Abstract
Objective
To investigate the correlation between endometriosis and mitochondrial DNA (mtDNA) D-loop single nucleotide polymorphisms (SNPs) and haplotype, as well as the predictive power of certain SNPs in reproductive outcomes in a Chinese Han population.
Methods
A case-control study was conducted in which 125 endometriosis patients and 124 controls were recruited from an academic fertility center. The entire 1124-bp D-loop region of mtDNA of whole blood samples from all subjects was amplified, sequenced, and compared with the revised Cambridge Reference Sequence (rCRS) to identify SNPs and haplotypes. The association between D-loop SNPs and embryo quality and clinical outcome following in vitro fertilization (IVF) was also assessed.
Results
A total of 321 polymorphisms were identified by sequencing, allowing comparison of the D-loop between endometriosis patients and controls. The frequency of the AC523–524 del, T16172C, and C16290T variants were significantly higher, while the frequency of polymorphisms T195C, 573XCins, 16036Gins, 16049Gins, T16140C, A16183C, T16189C, and 16193Cins were lower, in the endometriosis group compared with the control group (p < 0.05). Within the endometriosis group, the high-quality blastocyst rate in the 16,290T subgroup was significantly lower than that in the 16290C subgroup (p < 0.05). In the control group, 16519C carriers showed a lower rate of high-quality blastocyst development compared with 16519T (p < 0.05). In endometriosis patients clinical pregnancy rate was significantly lower in the 150T subgroup compared with the 150C subgroup (p < 0.05).
Discussion
Data confirms a correlation between D-loop polymorphisms and endometriosis. The polymorphisms AC523–524 del, T16172C, and C16290T are associated with increased risk of endometriosis, while T195C, 573XCins, 16036Gins, 16049Gins, T16140C, A16183C, T16189C, and 16193Cins are associated with decreased risk of endometriosis. In addition, C16290T and T16519C can be associated with poor quality blastocyst development in population with and without endometriosis, respectively and C150T can be a predictor of poor IVF outcome.
Electronic supplementary material
The online version of this article (10.1007/s10815-020-01853-z) contains supplementary material, which is available to authorized users.
Keywords: Endometriosis, Mitochondria, D-LOOP, Polymorphism, Haplotype
Introduction
Endometriosis is defined as the presence of endometrial tissue (either glands or stroma) outside the uterine cavity. It can invade a range of organs, among which the ovary and the uterosacral ligament are the most frequently involved. The incidence of this benign disorder is 5% in child-bearing women, and up to 50% in infertile women [1]. Many women suffering from endometriosis-related infertility choose to undergo in vitro fertilization (IVF). Dysmenorrhea, pelvic pain, menstrual disorders, infertility, and dyspareunia are common symptoms of endometriosis [2]. While there has been extensive research to investigate the mechanisms of the disease, the etiology and pathogenesis remain elusive [3, 4]. Four classical hypotheses exist: retrograde menstruation, endometrial stem cell implantation, Mullerian remnant abnormalities, and coelomic metaplasia. However, none can fully explain the pathogenesis and development of endometriosis [5].
A general consensus has formed that endometriosis is a polygenic and multifactorial disorder with familial aggregation [6, 7], and so recent studies have shifted attention to the genetic nature of the disease. Mutations in genes linked to inflammation, growth, and hormone synthesis have been implicated. Specific SNPs in both intronic and intergenic regions were linked to the development of endometriosis with different degrees of severity [8, 9]. Other studies have found that in patients with endometriosis, the menstrual reflux contains considerably more pro-inflammatory factors and inflammatory cells than controls, which can create adhesion sites and promote the attachment of endometrial cells to pelvic tissue [5]. Inflammation has been linked to the generation of reactive oxygen species (ROS) and oxidative damage, which arise due to mutations in mitochondrial DNA (mtDNA) [3]. Therefore, in addition to those in the nuclear genome, polymorphisms of mtDNA may also contribute considerably to the generation and development of endometriosis.
Mitochondria are semi-autonomous organelles playing an important role in ATP synthesis to provide energy for cells, regulation of cell differentiation, apoptosis, proliferation and metabolism [10]. Mitochondria primarily produce ATP through oxidative phosphorylation (OXPHOS), a process which includes an electron transport chain (ETC) within the mitochondrial matrix, wherein electrons interact with oxygen to form water. Protons are expelled across the mitochondrial inner membrane, establishing a proton motive force that allows protons to flow back into the matrix, allowing generation of ATP [11]. Although this process is efficient, a small number of electrons can escape the ETC and reduce oxygen to ROS such as superoxide, which can contribute to oxidative stress. Due to the lack of protective histones and DNA repair mechanism, mtDNA is more sensitive to ROS-induced damage than nuclear DNA.
The structure and composition of mtDNA is well documented: it is a closed circular molecule containing 37 genes responsible for encoding 13 proteins, 22 tRNAs, and 2 rRNAs [12, 13]. The only non-coding sequence is the displacement loop (D-loop) control region, which is rich in A–T bases and a hot spot for mutation. The start sites of heavy and light chain replication of mtDNA, as well as major promoters of transcription, are located in this region. It is therefore vital to the maintenance and function of the mitochondrial network. Studies have shown that SNPs in the mtDNA D-loop region can be of relevance to many gynecological diseases, but its association with endometriosis remains elusive [14–19].
Given the hypothesis that mtDNA polymorphisms increase the level of inflammation and so induce ROS, which have been implicated in the development of endometriosis, we investigated the association between endometriosis and D-loop polymorphisms, as well as with mitochondrial haplotypes. In addition, we evaluated the relationship between polymorphisms and embryo development and clinical pregnancy rate in endometriosis patients undergoing IVF.
Materials and methods
Patients
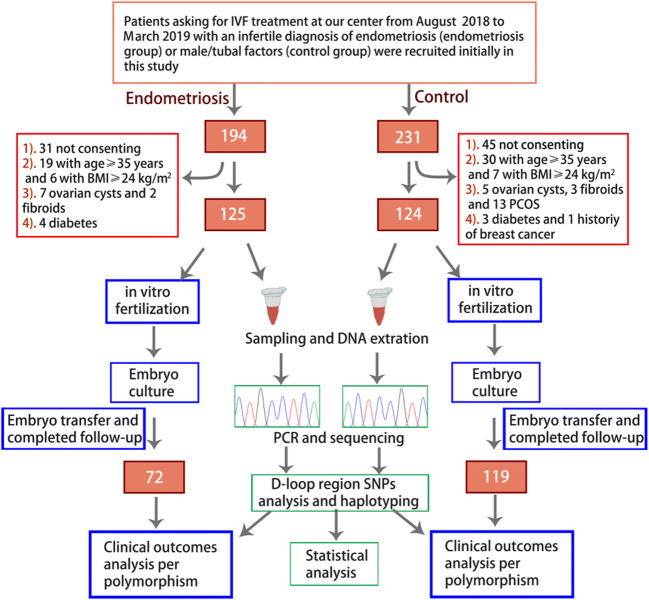
Cohorts of endometriosis patients and controls were recruited from the Reproductive Center of The First Affiliated Hospital of Anhui Medical University between August 2018 and March 2019. There were 194 and 231 subjects in the initial endometriosis and control group respectively. All endometriosis patients, encompassing mild- to severe-stage endometriosis, were surgically confirmed endometriosis cases by a prior laparoscopy. Women older than 35 or with other diseases of the reproductive system, such as polycystic ovarian syndrome, ovarian cysts, fibroids, or with disorders with a possible link to mtDNA polymorphisms, were excluded from the study. The control group was composed of females under 35 years old with infertility caused by male or tubal factors, with no evidence of endometriosis on laparoscopy. Individual matching of cohorts was performed with strict control for female age, body mass index (BMI), family economic situation, education level, and infertility duration in both groups. Routine physical examination was performed on all subjects. Disease history and living habits were investigated in detail to ensure confounders between the two groups had been eliminated and the data are comparable. Finally, a total of 125 and 124 patients in endometriosis and control group were included respectively. The process of clinical data collection and study design is shown in Fig. 1. All patients participating in this study signed informed consent and were approved by the Biomedical Ethics Board of Anhui Medical University (20180243).
Fig. 1.
Flowchart of patient gathering and study design
Genomic DNA extraction
Approximately 5 ml of peripheral blood was collected from subjects and stored at − 80 °C. Genomic DNA was extracted from the whole blood with the use of Magnetic Universal Genomic DNA Kit (Tiangen Biotech (Beijing) Co., Ltd). Absorbance of samples at 260/280 was measured with a Thermo Ultraviolet Spectrophotometer (NanoDrop 2000C), ensuring the ratio was between 1.8 and 2.0 in order to confirm a high purity of DNA samples. Samples were stored at − 20 °C.
Conditions for polymerase chain reaction and sequencing
The entire 1124 bp D-loop region of mtDNA was amplified by polymerase chain reaction (PCR) with one pair of primers: L15975F: CTCCACCATTAGCACCCAAAGC, H794R: AGGCTAAGCGTTTTGAGCTG. The 20 μl PCR reaction with the use of TAKARA LA Taq kit (Clontech), contained 10 μl mix, 2 μl primers, 7 μl ddH2O, and 1 μl DNA. PCR amplification conditions were as follows: 1 cycle at 95 °C for 3 min, followed by 30 cycles at 94 °C for 30 s, 54 °C for 30 s, 72 °C for 30 s, followed by 1 cycle at 72 °C for 10 min. Amplified products were detected by agarose gel electrophoresis and sent to Jiangsu Hongxun company for two-way Sanger sequencing using ABI 3730 sequencer. Sequencing results were spliced via SeqMan.
Mutational analysis and haplotyping of the D-loop region
Haplogrep 2 (https://haplogrep.i-med.ac.at/) was used to identify SNPs in sequencing results and served as a reference for haplotype classification. Two analysts were responsible for independently reading and interpreting the results.
Clinical outcomes analysis
We defined an embryo on day 5 or 6 with a grade of 4BB or above as a high-quality blastocyst according to the Gardner grading method [20]. Vaginal ultrasonography showing the pregnancy sac 30 days after embryo transfer was considered as clinical pregnancy. These two indicators were evaluated for each SNP individually in order to assess the predictive power of D-loop SNPs on embryo quality and clinical pregnancy rate in subjects.
Statistical analysis
Statistical analysis was performed using SPSS software version 23.0. Continuous variables were compared by Student’s t test and expressed as mean ± SD, or the Mann-Whitney U test and expressed as median and interquartile range. Categorical variables such as SNPs and endometriosis were evaluated by chi-square test (χ2) or Fisher’s test. Where possible, we performed logistic regression analysis to analyze the association between SNPs and endometriosis and expressed results as odds ratio (OR) and 95% confidence interval (CI). p < 0.05 was considered statistically significant.
Results
A total of 125 endometriosis and 124 control subjects were assessed in our study. Mean age ± SD was 29.22 ± 2.94 years in the study group and 29.47 ± 1.69 years in the control group, and BMI was 21.66 ± 2.78 kg/m2 in the study group compared with 21.65 ± 3.14 kg/m2 in the control group. Differences in baseline age (p = 0.424) and BMI (p = 0.987) were not significant, nor were other clinical characteristics of patients between the two groups except for the dosage of gonadotrophin (p = 0.035) (Table 1).
Table 1.
Baseline characteristics of subjects in the two groups
| Endometriosis (n = 125) | Controls (n = 124) | t/Z value | p value | |
|---|---|---|---|---|
| Age (years) | 29.22 ± 2.94 | 29.47 ± 1.69 | 0.802a | 0.424 |
| BMI (kg/m2) | 21.66 ± 2.78 | 21.65 ± 3.14 | 0.017a | 0.987 |
| Infertility duration (years) | 2.00 (1.50, 4.00) | 3.00 (2.00, 4.00) | 1.261b | 0.207 |
| FSH | 7.492 ± 2.40 | 7.75 ± 1.88 | 0.943a | 0.347 |
| Duration of Gn | 11.03 ± 2.08 | 11.55 ± 2.18 | 0.916a | 0.056 |
| Dosage of Gn | 2322.71 ± 863.66 | 2110.87 ± 7.3.25 | 2.123a | 0.035 |
aStudent’s t test, data are expressed as means ± standards; bMann-Whitney U test, data are expressed as median and interquartile range
BMI: body mass index; FSH: follicle-stimulating hormone; Gn: gonadotropin
Mutation in the D-loop region
The entire mitochondrial DNA D-loop region was successfully sequenced in all 249 subjects. Across both groups, 321 polymorphisms were identified in comparison to the rCRS, comprising 273 single nucleotide substitutions, 34 nucleotide insertions, and 14 nucleotide deletions. Only 11 sites were significantly different between the patient group and the control group, all of which were located in hypervariable regions (HV) with the D-loop. The frequencies of 8 polymorphisms (T195C, 573XCins, 16036Gins, 16049Gins, T16140C, A16183C, T16189C, 16193Cins) were significantly lower in the endometriosis group compared with the control group, while the other three (AC523–524 del, T16172C, C16290T) were significantly higher. We performed logistic regression analysis to analyze the association of these polymorphisms with endometriosis, excluding four sites (573XCins, 16036Gins, 16049Gins, and T16140C) owing to too few cases in the endometriosis group (Table 2 Fig. 2, and Table 3).
Table 2.
Significant mtDNA D-loop SNPs with a minor allele frequency > 5% in endometriosis and controls
| SNP | Frequency | OR (95% CI) | p value | |
|---|---|---|---|---|
| Endometriosis (n = 125) | Control (n = 124) | |||
| T195C | 6 (4.80%) | 17 (13.71%) | 0.317 (0.121–0.834) | 0.02 |
| AC523–524 del | 58 (46.40%) | 42 (33.87%) | 1.690 (1.013–2.819) | 0.044 |
| T16172C | 21 (16.80%) | 9 (7.26%) | 2.580 (1.131–5.886) | 0.024 |
| A16183C | 24 (19.20%) | 40 (32.26%) | 0.499 (0.279–0.894) | 0.019 |
| T16189C | 32 (25.60%) | 49 (39.52%) | 0.527 (0.307–0.903) | 0.02 |
| 16193C ins | 9 (7.20%) | 25 (20.16%) | 0.307 (0.137–0.689) | 0.004 |
| C16290T | 17 (13.60%) | 6 (4.84%) | 3.096 (1.178–8.139) | 0.022 |
Logistic regression analysis, data are expressed as n (%); SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence interval
Fig. 2.
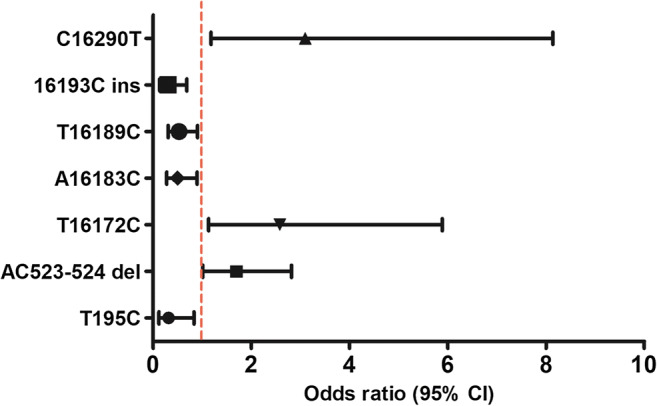
Forest plot of odds ratio of mtDNA D-loop SNPs with a minor allele frequency > 5% in endometriosis and controls
Table 3.
Significant mtDNA D-loop SNPs with a minor allele frequency < 5% in endometriosis or controls
| SNP | Frequency | p value | χ2 | |
|---|---|---|---|---|
| Endometriosis (n = 125) | Control (n = 124) | |||
| 573XC ins | 0 (0) | 6 (4.84%) | 0.038b | 4.311 |
| 16036G ins | 0 (0) | 17 (13.71%) | 0.000a | 18.393 |
| 16049G ins | 1 (0.8%) | 10 (8.06%) | 0.005a | 7.78 |
| T16140C | 2 (1.6%) | 10 (8.06%) | 0.017a | 5.671 |
Owing to the too few cases of patients in the patient group, chi-square test was performed without logistic regression analysis
achi-square test; bcontinuity correction, data are expressed as n (%)
It is well known that the incidence of endometriosis is significantly correlated with age and BMI. Therefore, for the polymorphisms mentioned above (excluding four with too few cases), multivariate regression analysis was performed for these two variables to eliminate the influence of confounding bias, even though matching of the age and BMI of our subjects was strictly controlled at the time of selection. After adjustment, the difference in frequencies of all polymorphisms between the two groups remained significant (Table 4 Fig. 3).
Table 4.
mtDNA D-loop SNPs with a minor allele frequency > 5% in endometriosis and controls after correction for age and BMI
| SNP | Adjusted OR(95% CI) | p value |
|---|---|---|
| T195C | 0.311 (0.118–0.821) | 0.018 |
| AC523–524 del | 1.739 (1.036–2.921) | 0.036 |
| T16172C | 2.737 (1.167–6.419) | 0.021 |
| A16183C | 0.481 (0.266–0.868) | 0.015 |
| T16189C | 0.512 (0.297–0.883) | 0.016 |
| 16193C ins | 0.313 (0.139–0.705) | 0.005 |
| C16290T | 2.953 (1.112–7.845) | 0.030 |
Multivariable logistic regression analysis adjusted for age and BMI; aOR: adjusted odds ratio
Fig. 3.
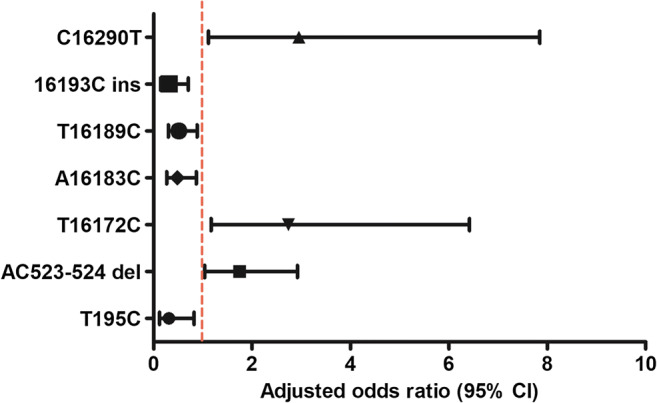
Forest plot of odds ratio of mtDNA D-loop SNPs with a minor allele frequency > 5% in endometriosis and controls after correction for age and BMI
Association between haplotype and endometriosis
We identified 153 different haplotypes across all subjects, but no significant differences in frequencies were observed between the endometriosis and control groups. The haplotypes F1a and F1a1 were present in 4 and 5 cases respectively in the endometriosis group but in no cases of the control group, but this did not reach statistical significance. We subsequently merged the corresponding haplotype subgroups and repeated our analysis. In the endometriosis group, the frequency of the D4 group was highest, followed by A, while in the control group the proportion of D4 and B4 haplotypes were dominant (Supplementary Table 1).
The association between polymorphism and clinical outcomes
Subjects in the endometriosis and control group were divided into subgroups according to the presence or absence of all 321 polymorphisms identified to assess the impact of polymorphisms on clinical outcomes. Two polymorphisms, C16290T and T16519C were associated with the development of fewer high-quality blastocysts in the endometriosis and control groups respectively. Endometriosis patients carrying 16290C showed a higher rate of high-quality blastocyst development compared with carriers of 16290T (46.15% vs 26.67%, p = 0.019) (Supplementary Table 2). In the control group, high-quality blastocysts were found at a higher rate in carriers of 16519T, compared with 16519C (50.00% vs 45.45%, p = 0.031) (Supplementary Table 3). After excluding patients without transferable embryos or not within a transfer cycle, a total of 72 and 119 cases were included in patients with or without endometriosis for clinical pregnancy outcome analysis. The polymorphism C150T was associated with a lower clinical pregnancy rate in endometriosis patients. Carriers of 150C showed a higher rate of clinical pregnancies post-IVF compared with 150T carriers (63.79% vs 28.57%, p = 0.017) (Supplementary Table 4). By contrast, no D-loop polymorphisms showed an association with clinical outcome in the control group. All patients who received a clinical pregnancy designation were currently in a state of ongoing pregnancy.
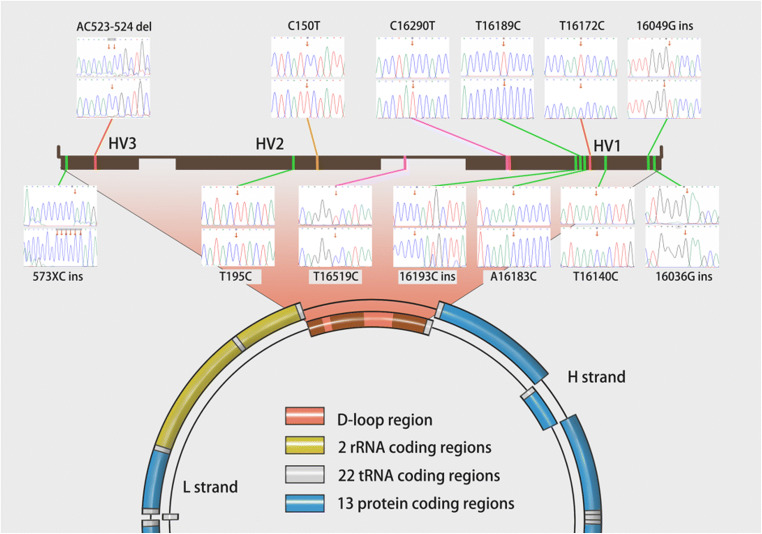
All significant polymorphisms are displayed in Fig. 4.
Fig. 4.
Structural overview of mtDNA and significant D-loop polymorphisms in this study. Polymorphic sites shown in red are positively correlated with endometriosis. Polymorphic sites shown in green are negatively correlated with endometriosis. Polymorphic sites shown in purple are associated with high-quality blastocyst development in endometriosis patients (C16290T) or controls (T16519C) (C16290T also shows a positive correlation with endometriosis). The polymorphic site shown in tan is associated with a higher rate of clinical pregnancy post-IVF in endometriosis patients. HV1/2/3: hypervariable region 1/2/3
Discussion
Endometriosis is a common disease in women of reproductive age suffering from infertility and is associated with inflammation and oxidative stress. The mtDNA D-loop region is responsible for regulating the transcription and replication of mtDNA, and so plays an indirect but important role in regulating levels of ROS production and oxidative stress. A range of relevant studies have demonstrated that polymorphisms in this control region can disrupt this regulation and increase the risk of diverse diseases such as diabetes and a wide spectrum of cancers [21–27].
In this study, we sequenced the entire D-loop region in 249 subjects and found a total of 13 polymorphisms with a significant association to either disease state, the development of high-quality blastocysts, or clinical pregnancy rates in IVF. Three polymorphisms appear to increase susceptibility to endometriosis. A point mutation, T16172C (located in HV1) was associated with susceptibility to endometriosis, with or without adjustment for the age and BMI of subjects. This represents the first reported association between T16172C and human reproductive diseases, and the first suggestion that variation at this site predisposes women to endometriosis. Another polymorphism we link to the risk of endometriosis in this study, C16290T (also belonging to HV1), has been identified as a risk factor for aggressive periodontitis and a predictor of post-operational survival in malignant fibrous histiocytoma [28, 29]. The AC523–524 del was shown to be significantly associated with an increased risk of endometriosis in our study and has also been reported to be related to the prognosis of operation in malignant fibrous histiocytoma [28].
Intriguingly, 8 other SNPs occurred more frequently in the control group than the endometriosis group, suggesting they might decrease the risk of endometriosis. Of these, 573XCins was also shown to be significantly associated with a decreased risk of endometriosis in another recent study [30]. However, because the frequency of this polymorphism is very small in our study we were not able to perform multivariate regression analysis and draw reliable conclusions. By the same token, the polymorphisms 16036Gins, 16049Gins, and T16140C were rare in the case group and so could not be included.
We found the T16189C polymorphism, a hot spot for mutation in the D-loop region, at a lower frequency in endometriosis patients compared with controls. This polymorphism has been associated with an increased risk of diverse human diseases, including endometriosis in Indian and Korean populations [31, 32]. However, it has also been reported that there is no difference in the incidence of this polymorphism between endometriosis patients and a control group in a Brazilian population. The Brazilian cohort consisted of patients from 4 different ethnic groups and includes women of more advanced age than ours [30]. It is clear that mtDNA haplotypes and polymorphisms are correlated with ethnic differences and geographical location, and so the heterogeneity of the Brazilian cohort may contribute to the discrepancy in reports of this polymorphism’s effect.
Of the other detected polymorphisms that we linked to a decreased risk of endometriosis, the polymorphism T195C has been reported to increase the risk of chronic kidney disease and colon cancer [26, 33]. The two remaining polymorphisms, A16183C and 16193Cins, have not been previously reported on, but are located on the termination associated sequence of H-strand synthesis [34], where mutations could have a profound impact on mitochondrial physiology. Although these polymorphisms appear to reduce the risk of endometriosis, a larger sample size may be necessary to draw convincing conclusions.
Our analysis of patient haplotype revealed that the haplotypes F1a and F1a1 were present in 4 and 5 cases of the endometriosis group, respectively, but in no cases of the control group. When we merged the corresponding haplotype subgroups, we found that the D4 and A haplotypes were most prevalent in the endometriosis group, while in the control group, the frequency of D4 and B4 was highest, though no differences were significant. Reports studying the phylogeography of Han Chinese have shown that the most frequent haplotypes in middle China, the broad geographical location of Anhui, are D and A [35, 36]. Our subjects reflect this distribution.
We used laboratorial and/or clinical outcomes to associate SNPs with IVF outcomes. A previous study has shown that oocyte quality is poor in patients with endometriosis [37]. Therefore, we speculated that mtDNA polymorphisms in endometriosis are also correlated with embryo quality, and/or clinical pregnancy rates in women with endometriosis.
Among endometriosis patients, we found the development of high-quality blastocysts occurred at a lower rate in the 16290T subgroup, compared with the 16290C subgroup. Interestingly, we also report 16290T to be a risk factor for endometriosis. Polymorphisms C527G and T16468C also appeared to be associated with embryo quality, however, the frequencies of these polymorphisms were so small that reliable conclusions cannot be drawn (data not shown). In the control group, the 16519C subgroup was associated with a lower rate of high-quality blastocyst compared with 16519T. The polymorphism T16519C was previously reported to be related to diverse disorders and considered to be a predictor for prognosis of pancreatic cancer [38, 39].
In addition, among endometriosis patients undergoing IVF, the 150C group showed a lower rate of clinical pregnancy compared with 150T. The C150T polymorphic site is in HV2 and has been associated with multitudinous disorders [17, 25, 33, 40]. Our results suggest these polymorphisms can be used as a predictor of embryo development or clinical pregnancy rate in patients with endometriosis (C16290T and C150T), or other causes of infertility (T16519C). This information may benefit the counseling of patients in IVF clinics.
The mechanisms underpinning the role of these polymorphisms in endometriosis are unclear. It may be that specific polymorphisms in the D-loop region cause a disorder in the regulation of the transcription and replication of mtDNA. This may lead to impaired OXPHOS function and ETC disruption due to decreased or dysfunctional mitochondrial proteins and RNAs, or reduced mtDNA copy number [28]. An aberrant ETC could lead to the release of large amounts of ROS production involved in the inflammatory mechanisms reported in endometriosis. Therefore, it is plausible that women carrying those polymorphisms may be more susceptible to endometriosis. Furthermore, endometriosis patients with certain nucleotide site variations manifesting less efficient OXPHOS or increasing oxidative stress may produce embryos with similar dysfunctions leading to the development of fewer high-quality embryos, and lower clinical pregnancy rates following IVF. Dedicated studies will be required to elucidate these mechanisms.
The main limitation of this study was the small number of samples. Consequently, when we conducted subgroup analysis in regard to haplotype, blastocyst development, or pregnancy rate, there was at times a large discrepancy in the sample size between the two subgroups, skewing statistical analysis. This is especially true when clinical pregnancy was assessed, as many patients were outside of the transfer cycle, leaving only 72 subjects in the endometriosis group. Even though some patients had transferable embryos, they had not entered the transfer cycle during data gathering. To draw more reliable conclusions, it is necessary to expand the sample size, especially to include more patients in a transfer cycle.
In conclusion, the present study demonstrated that the D-loop polymorphisms AC523–524 del, T16172C, and C16290T are associated with an increased risk of endometriosis after adjustment for age and BMI of patients, in Anhui Province of mid-east China. Polymorphisms T195C, 573XCins, 16036Gins, 16049Gins, T16140C, A16183C, T16189C, and 16193Cins are associated with decreased risk of endometriosis with varying degrees of confidence. Polymorphism C16290T is associated with poor quality blastocyst development in the endometriosis population while T16519C is associated with compromised blastocyst quality in infertile patients without endometriosis. In addition, polymorphism C150T impairs successful pregnancy in endometriosis patients undergoing IVF. This study is the first to evaluate the association between IVF outcomes and mtDNA D-loop region polymorphisms in patients with endometriosis. A larger sample size and well-designed studies are required to reach more reliable and convincing conclusions.
Electronic supplementary material
(DOCX 39 kb)
Acknowledgments
We are grateful to Kai Zong (Hefei Customs) for assisting in mtDNA sequencing and analysis.
Funding information
This study was supported by the National Key R&D Program of China (2017YFC1001300) and the National Natural Science Foundation of China (81601345, 81871216, 81771653).
Compliance with ethical standards
Competing interests
The authors declare that there are no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xinyuan Li and Dongmei Ji contributed equally to this work.
Contributor Information
Dongmei Ji, Email: cahsxjdm@aliyun.com.
Yunxia Cao, Email: caoyunxia6@126.com.
References
- 1.Govatati S, Tipirisetti NR, Perugu S, Kodati VL, Deenadayal M, Satti V, Bhanoori M, Shivaji S. Mitochondrial genome variations in advanced stage endometriosis: a study in south Indian population. PLoS One. 2012;7(7):e40668. doi: 10.1371/journal.pone.0040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creed J, Maggrah A, Reguly B, Harbottle A. Mitochondrial DNA deletions accurately detect endometriosis in symptomatic females of child-bearing age. Biomark Med. 2019;13(4):291–306. doi: 10.2217/bmm-2018-0419. [DOI] [PubMed] [Google Scholar]
- 3.Kao SH, Huang HC, Hsieh RH, Chen SC, Tsai MC, Tzeng CR. Oxidative damage and mitochondrial DNA mutations with endometriosis. Ann N Y Acad Sci. 2005;1042:186–194. doi: 10.1196/annals.1338.021. [DOI] [PubMed] [Google Scholar]
- 4.Van Langendonckt A, Casanas-Roux F, Donnez J. Oxidative stress and peritoneal endometriosis. Fertil Steril. 2002;77(5):861–870. doi: 10.1016/S0015-0282(02)02959-X. [DOI] [PubMed] [Google Scholar]
- 5.Vercellini P, Vigano P, Somigliana E, Fedele L. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10(5):261–275. doi: 10.1038/nrendo.2013.255. [DOI] [PubMed] [Google Scholar]
- 6.Greene AD, Lang SA, Kendziorski JA, Sroga-Rios JM, Herzog TJ, Burns KA. Endometriosis: where are we and where are we going? Reproduction. 2016;152(3):R63–R78. doi: 10.1530/REP-16-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanbo T, Fedorcsak P. Endometriosis-associated infertility: aspects of pathophysiological mechanisms and treatment options. Acta Obstet Gynecol Scand. 2017;96(6):659–667. doi: 10.1111/aogs.13082. [DOI] [PubMed] [Google Scholar]
- 8.Matalliotakis M, Zervou MI, Matalliotaki C, Rahmioglu N, Koumantakis G, Kalogiannidis I, Prapas I, Zondervan K, Spandidos DA, Matalliotakis I, Goulielmos GN. The role of gene polymorphisms in endometriosis. Mol Med Rep. 2017;16(5):5881–5886. doi: 10.3892/mmr.2017.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osinski M, Mostowska A, Wirstlein P, Wender-Ozegowska E, Jagodzinski PP, Szczepanska M. The assessment of GWAS - identified polymorphisms associated with infertility risk in polish women with endometriosis. Ginekol Pol. 2018;89(6):304–310. doi: 10.5603/GP.a2018.0052. [DOI] [PubMed] [Google Scholar]
- 10.Shu J, Xing L, Ding G, Luo Q, Liu X, Yan Q, Sheng J, Huang H. The effect of peritoneal fluid from patients with endometriosis on mitochondrial function and development of early mouse embryos. PLoS One. 2013;8(12):e82334. doi: 10.1371/journal.pone.0082334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang D, Hamasaki N. Alterations of mitochondrial DNA in common diseases and disease states: aging, neurodegeneration, heart failure, diabetes, and cancer. Curr Med Chem. 2005;12(4):429–441. doi: 10.2174/0929867053363081. [DOI] [PubMed] [Google Scholar]
- 12.Stefano GB, Bjenning C, Wang F, Wang N, Kream RM. Mitochondrial Heteroplasmy. Adv Exp Med Biol. 2017;982:577–594. doi: 10.1007/978-3-319-55330-6_30. [DOI] [PubMed] [Google Scholar]
- 13.Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. 2015;16(9):530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 14.Cheng M, Guo Z, Li H, Li Z, Li C, Geng C. Identification of sequence polymorphisms in the mitochondrial displacement loop as risk factors for sporadic and familial breast cancer. Tumour Biol. 2014;35(5):4773–4777. doi: 10.1007/s13277-014-1626-5. [DOI] [PubMed] [Google Scholar]
- 15.Kong D, Shi S, Li Y. Single nucleotide polymorphisms in the D-loop region of mitochondrial DNA are associated with epithelial ovarian cancer prognosis. Mitochondrial DNA. 2015;26(6):848–850. doi: 10.3109/19401736.2013.861425. [DOI] [PubMed] [Google Scholar]
- 16.Reddy TV, Govatati S, Deenadayal M, Sisinthy S, Bhanoori M. Impact of mitochondrial DNA copy number and displacement loop alterations on polycystic ovary syndrome risk in south Indian women. Mitochondrion. 2019;44:35–40. doi: 10.1016/j.mito.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Zhai K, Chang L, Zhang Q, Liu B, Wu Y. Mitochondrial C150T polymorphism increases the risk of cervical cancer and HPV infection. Mitochondrion. 2011;11(4):559–563. doi: 10.1016/j.mito.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Tipirisetti NR, Govatati S, Pullari P, Malempati S, Thupurani MK, Perugu S, Guruvaiah P, K LR, Digumarti RR, Nallanchakravarthula V, Bhanoori M, Satti V. Mitochondrial control region alterations and breast cancer risk: a study in south Indian population. PLoS One. 2014;9(1):e85363. doi: 10.1371/journal.pone.0085363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu VW, Wang Y, Yang HJ, Tsang PC, Ng TY, Wong LC, et al. Mitochondrial DNA variant 16189T>C is associated with susceptibility to endometrial cancer. Hum Mutat. 2003;22(2):173–174. doi: 10.1002/humu.10244. [DOI] [PubMed] [Google Scholar]
- 20.Gardner DK, Vella P, Lane M, Wagley L, Schlenker T, Schoolcraft WB. Culture and transfer of human blastocysts increases implantation rates and reduces the need for multiple embryo transfers. Fertil Steril. 1998;69(1):84–88. doi: 10.1016/S0015-0282(97)00438-X. [DOI] [PubMed] [Google Scholar]
- 21.Kumari T, Vachher M, Bansal S, Bamezai R, Kumar B. Meta-analysis of mitochondrial T16189C polymorphism for cancer and type 2 diabetes risk. Clin Chim Acta. 2018;482:136–143. doi: 10.1016/j.cca.2018.03.041. [DOI] [PubMed] [Google Scholar]
- 22.Govatati S, Saradamma B, Malempati S, Dasi D, Thupurani MK, Nagesh N, et al. Association of mitochondrial displacement loop polymorphisms with risk of colorectal cancer in south Indian population. Mitochondrial DNA A DNA Mapp Seq Anal. 2017;28(5):632–637. doi: 10.3109/24701394.2016.1160076. [DOI] [PubMed] [Google Scholar]
- 23.Hu WX, Ding CM, Li RJ, Fan HY, Guo ZJ, Liu W. Single nucleotide polymorphisms in the mitochondrial displacement loop and age-at-onset of non-small cell lung cancer. Genet Mol Res. 2015;14(1):2512–2517. doi: 10.4238/2015.March.30.9. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Deng B, Wang W, Jia Z, Liu X, Li N. Single nucleotide polymorphisms in the mitochondrial displacement loop region predict malignant melanoma outcome: a study in Chinese Han population. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(3):1812–1816. doi: 10.3109/19401736.2014.963824. [DOI] [PubMed] [Google Scholar]
- 25.Er LM, Wu ML, Gao Y, Wang SJ, Li Y. Identification of sequence polymorphisms in the displacement loop region of mitochondrial DNA as a risk factor for gastroenteropancreatic neuroendocrine neoplasm. J Clin Lab Anal. 2017;31(5):e22078. [DOI] [PMC free article] [PubMed]
- 26.Guo Z, Zhao S, Fan H, Du Y, Zhao Y, Wang G. Identification of sequence polymorphisms in the D-loop region of mitochondrial DNA as a risk factor for colon cancer. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(6):4244–4245. doi: 10.3109/19401736.2014.1003920. [DOI] [PubMed] [Google Scholar]
- 27.Bai Y, Guo Z, Xu J, Liu S, Zhang J, Cui L, Zhang H, Zhang S. Single nucleotide polymorphisms in the D-loop region of mitochondrial DNA is associated with renal cell carcinoma outcome. Mitochondrial DNA. 2015;26(2):224–226. doi: 10.3109/19401736.2013.825772. [DOI] [PubMed] [Google Scholar]
- 28.Xun J, Li Z, Feng J, Gao S, Yang H, Song X. Single nucleotide polymorphisms in the mitochondrial displacement loop region and outcome of malignant fibrous histiocytoma. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(1):177–181. doi: 10.3109/19401736.2013.879650. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Guo Y, Luan Q. Association of mitochondrial DNA displacement loop polymorphisms and aggressive periodontitis in a Chinese population: a pilot study. Mitochondrial DNA. 2015;26(3):389–395. doi: 10.3109/19401736.2013.840589. [DOI] [PubMed] [Google Scholar]
- 30.Andres MP, Cardena M, Fridman C, Podgaec S. Polymorphisms of mitochondrial DNA control region are associated to endometriosis. J Assist Reprod Genet. 2018;35(3):533–538. doi: 10.1007/s10815-017-1082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho S, Lee YM, Choi YS, Yang HI, Jeon YE, Lee KE, Lim KJ, Kim HY, Seo SK, Lee BS. Mitochondria DNA polymorphisms are associated with susceptibility to endometriosis. DNA Cell Biol. 2012;31(3):317–322. doi: 10.1089/dna.2011.1279. [DOI] [PubMed] [Google Scholar]
- 32.Govatati S, Deenadayal M, Shivaji S, Bhanoori M. Mitochondrial displacement loop alterations are associated with endometriosis. Fertil Steril. 2013;99(7):1980–6.e9. doi: 10.1016/j.fertnstert.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 33.Bai Y, Guo Z, Xu J, Zhang J, Cui L, Zhang H, Zhang S, Ai X. Association of sequence polymorphism in the mitochondrial D-loop with chronic kidney disease. Ren Fail. 2014;36(5):781–784. doi: 10.3109/0886022X.2014.890842. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88(1):41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- 35.Xu K, Hu S. Population data of mitochondrial DNA HVS-I and HVS-II sequences for 208 Henan Han Chinese. Leg Med (Tokyo) 2015;17(4):287–294. doi: 10.1016/j.legalmed.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet. 2002;70(3):635–651. doi: 10.1086/338999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu B, Guo N, Zhang XM, Shi W, Tong XH, Iqbal F, Liu YS. Oocyte quality is decreased in women with minimal or mild endometriosis. Sci Rep. 2015;5:10779. doi: 10.1038/srep10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navaglia F, Basso D, Fogar P, Sperti C, Greco E, Zambon CF, Stranges A, Falda A, Pizzi S, Parenti A, Pedrazzoli S, Plebani M. Mitochondrial DNA D-loop in pancreatic cancer: somatic mutations are epiphenomena while the germline 16519 T variant worsens metabolism and outcome. Am J Clin Pathol. 2006;126(4):593–601. doi: 10.1309/GQFCCJMH5KHNVX73. [DOI] [PubMed] [Google Scholar]
- 39.Zhou HY, Shu HY, Dai J, Li HC, Tang L, Wang HW, et al. Maternal genetic backgrounds contribute to the genetic susceptibility of tongue cancer patients in Hunan, central of China. Mitochondrial DNA A DNA Mapp Seq Anal. 2018;29(3):347–352. doi: 10.1080/24701394.2016.1278539. [DOI] [PubMed] [Google Scholar]
- 40.Guney O, Ak H, Atay S, Ozkaya AB, Aydin HH. Mitochondrial DNA polymorphisms associated with longevity in the Turkish population. Mitochondrion. 2014;17:7–13. doi: 10.1016/j.mito.2014.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 39 kb)