Abstract
Purpose
Changes in DNA methylation modifications have been associated with male infertility. With the development of assisted reproductive technologies (ARTs), abnormal DNA methylation in sperm, especially in imprinted genes, may impact the health of offspring and requires an in-depth study.
Methods
In this study, we collected abnormal human semen samples, including asthenospermic, oligospermic, oligoasthenospermic and deformed sperm, and investigated the methylation of imprinted genes by reduced representation bisulfite sequencing (RRBS) and bisulfite amplicon sequencing on the Illumina platform.
Results
The differentially methylated regions (DMRs) of imprinted genes, including H19, GNAS, MEG8 and SNRPN, were different in the abnormal semen groups. MEG8 DMR methylation in the asthenospermic group was significantly increased. Furthermore, higher methylation levels of MEG8, GNAS and SNRPN DMR in the oligospermic and oligoasthenospermic groups and a decrease in the H19 DMR methylation level in the oligospermic group were observed. However, the methylation levels of these regions varied greatly among the different semen samples and among individual sperm within the same semen sample. The SNP rs2525883 genotype in the H19 DMR affected DNA methylation. Moreover, DNA methylation levels differed in the abnormal semen groups in the non-imprinted genomic regions, including repetitive sequence DNA transposons and long/short interspersed nuclear elements (LINEs and SINEs).
Conclusion
Our study established that imprinted gene DMRs, such as H19, GNAS, SNRPN and MEG8, were differentially methylated in the abnormal semen groups with obvious inter- and intra-sample heterogeneities. These results suggest that special attention needs to be paid to possible epigenetic risks during reproduction.
Keywords: DNA methylation, Imprinted genes, Spermatozoa
Introduction
After a long period of differentiation, mature spermatozoa establish a unique epigenome [1]. As one of the most important epigenetic modifications, DNA methylation plays biological roles in genomic imprinting, reprogramming and stability; cellular differentiation; X-chromosome inactivation (XCI); transposon silencing; RNA splicing; and DNA repair [2–4]. Unlike genetic mutations, DNA methylation is highly susceptible to environmental cues and environmental insults, such as exposure to toxins, teratogens, diet (nutrient availability) and mental state (stress) [5–7].
Alterations in DNA methylation patterns are known to be associated with a variety of human diseases [5]. Aberrant DNA methylation changes in sperm have been shown to increase the risk of reproductive failures [8], to deregulate gene expression and to promote genomic instability [9, 10]. Moreover, specific alterations in germ line DNA methylation are associated with a variation in sperm morphology and motility, including asthenospermia, oligospermia and deformation [11–13]. These methylome defects can be transmitted to offspring and can affect the offspring’s susceptibility to disease [14]. More importantly, recent reports have indicated that changes in DNA methylation occur not only between different semen samples but also within the same semen sample [15]. This intra-sample heterogeneity of DNA methylation may be due to the presence of sperm populations with different epigenetic quality [16]. Assisted reproductive technologies (ARTs) are widely used, allowing infertile men to father their own children. However, ART carries a potential risk of transmission/induction of genetic/epigenetic alterations, particularly when more intrusive methodologies are applied, such as intra-cytoplasmatic sperm injection (ICSI) [17–19]. Indeed, an increased frequency of several diseases has been reported after ART, such as Angelman, Beckwith–Wiedemann, Silver–Russell and Prader–Willi syndromes [20–22]. Although these diseases are derived from different genetic causes, the role of sperm epigenetic aberrations has been proposed, igniting scientific interest about the relationship between genomic imprinting and fertility. ICSI and in vitro fertilization (IVF) routinely require the selection of gametes and the culture of embryos during the early stage of development when the genome is relatively vulnerable to external influences [20]. However, whether these syndromes arise from ART itself or from pre-existing epigenetic aberrations in gametes remains unclear. The so-called ‘imprinted genes’ escape epigenetic reprogramming after fertilization, leading to the maintenance of aberrant sperm DNA methylation in the developing embryo [23].
Genomic imprinting is an epigenetic phenomenon that describes parent-of-origin patterns of monoallelic gene expression [24]. This monoallelic expression is established by DNA methylation of CpG dinucleotides in the germ line, and these modifications then form germ line differentially methylated regions (gDMRs) after fertilization. These gDMRs act as imprinting control regions (ICRs) that generate extensive domains of imprinted chromatin, some of which span several megabases. The DNA methylation of imprinted gDMRs is reset with every reproductive cycle during gametogenesis [25, 26]. Ejaculated and mature sperm should be methylated in the paternal gDMRs (pgDMRs) but unmethylated in the maternal gDMRs (mgDMRs). In humans, over 100 genes are regulated by genomic imprinting, and many of these have critically important roles in early development [24]. The failure to correctly establish or maintain imprints causes rare but striking developmental imprinting disorders during childhood and, later in life, causes both metabolic and behavioural diseases [27–29]. In this context, several studies documented a strong association between idiopathic infertility and DNA methylation alterations in specific imprinted genes in spermatozoa from human men [30]. In particular, infertile and fertile men were compared, and an increased odds ratio of imprinting aberrations was demonstrated in two specifically imprinted genes, H19 and mesoderm-specific transcript (MEST) [31]. In addition, aberrant methylation on specific non-imprinted genes was found to be associated with infertility [32, 33]. All these data increased the scientific interest in the analysis of sperm DNA methylation as a valuable, non-invasive diagnostic marker of infertility.
Although largely debated in the literature, the link between epigenetics and sperm abnormalities has not been elucidated. High-throughput bisulfite sequencing is a new DNA methylation profiling method that can be applied to the study of genome-wide and gene-specific methylation levels with high accuracy. This study was designed to further evaluate the sperm DNA methylation patterns, particularly the methylation of imprinted genes, in abnormal semen samples by RRBS and bisulfite amplicon sequencing on the Illumina platform.
Methods and materials
Spermatozoa samples
Semen samples were collected from patients (age between 20 and 60 years) attending the Hospital of Shanghai Institute of Planned Parenthood Research for semen analysis from January 2019 to March 2019 (Table 1). After liquefaction at 37 °C for 30 min, semen parameters such as semen pH, volume, viscosity, liquefaction time, colour, sperm concentration, sperm rapid progressive motility, viability and sperm morphology were analysed for routine semen analysis using microcell slide and computer-assisted semen analysis (CASA, WLJY 9000, Weili New Century Science and Tech Dev, Beijing, China) according to World Health Organization guidelines [34]. The absence of leukocytes and other cells was confirmed by phase-contrast microscopic analysis of the sperm pellets. Finally, 8 normozoospermic, 16 asthenospermic, 3 oligospermic, 11 oligoasthenospermic and 2 morphologically deformed (with sperm deformity rate 40% and 55%) semen samples were collected from patients. These samples were stored at − 80 °C until processing. This study was approved by the Ethics Committee of Shanghai Institute of Planned Parenthood Research.
Table 1.
Sample parameters and global DNA methylation levels
| Normozoospermia | Asthenospermia | Oligospermia | Oligoasthenospermia | Deformity | |
|---|---|---|---|---|---|
| Number of samples | 8 | 16 | 3 | 11 | 2 |
| Age (year) | 30.00 ± 1.67 | 36.63 ± 2.48 | 32.67 ± 4.33 | 35.27 ± 1.95 | 36.50 ± 3.50 |
| Sperm concentration (× 106/ml) | 113.40 ± 21.95 | 53.14 ± 9.78* | 5.06 ± 1.30* | 8.79 ± 1.42* | 79.50 ± 24.90 |
| Total motility (%) | 66.51 ± 5.56 | 18.43 ± 2.78* | 45.10 ± 4.03 | 24.10 ± 2.57* | 52.10 ± 21.50 |
| Progressive motility (%) | 49.85 ± 4.07 | 12.45 ± 2.09* | 40.67 ± 4.81 | 19.24 ± 2.78* | 38.65 ± 22.15 |
| Nonprogressive motility (%) | 16.66 ± 2.81 | 5.98 ± 1.28* | 4.43 ± 1.79* | 4.04 ± 0.97* | 13.45 ± 0.65 |
| Sperm vitality (%) | 70.99 ± 3.52 | 38.82 ± 2.09* | 52.10 ± 7.13* | 43.06 ± 1.92* | 64.10 ± 16.10 |
| Number of samples analysed by RRBS | 8 | 4 | 3 | 2 | 2 |
| DNA methylation-whole genome (%) | 40.91 ± 2.41 | 37.84 ± 2.63 | 47.81 ± 4.67 | 40.56 ± 7.25 | 48.39 ± 6.90 |
| DNA methylation-gene body (%) | 40.19 ± 2.41 | 36.35 ± 2.67 | 46.43 ± 4.94 | 39.12 ± 7.53 | 47.32 ± 7.60 |
| DNA methylation-promoter (%) | 13.54 ± 1.22 | 11.05 ± 1.25 | 16.84 ± 3.07 | 12.66 ± 3.09 | 16.85 ± 4.54 |
| DNA methylation-CpG island (%) | 6.22 ± 0.22 | 5.30 ± 0.35* | 7.49 ± 1.23 | 6.21 ± 0.32 | 5.74 ± 0.40 |
| DNA methylation-3′-UTR (%) | 41.42 ± 2.24 | 38.44 ± 2.36 | 47.28 ± 4.61 | 40.40 ± 6.92 | 47.21 ± 6.94 |
| DNA methylation-5′-UTR (%) | 21.61 ± 2.15 | 17.80 ± 2.10 | 26.59 ± 4.58 | 20.10 ± 6.02 | 27.50 ± 7.37 |
| DNA methylation-exon (%) | 24.71 ± 1.49 | 20.11 ± 1.51 | 27.32 ± 4.14 | 22.83 ± 3.99 | 27.26 ± 6.02 |
| DNA methylation-intergenic (%) | 49.59 ± 2.29 | 51.82 ± 2.10 | 59.77 ± 2.50* | 55.07 ± 2.93 | 58.23 ± 1.61 |
| DNA methylation-intron (%) | 50.15 ± 2.31 | 47.31 ± 2.78 | 56.29 ± 4.15 | 49.71 ± 7.93 | 57.24 ± 6.28 |
*P value ≤ 0.05
RRBS, reduced representation bisulfite sequencing; UTR, untranslated region
DNA extraction
Sperm pellets were lysed in sperm lysis buffer (supplemented with proteinase K and DTT) in a 55 °C water bath for 2 h with intermittent mixing by inversion. Then, the DNA was extracted using phenol/chloroform extraction and ethanol precipitation.
Reduced representation bisulfite sequencing (RRBS)
Genomic DNA was used to perform RRBS. The standard protocol for RRBS library construction used here was described previously [35]. The library was 2 × 150 bp paired-end sequenced on a next-generation sequencing (NGS) HiSeq platform (Illumina, Inc., San Diego, CA, USA). The quality of the sequencing data was verified using ‘FastQC v.0.11.7’ package (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Adapter removal and quality trimming were performed using Cutadapt v1.9.1 (https://cutadapt.readthedocs.io/en/stable/). The reads were aligned to a reference genome (UCSC hg19) using Bismark v.0.19.0 (http://www.bioinformatics.babraham.ac.uk/projects/bismark/). The methylation level of each cytosine was calculated using the R package MethylKit (version 1.12.0) (https://bioconductor.org/packages/release/bioc/html/methylKit.html) [36].
Bisulfite amplicon sequencing
The DNA methylation levels of H19 DMR, GNAS XL DMR, MEG8 DMR and SNRPN DMR were analysed in samples via bisulfite amplification followed by NGS. A total of 500 ng of DNA was bisulfite treated using an EZ DNA Methylation Gold-Kit (Zymo Research Corp., Irvine, CA, USA). The bisulfite-converted DNA was used to amplify the candidate fragment with a TaKaRa EX Taq Hot Start Version Kit (TaKaRa Biotechnology Co., Ltd., Dalian, China). The PCR products were loaded on a 1.5% agarose gel for analysis, recovered for library construction and 2 × 300 bp paired-end sequenced using the MiSeq platform (Illumina, Inc., San Diego, CA, USA). The DNA methylation level of candidate fragments was determined by analysing the sequencing data. Haplotype analysis of sample methylation was performed using BiQ Analyzer HiMod (https://biq-analyzer-himod.bioinf.mpi-inf.mpg.de/). The primers for amplification were as follows: H19 DMR forward, 5′-AATAGTGTATTTTGGGGTGAA-3′, and H19 DMR reverse, 5′-AAAAACAATAAAATATCCCCATTCTT-3′; GNAS XL DMR forward, 5′-GTTGAAATTAGYGGAGGTATT-3′, and GNAS XL DMR reverse, 5′-CAAAAACTCCCACTACCCCA-3′; MEG8 DMR forward, 5′-GACAGGACAGCTCCCGGGACAGC-3′, and MEG8 reverse, 5′-AAACAATCTAAATTCTTCAAACACCA-3′; and SNRPN DMR forward, 5′-AGGYGTAAATAGGATTTGTTT-3′, and SNRPN DMR reverse, 5′-TCACTCTCCAAAAACATTATAATTCAA-3′.
Genotyping of the single nucleotide polymorphism (SNP) rs2525883
Genotyping of SNP rs2525883 within H19 DMR was performed by Sanger sequencing. DNA from 38 spermatozoa samples was used to amplify the DNA fragment surrounding rs2525883 with a TaKaRa EX Taq Hot Start Version Kit (TaKaRa Biotechnology Co., Ltd., Dalian, China). The PCR products were loaded on a 1.5% agarose gel for analysis and were recovered for Sanger sequencing. Genotypes of samples were determined by reading sequencing trace files. The primers for amplification were as follows: H19 DMR SNP forward, 5′-GGTGAATCAGACACATAGCC-3′, and H19 DMR SNP reverse, 5′-GGATGATGGGGATCTCGG-3′.
Statistical analysis
All data were analysed using the R program (version 3.3.2) (http://www.R-project.org/) and SPSS software (version 23.0). The samples included in this study were non-normally distributed (nonparametric) according to the value of the skewness test, Kurtosis test, Z value and Shapiro test. The independent sample t test (Mann–Whitney test) and Kruskal–Wallis test were used to compare the means of quantitative variables in addition to Spearman’s test to assess the correlation coefficient between methylation level and other parameters. The methylation levels between semen groups were further compared by using binary logistic regression models adjusted for age. The results in all of the aforementioned procedures were accepted as significant when the P value was less than or equal to 5% (P ≤ 0.05). Data are presented as the mean ± standard deviation (SD).
Results
Semen parameters and global DNA methylation levels
According to the World Health Organization guidelines [34], a total of 40 semen samples were collected and divided into five groups, including one normal group, normozoospermia (n = 8), and 4 abnormal groups: asthenospermia (n = 16), oligospermia (n = 3), oligoasthenospermia (n = 11) and deformed samples (n = 2) (Table 1). The sperm concentration, total motility and sperm viability of the samples from asthenospermia patients and oligoasthenospermia patients were significantly lower than those of the normozoospermia samples (P < 0.01). The sperm concentration and nonprogressive motility were significantly lower in samples from the oligospermic group than in samples from the control group (P < 0.001).
First, 19 semen samples from five groups were randomly selected for genome-wide methylation analysis by RRBS. The results showed that there was no significant difference in the global methylation levels of the genomes among the five groups, and the average DNA methylation levels of the samples were between 38 and 48%. Further, after classifying all CpG sites according to genomic location, methylation level analysis revealed that intergenic regions and intronic regions had the highest levels of DNA methylation, while CpG islands, promoter regions and 5′-UTR regions had lower levels of methylation. There was no significant difference among the groups, except for the asthenospermic group which had a lower methylation level at the CpG islands and the oligospermic group which had a higher methylation level at the intergenic regions than the normozoospermic group (Table 1).
Genome-wide methylation analysis of imprinted genes in normal and abnormal semen samples
Next, we performed an analysis of the methylation levels in the DMR regions of known imprinted genes [24]. In total, the methylation levels of the CpG sites within 64 imprinted DMR regions were detected in our RRBS. Most of these DMR regions were hypomethylated (mgDMRs), and only the IGF2-H19 and IG DMRs were hypermethylated (pgDMRs). We calculated the average methylation level of the CpG sites for each DMR region. By comparing all abnormal semen groups with the normal semen group, we found that aberrant methylation modifications existed in the DMRs of the imprinted genes H19, GNAS, SNRPN and MEG8 in the abnormal semen groups (Fig. 1a).
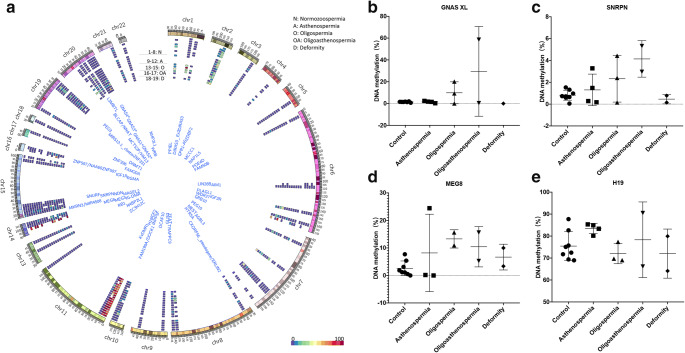
Fig. 1.
Methylation level of an imprinted gene DMR in semen samples analysed by RRBS (a). GNAS1*, GNAS; GNAS2*, NESP-AS/GNAS-AS1; GNAS3*, GNAS XL; GNAS4*, GNAS Ex1A. b–e The DNA methylation level of the DMR of GNSA XL (b), SNRPN (c), H19 (d) and MEG8 (e) in five groups. The bars represent the mean ± SD
Among the mgDMRs, the GNAS gene XL DMR was 1.47% methylated in normozoospermia but was 9.46% methylated in abnormal spermatozoa, 9.85% methylated in oligoasthenospermia and 29.50% methylated in oligospermia (Fig. 1b). The average methylation level of the DMR of the SNRPN gene was 0.82% in the normozoospermic group and 4.14% in the oligoasthenospermia group (Fig. 1c). In addition, the average methylation level of the DMR of the MEG8 gene was 2.61% in the normal group but was an average of 9.49% in the abnormal groups (8.20% in the asthenospermic group, 13.29% in the oligospermic group and 10.44% in the oligoasthenospermic group) (Fig. 1d). The average methylation of the DMR of the H19 gene on chromosome 11 was 79.92% in the normozoospermic group and decreased to an average of 77.31% in the abnormal groups. Notably, the methylation level in the asthenospermic group was 83.44%, which was higher than that in the control group (Fig. 1e).
We also found that the DMR methylation levels were strongly heterogeneous among abnormal semen samples, as large variations in methylation among different samples were identified. For example, the methylation level of GNAS XL DMR was extremely high in one sample from the oligospermic group and exceeded 50% (Fig. 1b). In SNRPN DMR, the methylation levels of some samples from the oligospermic group and oligoasthenospermic group were higher than those of the other samples (Fig. 1c). A similar phenomenon was also observed in the MEG8 DMR from the asthenospermic group: one abnormal sample displayed a high methylation level, while the other abnormal samples had methylation levels that were very similar to the level in the normal group (Fig. 1d).
Methylation validation of H19 DMR, GNAS XL DMR, SNRPN DMR and MEG8 DMR in normal and abnormal semen samples
Further, we validated the DNA methylation levels of H19 DMR, GNAS XL DMR and SNRPN DMR in all 38 semen samples using bisulfite amplicon sequencing. One oligospermic sample and one deformed sample in the RRBS experiment were not included in these 38 samples because their DNA was depleted. For the H19 DMR, the assay fragment was designed to cover a range of approximately 377 bp, from position 2021841 to 2022218 on chromosome 11, encompassing a total of 18 CpG sites. The results suggested that within this region, most of the sites fit the signature of hypermethylated pgDMR, with the exception of locus Chr11:2022019. The methylation levels of cytosine on Chr11:2022019 ranged from 60 to 90% among samples but did not show significant differences among the different groups. The Mann–Whitney test was used to compare Chr11:2021940 and Chr11:2021986 in the oligospermic group with those in the normozoospermic group, and a significant decrease in methylation levels from 98.18% and 97.61% to 90.90% and 95.39% was observed, respectively. In addition, Chr11:2022014 was methylated by 97.17% in the asthenospermic group, which was significantly lower than that in the normozoospermic group (98.44%) (Fig. 2a and d). However, these significant differences no longer existed after adjusting for age by binary logistic regression analysis.
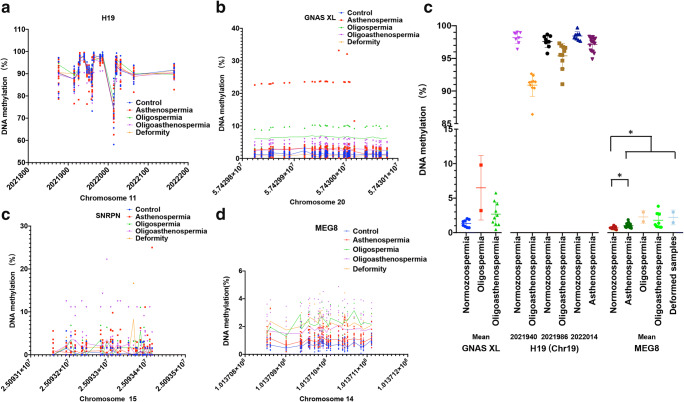
Fig. 2.
Validation of the methylation level of imprinted gene DMR in semen samples analysed by BSAS. a–c The methylation level of CpG sites in the DMR of H19 (a), GNAS XL (b), SNRPN (c) and MEG8 (d). The x-axis represents the chromosomal location. e Comparisons of the DNA methylation levels of DMR of GNSA XL and three CpG sites (Chr19:2021940, 2021986 and 2022014) from H19 DMR and MEG8 DMR in the semen groups. *P < 0.05 vs. normozoospermia group. Significance was determined using a binary logistic regression adjusted by age
To verify the methylation level of the XL DMR of the GNAS gene, we designed an amplicon fragment covering positions 57429801 to 57430101 of chromosome 20, containing a total of 30 CpG sites. The methylation levels of all loci were consistent with the characteristics of hypomethylated mgDMRs. Moreover, consistent with the RRBS results, the methylation levels of all CpGs in the oligospermic group were higher than those in the normozoospermic group, whereas the methylation levels of most CpG sites in the oligoasthenospermic group samples were also higher than those in the normozoospermic group samples. If the average methylation of this fragment was calculated, the average methylation was 6.50% in the oligospermic group and 2.68% in the oligoasthenospermic group; these levels were higher than those in the normal group (1.33%) (Fig. 2b and d). However, the increases in methylation in the abnormal groups were only significant in the Mann–Whitney test and not in the binary logistic regression model adjusted by age.
Validation experiments for the SNRPN DMR covered positions 25093077 to 25093438 of chromosome 15 and contained a total of 25 CpG sites. All sites were hypomethylated. Although consistent with the results of RRBS, the mean methylation value was increased in the oligospermic group compared with that in the normozoospermic group; this difference was 1.70% versus 0.42%, respectively, but was not statistically significant (Fig. 2c).
For the DMR of MEG8, an amplified fragment on the minus strand of chromosome 14 from 101371137 to 101370814 containing 23 CGs was designed. All sites were hypomethylated. The methylation levels were higher at most CG sites in the abnormal groups than those in the normal group. The mean methylation level of all sites was 0.70% in the normozoospermic group, while the mean levels of asthenospermia, oligospermia, oligoasthenospermia and deformed samples were 1.02%, 2.28%, 1.79% and 2.22%, respectively. The hypermethylation in all groups was significant by the Mann–Whitney test. After adjusting for age in the binary logistic regression, mean methylation levels of the combined abnormal samples and those of the asthenospermic samples were both significantly higher than those in the normal samples (Fig. 2d and e).
Inter-sample heterogeneities were still found in the DMR methylation validation assay. In the analysis of the GNAS XL DMR, one sample from the asthenospermic group had particularly high methylation levels, reaching an average of 18.24% and even more than 20% at multiple CpG sites. In addition, the average methylation level of one sample in the oligospermic group was 9.80%, which was much higher than the methylation level of most samples (Fig. 2b). In the SNRPN DMR methylation analysis results, although no statistically significant differences between groups were identified, individual samples still showed aberrantly increased methylation levels. For example, in one sample from the oligospermic group, the average methylation level reached 7.02% (Fig. 2c). The most interesting results were obtained from the H19 DMRs. Except for several sites, such as Chr11:2021940, Chr11:2021986 and Chr11:2022014, the methylation levels of the other sites were quite different among different samples, and the average methylation levels were mostly approximately 90%, which indicated that pgDMR was not fully methylated. However, the methylation levels of these sites were not significantly different among the different groups. In contrast, the methylation levels at several loci with small differences between samples were significantly different among groups, perhaps suggesting a more important biological function for these CpG sites (Fig. 2a).
Intra-sample heterogeneity analysis of DNA methylation in different spermatozoa from the same semen sample
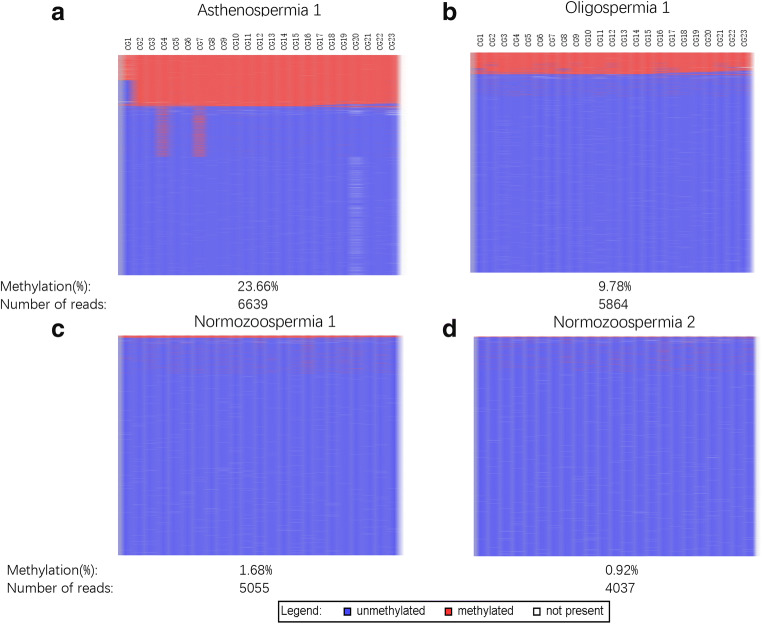
Because methylation abnormalities in the DMR fragments of the GNAS gene often occurred at multiple CpG sites simultaneously in our results, we further analysed the methylation sequencing results. By methylation haplotype analysis, we found that in one sample of the asthenospermic group, 23% of the reads, which should correspond to the sperm proportion, were aberrantly methylated on almost all CpG sites, and the other 77% of the reads remained unmethylated (Fig. 3a). This result indicated that approximately 23% of spermatozoa in this semen sample were aberrantly methylated, which suggested the presence of intra-sample heterogeneity. A similar result was observed in a sample from the oligospermic group, in which approximately 10% of the sperm were abnormally methylated at almost all CpG sites (Fig. 3b). Almost no aberrant methylation modifications were found in normal semen samples, except for a very small number of aberrantly methylated CpG sites, which were often linked (Fig. 3c and d). This result suggested that a small number of spermatozoa with aberrant methylation modifications in the GNAS XL DMR might exist even in semen samples with normal parameters.
Fig. 3.
Methylation analyses of GNAS XL DMR of BSAS of two normal controls (top a and b) and two semen samples from asthenospermic group (c) and oligoasthenospermic group (d). Mean methylation levels and number of reads are given below each pattern. Lines represent reads; columns represent CpG dinucleotides; blue squares represent unmethylated CpGs; red squares represent methylated CpGs; white squares represent missing sequence information
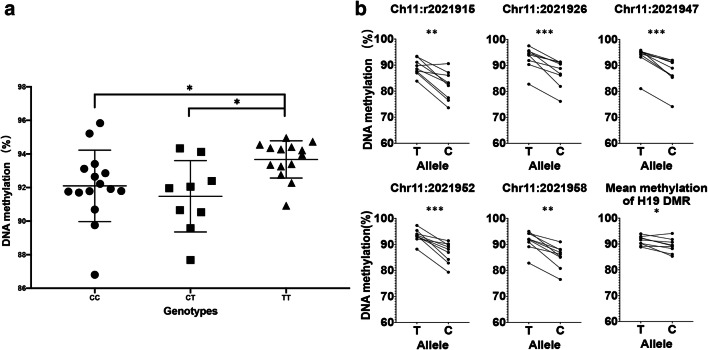
SNP effect on the methylation level of H19 DMR
A SNP (rs2525883) was included in the bisulfite amplicon sequencing (BSAS) fragment of the H19 DMR. The locus was highly polymorphic in 38 samples, with 15 CC, 9 CT and 14 TT genotypes. Interestingly, we found significant differences in the H19 DMR methylation levels between samples with different genotypes. Samples with the TT genotype had the highest mean methylation level of 93.67%, while samples with the CT and CC genotypes had mean methylation levels of 91.48% and 92.10%, respectively (Fig. 4a). Further, in nine CT heterozygous samples, allele-specific methylation levels were analysed, and the mean methylation level of the T allele was found to be significantly higher than that of the C allele, especially at CpG sites such as Chr11:2021915, Chr11:2021926, Chr11:2021947, Chr11:2021952 and Chr11:2021958 (Fig. 4b). Another interesting finding is that the T allele was presented only three times (20%) in the normozoospermic group, while it appeared 3 times (55%) in the abnormal groups, and this difference was significantly different (p-corrected of chi-square test = 0.033). Further investigations are needed to reveal the reasons and implications of this genetic-epigenetic interaction.
Fig. 4.
Methylation analyses of H19 DMR of BSAS according to the SNP rs2525883 genotype. a The mean methylation level of H19 DMR of the semen samples with different genotypes. b The allele-specific methylation level of 5 CpG sites and the mean of all CpG sites from H19 DMR of the semen samples with heterozygous genotype CT. Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Significance was determined using the Wilcoxon matched-pairs signed rank test
Aberrant DNA methylation analysis of non-imprinted gene regions
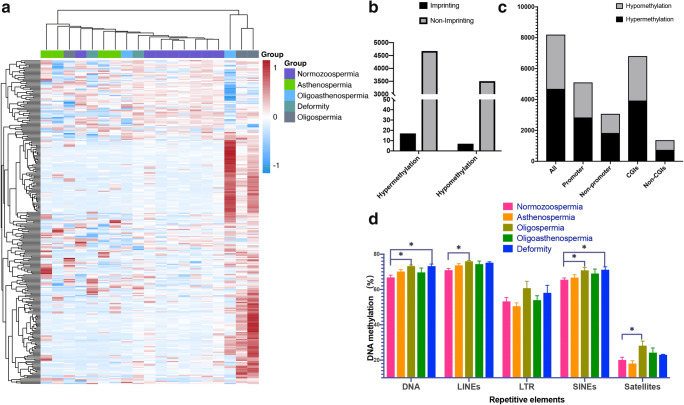
We also analysed the DNA methylation profiles of other genomic regions in the RRBS results. By comparing abnormal semen groups with the normal group using a p value threshold of less than 0.01, we found 10,429 differentially methylated sites and 8202 DMRs (one region per 1000 bp, with at least 5 CpG sites in one DMR and regional methylation as the average of methylation of all CpG sites within the region). Hierarchical clustering of DNA methylation data for the most 240 variable methylated regions highlighted the differences between abnormal semen groups and the normal group (Fig. 5a). Of these, 4691 regions were hypermethylated in abnormal semen samples, and 3511 regions were hypomethylated in abnormal semen samples. There were 24 (17 hypermethylated, 7 hypomethylated) regions with differential methylation modifications that mapped to imprinted gene DMR regions and 8178 (4674 hypermethylated, 3504 hypomethylated) that mapped to non-imprinted gene regions (Fig. 5b). In addition, there were 5106 DMRs in promoters and 6813 DMRs in CGIs (Fig. 5c). In the repetitive sequence regions, the methylation levels of DNA transposons, long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs) were significantly higher in the abnormal samples (especially in the oligospermic and deformed groups) than in the normal control samples (Fig. 5d).
Fig. 5.
DNA methylation patterns in normal and abnormal semen samples. a Unsupervised hierarchical clustering and heatmap including the 280 most DMRs (one region per 1000 bp, more than 5 CpG sites in one DMR, P < 0.01 and absolute methylation level differences > 10%) between normal and abnormal semen samples. Average methylation values are displayed from − 1 (blue) to 1 (red). b Distribution of differentially methylated regions relative to imprinting DMR. c Distribution of DMRs relative to the promoter and CGI. CGI, CpG islands. d The mean methylation level of the repetitive elements, including DNA transposons (DNA), long interspersed nuclear elements (LINEs), long terminal repeats (LTR), short interspersed nuclear elements (SINEs) and satellites. Data are presented as the mean ± SEM. *P < 0.05. Significance was determined using two-tailed Mann–Whitney test
Discussion
Abnormalities in sperm methylation may be associated with various male reproductive problems, but the conclusions remain unclear. In this study, we analysed and validated the DNA methylation profiles of abnormal semen samples at the genome-wide level by RRBS and bisulfite amplicon sequencing on the Illumina platform. The results showed that in the abnormal semen groups, some imprinted genes, including H19, GNAS, MEG8 and SNRPN, possibly had abnormal modifications of DNA methylation with great inter- and intra-sample heterogeneity. Some differentially methylated non-imprinting regions were also identified, including the hypermethylation of repetitive sequence DNA transposons, LINEs and SINEs, in abnormal groups. Another interesting result was that the genotypes of one SNP in H19 DMR affected nearby DNA methylation levels. Considering that these methylation modification abnormalities might escape the demethylation process of the zygote and thus be transmitted to the next generation, leading to related diseases, special attention needs to be paid to the possible epigenetic risks during the reproductive process, especially when ARTs are used.
ART can help infertile patients or patients with low fertility father their own children, though it may also allow sperm cells with genetic/epigenetic defects to escape natural selection and fertilize an egg. Therefore, clarifying what genetic/epigenetic defects may exist in sperm can help in the application of ART. Nonetheless, male infertility is a multifactorial and complex condition, and the genetic/epigenetic abnormalities associated with infertility are not entirely known [37]. Recently, epigenetic modifications, including DNA methylation, histone modifications and non-coding RNAs, have been suggested to be closely related to the aetiology of infertility in men [38]. The relationship between abnormal DNA methylation of sperm and male infertility as well as unfavourable reproductive phenotypes has been reported in several studies [6, 8, 16, 30, 32, 33], but the conclusions were not always consistent [17]. Meta-analyses have shown that aberrant methylation of imprinted genes H19, MEST or SNRPN in sperm increase the risk of male infertility [17, 31]. For other imprinting DMRs such as GNAS, MEG8, IGF2 and MEG3, a clear conclusion has not been drawn because of contradictory results or the lack of sufficient study [16, 17]. These inconsistent results might also be related to the different DNA methylation detection techniques used in each study. A semen sample is a mixture of spermatozoa, whereas the level of DNA methylation level in a sample is the average of the individual methylation levels of all spermatozoa. Methods with higher precision are required to identify some smaller differences in DNA methylation levels. Our study used the method of high-throughput bisulfite sequencing, which can provide highly accurate quantitative results because of the large number of sequencing reads. Our investigation confirmed that the imprinted gene DNA methylation levels were related to sperm phenotypes. We profiled the methylation patterns of more than 60 imprinted gene DMRs by RRBS and found that the imprinted regions of genes such as H19, GNAS, SNRPN and MEG8 may have aberrant methylation modifications in abnormal sperm.
In addition to imprinting DMRs, repetitive sequences can also escape waves of demethylation after fertilization [39]; thus, we analysed the DNA methylation levels of DNA transposons, LINEs and SINEs. Hypermethylation of these repetitive sequences in abnormal samples were found in our study; these results contradicted the results of some previous studies [13, 23, 40] and might have resulted from different sample selection strategies.
Another finding of our study was the presence of inter- and intra-sample epigenetic heterogeneity. Inter-sample heterogeneity was characterized by different semen samples with the same phenotype having different DNA methylation modifications, which might due to the complexity of the factors lead to infertility [37]. Intra-sample heterogeneity was characterized by abnormal samples containing spermatozoa with both normal and abnormal methylation modifications, which might be observed by analysis of haplotypes of DNA methylation on high-throughput bisulfite sequencing data. Similar findings have been reported in previous studies that also employed a high-throughput bisulfite sequencing approach, whereby DNA methylation heterogeneity was present in imprinted gene regions in oligoasthenoteratozoospermic (OAT) patients [16, 41]. In addition, sperm DNA methylation heterogeneity studies have shown that even in normal male semen, a small amount of DNA methylation variations may be present. These small amounts of heterogeneity affect the expression of some disease-related genes [42] and may also directly affect the health of the next generation.
Aberrant regulation of these four imprinted genes that we report is known to be responsible for various growth and behavioural syndromes [20, 22, 43, 44]. Loss of H19 methylation is associated with Beckwith–Wiedemann syndrome and Silver–Russell syndrome [20, 22], whereas the hypermethylation of SNRPN is associated with Angelman syndrome and Prader–Willi syndrome [22]. MEG8 DMR is also hypermethylated in individuals with Temple syndrome [43]. Defects in GNAS imprinting are associated with the imprinting disorder pseudohypoparathyroidism (PHP) [44]. Certainly, the transgenerational mechanisms of these imprinting DMRs and effects on the regulation of gene expression in the next generation require further investigation.
We included an SNP site in the H19 DMR analysis, and the results revealed that genotypes might affect methylation in this DMR region. SNP effects on methylation at adjacent CpG sites have been reported in several studies [45–47], but the SNP effects on methylation in the DMRs of imprinted genes in sperm are identified for the first time, and it is worth further investigation.
Our study has several limitations. First, due to the relatively small sample size together with the heterogeneity in DNA methylation, the study was more likely to provide some trends in change instead of the final conclusion. Second, millions of CG sites were sequenced in RRBS, but the coverage per site was relatively low, which likely caused false positives and false negatives in the statistics. In our study, except for the four imprinting DMRs, other results from RRBS require further validation by a more accurate assay before subsequent studies. Finally, many environmental factors and lifestyle habits could affect DNA methylation. Only age was included in our study. The relationship with other factors requires further study.
Conclusion
In this study, we evaluated aberrant DNA methylation modifications in sperm from abnormal semen samples by high-throughput sequencing. Our results validated the previously reported aberrant DNA methylation modification of the H19 gene in abnormal sperm and found that other imprinted genes, such as GNAS, SNRPN and MEG8, showed aberrant methylation modification. In addition, heterogeneities of DNA methylation modifications in inter- and intra- samples were identified in abnormal semen samples, and an SNP in the H19 DMR fragment affected the methylation levels. These results suggest that there are potential epigenetic risks in abnormal semen samples and that further investigation is required to determine if these risks will lead to health risks in offspring conceived via ART.
Authors’ contributions
All authors contributed to the study conception and design. The study was designed and supervised by Qihan Wu and Xin Wang. Sample collection and semen parameter analyses were performed by Yuhua Sun, Xing Feng and Xin Wang. Material preparation, data collection and analysis were performed by Wanhong He, Sufen Zhang, Minjie Xu, Jianfeng Dai, Xiaohua Ni and Qihan Wu. The first draft of the manuscript was written by Qihan Wu, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding information
This work was supported by the National Natural Science Foundation of China (grant numbers 81872103 and 81971917) and the Science and Technology Climbing Fund of SIPPR (grant numbers PD2017-2 and PD2017-4).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Medical Ethics Committee of Shanghai Institute of Planned Parenthood Research. All procedures performed in studies involving human participants were in accordance with the ethical standards of NHFPC Ethical Review of Human Biomedical Research (2010) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wanhong He and Υuhua Sun contributed equally to this work.
Contributor Information
Xin Wang, Email: wangxin51305@126.com.
Qihan Wu, Email: henrywuqh@hotmail.com.
References
- 1.Reik W, Surani MA. Germline and pluripotent stem cells. Cold Spring Harb Perspect Biol. 2015;7(11). [DOI] [PMC free article] [PubMed]
- 2.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20(3):320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ndlovu MN, Denis H, Fuks F. Exposing the DNA methylome iceberg. Trends Biochem Sci. 2011;36(7):381–387. doi: 10.1016/j.tibs.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liyanage VR, Jarmasz JS, Murugeshan N, Del Bigio MR, Rastegar M, Davie JR. DNA modifications: function and applications in normal and disease states. Biology (Basel) 2014;3(4):670–723. doi: 10.3390/biology3040670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laqqan M, Ahmed I, Yasin M, Hammadeh ME, Yassin M. Influence of variation in global sperm DNA methylation level on the expression level of protamine genes and human semen parameters. Andrologia. 2020;52(1):e13484. doi: 10.1111/and.13484. [DOI] [PubMed] [Google Scholar]
- 7.Siddeek B, Mauduit C, Simeoni U, Benahmed M. Sperm epigenome as a marker of environmental exposure and lifestyle, at the origin of diseases inheritance. Mutat Res. 2018;778:38–44. doi: 10.1016/j.mrrev.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins TG, Aston KI, Meyer TD, Hotaling JM, Shamsi MB, Johnstone EB, et al. Decreased fecundity and sperm DNA methylation patterns. Fertil Steril. 2016;105(1):51–7 e1–3. doi: 10.1016/j.fertnstert.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milekic MH, Xin Y, O'Donnell A, Kumar KK, Bradley-Moore M, Malaspina D, et al. Age-related sperm DNA methylation changes are transmitted to offspring and associated with abnormal behavior and dysregulated gene expression. Mol Psychiatry. 2015;20(8):995–1001. doi: 10.1038/mp.2014.84. [DOI] [PubMed] [Google Scholar]
- 10.Skinner MK, Guerrero-Bosagna C, Haque MM. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics. 2015;10(8):762–771. doi: 10.1080/15592294.2015.1062207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aston KI, Uren PJ, Jenkins TG, Horsager A, Cairns BR, Smith AD, et al. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril. 2015;104(6):1388–97 e1–5. doi: 10.1016/j.fertnstert.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 12.Du Y, Li M, Chen J, Duan Y, Wang X, Qiu Y, et al. Promoter targeted bisulfite sequencing reveals DNA methylation profiles associated with low sperm motility in asthenozoospermia. Hum Reprod. 2016;31(1):24–33. doi: 10.1093/humrep/dev283. [DOI] [PubMed] [Google Scholar]
- 13.Urdinguio RG, Bayon GF, Dmitrijeva M, Torano EG, Bravo C, Fraga MF, et al. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum Reprod. 2015;30(5):1014–1028. doi: 10.1093/humrep/dev053. [DOI] [PubMed] [Google Scholar]
- 14.Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A. 2014;111(5):1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins TG, Aston KI, Trost C, Farley J, Hotaling JM, Carrell DT. Intra-sample heterogeneity of sperm DNA methylation. Mol Hum Reprod. 2015;21(4):313–319. doi: 10.1093/molehr/gau115. [DOI] [PubMed] [Google Scholar]
- 16.Laurentino S, Borgmann J, Gromoll J. On the origin of sperm epigenetic heterogeneity. Reproduction. 2016;151(5):R71–R78. doi: 10.1530/REP-15-0436. [DOI] [PubMed] [Google Scholar]
- 17.Santi D, De Vincentis S, Magnani E, Spaggiari G. Impairment of sperm DNA methylation in male infertility: a meta-analytic study. Andrology. 2017;5(4):695–703. doi: 10.1111/andr.12379. [DOI] [PubMed] [Google Scholar]
- 18.Lou H, Le F, Hu M, Yang X, Li L, Wang L, Wang N, Gao H, Jin F. Aberrant DNA methylation of IGF2-H19 locus in human fetus and in spermatozoa from assisted reproductive technologies. Reprod Sci. 2019;26(7):997–1004. doi: 10.1177/1933719118802052. [DOI] [PubMed] [Google Scholar]
- 19.Choufani S, Turinsky AL, Melamed N, Greenblatt E, Brudno M, Berard A, Fraser WD, Weksberg R, Trasler J, Monnier P, et al. Impact of assisted reproduction, infertility, sex and paternal factors on the placental DNA methylome. Hum Mol Genet. 2019;28(3):372–385. doi: 10.1093/hmg/ddy321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72(1):156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortessis VK, Azadian M, Buxbaum J, Sanogo F, Song AY, Sriprasert I, Wei PC, Yu J, Chung K, Siegmund KD. Comprehensive meta-analysis reveals association between multiple imprinting disorders and conception by assisted reproductive technology. J Assist Reprod Genet. 2018;35(6):943–952. doi: 10.1007/s10815-018-1173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hattori H, Hiura H, Kitamura A, Miyauchi N, Kobayashi N, Takahashi S, Okae H, Kyono K, Kagami M, Ogata T, Arima T. Association of four imprinting disorders and ART. Clin Epigenetics. 2019;11(1):21. doi: 10.1186/s13148-019-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, Hahn T, et al. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011;5(2):60–69. doi: 10.1159/000323806. [DOI] [PubMed] [Google Scholar]
- 24.Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol. 2014;6(2). [DOI] [PMC free article] [PubMed]
- 25.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9(2):129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 27.Surani MA. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93(3):309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 28.Tilghman SM. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96(2):185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 29.Darbandi M, Darbandi S, Agarwal A, Baskaran S, Dutta S, Sengupta P, Khorram Khorshid HR, Esteves S, Gilany K, Hedayati M, Nobakht F, Akhondi MM, Lakpour N, Sadeghi MR. Reactive oxygen species-induced alterations in H19-Igf2 methylation patterns, seminal plasma metabolites, and semen quality. J Assist Reprod Genet. 2019;36(2):241–253. doi: 10.1007/s10815-018-1350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Q, Pan F, Yang J, Fu Z, Lu Y, Wu X, Han X, Chen M, Lu C, Xia Y, Wang X, Wu W. Idiopathic male infertility is strongly associated with aberrant DNA methylation of imprinted loci in sperm: a case-control study. Clin Epigenetics. 2018;10(1):134. doi: 10.1186/s13148-018-0568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klaver R, Gromoll J. Bringing epigenetics into the diagnostics of the andrology laboratory: challenges and perspectives. Asian J Androl. 2014;16(5):669–674. doi: 10.4103/1008-682X.125412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS One. 2007;2(12):e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sujit KM, Singh V, Trivedi S, Singh K, Gupta G, Rajender S. Increased DNA methylation in the spermatogenesis-associated (SPATA) genes correlates with infertility. Andrology. 2019. [DOI] [PubMed]
- 34.World Health Organization . WHO laboratory manual for the examination and processing of human semen. 5. Geneva: World Health Organization; 2010. p. xiv. [Google Scholar]
- 35.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6(4):468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 36.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuttelmann F, Gromoll J, Kliesch S. Genetics of male infertility. Urol A. 2008;47(12):1561–1562. doi: 10.1007/s00120-008-1804-4. [DOI] [PubMed] [Google Scholar]
- 38.Carrell DT. Epigenetics of the male gamete. Fertil Steril. 2012;97(2):267–274. doi: 10.1016/j.fertnstert.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 39.Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, et al. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161(6):1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson ME, Bleiziffer A, Tuttelmann F, Gromoll J, Wilkinson MF. Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum Mol Genet. 2014;23(1):12–23. doi: 10.1093/hmg/ddt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhtz J, Schneider E, El Hajj N, Zimmermann L, Fust O, Linek B, et al. Epigenetic heterogeneity of developmentally important genes in human sperm: implications for assisted reproduction outcome. Epigenetics. 2014;9(12):1648–1658. doi: 10.4161/15592294.2014.988063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, Petronis A. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79(1):67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beygo J, Kuchler A, Gillessen-Kaesbach G, Albrecht B, Eckle J, Eggermann T, Gellhaus A, Kanber D, Kordass U, Ludecke HJ, et al. New insights into the imprinted MEG8-DMR in 14q32 and clinical and molecular description of novel patients with Temple syndrome. Eur J Hum Genet. 2017;25(8):935–945. doi: 10.1038/ejhg.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zink F, Magnusdottir DN, Magnusson OT, Walker NJ, Morris TJ, Sigurdsson A, Halldorsson GH, Gudjonsson SA, Melsted P, Ingimundardottir H, Kristmundsdottir S, Alexandersson KF, Helgadottir A, Gudmundsson J, Rafnar T, Jonsdottir I, Holm H, Eyjolfsson GI, Sigurdardottir O, Olafsson I, Masson G, Gudbjartsson DF, Thorsteinsdottir U, Halldorsson BV, Stacey SN, Stefansson K. Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat Genet. 2018;50(11):1542–1552. doi: 10.1038/s41588-018-0232-7. [DOI] [PubMed] [Google Scholar]
- 45.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49(1):131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 46.Hannon E, Weedon M, Bray N, O'Donovan M, Mill J. Pleiotropic effects of trait-associated genetic variation on DNA methylation: utility for refining GWAS loci. Am J Hum Genet. 2017;100(6):954–959. doi: 10.1016/j.ajhg.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulz H, Ruppert AK, Herms S, Wolf C, Mirza-Schreiber N, Stegle O, Czamara D, Forstner AJ, Sivalingam S, Schoch S, Moebus S, Pütz B, Hillmer A, Fricker N, Vatter H, Müller-Myhsok B, Nöthen MM, Becker AJ, Hoffmann P, Sander T, Cichon S. Genome-wide mapping of genetic determinants influencing DNA methylation and gene expression in human hippocampus. Nat Commun. 2017;8(1):1511. doi: 10.1038/s41467-017-01818-4. [DOI] [PMC free article] [PubMed] [Google Scholar]