Abstract
The ubiquitin–proteasome system has the capacity to degrade polyubiquitinated proteins and plays an important role in many cellular processes. However, the role of Rpt3, a crucial proteasomal gene, has not been investigated in adult muscles in vivo. Herein, we generated skeletal-muscle-specific Rpt3 knockout mice, in which genetic inactivation of Rpt3 could be induced by doxycycline administration. The Rpt3-knockout mice showed a significant reduction by more than 90% in the expression of Rpt3 in adult muscles. Using this model, we found that proteasome dysfunction in adult muscles resulted in muscle wasting and a decrease in the myofiber size. Immunoblotting analysis showed that the amounts of ubiquitinated proteins were markedly higher in muscles of Rpt3-deficient mice than in those of the control mice. Analysis of the autophagy pathway in the Rpt3-deficient mice showed that the upregulation of LC3II, p62, Atg5, Atg7, and Beclin-1 in protein levels, which supposed to be compensatory proteolysis activation. Our results suggest that the proteasome inhibition in adult muscle severely deteriorates myofiber integrity and results in muscle atrophy.
Keywords: muscle homeostasis, Rpt3, ubiquitin proteasome system, muscle atrophy, adult skeletal muscle, sarcopenia
Introduction
Skeletal muscles make up approximately 40% of the total body weight, thus being the largest tissue in the body. Maintaining muscle homeostasis is essential for preserving the body’s integrity and daily function. The loss of skeletal muscle mass in humans at an older age, called sarcopenia, is a rapidly growing health issue worldwide (Vellas et al., 2013). Muscle impairment is associated with several diseases and ultimately leads to a poor quality of life. The process of proteolysis is important for the prevention of cellular dysfunction and disease progression. The regulation of skeletal muscle mass largely depends on protein synthesis and degradation processes, with the latter often associated with pathological conditions, rather than with normal cellular function. Therefore, understanding of the proteolytic system has important implications for normal maintenance of cells and tissues.
The ubiquitin–proteasome system (UPS) is the best-known cellular proteolytic system, which is responsible for degradation of the majority of misfolded or defective cellular proteins (Rock et al., 1994). Proteasomes are present in both the cytoplasm and nucleus and are enriched in nuclei of many proliferating eukaryotic cells (Enenkel, 2014; Pack et al., 2014). Also, the 26S proteasome has the capacity to degrade polyubiquitinated proteins and plays an important role in many cellular processes, such as proteostasis and transcriptional control (Durairaj and Kaiser, 2014; Gallagher et al., 2014). In skeletal muscles, excessive or defective activity of the UPS leads to detrimental effects on muscle homeostasis (Sandri et al., 2013). Therefore, protein degradation by the UPS needs to be precisely regulated to maintain muscle homeostasis.
The 26S proteasome is composed of one barrel-shaped proteolytic core complex (20S proteasome), capped at both ends with 19S regulatory complexes, which recognize ubiquitinated proteins (Baumeister et al., 1998; Tanaka, 2009). The 19S proteasome can be further divided into base and lid subcomplexes. The base has six different ATPase subunits, Rpt1–6. Rpt3, also known as PSMC4, is an essential subunit of the 26S proteasome and is required for the degradation of most proteasomal substrates. In particular, Rpt3-deficient mouse embryos die before implantation, owing to a defect in blastocyst development (Sakao et al., 2000), which indicates that Rpt3 plays essential roles in early embryogenesis and survival. Interestingly, Rpt3 mutations have been described in patients with Parkinson’s disease (Wahl et al., 2008). Furthermore, a previous study has revealed that phosphorylation of Rpt3 controls cell proliferation and tumorigenesis (Guo et al., 2016). To explore the organ- and cell-specific roles of the proteasome, we generated proteasome-deficient mice by targeting Rpt3 (Tashiro et al., 2012; Kitajima et al., 2014, 2018). In particular, we found that a conditional KO of Rpt3 in a motor neuron-specific manner resulted in locomotor dysfunction, accompanied by progressive motor neuron loss and gliosis (Tashiro et al., 2012). Moreover, we have also reported that proteasome dysfunction in Rpt3-deficient muscle stem cells impaired their ability to proliferate, survive, and differentiate, resulting in defective muscle regeneration (Kitajima et al., 2018).
Recently, we have also found that muscle-specific Rpt3-KO mice, which were generated using Cre recombinase under control of the myosin light chain 1 fast (Mlc1f, also known as Myl1) promoter (Bothe et al., 2000), exhibited proteasome insufficiency, leading to a muscle growth defect and an early death (Kitajima et al., 2014). Mlc1f transcripts are initially detected between E8.5 and E9.5 and are robustly expressed beginning at E10.5 (Mourkioti et al., 2008), suggesting that the Rpt3 gene in Mlc1f-Cre;Rpt3f/f pups can be knocked out during the embryonic stage. However, the effect of Rpt3 deficiency in the adult skeletal muscle is unknown. Here, we generated inducible muscle-specific Rpt3-KO mice to investigate the effect of proteasome insufficiency by the Rpt3 gene in adult skeletal muscle.
Materials and Methods
Mouse Strains
The Experimental Animal Care and Use Committee of the Kumamoto University approved the animal experimentation (Ref. No. A30-098). Rpt3-floxed mice (Tashiro et al., 2012; Kitajima et al., 2014) were crossed with ACTA1-rtTA;tetO-Cre mice (Rao and Monks, 2009) to generate ACTA1-rtTA;tetO-Cre; Rpt3f/f (mKO; muscle-specific Rpt3 knockout) mice. All mice used for these experiments were between 3 and 4 months of age.
Doxycycline (DOX) Treatments
As described in a previous study (Rao and Monks, 2009), solutions containing 2 mg/ml DOX (Sigma) or 5% sucrose in drinking water were prepared every third day and provided to the mice to voluntarily consume for 3 weeks to induce Cre-mediated excision. While the Dox solution was provided, the mice were not permitted to drink plain water.
Mouse Tissue Preparation
The body and wet muscle were weighed. The tibialis anterior, gastrocnemius, and soleus muscles were collected individually using standard dissection methods; cleared of excess fat, connective tissue, and tendons; and subjected to further preparation and analyses. The origins of the muscle samples (i.e., tibialis anterior, gastrocnemius, or soleus muscle) are described in each figure legend as relevant. Some portions of the muscles were frozen in isopentane and cooled with liquid nitrogen for histological and immunostaining analysis, and the other muscle portions were frozen directly in liquid nitrogen and stored at −80°C for RNA isolation or protein extraction.
Immunostaining
Immunohistochemistry of cryosections of muscle tissue was performed as described previously (Kitajima and Ono, 2018). In brief, cryosections of the muscle tissue (10-μm thickness) were cut from the middle portion of the muscle belly. These were fixed with 4% PFA, blocked with 5% goat serum or the M.O.M kit (Vector Laboratories) for 30 min at room temperature, and incubated with primary antibodies at 4°C overnight. All immunostained samples were visualized using appropriate species-specific Alexa Fluor 488 and/or 546 fluorescence conjugated secondary antibodies (Life Technologies). Samples were then observed using an Olympus fluorescence microscope IX83 (Olympus).
Measurement of Myofiber Diameter
For measurement of the myofiber diameter, immunostaining analysis was performed on the tibialis anterior muscle, as described in our previous study (Uchitomi et al., 2019). The primary antibodies were as follows: anti-type IIa myosin heavy chain (MyHC) (SC-71, 1:5) and anti-type IIb MyHC (BF-F3, 1:5) obtained from Deutsche Sammlung von Mikroorganismen. When stained with anti-type I MyHC antibody (BA-D5), no type I fibers were observed in the tibialis anterior muscle (data not shown). Thus, type IIa/IIb-unstained fibers were defined as type IIx fibers. Immunostained images were optimized globally and assembled into figures with Photoshop. The minimum fiber Feret’s diameter (Wang et al., 2019) was measured using ImageJ/Fiji software. Samples with significant staining artifacts were excluded from automated analyses.
Immunoblotting
Total protein lysates were extracted from the tibialis anterior muscle for immunoblotting analysis. In brief, the tibialis anterior muscle was crushed by a homogenizer, and the crushed solution was centrifuged for 15 min at 15,000 rpm and 4°C, and the supernatant was collected. We used the BCA method to determine protein concentrations. Then, the protein fractions were extracted with a reducing sample buffer containing 5% β-mercaptoethanol and complete protease inhibitor cocktail. The protein samples (20 μg per lane) were separated on a 10–20% gradient sodium dodecyl sulfate-polyacrylamide gels and subsequently transferred to polyvinylidene difluoride membranes. The membrane was then incubated with primary antibodies. Specific signals were detected using the enhanced chemiluminescence method. Densitometry was performed using ImageJ software (National Institute of Health).
Real-Time PCR
Total RNA was isolated using RNeasy (Qiagen). For real-time PCR, first-strand cDNA was synthesized using oligo-dT primers. The expression levels of selected genes were analyzed using the Bio-Rad CFX96 system according to the manufacturer’s instructions and quantitative PCR analysis was performed in triplicate using specific primers (Supplementary Table S1).
Proteasome Activity
As described in our previous study (Kitajima et al., 2014), proteasome activity was assessed using Proteasome-GloTM Assay kit (Promega) following the manufacturer’s instruction. The chymotrypsin-like activity assay was conducted using skeletal muscle homogenates in a total volume of 100 ml in opaque 96-well plates. For the assays, 100 mg of protein was added to assay buffer containing 20 mM Tris-HCl (pH 7.2), 0.1 mM EDTA, 5 mM ATP, 1 mM β-mercaptoethanol, 20% glycerol and 0.04% Nonidet P40. The proteasome reagent was added separately, and 30 min later, the luminescence was recorded as relative light units on a infinite F200 pro (Tecan). Each sample was measured in triplicate.
Antibodies
The following antibodies from Cell Signaling Technology were used: anti-GAPDH (Cat. No. 2118, 1:1000), Atg5 (Cat. No. 5840, 1:1000), Atg7 (Cat. No. 8558, 1:1000), Beclin1 (Cat. No. 3495, 1:1000), and LC3 (Cat. No. 3868, 1:200). Laminin (Cat. No. L9393, 1:100) and p62 (Cat. No. p0067, 1:1000) antibodies were obtained from Sigma. Type IIa myosin heavy chain (SC71, 1:5) and type IIb myosin heavy chain (BF-F3, 1:5) antibodies were purchased from Deutsche Sammlung von Mikroorganismen. We also used antibodies against LC3 (Novus, Cat. No. 100-2220, 1:300), ubiquitin (LifeSensors, Cat. No. VU101, 1:2000), and laminin α2 (Santa Cruz Biotechnology, Cat. No. 59854, 1:200).
Statistical Analysis
Statistical analyses were performed with GraphPad Prism8 to determine significant differences based on a two-tailed distribution using a Student’s t-test. P-values are indicated on each figure as <0.05 (∗), <0.01 (∗∗), <0.001 (∗∗∗) and <0.0001 (****). All error bars represent means ± SEM. n.s. represents statistically non-significant.
Results
Generation of Muscle-Specific Rpt3-KO Mice
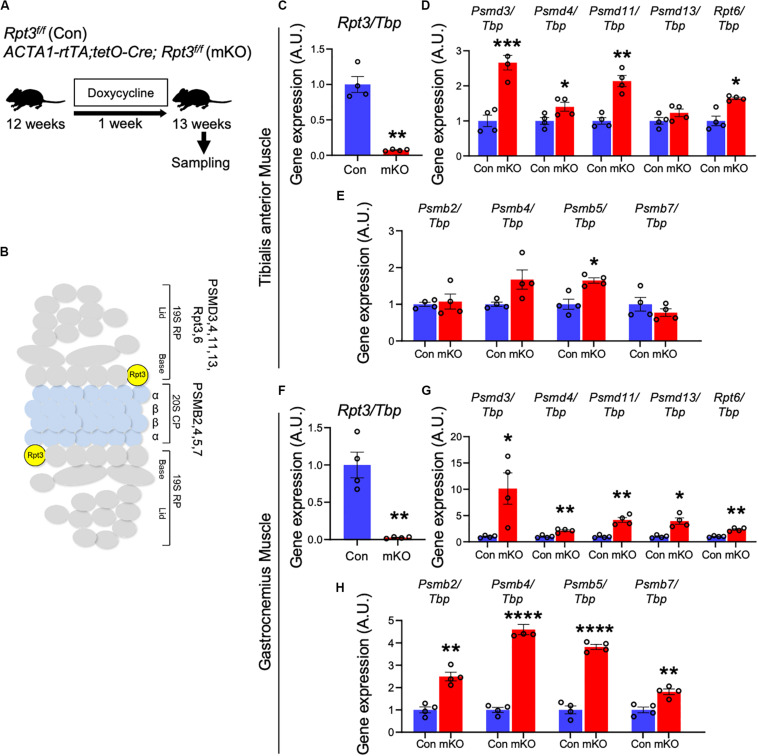
We crossed Rpt3-floxed (Rpt3f/f) mice (Tashiro et al., 2012; Kitajima et al., 2018) with a transgenic line expressing Cre recombinase under control of the skeletal muscle actin (ACTA1-rtTA;tetO-Cre) promoter (Rao and Monks, 2009) to generate muscle-specific Rpt3-KO mice. Genetic inactivation of Rpt3 was induced in adult ACTA1-rtTA;tetO-Cre;Rpt3f/f (mKO) mice by the administration of DOX in drinking water. DOX-treated Rpt3f/f littermates were used as wild-type control (Con) mice (Figure 1A). To determine the KO efficiency of the Rpt3 gene, we isolated the tibialis anterior muscles from Con and mKO mice after DOX treatment for 1 week (Figure 1A) and found that the Rpt3 expression was significantly lower (p < 0.001), by more than 90%, in the tibialis anterior muscles from the mKO mice than in those from the Con mice (Figures 1B,C). In our previously reported Mlc1f-Cre;Rpt3f/f KO mouse model, proteasome-related genes were upregulated (Kitajima et al., 2014). Therefore, we next investigated the expression of proteasome-related genes in the DOX-induced mKO mice. The expression of 19S proteasome-related genes, Psmd3, Psmd4, Psmd11, and Rpt6, was increased in the mKO mice compared to Con mice (Figures 1B,D), as was that of the Psmb5 gene, which are associated with the 20S proteasome (Figures 1B,E). Furthermore, we also examined gene expression in the gastrocnemius muscle, which is mainly composed of fast-type myofibers, as well as in the tibialis anterior muscle. mKO mice showed a significant decrease in the Rpt3 expression level and an increase in proteasome-related gene expression levels compared with those of Con mice (Figures 1F–H). By contrast, in the soleus muscle, which is mainly composed of slow-type myofibers, the Rpt3 gene expression level was reduced by about 50% in mKO mice compared to that of Con mice, whereas the expression levels of other proteasome-related genes were not significantly altered (Supplementary Figures S1A–C). In addition, Rpt3-deficient mice caused a significant increase in proteasome activity in the tibialis anterior and gastrocnemius muscles, but not in the soleus muscle (Supplementary Figure S2). These results confirmed the efficiency of the muscle-specific Rpt3 conditional KO in the tibialis anterior and gastrocnemius muscles.
FIGURE 1.
Generation of muscle-specific Rpt3-knockout mice. (A) Timeline of doxycycline (DOX) treatment. (B) The structure of the 26S proteasome. (C) Relative expression of Rpt3 mRNA in the tibialis anterior muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: **p < 0.01; n = 4 per group). (D) Relative mRNA expression of 19S proteasome genes (Psmd3, 4, 11, 13, and Rpt6) in the tibialis anterior muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: *p < 0.05, **p < 0.01, ***p < 0.001; n = 4 per group). (E) Relative mRNA expression of 20S proteasome genes (Psmb2, 4, 5, and 7) in the tibialis anterior muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: *p < 0.05; n = 4 per group). (F) Relative expression of Rpt3 mRNA in the gastrocnemius muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: **p < 0.01; n = 4 per group). (G) Relative mRNA expression of 19S proteasome genes (Psmd3, 4, 11, 13, and Rpt6) in the gastrocnemius muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: *p < 0.05, **p < 0.01; n = 4 per group). (H) Relative mRNA expression of 20S proteasome genes (Psmb2, 4, 5, and 7) in the gastrocnemius muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: **p < 0.01, ****p < 0.0001; n = 4 per group). Con indicates Rpt3f/f mice, and mKO indicates muscle-specific Rpt3-knockout mice (ACTA1-rtTA;tetO-Cre;Rpt3f/f). AU, arbitrary units.
Proteasome Inhibition Induces Muscle Atrophy
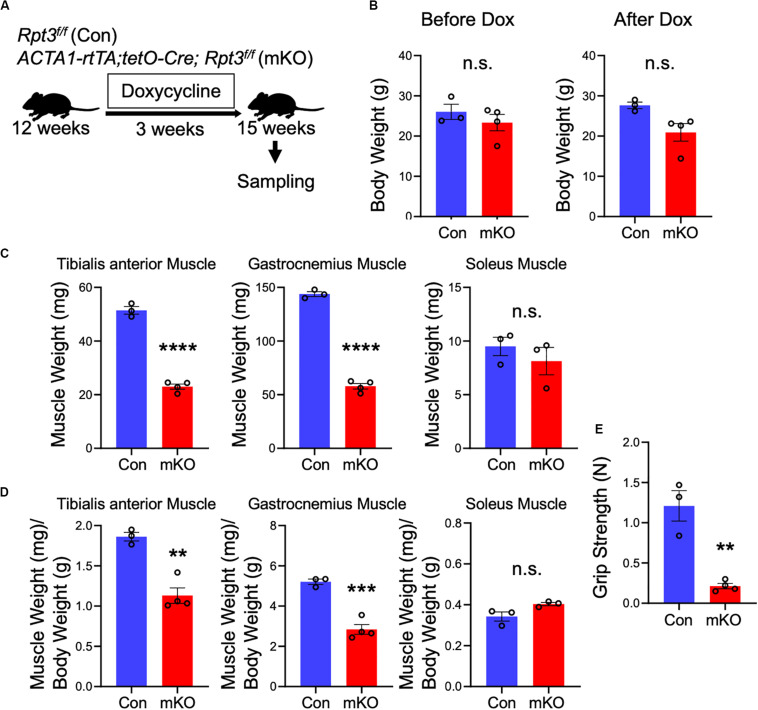
Next, we investigated whether Rpt3 deficiency can cause proteasome dysfunction in adult skeletal muscles, after mouse development is completed. The mice were sacrificed, and muscle weights were determined after DOX treatment for 3 weeks (Figure 2A). There were no differences in the body weights between the Con and mKO mice before and after DOX treatment (Figure 2B). As shown in Figure 1 and Supplementary Figure S1, the Rpt3 gene expression levels differed among the tibialis anterior, gastrocnemius, and soleus muscles; therefore, we examined the weight of each muscle. The absolute weights of the tibialis anterior and gastrocnemius muscles, but not the soleus muscles, were lower in the mKO mice (Figure 2C). Moreover, when muscle weights were evaluated per body weight, those of the tibialis anterior and gastrocnemius muscles, but not the soleus muscles, were lower in the mKO mice than in the Con mice (Figure 2D). Because proteasome-deficient mice exhibited muscle atrophy, we examined gene expression of MuRF1 and Atrogin1, which are markers of muscle atrophy. In the gastrocnemius muscle, but not in the tibialis anterior or soleus muscles, there was a significant increase in MuRF1 and Atrogin1 gene expression in mKO mice compared to Con mice (Supplementary Figure S3). In addition, the grip strength of the mKO mice was significantly lower than that of the Con mice (Figure 2E). These results suggest that post-developmental proteasome dysfunction induces muscle atrophy.
FIGURE 2.
Phenotypes of muscle-specific Rpt3-knockout mice. (A) Timeline of doxycycline (DOX) treatment. (B) Comparison of body weights between Con and mKO mice before and after DOX treatment. Data represent the means ± SEM (t-test: n.s., not significant; n = 3–4 per group). (C) Comparison of tibialis anterior, gastrocnemius, and soleus muscle weights between Con and mKO mice after DOX treatment. Data represent the means ± SEM (t-test: n.s., not significant, ****p < 0.0001; n = 3–4 per group). (D) Comparison of tibialis anterior, gastrocnemius, and soleus muscle weight-to-body weight ratios between Con and mKO mice after DOX treatment. Data represent the means ± SEM (t-test: n.s., not significant, **p < 0.01, ***p < 0.001; n = 3–4 per group). (E) Comparison of grip strength between Con and mKO mice. Data represent the means ± SEM (t-test: **p < 0.01; n = 3–4 per group). Con indicates Rpt3f/f mice, and mKO indicates muscle-specific Rpt3-knockout mice (ACTA1-rtTA;tetO-Cre;Rpt3f/f).
Muscle-Specific Rpt3-Deficient Mice Exhibit a Decrease in the Muscle Fiber Size
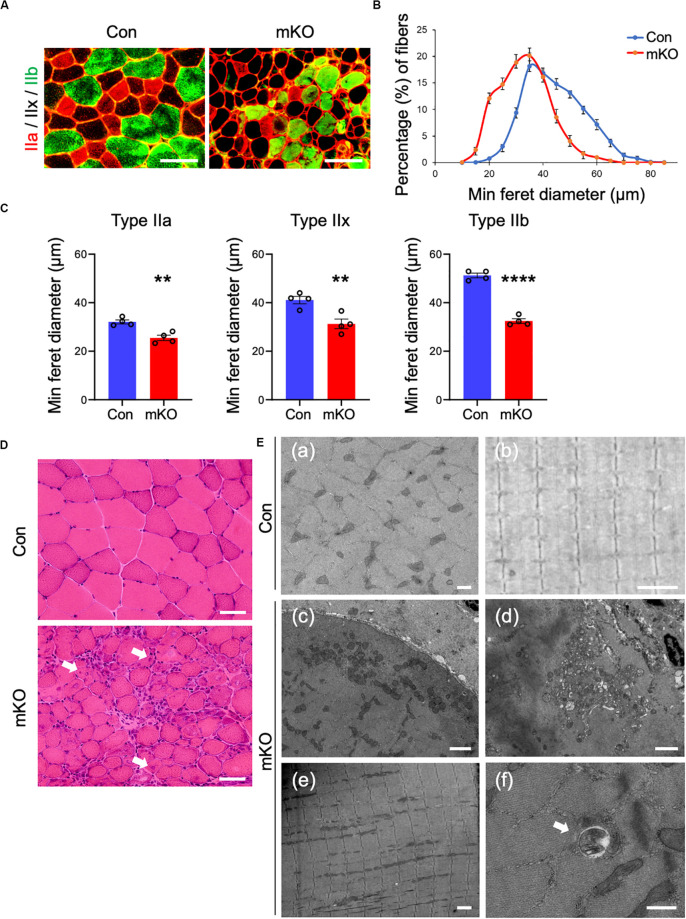
The fast-twitch fibers of the skeletal muscle are characterized by three types (type IIa, IIx, and IIb); type IIa fibers are oxidative, whereas type IIx and IIb fibers are mainly glycolytic. It has been reported that muscle atrophy varies with muscle fiber type (Wang and Pessin, 2013). Because the Rpt3-KO mice exhibited muscle atrophy, we next investigated the sizes of muscle fibers and fiber type in the Con and mKO mice. Immunofluorescence staining of cross-sections of the tibialis anterior muscles for myosin heavy chain type IIa/type IIb indicated that muscle fibers appeared smaller in the Rpt3-KO mice than in the Con mice (Figure 3A). Using a semi-automated measurement of the minimum Feret’s diameter of muscle cross-sections, we found that the myofiber sizes were smaller in the mKO mice than in the Con mice (Figure 3B). Moreover, we evaluated whether the extent of atrophy differed for different myofiber types and found that the sizes of type IIa, IIx, and IIb muscle fibers were significantly smaller in the mKO mice than in the Con mice (Figure 3C). Morphological observations revealed a general decrease in the sizes, along with the presence of central nuclei, in mKO myofibers (Figure 3D). Electron microscopy showed mitochondria and vacuolar structures in necrotising fibers in mKO mice (Figure 3E). Taken together, our data indicate that Rpt3-deficient mice exhibit decreased muscle fiber sizes.
FIGURE 3.
Muscle fiber sizes in muscle-specific Rpt3-deficient mice. (A) Immunohistochemical staining for myosin heavy chain type IIa (red), type IIb (green), and type IIx (unstained) in the tibialis anterior muscles of Con and mKO mice after doxycycline (DOX) treatment. Scale bar = 100 μm. (B) Distribution of muscle fiber minimal Feret’s diameters in the tibialis anterior muscles of Con and mKO mice. Data represent the means ± SEM (n = 4 per group). (C) Quantification of the minimal Feret’s diameter by the fiber type (IIa, IIx, and IIb) in the tibialis anterior muscles of Con and mKO mice. Data represent the means ± SEM (t-test: **p < 0.01, ****p < 0.0001; n = 4 per group). (D) H&E staining of the tibialis anterior muscles from Con and mKO mice. Arrows indicate the central nuclei in myofibers. Scale bar = 50 μm. (E) Electron micrographs of the tibialis anterior muscles from Con and mKO mice. (a) Cross-section of a muscle fiber. Scale bar = 500 nm. (b) Longitudinal section of a muscle fiber. Scale bar = 2 μm. (c) Cross-section of a muscle fiber. Normal mitochondria are located in the subsarcolemmal lesion. Scale bar = 2 μm. (d) Mitochondria and vacuolar structures in a necrotising fiber. Scale bar = 2 μm. (e) Longitudinal section of a muscle fiber. Scale bar = 2 μm. (f) Vacuolar structure in a non-necrotising fiber. Arrows indicate a vacuolar structure. Scale bar = 500 nm. Con indicates Rpt3f/f mice, and mKO indicates muscle-specific Rpt3-knockout mice (ACTA1-rtTA;tetO-Cre;Rpt3f/f).
Effects of Proteasome Dysfunction on the Proteolytic System
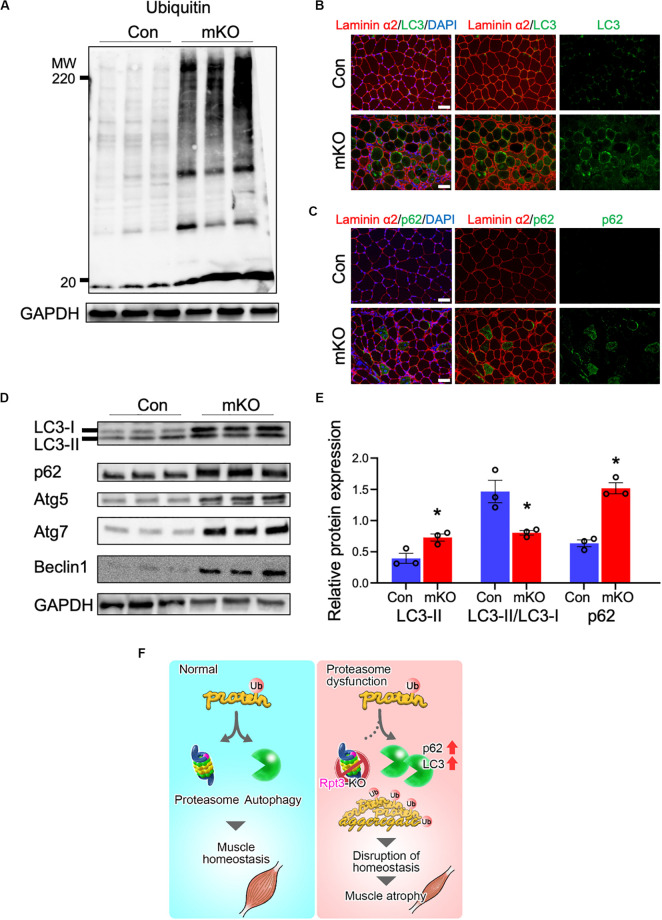
The UPS degrades most of long- and short-lived normal and abnormal intracellular proteins (Goldberg, 2003; Collins and Goldberg, 2017). Because most proteasomal substrates must be ubiquitinated before degradation (Schrader et al., 2009), we next analyzed ubiquitinated proteins in the tibialis anterior muscles by immunoblotting and, as expected, found that their amounts were markedly higher in Rpt3-KO mice than in the Con mice (Figure 4A).
FIGURE 4.
Effects of proteasome dysfunction on the proteolytic system. (A) Immunoblotting analysis of ubiquitinated proteins in the tibialis anterior muscles of Con and mKO mice after 3 weeks of doxycycline (DOX) treatment. GAPDH served as a loading control. (B) Immunostaining of cryosections of the tibialis anterior muscles of Con and mKO mice for laminin α2 (red), LC3 (green), and DAPI (blue) after 3 weeks of DOX treatment. Scale bar = 50 μm. (C) Immunostaining of cryosections of the tibialis anterior muscles of Con and mKO mice for laminin α2 (red), p62 (green), and DAPI (blue) after 3 weeks of DOX treatment. Scale bar = 50 μm. (D) Immunoblotting analysis of autophagy-related proteins in the tibialis anterior muscles of Con and mKO mice after 3 weeks of DOX treatment. (E) Quantification of LC3II and p62 proteins and the LC3II/LC3I ratio. Data represent the means ± SEM (t-test: *p < 0.05; n = 3 per group). (F) Graphical summary of the study. Con indicates Rpt3f/f mice, and mKO indicates muscle-specific Rpt3-knockout mice (ACTA1-rtTA;tetO-Cre;Rpt3f/f).
The autophagy pathway is also an important mechanism governing the degradation of proteins (Klionsky et al., 2016). LC3, which is a post-translational modifier, is required for autophagosome formation (Mizushima and Yoshimori, 2007). The p62 protein is involved in the aggregation of intracellular ubiquitin-associated proteins (Komatsu et al., 2007). Immunohistochemical analysis revealed higher levels of the LC3 and p62 proteins in the tibialis anterior muscles of the mKO mice than in those of the Con mice (Figures 4B,C). Next, autophagy-related proteins were evaluated by immunoblotting, and the data showed that the LC3II, p62, Atg5, Atg7, and Beclin-1 levels were higher and the LC3II/LC3I ratio showing autophagy flux was lower in the mKO mice than in the Con mice (Figures 4D,E). Taken together, these data suggest that proteasome-deficient mice had increased autophagosome formation but could not handle the accumulated proteins to be degraded.
Discussion
In this study, we demonstrated that proteasome dysfunction in adult skeletal muscles induced muscle atrophy in a mouse model. Ablation of Rpt3 in the skeletal muscle impaired proteasome-mediated proteolysis and led to muscle wasting and a decrease in the size of myofibers. In our previous study using Mlc1f-Cre;Rpt3f/f mice, we found that proteasome function is required for muscle growth (Kitajima et al., 2014). To better understand the role of the proteolytic system in adult skeletal muscles, a mouse model with post-developmental proteasome dysfunction was established.
Morphological observations in proteasome-deficient muscles showed myofiber degeneration, including the presence of central and abnormal myonuclei. The UPS degrades most of the normal and abnormal intracellular proteins (Goldberg, 2003; Collins and Goldberg, 2017). Especially in muscle, proteolysis by the UPS is a major mechanism involved in myofibrillar protein degradation (Attaix et al., 1998; Hasselgren et al., 2002). Furthermore, while examining proteasome activity during regeneration, we found that it increased (Kitajima et al., 2018), indicating that maintenance of muscle mass, involving muscle regeneration/degeneration, is associated with the activation of the machinery involved in protein degradation. In muscle, an association between proteasome abnormalities and muscle disease has also been reported (Fratta et al., 2005; Askanas and Engel, 2006). Therefore, further research related to muscle proteasome dysfunction and disease is needed, for which the mouse model developed in the present study could be useful. Interestingly, in mKO mice, proteasome-related genes other than Rpt3 were upregulated, similar to our previous observations in Mlc1f-Cre;Rpt3f/f mice (Kitajima et al., 2014). These findings reflect the adaptations in Rpt3-deficient muscle to eliminate misfolded proteins more efficiently.
There have been several reports on the relationship between proteasome function and longevity. Specifically, it has been reported that proteasome activity is reduced with aging in the brain (Zeng et al., 2005), liver (Hayashi and Goto, 1998; Dasuri et al., 2009), heart (Bulteau et al., 2002), and muscle (Ferrington et al., 2005). Overexpression of proteasome subunits in yeast and Caenorhabditis elegans results in increased proteasome activity and prolonged lifespan (Chen et al., 2006; Vilchez et al., 2012). On the contrary, decreased proteasome activity in flies and mice has been reported to be associated with a shorter lifespan (Tonoki et al., 2009; Tomaru et al., 2012). Investigating the influence of muscle proteasome dysfunction on longevity would be an interesting topic for further studies. Moreover, a previous study reported the enhancement of proteasome activity through inhibition of USP14, a proteasome-associated deubiquitinating enzyme (Lee et al., 2010). It will be interesting to determine whether the proteasome activators also reduce muscle atrophy in mKO mice.
In this study, the tibialis anterior and gastrocnemius muscles, which consist of fast-twitch fibers, were markedly atrophied in mice with proteasomal dysfunction, whereas the slow-twitch-fiber soleus muscle did not show atrophy. This may be due to differences in the knockout efficiency of Rpt3 gene expression. In fact, Rpt3 gene expression in the tibialis anterior and gastrocnemius muscles was suppressed by more than 90%, whereas it was suppressed by less than 50% in the soleus muscle. Furthermore, the levels of proteasome-associated genes were significantly increased in the tibialis anterior and gastrocnemius muscles, but not in the soleus muscle. Knockout of Rpt3 also caused a significant increase in proteasome activity in the tibialis anterior and gastrocnemius muscles. These results suggest that absence of Rpt3 in the tibialis anterior and gastrocnemius muscles induces a feedback increase in the expression levels of proteasome subunits. In the soleus muscle, the phenotypic difference from the tibialis anterior and gastrocnemius muscle may have been due to inadequate suppression of Rpt3 gene expression. Taken together, Rpt3 deficiency disturbed proper degradation of ubiquitinated proteins in proteasome and that seemed to result in compensatory upregulation of other proteasome-related genes. Because proteasome function declines with age (Bardag-Gorce et al., 1999; Ferrington et al., 2005), proteasome insufficiency may be partially related to sarcopenia which is an age-related loss of muscle mass and strength (Musaro and Scicchitano, 2019). Our mouse model may help to further understanding of the underlying mechanism of the decline in physical performance in the elderly.
Two muscle-specific E3 ubiquitin ligases, MuRF1 and Atrogin1, are thought to be key regulators of proteasomal proteolysis in skeletal muscle, especially under atrophy-inducing conditions (Cai et al., 2004; Sandri et al., 2004). In this study, when examining their gene expression in the gastrocnemius and tibialis anterior muscles that exhibited muscle atrophy, we found a significant increase in gene expression only in the gastrocnemius muscle. This difference in gene expression between the gastrocnemius and tibialis anterior muscles may be due to the timing of sampling. In fact, as shown in Figures 1D,E,G,H, proteasome-related genes were increased more in the gastrocnemius muscle compared to the tibialis anterior muscle. A study examining MuRF1 and Atrogin1 gene expression in several models of muscle atrophy has also reported that their gene expression is up-regulated at different time points (Bodine et al., 2001). In this study, an increase in MuRF1 and Atrogin1 gene expressions in the tibialis anterior muscle could be confirmed by examining the gene expression in several time courses.
Previous studies have shown that inhibition of the proteasome system induces autophagy in vivo and in vitro (Iwata et al., 2005; Pandey et al., 2007; Zhu et al., 2010). In this study, the Rpt3-deficient mice exhibited an increase in the expression levels of the p62 and LC3 proteins, suggesting the activation of autophagy. Immunostaining also detected apparent accumulation of p62 and LC3 in myofibers and elevation of the LC3II protein level in muscles of the mKO mice. As LC3II is a lipidated form of LC3 (Kabeya et al., 2000), these results indicated an increase in autophagosome formation. In a previous study, inhibition of the proteasome by specific silencing of a proteasome subunit also stimulated increases in Atg5 and Atg7 levels (Zhu et al., 2010). Consistently, the levels of Atg5, Atg7, and Beclin-1, which are involved in the formation of the isolation membrane, were elevated in the mKO muscle in this study. Thus, in adult muscle-specific proteasome-deficient mice, proteasome suppression caused an increase in autophagic protein degradation; however, this increase could not fully compensate for the loss of Rpt3 to maintain proteolytic system, resulting in skeletal muscle atrophy (Figure 4F).
Our results showed that the proteasome suppression in adult muscle severely deteriorated myofiber integrity and led to muscle atrophy. Although a further study is needed to understand the role of adult skeletal muscles in various physiological processes that involve the proteasome system, this work lays the foundation for understanding the regulation of proteolysis in adult skeletal muscles.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by The Experimental Animal Care and Use Committee of the Kumamoto University approved the animal experimentation (Ref. No. A30-098).
Author Contributions
YK designed the experiments, performed the experiments, interpreted the data, assembled the input data, and wrote the manuscript. NS, KY, RI, and MT performed the experiments. YT and RT produced the animals. MA and YO interpreted the data and assembled the input data. All authors discussed the results and implications and commented on the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Hinako Shigihara, Biomedical Research Core of Tohoku University Graduate School of Medicine for histological and electro-microscopic analysis.
Footnotes
Funding. This work was supported by the Grant-in-Aid for Scientific Research KAKENHI (15H05667, 16H05318, 18H03193, 18K07519, 18K17857, and 20H04078). This work was also supported by the Suzuken Memorial Foundation, and AMED (16bm0704010h0001, 18dk0310086, 18ek0109383h0001, and 19bm0704036h0001).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00859/full#supplementary-material
Expression of proteasome-related genes in the soleus muscles of Con and mKO mice after DOX treatment. (A) Relative expression of Rpt3 mRNA in the soleus muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (n = 4 per group). (B) Relative mRNA expression of 19S proteasome genes (Psmd3, 4, 11, 13, and Rpt6) in the soleus muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: ∗∗p < 0.01; n = 4 per group). (C) Relative mRNA expression of 20S proteasome genes (Psmb2, 4, 5, and 7) in the soleus muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: ∗p < 0.05; n = 4 per group). Con indicates Rpt3f/f mice, and mKO indicates muscle-specific Rpt3-knockout mice (ACTA1-rtTA;tetO-Cre;Rpt3f/f). AU, arbitrary units.
The chymotrypsin-like proteasome activities (relative to those in Con) in the soleus, tibialis anterior, and gastrocnemius muscles after DOX treatment are shown. Data represent means ± SEM (t-test: ∗p < 0.05, versus Con; n = 3–4 per group). IU, international units.
Relative expression of MuRF1 and Atrogin1 mRNA in the soleus, tibialis anterior, and gastrocnemius muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: ∗∗∗p < 0.001; n = 3–4 per group).
Primers for quantitative RT PCR.
References
- Askanas V., Engel W. K. (2006). Inclusion-body myositis: a myodegenerative conformational disorder associated with Abeta, protein misfolding, and proteasome inhibition. Neurology 66(2 Suppl. 1), S39–S48. [DOI] [PubMed] [Google Scholar]
- Attaix D., Aurousseau E., Combaret L., Kee A., Larbaud D., Ralliere C., et al. (1998). Ubiquitin-proteasome-dependent proteolysis in skeletal muscle. Reprod. Nutr. Dev. 38 153–165. 10.1051/rnd:19980202 [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F., Farout L., Veyrat-Durebex C., Briand Y., Briand M. (1999). Changes in 20S proteasome activity during ageing of the LOU rat. Mol. Biol. Rep. 26 89–93. 10.1023/a:1006968208077 [DOI] [PubMed] [Google Scholar]
- Baumeister W., Walz J., Zuhl F., Seemuller E. (1998). The proteasome: paradigm of a self-compartmentalizing protease. Cell 92 367–380. 10.1016/s0092-8674(00)80929-0 [DOI] [PubMed] [Google Scholar]
- Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., et al. (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294 1704–1708. 10.1126/science.1065874 [DOI] [PubMed] [Google Scholar]
- Bothe G. W., Haspel J. A., Smith C. L., Wiener H. H., Burden S. J. (2000). Selective expression of Cre recombinase in skeletal muscle fibers. Genesis 26 165–166. [DOI] [PubMed] [Google Scholar]
- Bulteau A. L., Szweda L. I., Friguet B. (2002). Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 397 298–304. 10.1006/abbi.2001.2663 [DOI] [PubMed] [Google Scholar]
- Cai D., Frantz J. D., Tawa N. E., Jr., Melendez P. A., Oh B. C. (2004). IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell 119 285–298. 10.1016/j.cell.2004.09.027 [DOI] [PubMed] [Google Scholar]
- Chen Q., Thorpe J., Dohmen J. R., Li F., Keller J. N. (2006). Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic. Biol. Med. 40 120–126. 10.1016/j.freeradbiomed.2005.08.048 [DOI] [PubMed] [Google Scholar]
- Collins G. A., Goldberg A. L. (2017). The logic of the 26S proteasome. Cell 169 792–806. 10.1016/j.cell.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasuri K., Nguyen A., Zhang L., Fernandez-Kim O. S., Bruce-Keller A. J., Blalock B. A., et al. (2009). Comparison of rat liver and brain proteasomes for oxidative stress-induced inactivation: influence of ageing and dietary restriction. Free Radic. Res. 43 28–36. 10.1080/10715760802534812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durairaj G., Kaiser P. (2014). The 26S proteasome and initiation of gene transcription. Biomolecules 4 827–847. 10.3390/biom4030827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C. (2014). Proteasome dynamics. Biochim. Biophys. Acta 1843 39–46. 10.1016/j.bbamcr.2013.03.023 [DOI] [PubMed] [Google Scholar]
- Ferrington D. A., Husom A. D., Thompson L. V. (2005). Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 19 644–646. 10.1096/fj.04-2578fje [DOI] [PubMed] [Google Scholar]
- Fratta P., Engel W. K., McFerrin J., Davies K. J., Lin S. W., Askanas V. (2005). Proteasome inhibition and aggresome formation in sporadic inclusion-body myositis and in amyloid-beta precursor protein-overexpressing cultured human muscle fibers. Am. J. Pathol. 167 517–526. 10.1016/s0002-9440(10)62994-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher P. S., Oeser M. L., Abraham A. C., Kaganovich D., Gardner R. G. (2014). Cellular maintenance of nuclear protein homeostasis. Cell Mol. Life Sci. 71 1865–1879. 10.1007/s00018-013-1530-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L. (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426 895–899. 10.1038/nature02263 [DOI] [PubMed] [Google Scholar]
- Guo X., Wang X., Wang Z., Banerjee S., Yang J., Huang L., et al. (2016). Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat. Cell Biol. 18 202–212. 10.1038/ncb3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselgren P. O., Wray C., Mammen J. (2002). Molecular regulation of muscle cachexia: it may be more than the proteasome. Biochem. Biophys. Res. Commun. 290 1–10. 10.1006/bbrc.2001.5849 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Goto S. (1998). Age-related changes in the 20S and 26S proteasome activities in the liver of male F344 rats. Mech. Ageing Dev. 102 55–66. 10.1016/s0047-6374(98)00011-6 [DOI] [PubMed] [Google Scholar]
- Iwata A., Riley B. E., Johnston J. A., Kopito R. R. (2005). HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280 40282–40292. 10.1074/jbc.M508786200 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., et al. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19 5720–5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima Y., Ono Y. (2018). Visualization of PAX7 protein dynamics in muscle satellite cells in a YFP knock-in-mouse line. Skelet. Muscle 8:26. 10.1186/s13395-018-0174-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima Y., Suzuki N., Nunomiya A., Osana S., Yoshioka K., Tashiro Y., et al. (2018). The ubiquitin-proteasome system is indispensable for the maintenance of muscle stem cells. Stem Cell Rep. 11 1523–1538. 10.1016/j.stemcr.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima Y., Tashiro Y., Suzuki N., Warita H., Kato M., Tateyama M., et al. (2014). Proteasome dysfunction induces muscle growth defects and protein aggregation. J. Cell Sci. 127(Pt 24), 5204–5217. 10.1242/jcs.150961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., Acevedo Arozena A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:222. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., et al. (2007). Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131 1149–1163. 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Lee B. H., Lee M. J., Park S., Oh D. C., Elsasser S., Chen P. C., et al. (2010). Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467 179–184. 10.1038/nature09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. (2007). How to interpret LC3 immunoblotting. Autophagy 3 542–545. 10.4161/auto.4600 [DOI] [PubMed] [Google Scholar]
- Mourkioti F., Slonimsky E., Huth M., Berno V., Rosenthal N. (2008). Analysis of CRE-mediated recombination driven by myosin light chain 1/3 regulatory elements in embryonic and adult skeletal muscle: a tool to study fiber specification. Genesis 46 424–430. 10.1002/dvg.20419 [DOI] [PubMed] [Google Scholar]
- Musaro A., Scicchitano B. M. (2019). Counteracting sarcopenia: the role of IGF-1 isoforms. Aging 11 3410–3411. 10.18632/aging.102027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack C. G., Yukii H., Toh-e A., Kudo T., Tsuchiya H., Kaiho A., et al. (2014). Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat. Commun. 5:3396. 10.1038/ncomms4396 [DOI] [PubMed] [Google Scholar]
- Pandey U. B., Nie Z., Batlevi Y., McCray B. A., Ritson G. P., Nedelsky N. B., et al. (2007). HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447 859–863. 10.1038/nature05853 [DOI] [PubMed] [Google Scholar]
- Rao P., Monks D. A. (2009). A tetracycline-inducible and skeletal muscle-specific Cre recombinase transgenic mouse. Dev. Neurobiol. 69 401–406. 10.1002/dneu.20714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., et al. (1994). Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78 761–771. 10.1016/s0092-8674(94)90462-6 [DOI] [PubMed] [Google Scholar]
- Sakao Y., Kawai T., Takeuchi O., Copeland N. G., Gilbert D. J., Jenkins N. A., et al. (2000). Mouse proteasomal ATPases Psmc3 and Psmc4: genomic organization and gene targeting. Genomics 67 1–7. 10.1006/geno.2000.6231 [DOI] [PubMed] [Google Scholar]
- Sandri M., Coletto L., Grumati P., Bonaldo P. (2013). Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J. Cell Sci. 126(Pt 23), 5325–5333. 10.1242/jcs.114041 [DOI] [PubMed] [Google Scholar]
- Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., et al. (2004). Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117 399–412. 10.1016/s0092-8674(04)00400-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader E. K., Harstad K. G., Matouschek A. (2009). Targeting proteins for degradation. Nat. Chem. Biol. 5 815–822. 10.1038/nchembio.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. (2009). The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85 12–36. 10.2183/pjab.85.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y., Urushitani M., Inoue H., Koike M., Uchiyama Y., Komatsu M., et al. (2012). Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J. Biol. Chem. 287 42984–42994. 10.1074/jbc.m112.417600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru U., Takahashi S., Ishizu A., Miyatake Y., Gohda A., Suzuki S., et al. (2012). Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am. J. Pathol. 180 963–972. 10.1016/j.ajpath.2011.11.012 [DOI] [PubMed] [Google Scholar]
- Tonoki A., Kuranaga E., Tomioka T., Hamazaki J., Murata S., Tanaka K., et al. (2009). Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell Biol. 29 1095–1106. 10.1128/MCB.01227-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchitomi R., Hatazawa Y., Senoo N., Yoshioka K., Fujita M., Shimizu T., et al. (2019). Metabolomic analysis of skeletal muscle in aged mice. Sci. Rep. 9:10425. 10.1038/s41598-019-46929-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellas B., Pahor M., Manini T., Rooks D., Guralnik J. M., Morley J., et al. (2013). Designing pharmaceutical trials for sarcopenia in frail older adults: EU/US Task Force recommendations. J. Nutr. Health Aging 17 612–618. 10.1007/s12603-013-0362-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D., Morantte I., Liu Z., Douglas P. M., Merkwirth C., Rodrigues A. P., et al. (2012). RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489 263–268. 10.1038/nature11315 [DOI] [PubMed] [Google Scholar]
- Wahl C., Kautzmann S., Krebiehl G., Strauss K., Woitalla D., Muller T., et al. (2008). A comprehensive genetic study of the proteasomal subunit S6 ATPase in German Parkinson’s disease patients. J. Neural Transm. 115 1141–1148. 10.1007/s00702-008-0054-3 [DOI] [PubMed] [Google Scholar]
- Wang Y., Pessin J. E. (2013). Mechanisms for fiber-type specificity of skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 16 243–250. 10.1097/MCO.0b013e328360272d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. X., Feige P., Brun C. E., Hekmatnejad B., Dumont N. A., Renaud J. M., et al. (2019). EGFR-aurka signaling rescues polarity and regeneration defects in dystrophin-deficient muscle stem cells by increasing asymmetric divisions. Cell Stem Cell 24 419–432. 10.1016/j.stem.2019.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B. Y., Medhurst A. D., Jackson M., Rose S., Jenner P. (2005). Proteasomal activity in brain differs between species and brain regions and changes with age. Mech. Ageing Dev. 126 760–766. 10.1016/j.mad.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Zhu K., Dunner K., Jr., McConkey D. J. (2010). Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene 29 451–462. 10.1038/onc.2009.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of proteasome-related genes in the soleus muscles of Con and mKO mice after DOX treatment. (A) Relative expression of Rpt3 mRNA in the soleus muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (n = 4 per group). (B) Relative mRNA expression of 19S proteasome genes (Psmd3, 4, 11, 13, and Rpt6) in the soleus muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: ∗∗p < 0.01; n = 4 per group). (C) Relative mRNA expression of 20S proteasome genes (Psmb2, 4, 5, and 7) in the soleus muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: ∗p < 0.05; n = 4 per group). Con indicates Rpt3f/f mice, and mKO indicates muscle-specific Rpt3-knockout mice (ACTA1-rtTA;tetO-Cre;Rpt3f/f). AU, arbitrary units.
The chymotrypsin-like proteasome activities (relative to those in Con) in the soleus, tibialis anterior, and gastrocnemius muscles after DOX treatment are shown. Data represent means ± SEM (t-test: ∗p < 0.05, versus Con; n = 3–4 per group). IU, international units.
Relative expression of MuRF1 and Atrogin1 mRNA in the soleus, tibialis anterior, and gastrocnemius muscles of Con and mKO mice after DOX treatment. The Tbp gene was used as an internal control. Data represent the means ± SEM (t-test: ∗∗∗p < 0.001; n = 3–4 per group).
Primers for quantitative RT PCR.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.