 | 1.000 | 1.000 |
| SPA upper | 18.2 ± 9.5 | 17.3 ± 13.1 | 15.0 ± 13.5 | 17.1 ± 30.8 | 1.000 | 1.000 |
| SPA lower | 13.8 ± 9.8 | 18.4 ± 10.5 | 14.0 ± 13.4 | 24.6 ± 36.5 | 1.000 | 1.000 |
| PPA | 8.2 ± 8.4 | 11.4 ± 9.3 | 8.3 ± 9.3 | 10.7 ± 9.3 | 1.000 | 1.000 |
| PPA upper | 9.1 ± 8.7 | 11.6 ± 8.0 | 7.2 ± 9.8 | 7.4 ± 12.0 | 0.508 | 0.486 |
| PPA lower | 6.9 ± 9.7 | 11.1 ± 11.8 | 10.1 ± 11.8 | 15.8 ± 11.8 | 0.646 | 0.646 |
| Controls | ||||||
| Entire | 8.9 ± 4.6 | 9.3 ± 5.7 | 13.3 ± 6.1 | 15.9 ± 7.0 | 0.084 | 0.034 |
| SPA | 10.8 ± 4.4 | 11.5 ± 5.6 | 15.3 ± 5.4 | 17.8 ± 6.6 | 0.061 | 0.040 |
| SPA upper | 14.8 ± 6.3 | 13.4 ± 6.2 | 19.3 ± 8.5 | 20.8 ± 9.4 | 0.104 | 0.070 |
| SPA lower | 6.7 ± 9.3 | 9.6 ± 10.7 | 11.4 ± 11.2 | 14.9 ± 12.2 | 0.187 | 0.166 |
| PPA | 4.3 ± 6.6 | 4.2 ± 8.1 | 8.2 ± 7.7 | 11.4 ± 8.7 | 0.209 | 0.030 |
| PPA upper | 5.2 ± 7.5 | 5.3 ± 7.7 | 11.5 ± 7.8 | 12.9 ± 9.6 | 0.059 | 0.012 |
| PPA lower | 3.2 ± 10.3 | 2.8 ± 11.5 | 4.9 ± 9.1 | 9.7 ± 9.4 | 0.665 | 0.200 |
| T2 | ||||||
| Low trypsin | ||||||
| entire | 5.5 ± 3.2 | 4.2 ± 5.6 | 2.4 ± 3.8 | 1.9 ± 7.2 | 0.064 | 0.237 |
| SPA | 5.0 ± 4.4 | 2.9 ± 7.7 | 1.4 ± 3.9 | − 0.2 ± 7.6 | 0.244 | 0.667 |
| SPA upper | 5.1 ± 5.7 | 1.4 ± 7.5 | 0.0 ± 5.5 | − 4.3 ± 8.8 | 0.020 | 0.076 |
| SPA lower | 4.9 ± 5.9 | 5.9 ± 12.2 | 3.6 ± 6.0 | 6.3 ± 8.7 | 1.000 | 1.000 |
| PPA | 6.2 ± 3.3 | 6.4 ± 4.9 | 4.1 ± 4.6 | 5.6 ± 9.1 | 0.244 | 0.667 |
| PPA upper | 8.0 ± 2.7 | 7.1 ± 5.1 | 4.9 ± 5.0 | 5.7 ± 9.3 | 0.042 | 0.466 |
| PPA lower | 4.2 ± 5.6 | 6.2 ± 6.1 | 2.1 ± 6.3 | 5.1 ± 10.8 | 0.842 | 0.842 |
| High trypsin | ||||||
| Entire | 1.1 ± 3.3 | − 0.3 ± 5.1 | − 1.5 ± 4.2 | − 6.2 ± 6.4 | 0.210 | 0.026 |
| SPA | − 0.2 ± 4.7 | − 2.9 ± 7.4 | − 3.0 ± 6.2 | − 9.9 ± 9.0 | 0.235 | 0.008 |
| SPA upper | − 0.5 ± 6.5 | − 7.4 ± 9.9 | − 4.9 ± 8.4 | − 15.2 ± 11.8 | 0.188 | 0.014 |
| SPA lower | 0.2 ± 4.7 | 3.7 ± 7.1 | 0.2 ± 7.7 | − 0.9 ± 10.8 | 0.998 | 0.126 |
| PPA | 3.5 ± 5.4 | 3.8 ± 5.1 | 0.9 ± 5.5 | 1.0 ± 6.8 | 0.468 | 0.468 |
| PPA upper | 4.7 ± 6.1 | 3.8 ± 7.0 | 1.3 ± 6.5 | 0.0 ± 7.8 | 0.328 | 0.328 |
| PPA lower | 1.7 ± 7.2 | 3.0 ± 6.1 | 0.2 ± 6.3 | 3.1 ± 7.4 | 1.000 | 1.000 |
| Controls | ||||||
| Entire | 0.2 ± 3.8 | − 2.9 ± 4.7 | − 1.1 ± 4.7 | − 2.8 ± 4.7 | 0.986 | 0.997 |
| SPA | − 0.5 ± 4.4 | − 3.7 ± 5.2 | − 2.1 ± 5.0 | − 4.4 ± 4.5 | 0.892 | 0.892 |
| SPA upper | 0.8 ± 3.3 | − 5.8 ± 4 | − 1.4 ± 5.5 | − 5.9 ± 4.9 | 0.366 | 0.961 |
| SPA lower | − 1.6 ± 6.1 | − 0.9 ± 8.1 | − 3.2 ± 5.6 | − 2.0 ± 8.9 | 1.000 | 1.000 |
| PPA | 2.4 ± 4.6 | − 0.2 ± 5.7 | 1.2 ± 5.0 | 0.5 ± 7.2 | 1.000 | 1.000 |
| PPA upper | 2.9 ± 5.1 | − 0.5 ± 5.2 | 3.6 ± 3.6 | 0.2 ± 7.9 | 1.000 | 1.000 |
| PPA lower | 0.5 ± 5.9 | − 1.1 ± 6.4 | 0.3 ± 8.1 | 1.4 ± 8.5 | 0.926 | 0.470 |
| T2* | ||||||
| Low trypsin | ||||||
| Entire | 8.3 ± 6.8 | 7.0 ± 6.1 | 5.0 ± 3.9 | 3.2 ± 4.9 | 0.136 | 0.136 |
| SPA | 8.2 ± 7.7 | 6.1 ± 8.4 | 3.9 ± 4.3 | 1.5 ± 5.1 | 0.310 | 0.310 |
| SPA upper | 5.1 ± 8.7 | 2.5 ± 8.6 | − 0.1 ± 7.1 | − 2.6 ± 7.0 | 0.028 | 0.061 |
| SPA lower | 12.7 ± 11 | 11.9 ± 11.4 | 10.7 ± 9.5 | 8.8 ± 6.0 | 0.868 | 0.868 |
| PPA | 8.4 ± 6.2 | 8.7 ± 4.5 | 6.6 ± 4.3 | 6.3 ± 7.6 | 0.310 | 0.310 |
| PPA upper | 8.6 ± 6.4 | 7.7 ± 4.8 | 5.4 ± 7.4 | 4.9 ± 8.8 | 0.002 | 0.253 |
| PPA lower | 8.8 ± 6.6 | 11.2 ± 5.7 | 7.9 ± 7.4 | 8.0 ± 10.0 | 0.806 | 0.806 |
| High trypsin | ||||||
| Entire | 3.6 ± 4.3 | 1.2 ± 6.1 | 2.7 ± 6.3 | − 8.2 ± 14.6 | 0.657 | 0.046 |
| SPA | 2.2 ± 5.4 | − 1.5 ± 8.1 | 1.6 ± 8.8 | − 11.6 ± 17.2 | 0.800 | 0.074 |
| SPA upper | 0.3 ± 7.4 | − 6.8 ± 10.3 | − 2.9 ± 11.5 | − 16.3 ± 18.4 | 0.327 | 0.056 |
| SPA lower | 5.3 ± 6.4 | 7.4 ± 9.7 | 7.2 ± 8.2 | − 4.1 ± 20.2 | 0.248 | 0.130 |
| PPA | 6.2 ± 5.3 | 6.2 ± 5.5 | 4.7 ± 6.3 | − 0.8 ± 13.1 | 0.463 | 0.088 |
| PPA upper | 4.7 ± 6.8 | 4.5 ± 9.1 | 2.2 ± 9.6 | − 1.3 ± 11.8 | 0.365 | 0.046 |
| PPA lower | 8.9 ± 8.7 | 8.3 ± 7.2 | 8.4 ± 7.7 | 1.0 ± 18.3 | 0.875 | 0.500 |
| Controls | ||||||
| Entire | 2.0 ± 5.3 | − 0.5 ± 6.1 | 0.1 ± 5.2 | − 2.3 ± 4.5 | 0.788 | 0.788 |
| SPA | 1.0 ± 5.8 | − 1.3 ± 6.5 | − 1.2 ± 5.6 | − 3.9 ± 4.5 | 0.552 | 0.552 |
| SPA upper | 0.8 ± 5.5 | − 3.3 ± 6.4 | − 2.7 ± 5.6 | − 7.2 ± 4.5 | 0.230 | 0.230 |
| SPA lower | 1.9 ± 7.2 | 1.1 ± 9.6 | 1.1 ± 6.8 | 0.8 ± 7.3 | 1.000 | 1.000 |
| PPA | 6.4 ± 10.6 | 3.0 ± 8.8 | 2.5 ± 4.6 | 0.9 ± 7.0 | 0.730 | 0.730 |
| PPA upper | 5.6 ± 10.2 | 0.8 ± 9.1 | 3.8 ± 5.3 | 0.2 ± 8.1 | 1.000 | 1.000 |
| PPA lower | 7.5 ± 20.3 | 6.2 ± 14.6 | 3.7 ± 8.4 | 3.2 ± 9.4 | 1.000 | 1.000 |
Pre- and post-exposure values of Δ1 and Δ2 were comparatively evaluated using paired Student's t-tests. Data are mean ± standard deviation [%]. Significant differences are indicated in bold type.
Generally, we observed substantial variability in response to loading in terms of absolute values and relative changes as indicated by high measures of statistical spread such as standard deviations and interquartile ranges throughout the study.
By trend, T1 at δ0 was consistently decreased in controls after incubation without enzyme exposure. After LT exposure, however, changes were variable with negligible decreases in the sub-pistonal area (SPA) and slight increases in the peri-pistonal area (PPA). In contrast, after HT exposure, we observed moderate, yet significant increases in the entire samples’ upper halves, both sub-pistonally (SPAupper [HT], Δ0 = 4.0 ± 3.3%, p = 0.004) and peri-pistonally (PPAupper [HT], Δ0 = 4.6 ± 3.2%, p = 0.004) as compared to slight and non-significant decreases in the lower halves (Table 1). Under loading, changes in T1 were dominated by decreases and -by and large- associated with strain, i.e. Δ1 < Δ2, particularly for the SPA and the upper halves. For the PPA, results differed with similar strain-related decreases in T1 in the upper sample halves (PPAupper) and ambiguous changes in the lower sample halves (PPAlower). In all samples, regardless of trypsin exposure, loading-induced changes in T1 demonstrated the following pattern of characteristic changes: Relative changes in the SPA were more pronounced than in the PPA and relative changes in the upper halves more pronounced than in the lower halves. Of note, these patterns were found for T1ρ and T2, too. Consequently, SPAupper and PPAlower displayed largest and smallest loading-induced decreases in T1, respectively. Overall, no significant differences between pre- and post-exposure relative changes in T1 were found in controls and after LT exposure. However, decreases in T1 were significantly larger after HT exposure in the SPAupper, both in response to moderate (SPAupper [LT], Δ1pre = − 9.6 ± 5.1%; Δ1post = − 15.1 ± 5.4%; p = 0.002) and strong loading (SPAupper [LT], Δ2pre = − 17.2 ± 6.5%; Δ2post = − 23.0 ± 5.8%; p = 0.003). In other regions-of-interest (ROIs), decreases in T1 were not significantly different (Table 2).
T1ρ at δ0 was consistently and significantly decreased in controls after incubation without enzyme exposure, e.g. (entire cartilage sample (ECS) [controls], Δ0 = − 12.1 ± 8.1%, p = 0.002), while hardly any changes were found after LT and HT exposure (Table 1). Even though dominated by considerable increases throughout, the T1ρ-associated responses to loading were characterized by substantial statistical variability. Consequently, we did not find any significant pre- vs. post-exposure differences in the loading-induced changes of T1ρ, regardless of trypsin exposure. If at all present, the association of relative changes with strain was only weak. While in controls, increases in T1ρ tended to be greater after incubation, no such trends were observed after LT and HT exposure (Table 2).
T2 at δ0 was significantly decreased after incubation without enzyme exposure in all ROIs of controls, similar to T1ρ (ECS [controls], Δ0 = − 12.4 ± 8.3%, p = 0.002), while after LT and HT exposure, T2 at δ0 was non-significantly increased (Table 1). Overall, T2 tended to decrease sub-pistonally and to increase peri-pistonally. While sub-pistonal changes were associated with strain, i.e. relative changes under moderate loading were smaller than under strong loading (Δ1 < Δ2), that was not the case for the largely ambiguous peri-pistonal changes. No significant differences in the pre- vs. post exposure loading responses were found in controls or after LT exposure, while after HT exposure, sub-pistonal decreases were significantly larger (SPA [HT], Δ2pre = − 2.9 ± 7.4%; Δ2post = − 9.9 ± 9.0%; p = 0.008). The significant sub-pistonal decreases were primarily driven by changes in the upper sample halves (Table 2).
Like T1ρ and T2, T2* at δ0 was substantially decreased in controls after incubation without enzyme exposure. Decreases in T2* at δ0 reached or tended towards significance in all ROIs of controls, e.g. (SPAupper [controls], Δ0 = − 7.8 ± 7.5%, p = 0.01). In contrast, T2* at δ0 was substantially increased after LT and HT exposure, e.g. (SPA [HT], Δ0 = 8.5 ± 8.6%, p = 0.01) (Table 1). Loading-induced changes in T2* were ambiguous without a clear pattern and seemingly not related to loading intensity. Even though characterized by large standard deviations, T2* tended to decrease sub-pistonally and to increase peri-pistonally. Significant pre- and post-exposure differences were only determined for the PPAlower after LT exposure (PPAlower [LT], Δ1pre = 8.6 ± 6.4%; Δ1post = 5.4 ± 7.4%; p = 0.002), while controls and HT exposed samples did not demonstrate significant differences. After HT exposure, we observed divergent increases in T2* under moderate loading alongside decreases under strong loading (Table 2).
Absolute values of T1, T1ρ, T2, and T2* (Supplementary Table S3) and T1, T1ρ, T2, and T2* maps (Figs. 1, 2 and 3, Supplementary Figure S1) were largely reflective of the above-mentioned changes. In controls (Fig. 1), no gross changes were noted after incubation without enzyme exposure. Pre- and post-exposure samples displayed characteristic decreases in the qMRI parameters as a function of tissue depth. Similarly, the response-to-loading patterns were not largely different with largest changes observed sub-pistonally. Peri-pistonal and deep tissue areas displayed least loading-induced changes.
After LT exposure (Fig. 2), slight changes were seen at δ0 that primarily involved the superficial tissue zone and sub-pistonal region, particularly in the T2 and T2* maps. Loading-induced changes tended to be more intense and widespread. In keeping with the pattern outlined above, deep and peri-pistonal areas underwent less changes than superficial and sub-pistonal areas.
After HT exposure (Fig. 3), the depth-wise stratification of the qMRI parameters became a lot clearer, particularly in the T2 and T2* maps. The tissue underwent substantial morphological deformation and flattening with increasing loading. Largest changes were noted for T1, where homogeneous decreases were observed sub-pistonally, and for T2 and T2*, where superficial increases were more marked. In other samples (Supplementary Figure S1), changes were considerably less pronounced. Increases of superficial zones at δ0 were accompanied by more intense and widespread loading-induced changes.
Cartilage sample heights at δ0, i.e. in the unloaded reference configuration, were slightly reduced in controls (from 2.77 ± 0.19 mm [pre exposure] to 2.66 ± 0.22 mm [post exposure], p = 0.016) and after LT exposure (from 2.59 ± 0.32 mm [pre] to 2.55 ± 0.31 mm [post], p = 0.373), while they were marginally increased after HT exposure (from 2.40 ± 0.41 mm [pre] to 2.43 ± 0.41 mm [post], p = 0.343). Additionally, serial measurements of sample heights at δ0, δ1, and δ2 indicated significant decreases in response to loading, irrespective of trypsin exposure (p < 0.001) (Supplementary Table S1). Correspondingly, pixel numbers, i.e. the number of pixels contained within the (sub-)segmentation outlines, were reflective of these changes. In response to loading, pixel numbers decreased significantly in all directly loaded regions-of-interest (ROIs), i.e. the sub-pistonal area (SPA), and in the upper sample halves of the peri-pistonal area (PPAupper), though significant only after trypsin exposure. Otherwise, not directly loaded regions, i.e. the entire peri-pistonal area (PPA), did not demonstrate significant pre- and post-exposure differences in pixel numbers (Supplementary Table S2).
Discussion
The most important findings of this study are that (1) proteoglycan depletion secondary to trypsin exposure affects human articular cartilage functionality and its imaging features and (2) the altered response-to-loading patterns demonstrate regional and zonal variability and may be best assessed using T2 and, to a lesser extent, T1 mapping.
In this pathomimetic in vitro model, trypsin was selected to induce depletion of proteoglycans and non-collagenous macromolecules because a solid body of scientific evidence details its effects on articular cartilage, thereby allowing comparative analyses24,25. As a serine protease, trypsin cleaves peptide bonds of proteoglycans and brings about their selective degradation as a function of concentration and exposure duration. Notably, trypsin-induced proteoglycan depletion is largely inconsistent and affected by the initial proteoglycan distribution and concentration, location within the joint, cartilage thickness, and surrounding medium. In bovine retropatellar cartilage, Moody et al. determined approximately 46% and 71% proteoglycan loss following exposure to 0.1 and 1.0 mg/mL trypsin for 2 h23. The trypsin concentrations in the present study were informed by these data and confirmed by other studies that used Safranin-O staining for reference purposes and demonstrated proteoglycan loss of 76% after exposure to 2.5 mg/mL trypsin over 3 h26 and of 51% after exposure to 0.1 mg/mL trypsin for 2 h27. Despite these studies’ differences in terms of species, trypsin type, and cartilage location, topography, and integrity, these studies are largely in line with one another and with our findings. Histologically, we found slight-to-moderate Safranin O de-staining of the superficial and transitional zones, confirming effective proteoglycan depletion as a function of concentration. Yet, at this point we can only speculate whether trypsin-induced degradation is specific (and exclusive) to proteoglycans. In contrast to the commonly held belief that -apart from specific collagenases- collagen fibrils are resistant to enzymatic cleavage28,29, previous studies have provided anecdotical evidence that human trypsin-2 is indeed capable of cleaving collagen-type II in human cartilage, while animal trypsin is not30. Consequently, in the absence of advanced biochemical reference methodology to prove or disprove trypsin specificity, the following discussion is based on the premise that porcine trypsin (as used in this study) did not affect other ECM constituents.
During physiological loading, intact articular cartilage is subjected to water redistribution, both within the tissue from loaded to less loaded areas and into and out of the tissue. Concomitantly, the ECM experiences condensation and deformation, relative increases in proteoglycan and collagen contents, and collagen fibre re-orientation31–33. These response-to-loading mechanisms may be reflected by the imaging characteristics of pre-exposure samples. For once, sample height decreased with loading. For another, the qMRI parameters underwent distinct changes. In this context, it is important to note that -with the exception of dGEMRIC (delayed gadolinium-enhanced MRI of cartilage) no qMRI parameter has exclusive specificity to any particular cartilage constituent. T1 decreased considerably and as a function of loading intensity with most pronounced decreases in the sub-pistonal upper sample halves. This is plausible in light of this marker’s primary predisposition to intra-tissue water34, resulting in longer T1 relaxation with higher fluid fractions35, while collagen or proteoglycans seem to contribute less36. T1ρ, however, increased under loading though not clearly related to strain. Interpretation is not straightforward as this parameter’s specificity profile remains unclear with water, collagen and proteoglycan content as well as collagen fibre orientation assumed to contribute to T1ρ characteristics37,38. Under loading, the deeper radial collagen fibers display fiber crimping31,39, which increases the percentage of fibers oriented at magic angle and reduces residual dipolar interaction, thereby increasing T1ρ (as well as T2) values. Moreover, recent literature reports have disproven the previously assumed proteoglycan specificity of T1ρ and qualified this association as weak35,40,41 so that loading-induced compositional changes may only be secondary. Consequently, T1ρ nowadays is considered a marker of the tissue’s macromolecular constitution with sensitivity to its solid and fluid constituents. Recently, positive correlations of T1ρ with fluid fraction and collagen fiber orientation, and negative correlations with collagen and proteoglycan content were reported35. Moreover, earlier studies by us and others have reported this parameter’s exquisite mechanosensitivity and loading-induced changes in T1ρ in a range of pre-clinical and clinical contexts17,21,22,42,43. T2 underwent divergent changes in response to loading, i.e. sub-pistonal decreases and peri-pistonal increases. While the former was associated with strain, the latter was not. As T2 is widely considered a marker of collagen fibre orientation and density, collagen content and network organisation, and intra-tissue water37,44, its discrepant changes most likely reflect the complex interplay of these constituents during loading and are largely in line with earlier literature data45,46. However, the relation between T2 and proteoglycan content remains unclear. Studying suspensions of chondroitin sulphate in experimental contexts, which is the most abundant sulphated glycosaminoglycan of cartilage proteoglycans47, Menezes et al. found an exponential association with higher T2 values indicating lower proteoglycan concentrations37. In the same study, a similar association was determined for T2 and collagen concentration. In light of the constitution of the ECM which is primarily made up of collagens (70%) and proteoglycans (20%)47, collagen is dominating -by far- over proteoglycan in both constituents’ contributions to T2 relaxation. These findings were later confirmed by other studies48,49. T2* was subject to ambiguous changes without a clear pattern or any obvious relation to strain. Susceptible to the magnetic susceptibility effects of inherently inhomogeneous cartilage, T2* supposedly reflects collagen microstructure rather than collagen content50,51. Like T2, T2* is sensitive towards water and its interactions with collagen, while its association with proteoglycans is still debated50. Earlier reports have reported an inverse relation, i.e. higher T2* values indicating lower proteoglycan and collagen contents35. However, the ambiguity of loading-induced changes in T2* does not allow solid conclusions to be made.
Substantial increases in T1, T2, and T2* were after HT exposure, mostly driven by changes in the upper sample halves. This is not surprising as proteoglycan depletion causes an influx of water52 and these three qMRI parameters are closely associated with hydration35,53. Moreover, the lower proteoglycan concentration itself probably contributes to these increase35,48. Further, the above-mentioned response-to-loading patterns were altered. First, significantly larger decreases in T1 and T2 were found after HT exposure, but only in the SPAupper (T1) or SPA (T2), respectively. It is important to note that the significant differences in T2 were not present in the unloaded configuration, thereby highlighting the potential value of cartilage functionality assessment and rendering T2 particularly suitable for assessing altered functionality after proteoglycan depletion. Otherwise, pre- and post-exposure changes in T1, T2 or T2* were not significantly different. This finding is plausible as, physiologically, proteoglycans pre-stress the collagen network by osmotic pressure54. Proteoglycan depletion, however, removes this pre-stress, thereby reducing mechanical properties, which renders the tissue more compliant and viscous, and subsequently facilitates water flow 29. These are the most likely imaging correlate of the pronounced decreases in T1 and T2. Therefore, proteoglycan depletion -one of the earliest signs of cartilage degeneration and OA6—compromises the load-bearing capacity of the superficial tissue (and surface), thereby promoting susceptibility to further damage to other tissue regions. Notably, the mechanical effects of trypsin are far more widespread than suggested by histological Safranin O de-staining. Griffin et al. determined trypsin-induced histological proteoglycan loss to a depth of up to 200 μm, while mechanical effects were noted to a depth of 500 μm29. Even when supplemented by more advanced reference measures such as optical absorbance measurements, proteoglycan concentration is variably affected by trypsin23, so that absolute quantification of proteoglycan content, let alone assessment of its integrity and functionality, may be challenging to realize.
Second, trypsin supposedly left the underlying collagen network’s microstructural architecture intact. Beyond histological referencing, this is evidenced by the fact that, overall, T2* and T1ρ, both parameters with sensitivity towards functional collagen properties35,44, were otherwise not significantly altered after trypsin exposure. Herein, it is worth realizing that T1ρ was not significantly affected by proteoglycan depletion, which is surprising, given this parameter’s sensitivity towards intra-tissue changes as detailed above. Most likely, these observations are due to the overpowering dominance of collagen as compared to proteoglycans in both constituents’ contribution to T1ρ relaxation37. Hence, proteoglycan depletion of the superficial and upper transitional tissue zones, i.e. those zones that display the lowest proteoglycan content anyway55, seems largely irrelevant to T1ρ characteristics, lending more evidence to the theory that proteoglycan content and T1ρ are largely unrelated.
Pre-exposure measurements under loading and incubation without enzyme exposure had significant effects on T1ρ, T2, and T2* that underwent significant decreases in controls. Probably, these decreases are related to the tissue’s stress relaxation after loading and an indication of ongoing ultrastructural recovery. The rate of stress relaxation is associated with strain and proceeds significantly slower after higher strains56. Consequently, full recovery may necessitate hours to days57 and may not have been completed. This, of course, limits repeatability and may be only partially remedied by intra-sample referencing. Incomplete ultrastructural recovery could also be a reason for the substantial standard deviations observed throughout the study. Regardless of the exact underlying cause, the tissue’s loading and cultivation history, if relevant, needs to be taken into account when interpreting these changes.
This study has several limitations. First, its in-vitro design limits generalizability to the in-vivo setting. Even though trypsin-2 has been detected in the synovial fluid of rheumatoid arthritis-affected human joints30, unphysiological trypsin concentrations as well as culture and loading conditions limit the in-vivo correspondence of trypsin-induced proteoglycan depletion as studied in vitro. Moreover, cartilage degeneration as the hallmark change of OA is characterized by numerous structural and compositional changes beyond proteoglycan depletion, i.e. disruption and disorganisation of the collagen network amongst others6, which are not emulated in our trypsin-induced experimental setup of isolated proteoglycan depletion. This again limits the generalizability to the in vivo setting. Beyond, proteoglycan depletion may not have been consistent throughout the samples. Following preparation, the bare sample shoulders and periphery might have been more susceptible to trypsin, which might have affected the PPA more than the SPA. Second, the study’s reference framework only included histology, which may be too coarse to detect finer compositional tissue alterations23 and could be supplemented by more advanced techniques such as micro-spectroscopy, biochemical assaying techniques, polarized light microscopy, dGEMRIC or solid-state NMR in the future. In particular, solid-state NMR allows the quantitative evaluation of cartilage and its organic constituents at the molecular level with potential prospects as an additional ultrastructural reference11. Alongside larger sample sizes, more refined references are a prerequisite for more in-depth functional analysis of proteoglycan depletion, for example as a function of gender or age. In addition, very-high-resolution scanning probe methodologies, i.e. atomic force microscopy, scanning electron microscopy or transmission electron microscopy, could provide further insights into cartilage structure, composition, and biomechanical properties for reference purposes to complement quantitative MRI measurements58. Third, despite histological quality checks, the tissue source of total joint replacements is a potential confounder. Beyond the extent appreciable histologically, samples may have been pre-degenerated and pre-inflamed, thereby potentially contributing to sample heterogeneity as signified by considerable measures of spread such as standard deviations. Nonetheless, given the stark differences between animal and human cartilage32,59, we intended to obtain clinically meaningful results and deliberately included human articular cartilage to study its functionality in reference to the sample’s histological tissue status, which partially remedies this situation. Our experimental imaging setup, i.e. the clinical 3.0 T MRI scanner and the ready-to-use mapping sequences with clinically applicable parameter settings are also reflective of this intention. Nonetheless, truly healthy cartilage from amputations, organ donor networks or body donors ought to be included in future studies. Fourth, histological mismatching of the cartilage sample and adjacent tissue may have decreased validity and representatively of histological results, in particular with regards to the variable degeneration in joints, compartments, and tissue regions. Fifth, trypsin exposure was only terminated by continuous washing with PBS, while no dedicated trypsin inhibitors were used. Consequently, traces of trypsin may have remained in the samples, bringing about stronger-than-intended proteoglycan-depletive effects. Yet, this source of variability was systematic after LT and HT exposure, thereby not affecting this study’s main outcomes. Sixth, pre-exposure response-to-loading patterns were quite heterogeneous, in particular for T1ρ and T2, which has been observed before18. Even though principally attributable to the small sample size, it is important to realize that the tissue loading response is not just related to cartilage sample integrity and its primary or secondary degradation, but also to other person-level factors such as age, gender, obesity, and genetics, as well as joint-level factors such as axis alignment, muscle strength and other factors that are reflective of abnormal loading60. These variables are beyond our experimental control and may affect the tissue’s loading responses in health and disease to a much larger extent than previously thought. As detailed above, larger sample sizes with alternative tissue sources and stricter inclusion criteria may help clarify the contributions of these factors to cartilage functionality. Seventh, for the sake of comparability with literature data, we only performed conventional mono-exponential fitting. Alternative fitting techniques such as bi-exponential relaxation of T261 help to quantify different water compartments in cartilage and may thus increase the specificity for altered hydration and functionality.
In conclusion, this study found that trypsin-induced proteoglycan depletion affects cartilage functionality as assessed by serial qMRI mapping and standardized pressure-controlled indentation loading. Dose-dependently, proteoglycan-depleted cartilage areas undergo substantially larger changes in T2 (and, to a lesser extent, T1) as an indication of the altered micromechanical environment secondary to deficient pre-stress of the collagen network. Simulating one of the earliest changes in cartilage degeneration, the close association between compositional deficit and altered functionality was not discernible from quantitative MR images in the unloaded configuration, thereby corroborating the diagnostic potential of imaging-based cartilage functionality assessment. Biomechanical imaging may provide a non-invasive means to study matrix changes in functional contexts and beyond mere composition and may perspectively advance joint and tissue diagnostics in patients.
Materials and methods
Study design
The present study was carried out as an intra-individual ex-vivo imaging study using human cartilage samples from the lateral femoral condyle that were harvested from total knee arthroplasties performed at the University Hospital Aachen. Prior to the study, local Institutional Review Board approval (Ethical Committee, RWTH Aachen, Germany, AZ-EK157/13) and individual written informed consent had been obtained. All methods were carried out in accordance with relevant guidelines and regulations.
Preparation of cartilage samples
Following resection during surgery, the surgical material was transferred to the laboratory in sterile cell culture medium, i.e. Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, Massachusetts, US) that contained 10% fetal calf serum (PAN-Biotech, Aidenbach, Germany), 100 μg/mL Gentamycin (GIBCO-BRL, Paisley, UK), 100 U/mL penicillin and streptomycin (Sigma-Aldrich, St. Louis, Missouri, US), and 1.25 μg/mL amphotericin B (GIBCO-BRL). Surgical material from a total of 29 patients (mean age: 67.4 ± 8.8 years; range, 53–86 years; 13 males, 16 females) was included. Patients were only included if they had primary OA of the knee joint that involved at least the medial compartment, but excluded if they had other bone and joint conditions such as rheumatoid arthritis, secondary OA, and historical findings suggestive of previous trauma or surgery to the index knee. To avoid sample pooling, only one sample was obtained from each patient.
The surgical material was kept refrigerated at 4 °C for a maximum of 24 h before samples were subject to the following standardized preparation steps. First, the material was evaluated macroscopically according to the Outerbridge classification62, the central lateral femoral condyle was identified, and only intact joint regions without any gross macroscopic damage, i.e. Outerbridge grade 0, were prepared. Second, the material was cut to osteochondral samples of standard square shape and size of 1.2 × 1.2 cm (width × length) using dedicated rongeurs and scalpels. Third, we prepared the samples to be as even and plane as possible by preserving the subchondral lamella and by removing the cancellous bone underneath. By doing so, we intended to prevent loading-induced compaction and heterogeneity in load distribution despite the considerable curvature of the femoral condyles. Fourth, for reference purposes, two notches at opposing sample sides were created via rongeur to define the mid-sagittal plane, while an additional third notch was used to define the sample centre as the intersection of the mid-sagittal plane and its perpendicular along the third notch (Fig. 4a). In line with the orientation of the native human knee joint, the sample’s midsagittal plane was aligned along the main magnetic field B0 within the sample box that was subsequently filled with PBS (phosphate-buffered solution, Sigma-Aldrich). Fifth, directly adjacent cartilage tissue with similarly grossly intact macroscopic appearance was prepared to determine the histological status of the sampled cartilage region before any intervention (see Histological analysis below).
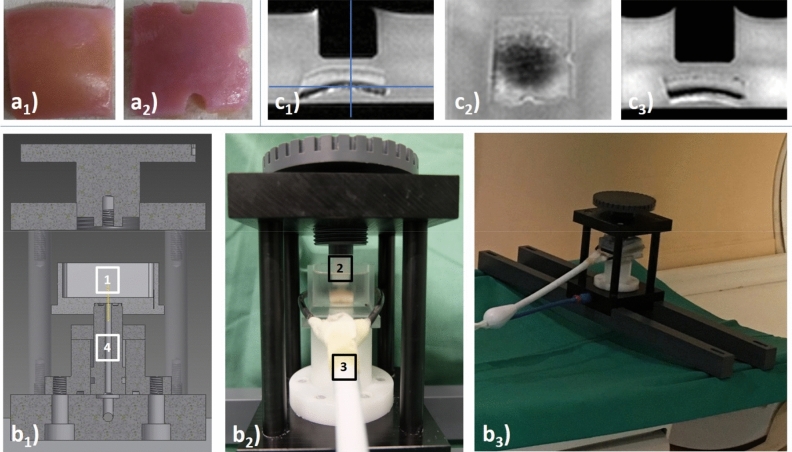
Figure 4.
Details of cartilage preparation steps and experimental setup. (a) Top view of a representative osteochondral sample of standard square shape and size of 1.2 × 1.2 cm before (a1) and after (a2) creation of three notches to define the mid-sagittal imaging plane (extending from the 12 to the 6 o ‘clock position) and the sample centre (as the intersection of the mid-sagittal imaging plane and its orthogonal along the 3 o ‘clock position). (b) Cross-sectional construction sketch (b1) and photographs (lateral [b2] and angulated [b3] views) of the pressure-controlled indentation loading device outside (b2) and inside the MRI scanner (b3). Centrally positioned within the sample box (1), the cartilage sample was upwardly displaced against the indentor piston (2) that was adjustable in height. MR imaging was performed using a dedicated receiver coil that circumferentially comprised the sample box (3). Even though laid out to generate substantially greater forces, the pneumatic mechanism (4) was used to apply forces of 15.1 N and 28.6 N in the context of this study. For more details on the device, the interested reader is referred to Truhn et al., JMBBM, 201964. (c) Sagittal (c1), axial (c2), and coronal (c3) Proton-Density-weighted images of a representative cartilage sample. Blue lines (in c1) indicate the respective heights of the axial (c2) and coronal (c3) images.
Prior to this study, sample size had been estimated by use of a dedicated online tool (https://www.statstodo.com). Based on earlier literature and our own experimental data17,18,43,63, we estimated a minimum sample size of 28 with a statistical power of 0.9, a probability of type-I error (α) of 0.05, a maximum inter-group difference of 0.6, an intra-group standard deviation of 0.5, and Cohen’s effect size model f2. Accordingly, we included a total of 29 macroscopically intact cartilage samples.
MRI-compatible device for standardized indentation loading
The device has been described and validated earlier64 and is presented synoptically in Fig. 4b. In brief, the setup consists of a loading unit inside the MRI scanner’s bore and an outside control unit. Both units are connected via standard pressure lines (Festo, Esslingen, Germany). Customized software routines implemented in LabVIEW software (v2017, National Instruments Corporation, Austin, US) are used to electronically actuate a high-precision valve (± 0.01 bar, Type: VPPM-6L-L-1-G18–0L6H-V1P-S1C1, Festo) via a digital-to-analogue converter (Multifunction I/O USB-6001, National Instruments Corporation). Maximum pressure (up to 4.69 bar) is provided by the hospital’s in-house pressure system and thus downregulated to pressures of 0.75 bar and 1.5 bar that are equivalent to forces of 15.1 N and 28.6 N, respectively64. These forces are approximately equivalent to the forces experienced in vivo during two-legged stance65. Once the desired pressure level is set, the pneumatic mechanism contained within the device’s framework displaces the firmly attached sample box upwards against the fixed and non-porous indentor piston (diameter: 10 mm, bevel: 1 mm) attached to the device’s upper frame. The centrally positioned cartilage sample thus experiences standardized indentation loading by control of pressure.
MRI measurements prior to exposure
Following preparation, pre-exposure MRI measurements were performed on a clinical 3.0 T MRI scanner (Achieva, Philips, Best, The Netherlands). A modified single channel receive-only prostate coil (BPX-30 disposable endorectal coil, Medrad/Bayer, Germany) without the inflatable balloon tip that circumferentially comprised the transparent sample box at the height of the cartilage layer was used for imaging. The scanner’s in-built body coil was used for the application of the radiofrequency pulses. Cartilage samples were centred underneath the indentor piston and attention was paid to align the piston’s undersurface with the sample’s surface.
Imaging of the cartilage samples was performed individually and serially, i.e. in the unloaded configuration (δ0) followed by two consecutive loading positions, i.e. at 0.75 bar (δ1) and at 1.5 bar (δ2). After scout views, Proton Density-weighted (PDw) sequences in all three planes (Fig. 4c) and T2*, T2, T1ρ, and T1 mapping sequences of the mid-sagittal plane were obtained in this order. Table 3 gives the detailed imaging protocol. Of note, the imaging protocol was completed for each individual sample and loading position. After setting pressure levels to 0.75 bar or 1.5 bar, we observed an equilibration period of 5 min prior to initiating the measurements. PDw sequences were assessed to confirm the absence of sample displacement and, thus, adequate loading prior to the acquisition of the mapping sequences. Moreover, PDw sequences were used to guide the mid-sagittal imaging plane along the mid-sagittal plane as defined by the notches. Per sample and loading position, scanner time was about 40 min, equaling 140 min per sample and measurement series (δ0–δ2). Imaging was performed at room temperature which was monitored during one measurement series (20.1 ± 0.6 °C).
Table 3.
Acquisition parameters of MR sequences. n/a—not applicable, ax—axial, sag—sagittal, cor—coronal, mid-sag—mid-sagittal. *Indicates the total duration of all three sequences.
| PDW | T1 | T1ρ | T2 | T2* | |
|---|---|---|---|---|---|
| Sequence type | Turbo-spin echo (2D) | Inversion-recovery (2D) | Spin-lock multi-gradient echo (3D) | Multi-spin echo (2D) | multi-gradient echo (2D) |
| Orientation | ax, sag, cor | mid-sag | sag | mid-sag | mid-sag |
| Repetition time (ms) | 1,500 | 3,000 | 30 | 1,500 | 700 |
| Echo time (ms) | 11.2 | 10.1 | 3.8 | n × 8.4 (n = 1–12) | 3.3 + n × 5.2 (n = 0–14) |
| Turbo spin-echo factor | 6 | 5 | 44 | 12 | 15 |
| Field of view (mm) | 62 × 62 | 62 × 62 | 52 × 52 | 52 × 52 | 52 × 52 |
| Acquisition matrix | 144 × 142 | 224 × 220 | 176 × 176 | 176 × 176 | 176 × 176 |
| Reconstruction matrix | 256 × 256 | 224 × 224 | 224 × 224 | 224 × 224 | 224 × 224 |
| Flip angle (°) | 90 | 90 | 11 | 90 | 90 |
| Number of signal averages | 2 | 1 | 4 | 2 | 3 |
| Slices | 10 | 1 | 7 | 1 | 1 |
| Slice Thickness / Gap (mm) | 1.0 / 0.5 | 2.0 / n/a | 3.2 | 2.0 / n/a | 2.0 / n/a |
| Inversion times (ms) | n/a | 150, 300, 500, 800, 1,000, 1,500 | n/a | n/a | n/a |
| Spin-lock durations (ms) | n/a | n/a | 0, 10, 20, 30, 40 | n/a | n/a |
| Duration (min:s) | 7:03 (*) | 7:12 | 14:30 | 4:29 | 03:08 |
Pathomimetic trypsin model
Following completion of the pre-exposure MRI measurement series, the cartilage samples were randomly allocated to one of three study arms, #1) trypsin at high concentration (1.0 mg/mL)—high trypsin (HT) exposure (n = 10), #2) trypsin at low concentration (0.1 mg/mL)—low trypsin (LT) exposure (n = 10), and #3) controls without any trypsin exposure (n = 9).
Practically, samples were individually suspended in 50 mL volume tubes (Falco Tube, Sarstedt, Nümbrecht, Germany) that contained.
for study arm #1 1.6 mL trypsin-Ethylenediaminetetraacetic acid (EDTA) solution (T4174, Sigma-Aldrich) and 6.4 mL sterile PBS to obtain the high trypsin concentration (1.0 mg/mL trypsin);
for study arm #2 0.16 mL trypsin–EDTA solution and 7.84 mL sterile PBS to obtain the low trypsin concentration (0.1 mg/mL trypsin);
for study arm #3 8 ml medium plus additives as detailed above to obtain controls.
Trypsin was obtained as a 10 × solution containing 5.0 g/L porcine trypsin (from porcine pancreas cells) and 2.0 g/L EDTA. Trypsin is commonly used in pathomimetic models of articular cartilage to induce proteoglycan depletion and the chosen concentrations of 0.1 mg/mL and 1.0 mg/mL are widely consented in the literature23–25. Even though these concentrations are unphysiological, they are commonly used to realize rapid proteoglycan degradation. By means of a tube rotator (VWR Tube Rotator, VWR, Amsterdam, The Netherlands), tubes were continuously rotated for 2 h while being incubated in a standard laboratory incubation unit at 37 °C, 5% CO2, and 21% O2. After incubation, proteolysis was halted by discarding the trypsin–EDTA solutions and by continuous washing with sterile PBS for 1 h, after which samples were replaced in medium + additives as above.
MRI measurements after exposure
Within 24 h after the pre-exposure MRI measurements, cartilage samples were imaged again in strict analogy to the first measurement conditions. While trying to align pre- and post-exposure framework conditions, we were particularly careful to realize identical sample orientation and position.
Image analysis and post processing
Cartilage sample height
Sample height was determined on the mid-sagittal PDw images for each cartilage sample and loading position individually. The first author (TH with 2 years of experience in musculoskeletal radiology) measured cartilage sample height at the sample centre and at an equidistance of 2 mm to both sides by use of the digital caliper tool of the in-house PACS (Picture Archiving and Communication System, Philips). With a step size of a single pixel, the caliper tool has a resolution of 0.24 × 0.24 mm.
Quantitative T1, T1ρ, T2 and T2* maps
First, image raw data were imported and respective time constants for each pixel of the mid-sagittal image were determined based on customized mono-exponential fitting routines implemented in Matlab (MatlabR2019a, Natick, USA) to generate spatially resolved quantitative T1, T1ρ, T2 and T2* maps as before17,20. Fit quality was checked using R2 statistics adjusted to the degrees of freedom and only pixel with R2 values > 0.95 were included. For T2 and T2* fitting, we only included echo times < 60 ms to decrease the effects of noise. Second, cartilage tissue was segmented by manual definition of the sample outlines. While considering the corresponding mid-sagittal PDw image, T2-weighted morphological image (of echo time 41.9 ms), and histological sections as reference, the first author (TH) performed the segmentations of each cartilage sample at each loading position. Segmentation was performed conservatively to decrease partial volume effects by only including pixels that safely lay inside the cartilage tissue and by excluding boundary pixels at the topmost (towards piston or medium) and lowermost layer (towards subchondral lamella). Third, the sample- and loading position-specific outlines were validated against the corresponding T1, T1ρ, T2 and T2* maps and quality checked for morphological correspondence by visual inspection (DT and SN, each with 8 years of experience in musculoskeletal radiology) (Fig. 5a,b). Fourth, using a customized algorithm implemented in Matlab, distinct regions-of-interest (ROIs) were automatically defined on the mid-sagittal images. Zonal ROIs were obtained by automatically partitioning the sample outlines into two equal layers based on pixel-wise measurements of cartilage sample height to halve the entire cartilage sample (ECS) in two halves of equal height, i.e. the superficial (‘upper’) and deep (‘lower’) halves. Regional ROIs were defined in relation to the indentor piston as the (i) sub-pistonal area (SPA, defined as interface between the piston’s undersurface and the sample’s surface of 8 mm width) and the ii) peri-pistonal area (PPA, defined as the tissue region that was located bilaterally adjacent to the SPA). Both bilateral PPA regions were merged, resulting in a total of seven distinct ROIs for downstream analyses: (1) entire cartilage sample (ECS), (2) SPA full thickness (SPA), (3) PPA full thickness (PPA), (4) SPA superficial half (SPAupper), (5) SPA deep half (SPAlower), (6) PPA superficial (PPAupper), and (7) PPA deep (PPAlower) (Fig. 5c).
Figure 5.
Details of image analysis, post processing, and histological referencing. (a) Mid-sagittal Proton Density-weighted images of a representative cartilage sample subject to increasing loading intensity, i.e. at the unloaded configuration (δ0) and under loading to 15.1 N (δ1), and 28.6 N (δ2). (b) Corresponding T2 maps overlaid onto the respective morphological images (TE 41.9 ms). Unit of scale on the right is [ms]. (c) Regional and zonal ROIs were defined as the upper and lower sample halves as well as the sub-pistonal (SPA) and peri-pistonal area (PPA). Consequently, seven ROIs were evaluated: (1) SPAupper, (2) SPAlower, (3) PPAupper, (4) PPAlower, (5) SPA full thickness (1 + 2), (6) PPA full thickness (3 + 4), (7) entire cartilage sample (ECS = 1 + 2 + 3 + 4). (d) Corresponding histological sections, stained with hematoxylin/eosin (d1,d3) and Safranin O (d2,d4), of the cartilage sample following incubation without enzyme exposure and pre- and post-exposure measurements (d1,d2) and of the adjacent cartilage tissue at the time of sample preparation (d3,d4). Same cartilage sample as in Fig. 4 (control).
Histological analysis
In line with standard procedures, the cartilage samples and the adjacent cartilage tissue were subject to histological processing and analysis. After simultaneous decalcification and fixation in Ossa fixona (Waldeck, Muenster, Germany), cartilage samples were sectioned along the mid-sagittal plane, while the adjacent tissue was sectioned parallel to it. Following sectioning, the material was embedded in paraffin, cut to 5-µm sections, stained with hematoxylin/eosin and Safranin O, and imaged using a standard light microscope (Leica DM/LM-P, Wetzlar, Germany) (Fig. 5d).
Histological sections were analysed by three blinded investigators experienced in musculoskeletal histopathology (TH [MD, 2 years of experience], MP [MD, 3 years of experience], SN [MD, fellowship trained, 11 years of experience]) who performed the histological evaluation individually. Adjacent cartilage tissue was assessed in terms of histological degeneration to quality check the included material. To this end, semi-quantitative grading of degeneration was performed according to the Mankin classification66 by assessment of tissue structure (score 0–6), cellularity (score 0–3), proteoglycan staining intensity (score 0–4), and tidemark integrity (score 0–1). Based on the resultant Mankin sum score (ranging from 0–14, i.e. no degeneration to most severe degeneration), the adjacent cartilage tissue was grouped into Mankin grades, i.e. MG 0 (Mankin sum scores 0–4), MG I (scores 5–8), MG II (scores 9–10), and MG III (scores 11–14)67. Only if initial scores were different between the investigators were individual sections discussed until consensus was established.
Cartilage samples following incubation with or without additional trypsin exposure were evaluated qualitatively with a focus on structural and compositional changes as compared to the adjacent tissue.
Statistical analysis
The first and last authors (TH, SN) carried out the statistical analyses using GraphPad Prism (Version 7.0; GraphPad, San Diego, CA, USA). As detailed above, δ0 refers to the unloaded reference configuration, while δ1 and δ2 refer to moderate and strong loading configurations, while the relative changes at δ1 or δ2 versus δ0 are referred to as Δ1 or Δ2. For a given qMRI parameter Tx, Δx was calculated as Δx = ((Txδx/Txδ0) − 1) * 100 [%]. Analogously, relative changes between the unloaded pre- and post-exposure configurations δ0post vs. δ0pre were defined as Δ0 and calculated as Δ0 = ((Txδ0post/Txδ0pre) − 1) * 100 [%]. Throughout, ROI-specific analyses were performed as a function of loading intensity and study arm. To evaluate loading-induced changes in the samples prior to as compared to after incubation with or without additional trypsin exposure, pair-wise comparisons of pre- and post-exposure measures, i.e. Δ1pre vs. Δ1post, and Δ2pre vs. Δ2post, were performed by paired Student's t tests. As absolute T1, T1ρ, T2, and T2* values were not assumed to be normally distributed, they were analysed using Friedman's test followed by Dunn's post-hoc test. To assess changes in the unloaded reference configuration because of incubation with or without additional trypsin exposure, δ0pre and δ0post were comparatively evaluated using the Wilcoxon matched pairs test. To assess loading-induced changes in the samples’ cross-sectional areas, sample heights and pixel numbers were analysed by repeated measures ANOVA. Supplementary Figure S2 graphically presents the most important comparisons.
Data are presented as median and interquartile range (for non-normally distributed data) or mean ± standard deviation (for normally distributed data). To account for the multiple comparisons performed in this exploratory study and control the family-wise error rate, Bonferroni-Holm correction was used. À priori, the level of significance was set to p ≤ 0.01 to contain the number of statistically significant, yet scientifically (likely) insignificant findings.
Supplementary information
Acknowledgements
This research project was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (NE 2136/3-1), the START Program of the Faculty of Medicine, RWTH Aachen, Germany, through means of grants (691702; 691905). We gratefully acknowledge Philips Healthcare for providing the T1ρ sequence and Dr. Quandel as well as Dr. Haberstroh and their team at Marien-Hospital Aachen for helping in sample provision and logistics.
Abbreviations
- ECM
Extracellular matrix
- ECS
Entire cartilage sample
- EDTA
Ethylenediaminetetraacetic acid
- HT
High concentration of trypsin
- LT
Low concentration of trypsin
- MG
Mankin Grade
- OA
Osteoarthritis
- PBS
Phosphate-buffered solution
- PDw
Proton Density-weighted
- PPA
Peri-pistonal area
- (q)MRI
(Quantitative) Magnetic Resonance Imaging
- ROI
Region-of-interest
- SPA
Sub-pistonal area
- Δ
Relative change
- δ0
Reference configuration
- δx
Measurement configuration x
Author contributions
T.H., J.S., and S.N. conceptualized and designed the study and are the guarantors of integrity of the entire study. T.H., J.S., M.P., D.T., and S.N. acquired the data. T.H., J.S., D.T., C.K., and S.N. analyzed and interpreted the data. T.H., D.B.A., M.K. and S.N. drafted the article. M.P., D.B.A., P.S., K.L., M.K., and D.T. provided the study materials, while T.H., J.S., and S.N. were responsible for the statistical analyses. S.N. obtained funding, while D.B.A., P.S., K.L., M.K., C.K. and D.T. provided administrative, technical, or logistic support. All authors critically revised the article for important intellectual content and finally approved the version to be submitted.
Funding
Open Access funding provided by Projekt DEAL.
Data availability
The datasets generated and analyzed in this study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tobias Hafner and Justus Schock.
Supplementary information
is available for this paper at 10.1038/s41598-020-72208-y.
References
- 1.Bortoluzzi A, Furini F, Scirè CA. Osteoarthritis and its management-epidemiology, nutritional aspects and environmental factors. Autoimmun. Rev. 2018;17(11):1097–1104. doi: 10.1016/j.autrev.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs, J., Kuhnert, R. & Scheidt-Nave, C. 12-Monats-Prävalenz von Arthrose in Deutschland (2017).
- 3.Wittenauer, R., Smith, L. & Aden, K. Background paper 6.12 osteoarthritis. World Health Organisation (2013).
- 4.Blüher M. Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019;15(5):288. doi: 10.1038/s41574-019-0176-8. [DOI] [PubMed] [Google Scholar]
- 5.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis. Rheum. 2012;64(6):1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saarakkala S, Julkunen P, Kiviranta P, Makitalo J, Jurvelin JS, Korhonen RK. Depth-wise progression of osteoarthritis in human articular cartilage: investigation of composition, structure and biomechanics. Osteoarthr. Cartil. 2010;18(1):73–81. doi: 10.1016/j.joca.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Marinetti A, Tessarolo F, Ventura L, Falzone A, Neri M, Piccoli F, Rigoni M, Masè M, Cortese F, Nollo G. Morphological MRI of knee cartilage: repeatability and reproducibility of damage evaluation and correlation with gross pathology examination. Eur. Radiol. 2020;30:1–10. doi: 10.1007/s00330-019-06627-5. [DOI] [PubMed] [Google Scholar]
- 8.Palmer AJ, Brown CP, McNally EG, Price AJ, Tracey I, Jezzard P, Carr AJ, Glyn-Jones S. Non-invasive imaging of cartilage in early osteoarthritis. Bone Jt. J. 2013;95-B(6):738–746. doi: 10.1302/0301-620X.95B6.31414. [DOI] [PubMed] [Google Scholar]
- 9.Link TM, Steinbach LS, Ghosh S, Ries M, Lu Y, Lane N, Majumdar S. Osteoarthritis: MR imaging findings in different stages of disease and correlation with clinical findings. Radiology. 2003;226(2):373–381. doi: 10.1148/radiol.2262012190. [DOI] [PubMed] [Google Scholar]
- 10.Guermazi A, Alizai H, Crema MD, Trattnig S, Regatte RR, Roemer FW. Compositional MRI techniques for evaluation of cartilage degeneration in osteoarthritis. Osteoarthr. Cartil. 2015;23(10):1639–1653. doi: 10.1016/j.joca.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 11.Singh C, Purusottam RN, Viswan A, Sinha N. Molecular level understanding of biological systems with high motional heterogeneity in its absolute native state. J. Phys. Chem. C. 2016;120(38):21871–21878. doi: 10.1021/acs.jpcc.6b08951. [DOI] [Google Scholar]
- 12.Xu J, Zhu P, Morris MD, Ramamoorthy A. Solid-state NMR spectroscopy provides atomic-level insights into the dehydration of cartilage. J. Phys. Chem. B. 2011;115(33):9948–9954. doi: 10.1021/jp205663z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horch RA, Gochberg DF, Nyman JS, Does MD. Non-invasive predictors of human cortical bone mechanical properties: T 2-discriminated 1 H NMR compared with high resolution X-ray. PLoS ONE. 2011;6(1):e16359. doi: 10.1371/journal.pone.0016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mroue KH, Viswan A, Sinha N, Ramamoorthy A (2017) Solid-state NMR spectroscopy: the magic wand to view bone at nanoscopic resolution. In: Annual Reports on NMR Spectroscopy, vol 92. Elsevier, pp 365–413
- 15.Neu CP. Functional imaging in OA: role of imaging in the evaluation of tissue biomechanics. Osteoarthr. Cartil. 2014;22(10):1349–1359. doi: 10.1016/j.joca.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jerban S, Chang EY, Du J. Magnetic resonance imaging (MRI) studies of knee joint under mechanical loading. Magn. Reson. Imaging. 2020;65:27–36. doi: 10.1016/j.mri.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nebelung S, Sondern B, Oehrl S, Tingart M, Rath B, Pufe T, Raith S, Fischer H, Kuhl C, Jahr H, Truhn D. Functional MR imaging mapping of human articular cartilage response to loading. Radiology. 2017;282(2):464–474. doi: 10.1148/radiol.2016160053. [DOI] [PubMed] [Google Scholar]
- 18.Truhn D, Sondern B, Oehrl S, Tingart M, Knobe M, Merhof D, Kuhl C, Thuring J, Nebelung S. Differentiation of human cartilage degeneration by functional MRI mapping-an ex vivo study. Eur. Radiol. 2019;29(12):6671–6681. doi: 10.1007/s00330-019-06283-9. [DOI] [PubMed] [Google Scholar]
- 19.Nebelung S, Post M, Knobe M, Shah D, Schleich C, Hitpass L, Kuhl C, Thuring J, Truhn D. Human articular cartilage mechanosensitivity is related to histological degeneration—a functional MRI study. Osteoarthr. Cartil. 2019;27(11):1711–1720. doi: 10.1016/j.joca.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Nebelung S, Post M, Knobe M, Tingart M, Emans P, Thuring J, Kuhl C, Truhn D. Detection of early-stage degeneration in human articular cartilage by multiparametric MR imaging mapping of tissue functionality. Sci. Rep. 2019;9(1):5895. doi: 10.1038/s41598-019-42543-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Souza RB, Kumar D, Calixto N, Singh J, Schooler J, Subburaj K, Li X, Link TM, Majumdar S. Response of knee cartilage T1rho and T2 relaxation times to in vivo mechanical loading in individuals with and without knee osteoarthritis. Osteoarthr. Cartil. 2014;22(10):1367–1376. doi: 10.1016/j.joca.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subburaj K, Souza RB, Stehling C, Wyman BT, Le Graverand-Gastineau MP, Link TM, Li X, Majumdar S. Association of MR relaxation and cartilage deformation in knee osteoarthritis. J. Orthop. Res. 2012;30(6):919–926. doi: 10.1002/jor.22031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moody HR, Brown CP, Bowden J, Crawford RW, McElwain D, Oloyede A. In vitro degradation of articular cartilage: does trypsin treatment produce consistent results? J. Anat. 2006;209(2):259–267. doi: 10.1111/j.1469-7580.2006.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukas VA, Fishbein KW, Reiter DA, Lin PC, Schneider E, Spencer RG. Sensitivity and specificity of univariate MRI analysis of experimentally degraded cartilage under clinical imaging conditions. J. Magn. Reson. Imaging JMRI. 2015;42(1):136–144. doi: 10.1002/jmri.24773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang N, Xia Y. Depth and orientational dependencies of MRI T2 and T1ρ sensitivities towards trypsin degradation and Gd-DTPA2−presence in articular cartilage at microscopic resolution. Magn. Reson. Imaging. 2012;30(3):361–370. doi: 10.1016/j.mri.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin L, Zheng Y, Leung C, Mak A, Choy W, Chan K. Ultrasound detection of trypsin-treated articular cartilage: its association with cartilaginous proteoglycans assessed by histological and biochemical methods. J. Bone Miner. Metab. 2002;20(5):281–287. doi: 10.1007/s007740200040. [DOI] [PubMed] [Google Scholar]
- 27.Duvvuri U, Kudchodkar S, Reddy R, Leigh JS. T(1rho) relaxation can assess longitudinal proteoglycan loss from articular cartilage in vitro. Osteoarthr. Cartil. 2002;10(11):838–844. doi: 10.1053/joca.2002.0826. [DOI] [PubMed] [Google Scholar]
- 28.Shingleton WD, Hodges DJ, Brick P, Cawston TE. Collagenase: a key enzyme in collagen turnover. Biochem. Cell Biol. 1996;74(6):759–775. doi: 10.1139/o96-083. [DOI] [PubMed] [Google Scholar]
- 29.Griffin DJ, Vicari J, Buckley MR, Silverberg JL, Cohen I, Bonassar LJ. Effects of enzymatic treatments on the depth-dependent viscoelastic shear properties of articular cartilage. J. Orthop. Res. 2014;32(12):1652–1657. doi: 10.1002/jor.22713. [DOI] [PubMed] [Google Scholar]
- 30.Stenman M, Ainola M, Valmu L, Bjartell A, Ma G, Stenman U-H, Sorsa T, Luukkainen R, Konttinen YT. Trypsin-2 degrades human type II collagen and is expressed and activated in mesenchymally transformed rheumatoid arthritis synovitis tissue. Am. J. Pathol. 2005;167(4):1119–1124. doi: 10.1016/S0002-9440(10)61200-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grunder W, Kanowski M, Wagner M, Werner A. Visualization of pressure distribution within loaded joint cartilage by application of angle-sensitive NMR microscopy. Magn. Reson. Med. 2000;43(6):884–891. doi: 10.1002/1522-2594(200006)43:6<884::AID-MRM15>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Grunder W. MRI assessment of cartilage ultrastructure. NMR Biomed. 2006;19(7):855–876. doi: 10.1002/nbm.1092. [DOI] [PubMed] [Google Scholar]
- 33.Nebelung S, Sondern B, Jahr H, Tingart M, Knobe M, Thuring J, Kuhl C, Truhn D. Non-invasive T1rho mapping of the human cartilage response to loading and unloading. Osteoarthr. Cartil. 2018;26(2):236–244. doi: 10.1016/j.joca.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Berberat JE, Nissi MJ, Jurvelin JS, Nieminen MT. Assessment of interstitial water content of articular cartilage with T1 relaxation. Magn. Reson. Imaging. 2009;27(5):727–732. doi: 10.1016/j.mri.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Thuring J, Linka K, Itskov M, Knobe M, Hitpass L, Kuhl C, Truhn D, Nebelung S. Multiparametric MRI and computational modelling in the assessment of human articular cartilage properties: a comprehensive approach. Biomed. Res. Int. 2018;2018:9460456. doi: 10.1155/2018/9460456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nissi MJ, Rieppo J, Toyras J, Laasanen MS, Kiviranta I, Nieminen MT, Jurvelin JS. Estimation of mechanical properties of articular cartilage with MRI - dGEMRIC, T2 and T1 imaging in different species with variable stages of maturation. Osteoarthr. Cartil. 2007;15(10):1141–1148. doi: 10.1016/j.joca.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 37.Menezes NM, Gray ML, Hartke JR, Burstein D. T2 and T1rho MRI in articular cartilage systems. Magn. Reson. Med. 2004;51(3):503–509. doi: 10.1002/mrm.10710. [DOI] [PubMed] [Google Scholar]
- 38.Link TM, Neumann J, Li X. Prestructural cartilage assessment using MRI. J. Magn. Reson. Imaging JMRI. 2017;45(4):949–965. doi: 10.1002/jmri.25554. [DOI] [PubMed] [Google Scholar]
- 39.Kääb MJ, Ito K, Clark JM, Notzli HP. Deformation of articular cartilage collagen structure under static and cyclic loading. J. Orthop. Res. 1998;16(6):743–751. doi: 10.1002/jor.1100160617. [DOI] [PubMed] [Google Scholar]
- 40.van Tiel J, Kotek G, Reijman M, Bos PK, Bron EE, Klein S, Nasserinejad K, van Osch GJ, Verhaar JA, Krestin GP, Weinans H, Oei EH. Is T1rho mapping an alternative to delayed gadolinium-enhanced mr imaging of cartilage in the assessment of sulphated glycosaminoglycan content in human osteoarthritic knees? An in Vivo Validation Study. Radiology. 2016;279(2):523–531. doi: 10.1148/radiol.2015150693. [DOI] [PubMed] [Google Scholar]
- 41.Wong CS, Yan CH, Gong NJ, Li T, Chan Q, Chu YC. Imaging biomarker with T1rho and T2 mappings in osteoarthritis—in vivo human articular cartilage study. Eur. J. Radiol. 2013;82(4):647–650. doi: 10.1016/j.ejrad.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 42.Hamada H, Nishii T, Tamura S, Tanaka H, Wakayama T, Sugano N. Comparison of load responsiveness of cartilage T1rho and T2 in porcine knee joints: an experimental loading MRI study. Osteoarthr. Cartil. 2015;23(10):1776–1779. doi: 10.1016/j.joca.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Pastrama MI, Ortiz AC, Zevenbergen L, Famaey N, Gsell W, Neu CP, Himmelreich U, Jonkers I. Combined enzymatic degradation of proteoglycans and collagen significantly alters intratissue strains in articular cartilage during cyclic compression. J. Mech. Behav. Biomed. Mater. 2019;98:383–394. doi: 10.1016/j.jmbbm.2019.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martín Noguerol T, Raya JG, Wessell DE, Vilanova JC, Rossi I, Luna A. Functional MRI for evaluation of hyaline cartilage extracelullar matrix, a physiopathological-based approach. Br. J. Radiol. 2019;92(1103):20190443. doi: 10.1259/bjr.20190443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liess C, Lusse S, Karger N, Heller M, Gluer CC. Detection of changes in cartilage water content using MRI T2-mapping in vivo. Osteoarthr. Cartil. 2002;10(12):907–913. doi: 10.1053/joca.2002.0847. [DOI] [PubMed] [Google Scholar]
- 46.Mosher TJ, Smith HE, Collins C, Liu Y, Hancy J, Dardzinski BJ, Smith MB. Change in knee cartilage T2 at MR imaging after running: a feasibility study. Radiology. 2005;234(1):245–249. doi: 10.1148/radiol.2341040041. [DOI] [PubMed] [Google Scholar]
- 47.Lohmander S. Proteoglycans of joint cartilage: structure, function, turnover and role as markers of joint disease. Bailliere's Clin. Rheumatol. 1988;2(1):37–62. doi: 10.1016/S0950-3579(88)80004-9. [DOI] [PubMed] [Google Scholar]
- 48.Linka K, Itskov M, Truhn D, Nebelung S, Thuring J. T2 MR imaging vs. computational modeling of human articular cartilage tissue functionality. J. Mech. Behav. Biomed. Mater. 2017;74:477–487. doi: 10.1016/j.jmbbm.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 49.Watrin-Pinzano A, Ruaud JP, Olivier P, Grossin L, Gonord P, Blum A, Netter P, Guillot G, Gillet P, Loeuille D. Effect of proteoglycan depletion on T2 mapping in rat patellar cartilage. Radiology. 2005;234(1):162–170. doi: 10.1148/radiol.2341030394. [DOI] [PubMed] [Google Scholar]
- 50.Hesper T, Hosalkar HS, Bittersohl D, Welsch GH, Krauspe R, Zilkens C, Bittersohl B. T2* mapping for articular cartilage assessment: principles, current applications, and future prospects. Skeletal Radiol. 2014;43(10):1429–1445. doi: 10.1007/s00256-014-1852-3. [DOI] [PubMed] [Google Scholar]
- 51.Williams AA, Titchenal MR, Do BH, Guha A, Chu CR. MRI UTE-T2* shows high incidence of cartilage subsurface matrix changes 2 years after ACL reconstruction. J. Orthop. Res. 2019;37(2):370–377. doi: 10.1002/jor.24110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q, Zheng YP, Leung G, Lam WL, Guo X, Lu HB, Qin L, Mak AF. Altered osmotic swelling behavior of proteoglycan-depleted bovine articular cartilage using high frequency ultrasound. Phys. Med. Biol. 2008;53(10):2537–2552. doi: 10.1088/0031-9155/53/10/006. [DOI] [PubMed] [Google Scholar]
- 53.Mosher TJ, Dardzinski BJ. Cartilage MRI T2 relaxation time mapping: overview and applications. Semin. Musculoskelet. Radiol. 2004;8(4):355–368. doi: 10.1055/s-2004-861764. [DOI] [PubMed] [Google Scholar]
- 54.Zernia G, Huster D. Collagen dynamics in articular cartilage under osmotic pressure. NMR Biomed. 2006;19(8):1010–1019. doi: 10.1002/nbm.1061. [DOI] [PubMed] [Google Scholar]
- 55.Wilson W, Huyghe JM, van Donkelaar CC. Depth-dependent compressive equilibrium properties of articular cartilage explained by its composition. Biomech. Model. Mechanobiol. 2007;6(1–2):43–53. doi: 10.1007/s10237-006-0044-z. [DOI] [PubMed] [Google Scholar]
- 56.June RK, Ly S, Fyhrie DP. Cartilage stress-relaxation proceeds slower at higher compressive strains. Arch. Biochem. Biophys. 2009;483(1):75–80. doi: 10.1016/j.abb.2008.11.029. [DOI] [PubMed] [Google Scholar]
- 57.Greaves L, Gilbart M, Yung A, Kozlowski P, Wilson D. Deformation and recovery of cartilage in the intact hip under physiological loads using 7 T MRI. J. Biomech. 2009;42(3):349–354. doi: 10.1016/j.jbiomech.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 58.Danalache M, Tiwari A, Sigwart V, Hofmann UK. Application of atomic force microscopy to detect early osteoarthritis. JoVE (J. Vis. Exp.) 2020;159:e61041. doi: 10.3791/61041. [DOI] [PubMed] [Google Scholar]
- 59.Taylor SD, Tsiridis E, Ingham E, Jin Z, Fisher J, Williams S. Comparison of human and animal femoral head chondral properties and geometries. Proc. Inst. Mech. Eng. [H] 2012;226(1):55–62. doi: 10.1177/0954411911428717. [DOI] [PubMed] [Google Scholar]
- 60.Neogi T, Zhang Y. Epidemiology of osteoarthritis. Rheum. Dis. Clin. 2013;39(1):1–19. doi: 10.1016/j.rdc.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharafi A, Chang G, Regatte RR. Biexponential T2 relaxation estimation of human knee cartilage in vivo at 3T. J. Magn. Reson. Imaging. 2018;47(3):809–819. doi: 10.1002/jmri.25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Outerbridge RE. The etiology of chondromalacia patellae. J Bone Joint Surg Br. 1961;43-B:752–757. doi: 10.1302/0301-620X.43B4.752. [DOI] [PubMed] [Google Scholar]
- 63.Collins AT, Hatcher CC, Kim SY, Ziemian SN, Spritzer CE, Guilak F, DeFrate LE, McNulty AL. Selective enzymatic digestion of proteoglycans and collagens alters cartilage T1rho and T2 relaxation times. Ann Biomed Eng. 2018 doi: 10.1007/s10439-018-02143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Truhn D, Brill N, Braun B, Merhof D, Kuhl C, Knobe M, Thüring J, Nebelung S. A multi-purpose force-controlled loading device for cartilage and meniscus functionality assessment using advanced MRI techniques. J. Mech. Behav. Biomed. Mater. 2020;101:103428. doi: 10.1016/j.jmbbm.2019.103428. [DOI] [PubMed] [Google Scholar]
- 65.Ahmed AM, Burke DL. In-vitro measurement of static pressure distribution in synovial joints–Part I: Tibial surface of the knee. J. Biomech. Eng. 1983;105(3):216–225. doi: 10.1115/1.3138409. [DOI] [PubMed] [Google Scholar]
- 66.Mankin HJ, Dorfman H, Lippiello L, Zarins A. Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips. II. Correlation of morphology with biochemical and metabolic data. J. Bone Joint Surg. Am. 1971;53(3):523–537. doi: 10.2106/00004623-197153030-00009. [DOI] [PubMed] [Google Scholar]
- 67.Gahunia HK, Babyn P, Lemaire C, Kessler MJ, Pritzker KP. Osteoarthritis staging: comparison between magnetic resonance imaging, gross pathology and histopathology in the rhesus macaque. Osteoarthr. Cartil. 1995;3(3):169–180. doi: 10.1016/S1063-4584(05)80051-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed in this study are available from the corresponding author on reasonable request.