Abstract
Purpose
miRNAs have been suggested as biomarkers of embryo viability; however, findings from preliminary studies are divergent. Furthermore, the presence of other types of small RNA molecules remains to be investigated. The purpose of this study was to perform a comprehensive analysis of small non-coding RNA levels in spent and unconditioned embryo culture media, along with miRNA levels in blastocoelic fluid samples from human embryos.
Methods
miRNAs in unconditioned culture medium from 3 different manufacturers, along with miRNA from day 5 conditioned culture medium, control medium, and corresponding blastocoel fluid from 10 human blastocysts were analyzed with array-based q-PCR analysis. Subsequently, deep sequencing of total and small RNA in day 5 spent culture medium from 5 human blastocysts and corresponding controls was performed.
Results
In spite of using state-of-the-art sensitive detection methods, no miRNAs were found to be reliably present in the spent culture medium or the blastocoel fluid. Ct values were above the recommended limit for detection in the array-based analysis, a finding that was confirmed by deep sequencing. The majority of miRNAs identified by deep sequencing were expressed in all samples including control media and seem to originate from sources other than conditioned IVF media.
Conclusions
Our findings question the use of miRNAs as a reliable biomarker and highlight the need for a critical methodological approach in miRNA studies. Interestingly, tiRNA fragments appear to be overexpressed in conditioned IVF media samples and could potentially be a novel biomarker worthy of investigation.
Keywords: miRNA, tiRNA, Biomarker, Culture medium, ART
Introduction
miRNAs are small, non-coding RNA molecules that have attracted attention in several areas of medicine, including assisted reproduction. miRNAs act by binding mRNA targets to repress their expression and one miRNA can target several different mRNAs [1]. In addition to regulating intracellular gene expression, miRNAs can be packaged into exosomes and released into the extracellular environment. In there, they can be taken up by recipient cells and modulate their gene expression [2]. As such, the discovery that miRNAs are secreted by human pre-implantation embryos [3, 4] and the endometrium [5] has led to the suggestion that miRNAs could play a role in embryo-endometrium crosstalk and be active participants in the implantation process [6]. Another types of small RNA species found both inside cells and in extracellular vesicles are fragmented tRNAs (referred to as tiRNAs, tsRNAs, or tRNA halves). Fragmented tRNAs are 35 nt molecules produced by cleavage of the full length tRNAs and have been implicated in numerous processes, including translational control (PMID 26463210). Similar to miRNAs, tiRNAs could serve as potential biomarkers.
Spent culture media from in vitro culture of embryos are readily accessible for non-invasive analysis of secreted molecules that may reflect the embryo viability and implantation potential. Analysis of spent culture media as a way to guide embryo selection would represent an attractive alternative to invasive methods such as PGT-A, as well as an important complement to existing morphological embryo assessment. So far, several strategies including measurement of amino acid turnover, glucose uptake, oxygen consumption, protein composition, and mapping of the entire metabolome have been investigated with limited clinical success [7–12]. More recently, miRNA secretion was suggested as a potential marker of embryo viability [4, 13]. Because miRNAs show high extracellular stability and—in contrast to proteins and metabolites—can be amplified by the detection assay, they offer several advantages over previously investigated biomarkers. However, previous studies show that analysis of culture media for embryo selection is complicated by technical limitations for detection of small molecular changes, and by the concealment of media composition where undeclared molecules, such as proteins, are present in the unconditioned culture media [14]. So far, published studies suggest that similar to proteins, miRNAs are present in the media before embryo exposure [4, 15]. This potentially introduces an important source of error in biomarker studies. Therefore, the purpose of the present study was to perform a comprehensive analysis of soluble RNA levels in both spent and unconditioned embryo culture media, as well as miRNA in blastocoelic fluid samples.
Materials and methods
Design
The study was designed as a three separate experiments (Table 1). First, unconditioned culture media from 3 different manufactures were analyzed for miRNA expression with an array-based q-PCR analysis. Second, day 5 spent culture media, control media, and blastocoelic fluid from human blastocysts were analyzed using an identical array platform. Finally, day 5 spent media samples and corresponding unconditioned control samples were analyzed by deep sequencing for both small RNA and total RNA fractions. The study was approved by the Ethics committee in Gothenburg (Dnr: 066-15), by the Central Denmark Region Committees on Biomedical Research Ethics and the Danish Data Protection Agency.
Table 1.
Overview of study design
| Analysis | Sample collection | Samples analyzed | |
|---|---|---|---|
| (1) Unconditioned culture media | Array-q-pcr |
(1) Sydney IVF Blastocyst medium K-SIBM-20 (2) Sydney IVF Blastocyst medium K-SIBM-50 (3) G2 (protein free) Vitrolife (4) GTL Vitrolife |
n = 16 (4 from each container) |
| (2) miRNA in culture media and blastocoel | Array-q-pcr | 10 donated surplus blastocysts |
Culture media (n = 10) Blastocoel fluid (n = 7) Corresponding media controls (n = 3) MOPS media (n = 1) |
| (3) Total small RNA in culture media | Deep sequencing | 5 embryos used for cryopreservation or transfer |
Culture media (n = 5) Corresponding media controls (n = 4) PBS controls (n = 2) |
Media
For the first experiment, four samples of unconditioned culture media were drawn from each of the following four unopened containers (n = 16): Sydney IVF Blastocyst medium K-SIBM-20 lot S20319 (1–4) and K-SIBM-50 lot S20380 (5–8); G2 (protein free) Vitrolife 30 ml, ref. 10131, lot 505393 (9–12); and GTL Vitrolife 30 ml, ref. 10145, lot 022304 (10–16). Two vials of identical culture media (Sydney IVF Blastocyst medium) were included to evaluate batch to batch variation. Protein-free G2 medium was included to assess potential contribution from added proteins in the other media.
For the second experiment, analyzing spent culture media and blastocoel fluid (Table 1), we profiled miRNA from embryo culture medium and corresponding blastocoelic fluid collected from 10 donated blastocysts. For technical reasons, no blastocoel fluid was collected from two blastocysts, leaving a total of 18 samples (10 culture media, 8 blastocoelic fluids) for analysis. Corresponding blank culture media from the same lot cultured alongside the embryos and MOPS-buffered media (G-gamete, Vitrolife AB, Sweden) were included as negative controls for the spent culture medium and blastocoel fluid, respectively.
For the sequencing analysis, spent culture media was collected on day 5 from 5 additional human blastocysts. Corresponding blank culture media from the same lot cultured alongside the embryos and PBS media were included as negative controls (Table 1).
Embryo culture and collection of blastocoelic fluid
Embryos cryopreserved on day 2 of development and close to the maximum time allowed for storage in Sweden (5 years) were donated to this study by patients giving their informed consent. Embryos with a 100% cell survival were transferred into individual droplets of 25 μl of GTL (ref. 10145, Vitrolife AB, Sweden) for culture to the blastocyst stage. On day 5, blastocoelic fluid was aspirated from all good-quality blastocysts (Gardner score ≥ 3BB) [16] according to the method described by Gianaroli et al. [17]. An ICSI pipette (TPC, ref. LICR-BA21TL0.6, Australia) was used to penetrate the boundary between two trophectoderm cells and enter the blastocoelic cavity. Care was taken to avoid aspiration of any cellular material inside the cavity during collection at the same time as attempting to extract a maximum volume of fluid. The sample volume was too small to measure. The samples were immediately transferred into a 0.2-ml PCR tube (ref. 0030124332, Eppendorf, Germany) kept on ice without the addition of any buffer. Simultaneously, all available (20 μl) spent embryo culture media from these blastocysts and the corresponding control media where no embryo was present were collected from the same culture dish. All samples were immediately stored at − 80 °C until analysis.
For the sequencing analysis of spent culture media, women from the Fertility Clinic, Regional Hospital Horsens, were eligible for inclusion if single embryo transfer was planned. The women gave informed consent to RNA analysis of spent culture medium on day 5. Ovarian stimulation, oocyte retrieval, and fertilization using IVF or ICIS were performed according to standard procedures. Embryos were cultured in SAGE 1 step medium in individual wells until day 5. Control media from the same batch were placed alongside the embryo. All available spent culture medium and corresponding control media were collected individually on day 5 from all embryos that were selected for transfer or cryopreservation and stored in individually labeled vials at – 80 °C until analysis.
miRNA extraction, reverse transcription, pre-amplification, and array PCR
All steps for extraction, reverse transcription, pre-amplification, and array PCR were performed according to the manufacturer’s protocol. All amplifications were performed on the Roche LightCycler 480. The miRNeasy Serum/Plasma kit designed for purification of miRNA from small volumes was used for extraction (Qiagen, cat. no. 217184), using a minimum of 12 μl for elution. Before purification, 1 μl MS2 carrier RNA (Roche, cat. no. 10165948001) was added to each sample for maximum recovery. Adding carrier RNA improves precipitation of RNA and ensures that minimal RNA is lost during processing. Samples were spiked with 3.5 μl C. elegans miR-39 miRNA mimic (miRNeasy Serum/Plasma Spike-In Control, 1.6 × 108 copies/μl (cat. no. 219610), Qiagen). For analysis of unconditioned culture media, a maximum input of 200 μl was used. For the embryo culture media and blastocoel fluid samples, all available media (~ 20 μl) were used to ensure extraction of all available miRNA. RNAse-free water was added to a final volume of 50 μl. According to protocol, 5 μl was taken from the eluate for reverse transcription of the extracted miRNA into cDNA using the miScript II RT kit (Qiagen) in a 10 μl reaction using the 5× miScript HiSpec Buffer which selectively converts miRNA and certain small nuclear RNAs. After dilution 1:5, 5 μl cDNA was taken for a pathway-focused 96-plex pre-amplification using the miScript PreAMP PCR kit (Qiagen) (activation 95 °C for 15 min followed by 12 cycles of 94 °C for 30 s, 60 °C for 3 min for amplification) followed by the minimal possible dilution of 1:20. The pre-amplification step lowers Ct cutoff values for positive calls to 30. The cutoff Ct values are lower than those used for non-pre-amplified cDNA, where a cutoff Ct value of 35 is used. This decrease is due to the additional 12 PCR cycles performed during pre-amplification, a strategy that has been experimentally determined by the manufacturer (miScript PreAmp Handbook, Qiagen) to have no effect on sensitivity. The quality control experiments were performed using the miScript miRNA QC PCR Array (positive control CtPPC 19 ± 2. Reverse transcription control: miRTC ΔCt = AVG CtmiRTC – AVG CtPPC < 2 miRTC 17 ± 3). The pre-amplified products were profiled without further dilution using the pathway-focused miScript miRNA PCR array plate F from Human Cell Development and Differentiation (Qiagen), which detects 84 individual miRNAs. Cycling conditions were 15 min at 95 °C followed by 40 cycles of 94 C for 15 s, 55 C for 30 s, and 70 °C for 30 s.
RNA extraction, library preparation, and deep sequencing of total and small RNA
Five spent media samples, 4 corresponding controls, and 2 PBS-negative controls were purified prepared for total RNA sequencing using SMARTer® Stranded Total RNA-Seq Kit v2-Pico Input Mammalian (96 reactions, 634413, Takara) following the manufacturer’s protocol. Lysis buffer was added before purification to release RNA from exosomes. Ribosomal RNA was removed after the first round of amplification. The workflow used in this kit takes advantage of a novel technology allowing removal of ribosomal cDNA (cDNA fragments originating from rRNA molecules) after cDNA synthesis using probes specific to mammalian rRNA. The probes target ribosomal RNA and mitochondrial rRNA sequences; however, the mitochondrial R-probes are derived from the human mitochondrial genome and are therefore strictly human-specific. The rRNA depletion method used in this kit makes it especially well-suited for working with very small quantities of total RNA. Prepared libraries were sequenced as 100 bp paired end on an Illumina HiSeq 4000 sequencer.
For small RNA sequencing, libraries were prepared using the QIAseq RNA Library Prep kit (Qiagen). One sample was used for an unsuccessful analysis of miRNA with nanostring, which left 4 spent media samples, 4 corresponding controls, and 2 PBS controls for small RNA analysis. The finished libraries were quality controlled using an Agilent Bioanalyzer 2100 and quantified by use of q-PCR. Libraries were pooled and sequenced on an Illumina NextSeq500 sequencer.
Data analysis
For the q-PCR array experiments, data analysis was performed using the web-based data analysis tool GeneGlobe provided by Qiagen and the statistical package STATA for Mac, version 14.0 (StataCorp, USA). The spiked-in C. elegans miR-39 miRNA mimic was used for calibration. Ct > 30 were considered negative calls. Only Ct values < 27 were categorized as reliable detection. The threshold of Ct values < 27 is given by the manufacturer and represents the level above which any detection should be interpreted with caution.
For the sequencing analysis, raw data was filtered and trimmed using trimgalore (v 0.4.1). Tophat2 (v2.1.1) was used to map the filtered reads to human genome hg 19 guided by Gencode annotation release 29. The feature counts from the Subread package (v 1.6.1) was used to quantify the number of reads mapping to each gene in Gencode annotation release 29, containing both mRNA and various lncRNA genes. Filtered reads were first mapped to human tiRNA sequences. If mapping was not possible, the reads were mapped to human miRNA sequences. Reads not mapping to miRNAs were mapped to other relevant transcriptomes. Remaining reads still not mapping were mapped to the human genome (hg19). This was done to assay which RNA species were sequenced in the study. The miRNA mapping reads were deduplicated, meaning identical reads with identical unique molecular identifiers were collapsed to a single read. This is done to remove PCR duplicates and has an enormous impact on very low input samples as PCR bias increases with low input given the extra PCR cycles needed to attain viable sequencing libraries. The deduplicated miRNA read counts were subjected to differential expression analysis using DESeq2 in R.
Results
Unconditioned media
Samples from unconditioned media were analyzed by RT-q-PCR. The resulting CtPPC value was 19 ± 2 for all 16 samples as required, confirming that the RNA was of sufficiently high quality, the cycling program was run correctly, and the thresholds were correctly defined (miRTC ΔCt (AVG CtmiRTC – AVG CtPPC) < 2) for all samples except samples 1 (2.44) and 2 (2.41), which were excluded, confirming that there was no inhibition of the reverse transcription reaction in the remaining samples. The spike-in (C. elegans miR-39 miRNA mimic) was detected equally in all samples.
Four miRNAs (hsa-miR-92a-3p, hsa-miR-124-3p, hsa-miR-142-5p, and hsa-miR-375) out of the 84 candidates tested in the array were detected consistently in all 14 samples. The Ct values are listed in Table 2. Notably, the 4 miRNAs listed were detected in all samples in all 3 types of media with no clear difference in Ct values. The 6 endogenous reference genes (SNORD61, SNORD68, SNORD72, SNORD95, SNORD96A, and RNU6-6P) were not consistently detected. However, SNORD68 was detected in 3 out of 14 samples (1 sample G2, 2 samples GTL) while SNORD61 and SNORD95 were detected in 1 sample (GTL). miRNA profiling by RT-q-PCR was also performed in spent culture media (n = 10) and corresponding blastocoel fluid (n = 8) from 10 blastocysts. All samples passed the QC step (CtPPC 19 ± 2 and miRTC ΔCt (AVG CtmiRTC – AVG CtPPC) < 2), except for one blastocoel fluid sample (that was subsequently excluded). The spike-in (C. elegans miR-39 miRNA mimic) was detected equally in all samples. Apart from hsa-miR-124-3p, no miRNA was consistently present in all samples. Similar to the unconditioned media, hsa-miR-124-3p was detected in all samples including the 3 control media samples (Table 3). Surprisingly, hsa-miR-92a-3p was detected in all control media samples, but only in 3 out of 10 spent media samples. The hsa-miR-142-5p and hsa-miR-375, which were consistently detected in the unconditioned media, were not present in the control or in the spent media samples. In contrast, hsa-miR-196a-5p and hsa-miR-214-3p, which were not detected in the unconditioned media, were present in all 3 control media samples and in 7 out of 10 and 6 out of 10 spent media samples, respectively. Similar to what was seen in the media analysis, hsa-miR-124-3p was detected in all blastocoel fluid samples including the control. Furthermore, hsa-miR-214-3p and hsa-miR-196a-5p were detected in the control samples and in 6 out of 7 and all 7 blastocoel fluid samples, respectively, and hsa-miR-92a-3p, which was detected consistently in the unconditioned media, was detected in 5 out of 7 blastocoel fluid samples and in the control sample. None of the other miRNAs was detected in more than one sample. Only hsa-miR-124-3p, showed Ct values < 27 and was therefore categorized as being reliably detected (Table 4). Of note, hsa-miR-124-3p was found consistently in unconditioned media, blastocoel fluid samples, spent media samples, and controls and thus likely represents a false positive call.
Table 2.
Absolute Ct values for miRNAs detected in unconditioned media
| miRNA | Sydney IVF Blastocyst medium (lot S20319) (n = 2) | Sydney IVF Blastocyst medium (lot S20380) (n = 4) | G2 (protein-free) (n = 4) | G-TL (n = 4) |
|---|---|---|---|---|
| hsa-miR-92a-3p | 23.0 ± 0.04 (23.0; 23.02) | 22.7 ± 0.18 (22.5; 22.9) | 23.5 ± 53 (23.0; 24;2) | 24.0 ± 0.59 (24.7; 23;5) |
| hsa-miR-124-3p | 28.1 ± 0.33 (27.9; 28.3) | 28.1 ± 0.58 (27.5; 28.9) | 28.4 ± 0.29 (28.1; 28.7) | 28.1 ± 0.34 (27.9; 28.6) |
| hsa-miR-142-5p | 28.9 ± 0.23 (28.7; 29.0) | 28.5 ± 1.7 (26.2; 29.8) | 28.5 ± 0.61 (27.9; 29.1) | 28.0 ± 1.2 (26.9; 29.6) |
| hsa-miR-375 | 28,4 ± 0.93 (27.8; 29.1) | 27,7 ± 0.57 (27.1;28.2) | 27,2 ± 0.51 (26.7; 27.9) | 27,6 ± 0.18 (27.3; 27.7) |
Ct values are listed as mean ± SD (range). Ct values were calibrated to the spiked-in miR-39 miRNA mimic
Table 3.
miRNAs detected (Ct < 30) displayed as number of samples, where the miRNA was detected
| miRNA/SnoRNA | Control media (n = 3) | Control MOPS-buffered media (n = 1) | Spent media (n = 10) | Blastocoel fluid (n = 7) |
|---|---|---|---|---|
| hsa-miR-92a-3p | 3 | 1 | 3 | 5 |
| hsa-miR-124-3p | 3 | 1 | 10 | 7 |
| hsa-miR-196a-5p | 3 | 1 | 7 | 7 |
| hsa-miR-214-3p | 3 | 1 | 6 | 6 |
| hsa-miR-22-3p | 0 | 1 | 3 | 0 |
| hsa-miR-142-5p | 0 | 0 | 0 | 0 |
| hsa-miR-375 | 0 | 1 | 0 | 1 |
| hsa-miR-10a-5p | 0 | 0 | 0 | 1 |
| hsa-miR-100-5p | 0 | 0 | 0 | 1 |
| hsa-miR-137 | 0 | 0 | 1 | 0 |
| hsa-miR-15a-5p | 0 | 1 | 0 | 1 |
| hsa-miR-99a-5p | 0 | 0 | 0 | 1 |
| hsa-miR-23b-3p | 0 | 0 | 1 | 0 |
| hsa-let-7c-5p | 0 | 0 | 0 | 1 |
| hsa-let-7a-5p | 0 | 0 | 0 | 1 |
| hsa-let-7b-5p | 1 | 0 | 0 | 0 |
| SNORD68 | 0 | 1 | 0 | 0 |
Control media were from the same batch as the culture media and cultured alongside the embryos. Several embryos could share a control if from the same batch and cultured the same day. MOPS-buffered media was taken directly from the bottle
Table 4.
Average Ct values for miRNAs that were detected in > 3 samples (MOPS-buffered media not included as n = 1)
| miRNA | Control media | Spent media | Blastocoel fluid |
|---|---|---|---|
| hsa-miR-92a-3p | 29.1 ± 0.23 | 29.2 ± 0.61 | 29.9 ± 1.77 |
| hsa-miR-124-3p | 25.9 ± 0.28 | 27.0 ± 1.1 | 24.5 ± 0.60 |
| hsa-miR-196a-5p | 27.3 ± 0.57 | 28.1 ± 1.4 | 28.4 ± 1.1 |
| hsa-miR-214-3p | 28.1 ± 0.89 | 28.2 ± 1.3 | 27.7 ± 0.80 |
Deep sequencing of spent culture media
To allow a more thorough and sensitive analysis of the small RNA content in media and blastocoel fluid, we conducted next-generation sequencing of the different samples, using libraries for both total RNA and small RNA. The number of filtered reads for total and small RNA is shown in Table 5. Gene annotation for total RNA mapped only 3% of the reads to gene regions in spent culture media and control and only 2% in PBS samples mapped to gene regions, which does not allow for differential gene expression analysis. The majority of reads generated in the total RNA-seq were simple repeats.
Table 5.
Filtered reads/sample of total RNA-seq and small RNA-seq
| Sample | Total RNA-seq total reads (mio) |
| Conditioned media | |
| 1 | 54.5 |
| 2 | 52.8 |
| 3 | 43.8 |
| 4 | 86.6 |
| 5 | 50.0 |
| Unconditioned media | |
| 1 | 55.6 |
| 2 | 58.1 |
| 3 | 53.3 |
| 4 | 44.6 |
| PBS | |
| 1 | 35.9 |
| 2 | 62.7 |
| Sample | Small RNA-seq total reads (mio) |
| Conditioned media | |
| 1 | 35.0 |
| 2 | 42.8 |
| 3 | 34.3 |
| 4 | 43.5 |
| Unconditioned media | |
| 1 | 38.2 |
| 2 | 38.2 |
| 3 | 37.6 |
| PBS | |
| 1 | 35.6 |
| 2 | 37.3 |
For the small RNA analysis, human annotated miRNAs constituted from 0.1 to 1.4% of the mapped reads and 40% of these reads mapped to the human genome (Table 6). Most RNA species were present at comparable levels in all samples with a possible exception for tiRNA fragments, which were generally more abundant in conditioned media samples (Fig. 1). No miRNAs had adjusted p values below 0.05 when comparing conditioned to unconditioned media. In contrast, tiRNAs derived from 5 different mature tiRNA were differentially expressed when comparing conditioned to unconditioned media (adjusted p values < 0.05) (Figs. 1 and 2). There was no difference in tiRNA fragment levels between PBS controls and unconditioned media.
Table 6.
small RNA mapping
| Samples | Human miRNAs | Other miRNAs | tRNA | Pre-tRNA | Other small RNAs | piRNA | rRNA | mRNA | Human genome |
|---|---|---|---|---|---|---|---|---|---|
| Conditioned media | |||||||||
| 1 | 0.5% | 0.1% | 5.7% | 0.1% | 0.3% | 29.0% | 0.7% | 6.1% | 37.6% |
| 2 | 0.2% | 0.0% | 1.2% | 0.0% | 0.2% | 30.2% | 22.4% | 6.6% | 39.2% |
| 3 | 0.6% | 0.0% | 1.4% | 0.0% | 0.2% | 31.6% | 17.7% | 7.1% | 41.3% |
| 4 | 1.0% | 0.3% | 4.5% | 0.1% | 0.4% | 30.3% | 22.3% | 6.2% | 34.9% |
| Control media | |||||||||
| 1 | 0.5% | 0.1% | 0.4% | 0.0% | 0.2% | 29.9% | 21.6% | 6.5% | 40.8% |
| 2 | 0.2% | 0.3% | 0.6% | 0.0% | 0.2% | 30.7% | 20.7% | 7.0% | 40.3% |
| 3 | 1.4% | 0.1% | 2.2% | 0.1% | 0.3% | 28.5% | 21.2% | 6.7% | 39.4% |
| PBS | 0.0% | 0.5% | 0.0% | 0.1% | 28.3% | 22.3% | 7.5% | 41.1% | |
| 1 | 0.9% | 0.0% | 0.6% | 0.1% | 0.3% | 31.6% | 16.7% | 7.2% | 42.6% |
| 2 | 0.1% | 0.0% | 0.5% | 0.0% | 0.1% | 28.3% | 22.3% | 7.5% | 41.1% |
Fig. 1.
Volcano plot of conditioned vs unconditioned media. Plotted are –log10 (p values) vs log2(fold change). Red means < 0.05 p value. Vertical lines indicate ± 1 log2 (fold change). The gene names for the 10 most significant tRNAs are indicated
Fig. 2.
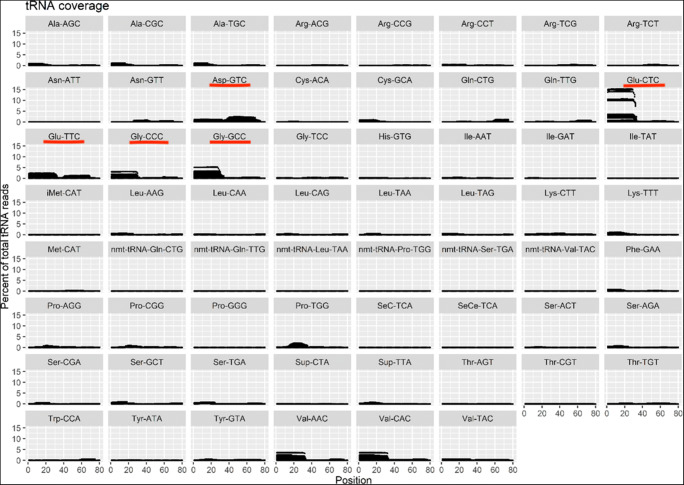
tiRNA map. The tiRNAs found to be significantly upregulated in conditioned media samples are underlined in red
Discussion
miRNA profiling of spent culture media has been suggested as a novel strategy for assessing embryo viability. In the present study, we have performed a basic methodological background analysis of the presence of miRNAs in unconditioned culture media from different manufacturers and batches and compared the results to spent embryo culture media. In contrast to previously published studies, we found no miRNAs that were reliably present in the unconditioned or spent culture medium, as Ct values were above the recommended limit for detection or the miRNA was detected in all samples including negative controls, thus representing false positives. A subsequent sequencing of conditioned and unconditioned culture media confirmed this interpretation. Our findings suggest that if miRNAs are present in the culture medium, the detected levels can only be counted as a few molecules with a concomitant high risk of random variation, which represents a plausible explanation of the high degree of discrepancy in results between studies and challenges in the use of miRNA as a useful clinical biomarker.
Recent studies have reported that miRNAs are present in embryo culture media and suggested specific miRNAs to be associated with implantation [4, 13]. Concomitantly, the presence of several miRNAs has been reported in culture media unexposed to embryos [4, 15], which presents an important source of error with potential biological implications for the developing embryo. However, there is very little agreement between different studies about the specific miRNAs being present in unconditioned culture media. This inconsistency may in principle be explained by true differences between different culture media used in the studies, or by different approaches for sample handling and data analysis or merely represent random findings. Two highly plausible explanations for the discrepancies between our findings and previous studies using array q-PCR are the choice of Ct cutoff values after pre-amplification and the data normalization strategy.
A fundamental, inherent limitation of all culture media studies is that the starting material is limited. For detection of DNA or RNA, this is often overcome by introducing a pre-amplification step. Targeted pre-amplification substantially increases the amount of genetic material of interest and is therefore highly useful for samples with a low amount of RNA as the sensitivity of gene detection is substantially lowered without introducing bias in the gene expression analysis [18]. Pre-amplification typically results in a 1000–4000-fold amplification which means that miRNA with considerably lower Ct values is classified as being detected compared with non-pre-amplified material. This in turn implicates that the number of miRNA molecules that give rise to a specific Ct value is fewer compared with non-amplified material. Detection of a limited number of miRNA molecules will invariably increase the stochastic noise in the system. As a result, lower cutoff Ct values for detection must be applied for pre-amplified compared with non-amplified material to avoid non-reproducible and false positive calls. When applying a pre-amplification step, it is crucial to adjust cutoff Ct values according to the manufacturer’s instructions. In general, high Ct values after pre-amplification may represent as little as one molecule of RNA. High Ct values must therefore be interpreted with caution and subsequently validated. Studies reporting the presence of miRNA in spent culture media have used Ct values of 38 [4] and 35 [13] as cutoff values after pre-amplification, i.e., much higher Ct values than the present study. Although Capalbo et al. subsequently tested the presence of 7 miRNA expressed in the spent media of 5 samples with RT-q-PCR, the Ct values were generally high (> 30) (for reference, see supplemental Table 5 in Capalbo et al. 2016). Five of those 7 miRNAs were covered by our assay (including miR-20a and miR-30, which were reported to be differentially expressed in implanted and non-implanted embryos), but none of them was detected above background levels in our study. In the study of Rosenbluth et al., only 2 out of 10 miRNAs detected were confirmed by single-assay PCR to be present solely in the spent media samples; however, none of these (miR-372 and miR-191) was covered by our assay. Battaglia et al. (2019) reported the presence of 89 out of 384 tested miRNAs in blastocoel fluid from 9 blastocysts [19]. The Ct cutoff value was 35 after pre-amplification, thus introducing a high risk of false positive calls, but no absolute Ct values were reported making a direct comparison difficult. Validation was performed using digital droplet PCR of 3 miRNAs.
The present study used a recommended Ct cutoff value of 30 for positive calls in the array analysis, which might explain the disagreement with previous studies. Apart from one miRNA (hsa-miR-124-3p), all Ct values were above 27, which is the limit above which any detection should be interpreted with caution according to the manufacturer’s recommendation. Of note, this particular miRNA was detected consistently across all samples and likely represents a false positive call. We therefore conclude that the presence of miRNAs in the culture media is uncertain. The negligible amount of miRNA was confirmed by deep sequencing, further strengthening the interpretation of minute amounts. Our findings suggest that if any miRNAs are present, they are counted only as a few molecules with a concomitant high risk of random variation, which represents a plausible explanation of the high degree of discrepancy in results between studies, and severely limits the use as a biomarker.
Sanchez-Ribas et al. used next-generation sequencing to detect several miRNAs in both spent and blank culture media, and found that only two miRNAs (miR-181b-5p and miR-191-5p) showed sufficiently high expression to candidate as biomarkers. The expression levels did, however, not differ between spent and control media, limiting the potential use as a biomarker. This is in line with our findings. Of note, media sampling was performed on day 3, where the number of embryo cells is substantially lower than on day 5/6 and before the embryonic genetic activation [20]. It has previously been suggested that miRNAs could be detected only after blastulation [13].
Therefore, it could be argued that miRNAs would have been detected if the embryos had been cultured until day 5/6. However, in the present study, where embryos were cultured until after blastulation, no miRNAs were reliably detected,
Another important methodological consideration with tremendous impact on the interpretation of data is the choice of strategy for data normalization and analysis. The aim of normalization is to even out differences due to sampling and quality of the samples. In most studies, the Ct values of target miRNA are normalized to one or more stably expressed endogenous miRNA (reference/housekeeping gene) from the same sample (ΔCt). Any variation in expression of the reference RNA will obscure real changes and produce artifactual changes. A prerequisite for a meaningful data interpretation requires selection of reference genes that are stably expressed with little variation, which is not always achievable [21]. An alternative approach is a strategy that normalizes miRNA expression to the mean expression value of all miRNAs in the samples, called global normalization. An adequate application of this strategy performs better than reference genes, in particular if the preferred reference gene is variably expressed [22]. However, the global normalization strategy is valid only if a large number of genes are expressed and analyzed. Our study showed that the endogenous reference genes included in the standard q-PCR assay, including snRNA U6, which was also used as an endogenous reference gene in previous studies [4], were undetectable. This is in line with the study from Capalbo et al., who reported a non-consistent detection of the putative endogenous reference genes in spent culture media. As a consequence, the authors adapted a global normalization strategy [13]. However, as only 16% of the miRNAs tested were detected in the medium with a mean Ct value of 29.9 ± 2.7, i.e., close to the cutoff for detection, this strategy is questionable. Similarly, Battaglia et al. used global normalization, although only 89/384 (23%) miRNAs were detected by the assay using a high cutoff. In our study, we found no miRNAs that were reliably present in the blastocoelic fluid or the spent culture medium using the recommended Ct value as cutoff for detection for the specific assay and a global normalization was therefore not possible.
In summary, our study confirms existing knowledge that high Ct values after pre-amplification may either be false positive calls or represent very few molecules with a concomitant high risk of random variation. Furthermore, it is known that inappropriate choice and validation of normalizer will obscure real changes and produce artefactual changes. We consider these two important methodological steps a potential explanation for discrepancies between studies.
Whenever a negative result is reported, the question arises, whether the methodology used was insufficient. A limitation to the use of microarray plates is that only miRNA with known sequences included in the plates are detected. The array plates used in the present study allowed for detection of 84 individual miRNAs on a pre-manufactured plate covering human cell development. However, array plates also require larger input material than individual miRNA assays, which implies that if more miRNAs are included on the plate, the more input material is needed. In these types of studies, the starting material is inherently low. We therefore chose a conservative approach using plate with fewer sequences compared with other available options. Pre-processing of samples is an additional important methodological consideration and a possible explanation for negative results. Several other studies, where miRNA has been detected, such as Battaglia et al., have in contrast to our study left out the RNA purification step, thus minimizing loss of material. In the present study, MS2 carrier RNA was added to each sample before purification prior to array q-PCR to maximize recovery. While we can never definitively rule out that miRNAs could have been lost during processing of the samples, the internal controls including the analysis of the spiked-in C, 39 mimic suggested that miRNA was recovered, RT-PCR was performed with no inhibition, the cycling program correctly run, and the thresholds correctly defined. We therefore consider our results reliable. As noted, previous studies have used higher Ct threshold values for detection which complicates direct comparisons. However, the subsequent deep sequencing of an additional set of culture media, where no miRNAs were detected, supports our interpretation.
The fact that we analyzed single embryo samples rather than pooled samples represents a potential explanation for the lack of detected miRNA. The study design was, however, chosen to evaluate the potential use of miRNA as a reliable biomarker for individual embryo viability rather than identifying potential biological roles of miRNA in signaling pathways.
Although the number of embryos tested is small, the strength lies in the stringency of the methodology, which includes testing of blank culture media from different manufacturers, corresponding controls for each sample, and the use of two different platforms (array and NGS), with identical results.
While miRNAs were either unchanged or undetected in the deep sequencing data, we found that 7 different tiRNA fragments were more abundant in media that had been used to culture embryos compared with the unconditioned media (adjusted p values < 0.05). This could reflect a stress response in the embryos, as stress has been linked to the formation of tiRNAs. Previous work has suggested that tiRNA fragments are secreted as non-vesicle-bound RNPs, rather than packaged in exosomes, but this RNP form appears sufficiently stable to allow the detection of tiRNA fragments in conditioned cell media [23]. To our knowledge, the present study provides the first indication that cultured embryos can release tiRNAs and future work will be required to test if these RNAs play a functional role in development or can serve as biomarkers for embryo status.
In conclusion, the present study found no miRNAs that were reliably present in the unconditioned or the spent culture medium by using array q-PCR or deep sequencing. It seems reasonable to conclude that if miRNAs are indeed present in the culture medium, the amount is minute. Although this does not exclude a biological role for miRNAs in the embryo-endometrium crosstalk, it questions the use of miRNAs as a biomarker in the near future and highlights the need for a critical methodological approach in biomarker studies.
Acknowledgments
Anne Færch Nielsen is thanked for critical reading of the manuscript. Anette Gabrielsen is thanked for assistance in collecting spent media at Regional Hospital Horsens.
Authors’ contribution
KK, KL, and AA designed the study. KL and AA designed and acquired samples for the spent culture media analysis. AA performed the experiments on surplus embryos embryos. UBK took part in study design and data acquisition at the region Hospital Horsens. YY designed and performed the deep sequencing experiments. JK designed the deep sequencing experiments and performed the interpretation of the sequencing analysis. BS contributed substantially to the acquisition of data and interpretation of the miRNA analysis. JI, TH and CH contributed substantially to the interpretation of data. KK interpreted the miRNA data and wrote the first draft. All authors critically reviewed and approved the final version of the manuscript.
Funding information
This work was supported by unrestricted grants from Vitrolife (Grant for the development of ART), the Hjalmar Svensson Foundation, VP legacy on recommendation from Novo Nordisk, and the Danish Council for Independent Research Medical Sciences.
Data availability
All data will be made accessible on request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The study was approved by the Ethics committee in Gothenburg (Dnr: 066-15), by the Central Denmark Region Committees on Biomedical Research Ethics and the Danish Data Protection Agency.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 3.Rosenbluth EM, et al. MicroRNA expression in the human blastocyst. Fertil Steril. 2013;99(3):855–861.e3. doi: 10.1016/j.fertnstert.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbluth EM, et al. Human embryos secrete microRNAs into culture media--a potential biomarker for implantation. Fertil Steril. 2014;101(5):1493–1500. doi: 10.1016/j.fertnstert.2014.01.058. [DOI] [PubMed] [Google Scholar]
- 5.Ng YH, et al. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS One. 2013;8(3):e58502. doi: 10.1371/journal.pone.0058502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galliano D, Pellicer A. MicroRNA and implantation. Fertil Steril. 2014;101(6):1531–1544. doi: 10.1016/j.fertnstert.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Katz-Jaffe MG, Gardner DK, Schoolcraft WB. Proteomic analysis of individual human embryos to identify novel biomarkers of development and viability. Fertil Steril. 2006;85(1):101–107. doi: 10.1016/j.fertnstert.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Hardarson T, et al. Non-invasive metabolomic profiling of day 2 and 5 embryo culture medium: a prospective randomized trial. Hum Reprod. 2012;27(1):89–96. doi: 10.1093/humrep/der373. [DOI] [PubMed] [Google Scholar]
- 9.Gardner DK, et al. Glucose consumption of single post-compaction human embryos is predictive of embryo sex and live birth outcome. Hum Reprod. 2011;26(8):1981–1986. doi: 10.1093/humrep/der143. [DOI] [PubMed] [Google Scholar]
- 10.Brison DR, et al. Identification of viable embryos in IVF by non-invasive measurement of amino acid turnover. Hum Reprod. 2004;19(10):2319–2324. doi: 10.1093/humrep/deh409. [DOI] [PubMed] [Google Scholar]
- 11.Tejera A, et al. Time-dependent O2 consumption patterns determined optimal time ranges for selecting viable human embryos. Fertil Steril. 2012;98(4):849–57 e1–3. doi: 10.1016/j.fertnstert.2012.06.040. [DOI] [PubMed] [Google Scholar]
- 12.Kirkegaard K, et al. Nuclear magnetic resonance metabolomic profiling of day 3 and 5 embryo culture medium does not predict pregnancy outcome in good prognosis patients: a prospective cohort study on single transferred embryos. Hum Reprod. 2014;29(11):2413–2420. doi: 10.1093/humrep/deu236. [DOI] [PubMed] [Google Scholar]
- 13.Capalbo A, et al. MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil Steril. 2016;105(1):225–35.e1–3. doi: 10.1016/j.fertnstert.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Dyrlund TF, et al. Unconditioned commercial embryo culture media contain a large variety of non-declared proteins: a comprehensive proteomics analysis. Hum Reprod. 2014;29(11):2421–2430. doi: 10.1093/humrep/deu220. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Ribas I, et al. NGS analysis of human embryo culture media reveals miRNAs of extra embryonic origin. Reprod Sci. 2018;1933719118766252. 10.1177/1933719118766252. [DOI] [PubMed]
- 16.Gardner DK, Schoolcraft W. In virto culture of human blastocyst. In: Janson R, Mortimer D, editors. Towards reproductive certainty: infertility and genetics beyond. Carnforth: Parthenon Press; 1999. pp. 378–388. [Google Scholar]
- 17.Gianaroli L, et al. Blastocentesis: a source of DNA for preimplantation genetic testing. Results from a pilot study. Fertil Steril. 2014;102(6):1692–1699.e6. doi: 10.1016/j.fertnstert.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 18.Zeka F, et al. Straightforward and sensitive RT-qPCR based gene expression analysis of FFPE samples. Sci Rep. 2016;6:21418. doi: 10.1038/srep21418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battaglia R, et al. Identification of extracellular vesicles and characterization of miRNA expression profiles in human blastocoel fluid. Sci Rep. 2019;9(1):84. doi: 10.1038/s41598-018-36452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, Pera RAR. The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet. 2004;13(14):1461–1470. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- 21.Schwarzenbach H, et al. Data normalization strategies for MicroRNA quantification. Clin Chem. 2015;61(11):1333–1342. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mestdagh P, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyuris A, et al. Physical and molecular landscapes of mouse glioma extracellular vesicles define heterogeneity. Cell Rep. 2019;27(13):3972–3987.e6. doi: 10.1016/j.celrep.2019.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data will be made accessible on request.