Abstract
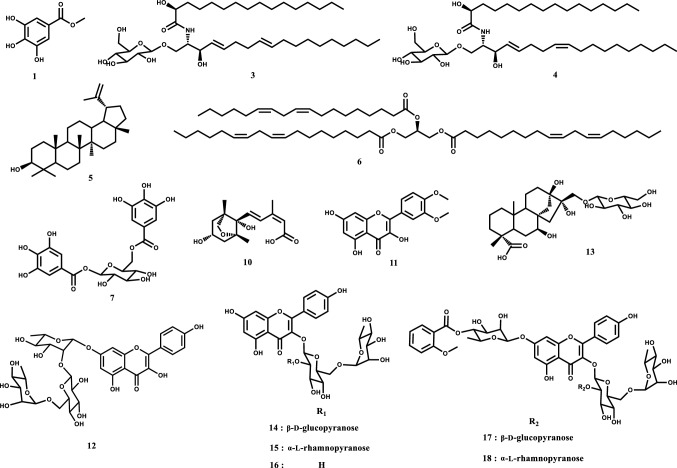
Eighteen compounds including new caryophyllene-type sesquiterpene and flavonol tetraglycoside were purified and isolated from sword beans (Canavalia gladiata). Two new compounds, (Z,1R,7S,9S)-7-hydroxy-11,11-dimethyl-8-methylenebicyclo[7.2.0]undec-4-ene-4-carboxylic acid (2) and kaempferol-7-O-α-l-dirhamnopyranosyl(1 → 2;1 → 6)-O-β-d-glucopyranosyl(1 → 2)-O-α-l-rhamnopyranoside (9), were identified. Other known compounds including methyl gallate (1), (2S,3S,4E,8E)-2-aminooctadeca-4,8-diene-1,3-diol 1-O-β-d-glucopyranoside (3), (2S,3S,4E,8Z)-2-aminooctadeca-4,8-diene-1,3-diol 1-O-β-d-glucopyranoside (4), lupeol (5), trilinolein (6), 1,6-di-O-galloyl β-d-glucopyranoside (7), N-(2-methoxybenzoyl)homoserine (8), dihydrophaseic acid (10), dillenetin (11), kaempferol-7-O-[2-O-β-d-glucopyranosyl-6-O-α-l-rhamnopyranosyl]-α-l-rhamnopyranoside (12), canavalioside (13), kaempferol-3-O-[2-O-β-d-glucopyranosyl-6-O-α-l-rhamnopyranosyl]-β-d-glucopyranoside (14), kaempferol-3-O-(2,6-O-α-l-dirhamnopyranosyl)-β-d-glucopyranoside (15), kaempferol-3-O-rutinoside (16), gladiatoside A1 (17), and gladiatoside B1 (18) were identified. The chemical structures of these compounds were determined by ESI–MS and NMR analyses.
Electronic supplementary material
The online version of this article (10.1007/s10068-020-00794-8) contains supplementary material, which is available to authorized users.
Keywords: Canavalia galidata, Sword bean, Flavonoid tetraglycoside, Sesquiterpenoic acid, Cerebioside
Introduction
Sword bean (Canavalia gladiata) belongs to the legume family, and it is widely cultivated in Asia and Africa (Lee and Jeong, 2005). The green fruit of sword bean is generally used as a domestic vegetable, and a ripe seed is consumed as cooked beans (Chang et al., 2011; Ekanayake et al., 2007). The bean has been used in traditional Chinese medicine for the treatment of purulent inflammation, such as sinusitis, hemorrhoids, and boils (Kim et al., 2012). Sword bean has been demonstrated to have several biological effects, including anti-oxidant (Gan et al., 2016), anti-bacterial (Lee and Jeong, 2005; Cho et al., 2000), anti-gastric inflammatory (Kim et al., 2013a, b), attenuation of bowel disease (Ji et al., 2018), and antiangiogenic activity (Jeon et al., 2005). Interestingly, sword bean does not contain isoflavones, such as daidzin, genistin, daidzein, and genistein, which are common bioactive compounds in soybean and black bean (Cho et al., 1999). Moreover, sword bean has high total phenolic and flavonoid contents, which are similar to those of soybean and black bean (Gan et al., 2018). Understanding the chemical constituents of sword bean is very important to assess its quality as a food material. Bioactive compounds including gallic acid derivative, ent-kaurane-type diterpene glycoside (canavalioside), and various flavonol glycosides (gladiatosides) have been found in sword beans (Kim et al., 2013a, b; Lee and Jeong, 2005; Murakami et al., 2000). However, information on the chemical constituents of the sword bean is limited. We purified and isolated 18 compounds, including new caryophyllene-type sesquiterpene and flavonol tetraglycoside from sword bean. In this study, we demonstrated the structural determination of new caryophyllene-type sesquiterpene and flavonol tetraglycoside found in sword beans.
Materials and methods
General experimental procedures
Nuclear magnetic resonance (NMR) spectra were recorded using unitINOVA 500 and 600 spectrometers (Varian, Walnut Creek, CA). Deuterated methanol (CD3OD), deuterated chloroform (CDCl3), and deuterated pyridine (Acros Organics, Morris Plains, NJ, USA) containing tetramethylsilane as an analytical solvent was used. The mass spectra were acquired on a Synapt G2 HDMS electrospray ion source (ESI) quadrupole time-of-flight (Q-TOF) mass spectrometer (Waters, Milford, MA). UV spectra were obtained by a Shimadzu Mini-1240 UV/VIS spectrometer (Shimadzu, Kyoto, Japan). The optical rotations were measured by a A.KRÜSS Optronic P8000 Automatic Polarimeter (A.KRÜSS, Berlin, Germany). Silica gel (Kieselgel 60, 70-230 mesh, Merck, Darmstadt, Germany), Amberlite XAD-2 (70-230 mesh, Merck), and ODS (70-230 mesh, Merck) for column chromatography were used to purify the compounds from the sword beans. The compounds were purified and isolated using a high-performance liquid chromatography (HPLC) instrument (LC-20A, Shimadzu, Kyoto, Japan) equipped with Silica-60 (5 μm, 7.8 × 250 mm, Shimadzu), Shim-pack Prep-ODS (H) Kit (5 μm, 20 × 250 mm, Shimadzu), Intersil ODS-3 (5 μm, 7.6 × 250 mm, Shimadzu), and ODS-80Ts (TSK-gel, 4.6 × 250 mm, Tosoh, Tokyo, Japan)]. The eluents were monitored at 210 and 254 nm. Thin-layer chromatography (TLC) was carried out using silica gel TLC plates (silica gel 60 F254, 0.25 mm thickness, Merck) and developed using a mixture of EtOAc/CHCl3/acetic acid = 3:1:2 (v/v). The spots were spayed by a 200 μM DPPH ethanol solution to screen antioxidative compounds. The methanol (MeOH), acetonitrile (MeCN), n-hexane, 2-propanol, and ethanol (EtOH) solutions used for analyses were of HPLC grade and were purchased from Fisher Scientific Korea Ltd. (Seoul, Korea). Acetic acid (AcOH) and trifluoroacetic acid (TFA) were obtained from Sigma-Aldrich Chemical Co. (St. Louis, Mo, USA).
Plant material
Sword beans (Canavalia gladiata, Jacq. DC) were harvested in September, in Gwang-ju, South Korea. This bean was identified by Prof. Sang-Hyun Lee, Laboratory of Pomology, College of Agriculture and Life Science, Chonnam National University. A voucher sample (no. JNU CG 20150931) was deposited in the laboratory herbarium. The fresh white sword beans were separated from the pod and stored at –70°C before using.
Extraction and solvent fractionation
The fresh sword bean (5 kg) was homogenized with MeOH (20 L) using a homogenizer (Ultra-Turrax T50, IKA, Seoul, Korea). After extraction for 1 day at room temperature, the mixture was filtered under a vacuum using No. 2 filter paper (Whatman, Maidstone, England). The residue was repeatedly extracted by adding MeOH (15 L). The MeOH extracts were combined and concentrated using a vacuum evaporator at 37 °C. The MeOH extracts (206.5 g) were suspended in 2 L of distilled water and solvent fractionated with n-hexane (2 L, 3 times), followed by CHCl3 (2 L, 3 times), EtOAc (2 L, 3 times), and water-saturated n-butanol (BuOH) (2 L, 3 times).
Purification of 1–6 from n-hexane and CHCl3 fraction
The combined n-hexane and CHCl3 fraction (21.7 g) was fractionated by silica-gel column (5 × 82 cm) chromatography and eluted with n-hexane/EtOAc/MeOH (10:0:0, 8:2:0, 6:4:0, 4:6:0, 2:8:0, 0:10:0, 0:8:2, 0:6:4, 0:4:6, 0:2:8, 0:0:10, v/v, step-wise system, each 2.6 L). Forty fractions (H1 ~ H40) were grouped by the TLC analysis. Fraction H25 (424.5 mg) was subjected to ODS-HPLC [column, Shim-pack Prep-ODS (H) KIT; flow rate, 9.0 mL/min; linear gradient elution of 20% MeOH (pH 3.0) containing TFA (A) and 100% MeOH (B), 0% B for initial → 50% B for 25 min → 50% B for 40 min; detection, 254 nm] to afford two compounds (1, tR 18.43 min, 23.3 mg; 2, tR 37.89 min, 1.6 mg) and the mixture of 3 and 4. Fraction H3 (6.0 g) was injected onto normal phase-HPLC [column, silica-60; flow rate, 5.0 mL/min; linear gradient elution of n-hexane (C) and n-hexane/2-propanol/EtOH = 8:4:1 v/v (D), initial 0% D → 100% D for 25 min; detection, 210 nm] to obtain the mixture of 5 and 6 (tR 15.3 min, 60.2 mg).
Purification of 7–18 from BuOH fraction
The BuOH fraction (13.0 g) was separated by Amberlite XAD-2 column (5 × 82 cm) chromatography and eluted with H2O/MeOH (10:0, 8:2, 6:4, 4:6, 2:8, 0:10, v/v, step-wise system, each 2.4 L). Nineteen fractions (BA-BP) were grouped by the TLC analysis. Fraction BI (424.5 mg) was separated by ODS column chromatography (3.2 × 95 cm) and eluted with H2O/MeOH (8:2, 6:4, 4:6, 2:8, 0:10, v/v, step-wise system, each 1.2 L) to obtain 24 subfractions (BI1-BI24). Compound 7 (tR 20.0 min, 1.0 g) was isolated from subfraction BI6 (1.13 g) by ODS-HPLC [column, Intersil ODS-3; flow rate, 1.0 mL/min; linear gradient elution of 20% MeOH (E) and 40% MeOH (F), initial 0% F → 30% F for 20 min → 100% F for 40 min; detection, 210 and 280 nm]. The subfractions, BI9, BI11, BI14, BI17, and BI19, were purified by ODS-HPLC [column, Intersil ODS-3; flow rate, 5.0 mL/min; detection, 210 nm] with their different elutions. Compounds 8 (tR 8.1 min, 3.0 mg) and 9 (tR 21.4 min, 19.4 mg) were separated from subfraction BI9 (182.6 mg) by ODS-HPLC [linear gradient elution of 20% MeOH (G) and 35% MeOH (H), initial 0% H → 30% G for 25 min → 100% G for 40 min]. Compounds 10 (tR 13.9 min, 1.0 mg), 11 (tR 23.2 min, 3.5 mg), and 12 (tR 28.8 min, 12.0 mg) were separated from subfraction BI11 (29.2 mg) by ODS-HPLC [linear gradient elution of 20% MeOH (I) and 30% MeOH (J), initial 0% J → 70% J for 25 min → 70% J for 35 min]. The mixture of 13 and 14 (tR 11.0 min, 12.0 mg) was separated from subfraction BI14 (56.4 mg) by ODS-HPLC [linear gradient elution of 20% MeOH (K) and 50% MeOH (L), initial 0% L → 70% L for 25 min → 70% L for 35 min]. Compound 15 (tR 12.5 min, 6.0 mg) was separated from subfraction BI17 (71.4 mg) by ODS-HPLC [linear gradient elution of 35% MeOH (M) and 40% MeOH (N), initial 0% N → 100% N for 30 min → 100% L for 40 min]. Compounds 16 (tR 14.2 min, 5.0 mg), 17 (tR 19.7 min, 3.0 mg), and 18 (tR 21.8 min, 4.0 mg) were separated from subfraction BI19 (20.9 mg) by ODS-HPLC [linear gradient elution of 35% MeOH (O) and 60% MeOH (P), initial 0% P → 100% N for 40 min → 100% P for 50 min].
Compound 1: white powder; 1H-NMR (500 MHz, CD3OD) δ 7.04 (2H, s, H-2 and H-6), 3.81 (3H, s, -OCH3); ESI–MS (negative) m/z 183.0 [M – H]−.
Compound 2: white powder; 1H- and 13C-NMR data are shown in Table 1. HR–ESI–MS m/z 249.1493 [M – H]– (calculated for C15H21O3, m/z 249.1493, +0.2 mDa).
Table 1.
1H- and 13C- NMR spectral data of 2 in CD3OD
| Carbon | δH (int., mult., J in Hz) | δC |
|---|---|---|
| 1 | 1.75 (1H, m) | 55.1 |
| 2a | 1.81 (1H, m) | 28.1 |
| 2b | 1.66 (1H, m) | |
| 3a | 2.57 (1H, m) | 24.3 |
| 3b | 2.33 (1H, m) | |
| 4 | – | 135.5 |
| 5 | 6.69 (1H, dd, 9.0, 5.5) | 139.5 |
| 6a | 2.72–2.66 (1H, m) | 36.0 |
| 6b | 2.53 (1H, m) | |
| 7 | 4.40 (1H, dd, 7.5, 4.5) | 76.2 |
| 8 | – | 159.0 |
| 9 | 2.77 (1H, td, 7.0, 7.5) | 33.6 |
| 10a | 1.69 (1H, m) | 41.9 |
| 10b | 1.80 (1H, m) | |
| 11 | – | 33.8 |
| 12 | – | 171.7 |
| 13a | 5.02 (1H, br. s) | 113.0 |
| 13b | 5.07 (1H, br. s) | |
| 14 | 1.00 (3H, s) | 24.3 |
| 15 | 1.03 (3H, s) | 30.8 |
Compound 3: white powder; 1H-NMR (500 MHz, pyridine-d5, TMS) δ 8.40 (1H, d, J = 8.5 Hz, NH), 7.59 (1H, br. s, C-2′-OH), 6.90 (1H, br. s, C-3-OH), 6.00 (1H, dd, J = 15.5, 6.0 Hz, H-4), 5.96 (1H, dt, J = 15.5, 6.0 Hz, H-5), 5.50 (2H, m, H-8 and H-9), 4.91 (1H, d, J = 7.7 Hz, H-1′′), 4.82 (1H, m, H-2), 4.70 (1H, dd, J = 10.5, 5.5 Hz, H-1a), 4.53 (1H, m, H-1b), 4.52 (1H, dd, J = 11.5, 3.0 Hz, H-2′), 4.50 (1H, dd, J = 11.8, 4.0 Hz, H-6′′a), 4.40 (1H, dd, J = 11.8, 5.5 Hz, H-6′′b), 4.21 (2H, s, H-3′′ and H-4′′), 4.01 (1H, t, J = 7.5 Hz, H-2′′), 3.90 (1H, m, H-5′′), 2.20 (2H, m, H-6a), 2.09 (2H, m, H-9), 1.86 (1H, m, H-3′a), 1.80 (2H, m, H-10), 1.74 (1H, m, H-4′a), 1.32–1.45 (36H, m, CH2), 0.88 (3H, t, J = 7.5 Hz, H-18, CH3); 13C-NMR (125 MHz, pyridine-d5, TMS) δ 176.0 (C-1′), 132.5 (C-5), 132.4 (C-4), 131.0 (C-9), 129.7 (C-8), 106.0 (C-1′′), 78.9 (C-5′′), 78.8 (C-3′′), 75.5 (C-2′′), 72.8 (C-2′), 72.6 (C-3), 71.8 (C-4′′), 70.5 (C-1), 63.0 (C-6′′), 54.9 (C-2), 36.0 (C-3′), 33.2 (C-6), 32.5 (C-16), 30.0–30.4 (C-11–C-15), 27.9 (C-10), 27.8 (C-7), 26.3 (C-4′), 23.3 (C-17), 23.3–32.5 (C-5′–C-17′), 14.6 (C-18); ESI–MS (negative) m/z 712.60 [M – H]−.
Compound 4: white powder; 1H-NMR (500 MHz, pyridine-d5, TMS) δ 8.40 (1H, d, J = 8.7 Hz, NH), 7.59 (1H, br. s, C-2′-OH), 6.90 (1H, br. s, C-3-OH), 6.00 (1H, dd, J = 15.0, 6.0 Hz, H-4), 5.96 (1H, dt, J = 15.0, 6.0 Hz, H-5), 5.50 (2H, m, H-8 and H-9), 4.91 (1H, d, J = 7.7 Hz, H-1′′), 4.82 (1H, m, H-2), 4.70 (1H, dd, J = 10.0, 6.0 Hz, H-1a), 4.53 (1H, m, H-1b), 4.52 (1H, dd, J = 11.5, 2.5 Hz, H-2′), 4.50 (1H, dd, J = 11.8, 2.5 Hz, H-6′′a), 4.40 (1H, dd, J = 11.8, 5.5 Hz, H-6′′b), 4.21 (2H, s, H-3′′ and H-4′′), 4.01 (1H, t, J = 7.5 Hz, H-2′′), 3.90 (1H, m, H-5′′), 2.20 (3H, m, H-6a and H-7), 2.04 (1H, m, H-3′a), 1.80 (2H, m, H-10), 1.74 (1H, m, H-4′a), 1.32–1.45 (36H, m, CH2), 0.88 (3H, t, J = 15.0 Hz, H-18); 13C-NMR (125 MHz, CD3OD, TMS) δ 176.0 (C-1′), 132.5 (C-5), 132.4 (C-4), 131.0 (C-9), 129.7 (C-8), 106.0 (C-1′′), 78.9 (C-5′′), 78.8 (C-3′′), 75.5 (C-2′′), 72.8 (C-2′), 72.6 (C-3), 71.8 (C-4′′), 70.5 (C-1), 63.0 (C-6′′), 54.9 (C-2), 36.0 (C-3′), 33.2 (C-6), 32.5 (C-16), 30.0–30.4 (C-11–C-15), 27.9 (C-10), 27.8 (C-7), 26.3 (C-4′), 23.3 (C-17), 23.3–32.5 (C-5′–C-17′), 14.6 (C-18 and C-18′); ESI–MS (negative) m/z 712.60 [M – H]−.
Compound 5: white powder; 1H-NMR (500 MHz, CDCl3, TMS) δ 4.69 (1H, s, H-29a), 4.56 (1H, s, H-29b), 3.18 (1H, dd, J = 11.5, 5.0 Hz, H-3), 1.68 (3H, s, H-30), 1.02 (3H, s, H-26), 0.96 (3H, s, H-23), 0.94 (3H, s, H-27), 0.82 (3H, s, H-25), 0.78 (1H, s, H-28), 0.76 (3H, s, H-24); 13C-NMR (125 MHz, CDCl3, TMS) δ 150.96 (C-20), 109.31 (C-29), 78.99 (C-3), 55.30 (C-4), 50.44 (C-9), 48.30 (C-18), 47.98 (C-19), 43.00 (C-17), 42.83 (C-14), 40.83 (C-8), 40.00 (C-22), 38.86 (C-4), 38.71 (C-1), 38.05 (C-13), 37.17 (C-10), 35.58 (C-16), 34.28 (C-7), 29.85 (C-21), 27.98 (C-23), 27.44 (C-2), 27.30 (C-15), 25.14 (C-12), 20.93 (C-11), 18.32 (C-6), 19.30 (C-30), 18.00 (C-28), 16.11 (C-25), 15.97 (C-26), 15.36 (C-24), 14.54 (C-27); ESI–MS (negative) m/z 425.30 [M – H]−.
Compound 6: white powder; 13C-NMR (125 MHz, CDCl3, TMS) δ 173.27 (C-1′ and C-1′′′), 172.82 (C-1′′), 130.21 (C-9′′ and C-13′′), 130.19 (C-9′ and C-13′), 130.00 (C-9′′′ and C-13′′′), 128.07 (C-10′ and C-12′), 128.05 (C-10′′ and C-12′′), 127.88 (C-10′′′ and C-12′′′), 68.87 (C-1 and C-3), 62.09 (C-2), 34.19 (C-2′′), 34.02 (C-2′ and C-2′′′), 31.52 (C-16′, C-16′′ and C-16′′′), 29.65 (C-4′ and C-7′), 29.62 (C-4′′, C-4′′′, C-7′′, and C-7′′′), 29.36, (C-15′, C-15′′, and C-15′′′), 29.27 (C-5′), 29.20 (C-5′′ and C-5′′′), 29.19 (C-8′, C-8′′, and C-8′′′), 29.18 (C-6′), 29.11 (C-6′′ and C-6′′′), 29.08 (C-4′), 29.05 (C-4′′ and C-4′′′), 27.19 (C-14′, C-14′′, and C-14′′′), 25.63 (C-11′, C-11′′, and C-11′′′), 24.88 (C-3′), 24.86 (C-3′′ and C-3′′′), 22.57 (C-17′, C-17′′, and C-17′′′), 14.07 (C-18′, C-18′′, and C-18′′′); ESI–MS (positive) m/z 879.35 [M + H]+.
Compound 7: white powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 7.14 (2H, s, H-2′′ and H-6′′), 7.09 (2H, s, H-2′ and H-6′), 5.68 (1H, d, J = 8.0 Hz, H-1), 4.55 (1H, dd, J = 12.0 2.0 Hz, H-6a), 4.40 (1H, dd, J = 12.0, 5.0 Hz, H-6b), 3.73 (1H, m, H-5), 3.49–3.53 (3H, m, H-2, H-3, and H-4); ESI–MS (positive) m/z 507.08 [M + Na]+.
Compound 8: white powder; –1.9 (c 0.006, MeOH); UV (CH3OH) λmax (nm) (log ε): 285 (4.49); 1H- and 13C-NMR data are shown in Table 2.; HR–ESI–MS m/z 254.0968 [M + H]+ (calculated for C12H16N1O5, m/z 254.0950, +1.8 mDa).
Table 2.
1H- and 13C- NMR spectral data of 8 in CD3OD
| Carbon | δH (int., mult., J in Hz) | δC |
|---|---|---|
| 1 | – | 120.9 |
| 2 | – | 158.1 |
| 3 | 7.16 (1H, br. d, 8.0) | 111.6 |
| 4 | 7.51 (1H, dt, 8.0, 2.0) | 133.0 |
| 5 | 7.06 (1H, dt, 8.0, 1.0) | 120.6 |
| 6 | 8.02 (1H, dd, 8.0, 2.0) | 130.9 |
| 7 | – | 165.2 |
| –OCH3 | 4.01 (3H, s) | 55.2 |
| 1′ | 176.9 | |
| 2′ | 4.57 (1H, t, 6.0) | 52.7 |
| 3′ | 2.19–2.33 (2H, m) | 31.2 |
| 4′ | 3.98–4.06 (2H, m) | 73.5 |
Compound 9: white powder; –3.7 (c 0.02, MeOH); UV (CH3OH) λmax (nm) (log ε): 265 (4.50) and 348 (4.33); 1H- and 13C-NMR data are shown in Table 3; ESI–MS (positive) m/z 925.2595 [M + Na]+ (calculated for C39H50O24Na, m/z 925.2590, −0.5 mDa).
Table 3.
1H- and 13C- NMR spectral data of 9 in CD3OD
| Carbon | δH (int., mult., J in Hz) | δC |
|---|---|---|
| 2 | – | 157.79 |
| 3 | – | 133.31 |
| 4 | – | 178.12 |
| 5 | – | 161.52 |
| 6 | 6.47 (1H, d, 2.0) | 98.99 |
| 7 | – | 161.98 |
| 8 | 6.73 (1H, d, 2.0) | 94.19 |
| 9 | – | 156.52 |
| 10 | – | 106.07 |
| 1′ | – | 121.36 |
| 2′, 6′ | 8.10 (2H, d, 9.0) | 130.93 |
| 3′, 5′ | 6.91 (2H, d, 9.0) | 114.83 |
| 4′ | – | 160.12 |
| 1′′ | 5.57 (1H, d, 1.5) | 98.49 |
| 2′′ | 4.03 (1H, dd, 3.5, 1.5) | 70.29 |
| 3′′ | 3.84 (1H, dd, 9.0, 3.5) | 70.83 |
| 4′′ | 3.49 (1H, dt, 9.0, 7.0) | 72.20 |
| 5′′ | 3.61 (1H, m) | 69.85 |
| 6′′ | 1.27 (1H, d, 6.0) | 16.69 |
| 1′′′ | 5.65 (1H, d, 7.5) | 99.42 |
| 2′′′ | 3.95 (1H, dd, 9.5, 7.5) | 76.15 |
| 3′′′ | 3.71 (1H, dd, 9.5, 8.0) | 74.31 |
| 4′′′ | 3.79 (1H, dd, 8.0, 8.0) | 69.29 |
| 5′′′ | 3.66 (1H, m) | 73.96 |
| 6′′′a | 3.73 (1H, dd, 11.5, 6.0) | 65.74 |
| 6′′′b | 3.46 (1H, dd, 11.5, 1.5) | |
| 1′′′′ | 5.23 (1H, d, 1.5) | 101.20 |
| 2′′′′ | 4.01 (1H, dd, 3.5, 1.5) | 71.00 |
| 3′′′′ | 3.80 (1H, dd, 9.5, 3.5) | 70.92 |
| 4′′′′ | 3.27 (1H, dd, 9.5, 9.5) | 72.47 |
| 5′′′′ | 3.47 (1H, m) | 68.27 |
| 6′′′′ | 1.18 (1H, d, 6.0) | 16.54 |
| 1′′′′′ | 4.53 (1H, d, 1.5) | 100.42 |
| 2′′′′′ | 3.55 (1H, dd, 3.5, 1.5) | 70.66 |
| 3′′′′′ | 3.48 (1H, dd, 9.5, 3.5) | 70.83 |
| 4′′′′′ | 3.35 (1H, dd, 9.5, 9.5) | 72.64 |
| 5′′′′′ | 4.06 (1H, m) | 68.42 |
| 6′′′′′ | 0.98 (1H, d, 6.5) | 16.13 |
Compound 10: white powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 8.01 (1H, d, J = 16.0 Hz, H-2′), 6.55 (1H, d, J = 16.0 Hz, H-1′), 5.77 (2H, s, H-4′), 4.12 (1H, m, H-4), 3.81 (1H, dd, J = 7.3, 2.5 Hz, H-8a), 3.72 (1H, br. d, J = 7.3 Hz, H-8b), 2.09 (3H, s, H-6′), 2.05 (2H, br. d, J = 10.3 Hz, H-5a), 1.86 (1H, ddd, J = 13.4, 7.0, 1.7 Hz, H-3a), 1.75 (1H, dd, J = 13.8, 10.3 Hz, H-5b), 1.68 (1H, ddd, J = 13.4, 11.0, 1.7 Hz, H-3b), 1.16 (3H, s, H-9), 0.94 (3H, s, H-7); 13C-NMR (125 MHz, CD3OD, TMS) δ 168.16 (C-5′), 150.11 (C-3′), 133.78 (C-1′), 130.37 (C-2′), 117.77 (C-4′), 86.39 (C-6), 81.83 (C-1), 75.85 (C-8), 64.59 (C-4), 48.21 (C-2), 44.58 (C-5), 43.12 (C-3), 19.83 (C-6′), 18.23 (C-9), 14.93 (C-7); HR–ESI–MS m/z 305.1382 [M + Na]− (calculated for C15H22O5Na, m/z 305.1365, +1.7 mDa).
Compound 11: yellow powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 8.11 (1H, br. s, H-2′), 8.10 (1H, br. d, J = 9.0 Hz, H-6′), 6.91 (1H, d, J = 9.0 Hz, H-5′), 6.80 (1H, d, J = 2.5 Hz, H-8), 6.74 (1H, d, J = 2.5 Hz, H-6), 3.82 (3H, s, H-7′), 3.81 (3H, s, H-8′); 13C-NMR (125 MHz, CD3OD, TMS) δ 182.83 (C-4), 163.90 (C-5), 157.95 (C-7), 157.79 (C-9), 156.56 (C-2), 151.23 (C-3′), 147.54 (C-4′), 128.93 (C-3), 122.84 (C-1′), 121.38 (C-6′), 116.37 (C-2′), 110.41 (C-5′), 105.11 (C-10), 103.59 (C-6), 99.41 (C-8); ESI–MS (positive) m/z 353.27 [M + Na]+.
Compound 12: yellow powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 8.13 (2H, br. d, J = 9.0 Hz, H-2′ and H-6′), 6.91 (2H, br. d, J = 9.0 Hz, H-3′ and H-5′), 6.74 (1H, d, J = 2.3 Hz, H-8), 6.47 (1H, d, J = 2.3 Hz, H- 6), 5.64 (1H, d, J = 8.0 Hz, H-1′′′), 5.57 (1H, d, J = 1.5 Hz, H-1′′), 4.52 (1H, d, J = 1.5 Hz, H-1′′′′), 4.06 (1H, m, H-5′′′), 4.03 (1H, dd, J = 5.0, 1.5 Hz, H-2′′), 3.95–3.27 (H-2′′–H-5′′, H-2′′′–H-6′′′, and H-2′′′′–H-5′′′′), 1.27 (3H, d, J = 6.0 Hz, H-6′′), 1.18 (3H, d, J = 6.0 Hz, H-6′′′′); 13C-NMR (125 MHz, CD3OD, TMS) δ 178.12 (C-4), 161.98 (C-7), 161.52 (C-5), 160.12 (C-4′), 157.79 (C-2), 156.52 (C-9), 133.31 (C-3), 130.93 (C-2′ and C-6′) 121.36 (C-1′), 114.83 (C-3′ and C-5′), 106.07 (C-10), 101.20 (C-1′′′′), 99.42 (C-1′′′), 98.99 (C-6), 98.49 (C-1′′), 94.19 (C-8), 76.15 (C-2′′′), 74.31 (C-3′′′), 73.96 (C-5′′′), 72.64 (C-4′′′′), 72.20 (C-4′′), 71.00 (C-2′′′′), 70.92 (C-3′′′′), 70.66 (C-3′′), 70.29 (C-2′′), 69.85 (C-5′′), 69.29 (C-4′′′), 68.42 (5′′′′), 65.74 (C-6′′′), 16.69 (6′′), 16.13 (6′′′′); ESI–MS (positive) m/z 741.20 [M + H]+.
Compound 13: white powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 4.31 (1H, d, J = 7.0 Hz, H-1′), 4.26 (1H, dd, J = 11.5, 5.5 Hz, H-6), 4.11 (1H, dd, J = 8.0, 6.0 Hz, H-3′), 3.82 (1H, dd, J = 10.0, 2.5 Hz, H-6′b), 3.71 (1H, dd, J = 10.0, 5.5 Hz, H-6′a), 3.37 (1H, m, H-5′), 3.24 (1H, dd, J = 8.0, 6.0 Hz, H-2′), 4.10 (1H, d, J = 5.5 Hz, H-7), 3.61 (1H, d, J = 11.0 Hz, H-17b), 3.50 (1H, d, J = 5.0 Hz, H-17a), 3.49 (1H, dd, J = 8.0, 5.6 Hz, H-4′), 3.39 (1H, d, J = 2.0 Hz, H-7), 2.12 (1H, d, J = 12.5 Hz, H-15b), 2.11 (1H, s, H-13), 2.10 (1H, m, H-2b), 1.93 (1H, m, H-3b), 1.88 (1H, m, H-12b), 1.87 (1H, m, H-5), 1.86 (1H, m, H-11b), 1.85 (1H, d, J = 12.5 Hz, H-15a), 1.84 (1H, m, H-14b), 1.83 (1H, m, H-1b), 1.76 (1H, m, H-3a), 1,73 (1H, m, H-14a), 1,67 (1H, m, H-11a), 1,65 (1H, m, H-12a), 1,47 (1H, m, H-9), 1,39 (1H, m, H-2a), 0.88 (1H, m, H-1a), 1.45 (3H, s, H-18), 1.00 (3H, s, H-20); 13C-NMR (125 MHz, CD3OD, TMS) δ 179.7 (C-19), 104.6 (C-1′), 81.2 (C-7), 80.6 (C-13), 79.8 (C-3′ and C-5′), 77.4 (C-16), 77.3 (C-17), 74.7 (C-2′), 72.4 (C-6), 71.0 (C-4′), 63.5 (C-6′), 50.8 (C-5), 49.1 (C-15), 48.6 (C-9), 45.3 (C-8), 43.9 (C-4,14), 42.4 (C-10), 40.7 (C-1), 40.5 (C-3), 34.7 (C-12), 32.4 (C-18), 19.7 (C-2, 11), 16.9 (C-20); m/z 547.28 [M + H]+.
Compound 14: yellow powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 8.09 (2H, br. d, J = 9.0 Hz, H-2′ and H-6′), 6.91 (2H, br.d, J = 9.0 Hz, H-3′ and H-5′), 6.41 (1H, d, J = 2.0 Hz, H-8), 6.21 (1H, d, J = 2.0 Hz, H-6), 5.25 (1H, d, J = 7.5 Hz, H-1′′), 4.77 (1H, d, J = 7.8 Hz, H-1′′′), 4.50 (1H, d, J = 1.5 Hz, H-1′′′′), 4.28–3.27 (H-2′′–H-6′′, H-2′′–H-6′′′, and H-2′′′′–H-5′′′′), 1.18 (3H, d, J = 6.5 Hz, H-6′′′′); 13C-NMR (125 MHz, CD3OD, TMS) δ 180.60 (C-4), 164.50 (C-7), 161.61 (C-5), 160.18 (C-4′), 157.79 (C-2), 157.06 (C-9), 133.47 (C-3), 131.08 (C-2′ and C-6′) 121.26 (C-1′), 114.90 (C-3′ and C-5′), 104.25 (C-10), 103.24 (C-1′′′), 100.43 (C-1′′), 100.37 (C-1′′′′), 98.54 (C-6), 93.44 (C-8), 78.41 (C-2′′), 76.61 (C-5′′′), 76.42 (C-2′′′), 73.91 (C-3′′), 73.73 (C-5′′), 73.72 (C-3′′′), 72.44 (C-4′′′′), 71.06 (C-4′′′), 70.65 (C-2′′′′), 70.13 (C-4′′), 69.84 (3′′′′), 68.29 (C-5′′′′), 65.72 (C-6′′), 61.18 (6′′′), 16.54 (6′′′′); ESI–MS (positive) m/z 779.20 [M + Na]+.
Compound 15: yellow powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 8.07 (2H, br. d, J = 9.0 Hz, H-2′ and H-6′), 6.90 (2H, br. d, J = 9.0 Hz, H-3′ and H-5′), 6.39 (1H, d, J = 2.3 Hz, H-8), 6.19 (1H, d, J = 2.3 Hz, H- 6), 5.62 (1H, d, J = 8.0 Hz, H-1′′), 5.22 (1H, d, J = 1.5 Hz, H-1′′′), 4.53 (1H, d, J = 1.5 Hz, H-1′′′′), 4.07–3.32 (H-2′′–H-6′′, H-2′′′–H-5′′′, and H-2′′′′–H-5′′′′), 1.18 (3H, d, J = 6.0 Hz, H-6′′′′), 0.98 (3H, d, J = 6.0 Hz, H-6′′′); 13C-NMR (125 MHz, CD3OD, TMS) δ 177.99 (C-4), 164.33 (C-7), 161.72 (C-5), 159.87 (C-4′), 157.24 (C-2), 157.00 (C-9), 133.02 (C-3), 130.79 (C-2′ and C-6′) 121.62 (C-1′), 114.76 (C-3′ and C-5′), 104.42 (C-10), 101.19 (C-1′), 100.42 (C-1′′′′), 99.44 (C-1′′), 98.43 (C-6), 93.27 (C-8), 76.13 (C-2′′), 74.30 (C-3′′), 73.87 (C-5′′), 72.66 (C-4′′′), 72.66 (C-4′′′), 72.46 (C-4′′′′), 71.00 (C-2′′′), 70.92 (C-3′′′), 70.86 (C-3′′′′), 70.67 (C-2′′′′), 69.29 (C-4′′), 68.42 (5′′′), 68.28 (C-5′′′′), 65.69 (C-6′′), 16.54 (C-6′′′′), 16.12 (C-6′′′); ESI–MS (negative) m/z 739.30 [M–H]−.
Compound 16: yellow powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 8.11 (2H, br. d, J = 9.0 Hz, H-2′ and H-6′), 6.90 (2H, br. d, J = 9.0 Hz, H-3′ and H-5′), 6.40 (1H, d, J = 2.3 Hz, H-8), 6.23 (1H, d, J = 2.3 Hz, H-6), 5.05 (1H, d, J = 8.0 Hz, H-1′′), 4.52 (1H, d, J = 1.5 Hz, H-1′′′), 3.87–3.28 (H-2′′–H-6′′ and H-2′′′–H-5′′′), 1.19 (3H, d, J = 6.1 Hz, H-6′′′); ESI–MS (negative) m/z 593.20 [M–H]−.
Compound 17: yellow powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 8.11 (2H, br. d, J = 9.0 Hz, H-2′ and H-6′), 7.83 (1H, dd, J = 7.5, 1.7 Hz, H-6′′′′′′), 7.55 (1H, ddd, J = 8.5, 7.0, 1.7 Hz, H-4′′′′′′), 7.15 (1H, dd, J = 7.3, 1.7 Hz, H-3′′′′′′), 7.03 (1H, ddd, J = 8.5, 7.3, 1.7 Hz, H-5′′′′′′), 6.91 (2H, br.d, J = 9.0 Hz, H-3′ and H-5′), 6.73 (1H, d, J = 2.0 Hz, H-8), 6.48 (1H, d, J = 2.5 Hz, H-6), 5.66 (1H, d, J = 1.5 Hz, H-1′′′′′), 5.57 (1H, d, J = 1.5 Hz, H-1′′′′′), 5.25 (1H, d, J = 7.5 Hz, H-1′′), 4.77 (1H, d, J = 7.8 Hz, H-1′′′), 4.50 (1H, d, J = 1.5 Hz, H-1′′′′), 4.28–3.27 (H-2′′–H-6′′, H-2′′′–H-6′′′, H-2′′′′–H-5′′′′, and H-2′′′′′–H-5′′′′′), 1.27 (3H, d, J = 6.0 Hz, H-6′′′′′), 1.18 (3H, d, J = 6.5 Hz, H-6′′′′); 13C-NMR (125 MHz, CD3OD, TMS) δ 178.0 (C-4), 165.0 (C-7′′′′′′), 161.7 (C-7), 160.6 (C-4′), 158.2 (C-1′′′′′′), 156.8 (C-2), 156.3 (C-9), 134.1 (C-5′′′′′′), 133.6 (C-3), 131.7 (C-2′ and C-6′), 131.6 (C-3′′′′′′), 121.1 (C-1′′′′′′), 120.6 (C-1′), 120.3 (C-4′′′′′′), 115.7 (C-3′ and C-5′), 113.0 (C-6′′′′′′), 106.0 (C-10), 104.6 (C-1′′′ and C-1′′′′), 99.8 (C-6), 98.9 (C-1′′), 98.7 (C-1′′′′′), 95.2 (C-8), 80.6 (C-2′′), 77.0 (C-5′′′), 76.7 (C-3′′′), 74.7 (C-2′′′), 74.3 (C-4′′′′′), 74.1 (C-5′′), 73.6 (C-3′′), 72.4 (C-4′′′′), 70.8 (C-2′′′′), 70.7 (C-3′′′′), 70.1 (C-2′′′′′), 69.2 (C-4′′), 68.6 (C-5′′′′), 68.1 (C-3′′′′′), 67.8 (C-4′′′ and C-5′′′′′), 65.1 (C-6′′), 61.2 (C-6′′′), 56.2 (-OCH3) 18.4 (C-6′′′′ and C-6′′′′′); ESI–MS (positive) m/z 1059.30 [M + Na]+.
Compound 18: yellow powder; 1H-NMR (500 MHz, CD3OD, TMS) δ 8.11 (2H, br. d, J = 9.0 Hz, H-2′ and H-6′), 7.83 (1H, dd, J = 7.5, 1.7 Hz, H-6′′′′′′), 7.55 (1H, J = ddd, 8.5, 7.5, 1.7 Hz, H-4′′′′′′), 7.15 (1H, dd, J = 8.0, 2.0 Hz, H-3′′′′′′), 7.03 (1H, ddd, J = 8.5, 7.5, 1.7 Hz, H-5′′′′′′), 6.91 (2H, br. d, J = 9.0 Hz, H-3′ and H-5′), 6.73 (1H, d, J = 2.3 Hz, H-8), 6.48 (1H, d, J = 2.3 Hz, H-6), 5.62 (1H, d, J = 8.0 Hz, H-1′′), 5.57 (1H, d, J = 1.5 Hz, H-1′′′′′), 5.22 (1H, d, J = 1.5 Hz, H-1′′′), 4.53 (1H, d, J = 1.5 Hz, H-1′′′′), 4.07 (1H, m, H-5′′′), 4.03–3.32 (H-2′′–H-6′′, H-2′′′–H-5′′′, H-2′′′′–H-5′′′′, and H-2′′′′′–H-5′′′′′), 1.27 (3H, d, J = 6.0 Hz, H-6′′′′′), 1.18 (3H, d, J = 6.0 Hz, H-6′′′′), 0.98 (3H, d, J = 6.0 Hz, H-6′′′); 13C-NMR (125 MHz, CD3OD, TMS) δ 177.9 (C-4), 165.0 (C-7′′′′′′), 161.4 (C-7), 158.9 (C-4′), 158.2 (C-2′′′′′′), 156.4 (C-2), 155.9 (C-9), 134.1 (C-5′′′′′′), 133.4 (C-3), 131.4 (C-2′ and C-6′), 131.6 (C-3′′′′′′), 121.1 (C-1′′′′′′), 120.6 (C-1′), 120.3 (C-4′′′′′′), 115.6 (C-3′ and C-5′), 113.0 (C-6′′′′′′), 106.2 (C-10), 100.4 (C-1′′′), 100.0 (C-1′′′′), 99.4 (C-6), 98.7 (C-1′′), 98.1 (C-1′′′′′), 95.3 (C-8), 75.3 (C-2′′), 74.2 (C-4′′′′′), 73.8 (C-5′′), 73.7 (C-4′′′′′), 71.9 (C-4′′′′), 70.5 (C-2′′′′), 70.3 (C-3′′′′), 70.6 (C-2′′′ and C-3′′′), 70.1 (C-2′′′′′), 69.2 (C-4′′), 68.7 (C-5′′′′), 68.2 (C-5′′′), 68.1 (C-3′′′′′), 67.8 (C-4′′′ and C-5′′′′′), 65.1 (C-6′′), 61.2 (C-6′′′), 56.2 (-OCH3), 18.4 (C-6′′′′ and C-6′′′′′); ESI–MS (positive) m/z 1043.32 [M + Na] +.
Determination of DPPH radical-scavenging activity
The DPPH free radical-scavenging activities of the isolated 12 compounds except for 3–6, 13, and 14, which were finally purified as mixtures, and ferulic acid, as a positive control, were determined. In brief, 100 μL of 0.5 mM compound (final concentration, 50 μM) in EtOH was mixed to the 900 μL of DPPH radical EtOH solution (final concentration, 100 μM). The mixture was incubated for 20 min in darkness and the absorbance was measured at 517 nm. The DPPH radical-scavenging activities of the isolated 12 compounds and ferulic acid were calculated as the decreased percentage for the absorbance of sample solution to control. The DPPH radical-scavenging activity was expressed as the mean ± standard deviation (n = 3) using (SPSS, Chicago, IL, USA) 20.0 package programs. Statistical differences were measured by one-way analysis of variance followed by Duncan’s multiple-range test. The significance was considered in p < 0.05.
Results and discussion
DPPH free radical-scavenging activities of solvent-fractionated fractions of sword bean MeOH extracts
The MeOH extracts of the fresh sword bean were partitioned to obtain n-hexane, CHCl3, EtOAc, and BuOH fractions. These fractions were subjected to TLC analysis and screened as antioxidative compounds by spraying of DPPH solution. Various antioxidative compounds were observed in the n-hexane, CHCl3, and BuOH fractions. The n-hexane and CHCl3 factions exhibited similar patterns of antioxidative compounds detected on their TLC plates (data not shown).
Structural determination of 1–18 from sword bean MeOH extracts
The chemical constituents were separated from the n-hexane/CHCl3 and BuOH fractions of the fresh sword bean MeOH extracts. Briefly, 6 compounds were purified and isolated from the n-hexane/CHCl3 fraction by silica-gel column chromatography and HPLC. Additionally, 12 compounds were purified and isolated from the BuOH fraction by separation tools using Amberlite XAD-2, ODS, and ODS-HPLC.
The structures of the 18 compounds were determined by MS and NMR experiments. Among them, the 15 known compounds were methyl gallate (1) (Hussain et al., 1979), (2S,3S,4E,8E)-2-aminooctadeca-4,8-diene-1,3-diol 1-O-β-d-glucopyranoside (3) (Cateni et al., 2008), (2S,3S,4E,8Z)-2-aminooctadeca-4,8-diene-1,3-diol 1-O-β-d-glucopyranoside (4) (Cateni et al., 2008), lupeol (5) (Cho et al., 2013), trilinolein (6) (Fierro et al., 2012), 1,6-di-O-galloyl β-d-glucopyranoside (7) (Kim et al., 2013a, b), dihydrophaseic acid (10) (Rho and Yoon, 2017), dillenetin (11) (Megawai and Fajriah, 2013), kaempferol-7-O-[2-O-β-d-glucopyranosyl-6-O-α-l-rhamnopyranosyl]-α-l-rhamnopyranoside (12) (Okoye et al., 2012), canavalioside (13) (Murakami et al., 2000), kaempferol-3-O-[2-O-β-d-glucopyranosyl-6-O-α-l-rhamnopyranosyl]-β-d-glucopyranoside (14) (Chen et al., 2009), kaempferol-3-O-(2,6-O-α-l-dirhamnopyranosyl)-β-d-glucopyranoside (15) (Clarkson et al., 2005), kaempferol-3-O-rutinoside (16) (Clarkson et al., 2005), gladiatoside A1 (17) (Murakami et al., 2000), and gladiatoside B1 (18) (Murakami et al., 2000) (Fig. 1). We confirmed that 3 and 4, 5 and 6, and 13 and 15 were purified respectively as mixtures, based on their 1H- and 13C-NMR spectra. The structures of cerebrosides (3 and 4), terpene derivatives (10 and 13), and kaempferol derivative and its glycosides (11, 12, and 14–18) were determined by ESI–MS, 1H-NMR, 13C-NMR, HSQC, 1H-1H COSY, and HMBC experiments. Compounds 2 and 9 were elucidated as new compounds. Compound 8 was isolated for the first time in nature; consequently, no report on its MS and NMR spectroscopic data exists. Therefore, herein, we described the structural determination of 2, 8, and 9.
Fig. 1.
Structures of the isolated compounds, 1, 3–7, and 10–18, from sword beans
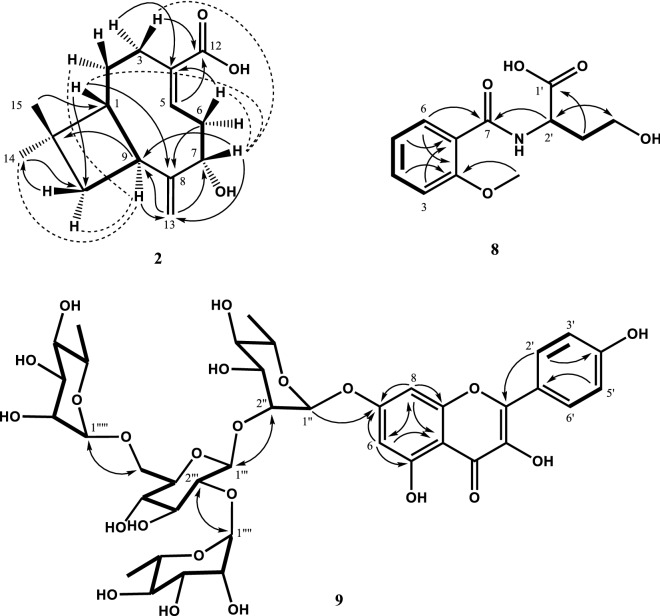
The HR-ESI–MS (negative) spectrum of 2 showed a quasi-molecular ion peak at m/z 249.1493 [M – H]− (C15H21O3), indicating that its molecular formula was C15H22O3 (MW 250). The 1H-NMR spectrum exhibited two singlet exomethylene proton signals at δ 5.07 (H-13b) and 5.02 (H-13a), as well as one olefinic double bond proton signal at δ 6.69 (H-5) (Table 1). Additionally, an oxygenated methine proton signal at δ 4.40 (H-7), five methylene proton signals at δ 1.66-2.77 (H-2, H-3, H-6, H-9, and H-10), and two methyl group signals at δ 1.03 (H-15) and 1.00 (H-14) were observed in the 1H-NMR spectrum. The 13C-NMR spectrum of 2 contained 15 carbon signals, including a carbonyl carbon at δ 171.7 (C-12), four olefinic double bond carbons at δ 159.0 (C-8), 139.5 (C-5), 135.5 (C-4), and 113.0 (C-13), one quaternary carbon at 33.8 (C-11), four methylene carbons at δ 41.9 (C-10), 36.0 (C-6), 28.1 (C-2), and 24.3 (C-3), and two methyl carbons at δ 30.8 (C-15) and 24.3 (C-14). From the MS and 1D-NMR results, 2 was suggested to be caryophyllene-type sesquiterpene that is composed of cyclobutane and cyclononene rings. The accurate structure of compound 2 could be determined by 1H-1H COSY, HSQC, and HMBC experiments. The 1H-1H COSY correlations (bold lines) of H-1/H-2, H-1/H-9, H-2/H-3, H-5/H-6, and H-6/H-7 and the HMBC correlations of δ 1.75 (H-1) to 159.0 (C-8), δ 2.53 (H-6b) to 135.5 (C-4) and 159.0 (C-8), and δ 1.81 (H-2a) to 135.5 (C-4) established a connection of the cyclononene ring in 2 (Fig. 2). Additionally, the 1H-1H COSY correlations (bold lines) of H-1/H-9 and H-9/H-10 and the HMBC correlations of δ 1.75 (H-1) to 41.9 (C-10), 24.3 (C-14), and 30.8 (C-15), δ 2.77 (H-9) to 33.8 (C-11), and δ 1.00 (H-14) and 1.03 (H-15) to 55.1 (C-1) indicated that the cyclobutane ring fused to C-1 and C-9 of the cyclononene ring and two methyl groups coupled with C-11 of the cyclobutane ring. The exomethylene group positioned at C-8 (δ 159.0) and the hydroxyl group positioned at C-7 (δ 76.2) were confirmed by the HMBC correlations of δ 5.07 (H-13b) and 5.02 (H-13a) to 76.2 (C-7) and 33.6 (C-9), and δ 2.77 (H-9) and 4.40 (H-7) to 113.0 (C-13). The carboxylic acid group positioned at C-4 (δ 135.5) was also confirmed by the HMBC correlations of δ 2.33 (H-3b) and 6.69 (H-5) to 171.7 (C-12). Compound 2 was suggested to be 7-hydroxy-11,11-dimethyl-8-methylenebicyclo[7.2.0]undec-4-ene-4-carboxylic acid, which is the similar structure of lychnopholic acid identified from Lychnophora martiana (Raffauf et al., 1987; Vichewski et al., 1980). The relative stereochemistry of compound 2 was established, based on the nuclear Overhauser effect (NOE) experiment and comparison of 13C-NMR data reported previously in literature (Raffauf et al., 1987; Vichewski et al., 1980). H-1 at δ 1.75, H-3a at δ 2.57, H-5 at δ 6.69, H-13a at 5.02, and H-6a at δ 2.72–2.66 were enhanced by irradiation of H-7 at δ 4.40, suggesting that H-1 and H-7 have α-orientation and hydroxyl group of C-7 has β-orientation (Fig. 2). The β-orientation of H-9 and C-14 was deduced from enhancement of H-14 at δ 1.00, H-10b at δ 1.80, and H-2b at δ 1.66 by irradiation of H-9 at δ 2.77. Therefore, the structure of compound 2 was determined as (1R,7S,9S)-7-hydroxy-11,11-dimethyl-8-methylenebicyclo[7.2.0]undec-4-ene-4-carboxylic acid (caryophyllenic acid) (Fig. 2).
Fig. 2.
Structures, 1H-1H COSY (bold line), NOE (dash line), and HMBC (arrows) correlations of 2, 8, and 9
The molecular formula (C12H15N1O5, MW 253) of 8 was established by observation of quasi-molecular ion peak at m/z 254.0968 [M + H]+ (calculated for C12H16N1O5, +1.8 mDa) by the HR–ESI–MS analysis. The 1H-NMR (500 MHz, CD3OD, TMS) spectrum of 8 showed four proton signals corresponding to ortho-substituted benzene ring at δ 8.02 (1H, dt, J = 8.0, 2.0 Hz, H-6), 7.51 (1H, dt, J = 8.0, 2.0 Hz, H-4), 7.16 (1H, br. d, J = 8.0 Hz, H-3), and 7.06 (1H, dt, J = 8.0, 1.0 Hz, H-5), and a methoxy proton signal at δ 4.01 (3H, s) (Table 2). Additionally, a nitrogenated methine proton signal was observed at δ 4.57 (1H, t, J = 6.0 Hz, H-2′), an oxygenated methylene proton signal at δ 3.98–4.06 (2H, m, H-4′), and a methylene proton signal at δ 2.19–2.33 (2H, m, H-3′) that were confirmed by the 13C-NMR and HSQC spectra. The 13C-NMR (125 MHz, CD3OD, TMS) spectrum showed 12 carbon signals, including two carbonyl carbons at δ 176.9 (C-1′) and 165.2 (C-7) (Table 2). The presence of 2-methoxybenzoic acid was established by 1H-1H COSY correlations of H-3/H-4, H-4/H-5, and H-5/H-6 and HMBC correlations of δ 8.02 (H-6) to 165.2 (C-7) and δ 7.51 (H-4) and 4.01 (-OCH3) to 158.1 (C-2) (Fig. 2). Homoserine moiety was confirmed by the 1H-1H COSY correlations of H-2′/H-3′ and H-3′/H-4′ and HMBC correlations of δ 4.57 (H-2′) and 2.19–2.33 (H-3′) to 176.9 (C-1′). In particular, the correlation of δ 4.57 (H-2′) to 165.2 (C-7) was observed in the HMBC experiment, indicating that 2-methoxybenzoic acid was amidified with the C-2 of homoserine. Therefore, the planar structure of 8 was determined as N-(2-methoxybenzoyl)homoserine (Fig. 2). Further investigation of the absolute stereochemical structure of 8 is required.
The molecular formula (C39H50O23, MW 908) of 9 was determined by analyzing the quasi-molecular ion peak at m/z 925.2595 [M + H]+ by the HR–ESI–MS (positive) spectrum. This result was supported by the 1H-NMR, 13C-NMR, and HSQC spectra. From the MS and 1D-NMR spectra, 9 was suggested to be flavonoid tetraglycoside. The presence of a kaempferol moiety was suggested by two p-substituted benzene ring proton signals at δ 8.10 (2H, d, J = 9.0 Hz, H-2′ and H-6′) and 6.91 (2H, d, J = 9.0 Hz, H-3′ and H-5′) and two meta-coupling benzene ring proton signals at δ 6.73 (1H, d, J = 2.0 Hz, H-8), and 6.47 (1H, d, J = 2.0 Hz, H-6) in the 1H-NMR (500 MHz, CD3OD, TMS) spectrum (Table 4). The presence of the kaempferol moiety was further supported by its related 15 carbon signals, including one carbonyl carbon at δ 178.12 (C-4) and four oxygenated quaternary carbons at δ 161.98–156.52. The presence of one β-d-glucopyranose and three α-l-rhamnopyranose was suggested by four anomeric proton signals at δ 5.65 (1H, d, J = 7.5 Hz, H-1′′′), 5.57 (1H, d, J = 1.5 Hz, H-1′′), 5.23 (1H, d, J = 1.5 Hz, H-1′′′′), and 4.53 (1H, d, J = 1.5 Hz, H-1′′′′′), and three methyl proton signals at δ 1.27 (3H, d, J = 6.0 Hz, H-6′′), 1.18 (3H, d, J = 6.0 Hz, H-6′′′′), and 0.98 (3H, d, J = 6.5 Hz, H-6′′′′′) observed in the 1H-NMR spectrum of 9. These glycoses were also confirmed by their 1H-1H COSY correlations and coupling constant values in the 1H-NMR spectrum. From the HMBC spectrum of 9, it was possible to establish a connection with kaempferol tetraglycoside. In particular, the HMBC correlations of δ 5.57 (H-1′′) to 161.98 (C-7) established the C-1 of rhamnose coupled with the C-7 position of kaempferol (Fig. 2). The HMBC correlations of δ 4.03 (H-2′′) to 99.42 (C-1′′′), and δ 5.65 (H-1′′′) to 70.29 (C-2′′) established that the C-1 of glucose was connected to the C-2 position of rhamnose in kaempferol 7-O-rhamnoside. The HMBC correlations of δ 3.95 (H-2′′′) to 101.20 (C-1′′′′), δ 5.23 (H-1′′′′) to 76.15 (C-2′′′), δ 3.73 (H-6′′′a) to 100.42 (C-1′′′′′), and δ 4.53 (H-1′′′′′) to 65.74 (C-6′′′) established that two rhamnopyranoses were etherified to the C-2 and C-6 positions of glucose. Therefore, compound 9 was determined to be kaempferol-7-O-α-l-dirhamnopyranosyl(1 → 2;1 → 6)-O-β-d-glucopyranosyl(1 → 2)-O-α-l-rhamnopyranoside (Fig. 2).
Table 4.
DPPH radical-scavenging activities of the isolated compounds from sword beans
| Compounds | DPPH radical-scavenging activities (%) |
|---|---|
| 1 | 85.6 ± 3.6a |
| 2 | 4.1 ± 1.7e |
| 7 | 90.6 ± 5.5a |
| 8 | 10.2 ± 2.1d |
| 9 | 15.9 ± 2.4c |
| 10 | 10.8 ± 2.2d |
| 11 | 9.3 ± 2.4d |
| 12 | 15.6 ± 3.2c |
| 15 | 14.4 ± 2.6c |
| 16 | 14.1 ± 1.4c |
| 17 | 13.2 ± 2.8c |
| 18 | 13.8 ± 2.5c |
| Ferulic acid | 71.3 ± 2.3b |
Ferulic acid was used as positive control. The isolated compounds except for 3–6, 13, and 14 were assayed at the concentration of 50 μM. Values are expressed as mean ± SD (n = 3)
a–eResults with a different letter differ significantly (p < 0.05)
DPPH free radical-scavenging activities of the isolated compounds
In this experiment, compounds 3–6, 13, and 14 were excluded because these compounds were finally purified as mixtures. The DPPH free radical-scavenging activities of 12 compounds of the isolated 18 compounds and ferulic acid at the same concentration (50 μM) were evaluated (Table 4). As expected, two galloyl derivatives (1 and 7) showed the highest DPPH free radical-scavenging activities. These activities were very higher in comparison to that of ferulic acid, which is used as a positive control. However, the free radical-scavenging activities of flavonol glycosides 9 and 12–18 were similar and lowered in compared to that of ferulic acid. Other compounds including new compounds 2 and 8 were very low DPPH free radical-scavenging activities, although these activities in TLC analysis during purification and isolation were observed. These observations indicated that two galloyl derivatives (1 and 7) including the phenolic compounds identified in this study may be responsible for the antioxidative activity of sword bean.
In this study, we demonstrated the presence of 18 compounds, including two galloyl derivatives (1 and 7), trilinolein (6), two cerebrosides (3 and 4), three terpene derivatives (2, 5, 10, and 13), salicyloyl derivative (8), and eight kaempferol derivatives and their glycosides (9, 11, 12, and 14–18) in sword beans. Among them, caryophyllene-type sesquiterpene (2) and flavonol tetraglycoside (9) were elucidated as new compounds. The occurrence of two galloyl derivatives (1 and 7), canavalioside (13), and salicyloyl kaempferol glycosides (17 and 18) in sword bean (Kim et al., 2013a, b; Lee and Jeong, 2005; Murakami et al., 2000) has been previously reported. Contrarily, compounds 3–8, 10–12, 14, and 15 were newly identified in this plant.
Compound 2 is a caryophyllene-type sesquiterpene like β-caryophyllene, β-caryophyllene oxide, and humulene, which are constituents of essential oil in plants and exert anticancer, antioxidant, and antimicrobial activities (Dahham et al., 2015; Fidyt et al., 2016). Phenolic compounds including two galloyl derivatives (1 and 7) and eight kaempferol glycosides (9, 11, 12, and 14–18) are widely distributed in plant kingdoms and are also known as biological active compounds that have anticancer, antioxidant, and anti-inflammatory effects (Kumar and Pandey, 2013; Tanase, 2019). In this study, we confirmed two galloyl derivatives (1 and 7) exerting strong DPPH free radical-scavenging activities. Additionally, cerebrosides (3 and 4) inhibited the growth of bacteria and Candida species (Cateni et al., 2008). Trilinolein (6) has been demonstrated to have anti-ischemic, antiarrhythmic, and antioxidant properties (Chan et al., 2005). The chemical constituents found in this study might offer valuable information for evaluating the health benefits and food quality of sword beans and for selecting biomarker compounds in food processing.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea government (Ministry of Science and ICT, MSIT, No. NRF-2017R1D1A1B03034520).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hyun Jeong An, Email: hyunjeon5722@naver.com.
Eun Hee Kim, Email: ekdmacc@hanmail.net.
Hyoung Jae Lee, Email: ableleehj@gmail.com.
Jeong-Yong Cho, Email: jyongcho17@jnu.ac.kr.
Jae-Hak Moon, Email: nutrmoon@jnu.ac.kr.
References
- Cateni F, Zilic J, Zacchigna M. Isolation and structure elucidation of cerebrosides from Euphorbia platyphyllos L. Sci. Pham. 2008;76:451–469. doi: 10.3797/scipharm.0805-03. [DOI] [Google Scholar]
- Chan P, Kao PF, Tomlinson B. Cardiovascular effects of trilinolein, a natural triglyceride isolated from the herb Sanchi (Panax notoginseng) Acta Cardiol. Sin. 2005;21:71–76. [Google Scholar]
- Chang MI, Kim JY, Kim SJ, Baek SH. Effect of sword bean chunggukjang addition on quality of kochujang. J. Korean Soc. Food Sci. Nutr. 2011;40:1292–1299. doi: 10.3746/jkfn.2011.40.9.1292. [DOI] [Google Scholar]
- Chen JH, Liau BC, Jong TT, Chang CMJ. Extraction and purification of flavanone glycosides and kaemferol glycosides from defatted Camellia oleifera seeds by salting-out using hydrophilic isopropanol. Sep. Purif. Technol. 2009;67:31–37. doi: 10.1016/j.seppur.2009.02.020. [DOI] [Google Scholar]
- Cho JY, Kim CM, Lee HJ, Lee SH, Cho JA, Kim WS, Park KH, Moon JH. Caffeoyl triterpenes from pear (Pyrus pyrifolia Nakai) fruit peels and their antioxidative activities against oxidation of rat blood plasma. J. Agric. Food Chem. 2013;61:4563–4569. doi: 10.1021/jf400524b. [DOI] [PubMed] [Google Scholar]
- Cho YS, Bae YI, Shim KH. Chemical components in different parts of Korean sword bean (Canavalia gladiata) Korean J. Postharv. Sci. Technol. 1999;6:475–480. [Google Scholar]
- Cho YS, Seo KI, Shim KH. Antimicrobial activities of Korean sword bean (Canavalia gladiata) extracts. Korean J. Food Preserv. 2000;7:113–116. [Google Scholar]
- Clarkson C, Staerk D, Hansen SH, Jaroszewski JW. Hyphenation of solid-phase extraction with liquid chromatography and nuclear magnetic resonance: Application of HPLC-DAD-SPE-NMR to identification of constituents of Kanahia laniflora. Anal. Chem. 2005;77:3547–3553. doi: 10.1021/ac050212k. [DOI] [PubMed] [Google Scholar]
- Dahham SS, Tabana YM, Iqbal MA, Ahamed MB, Ezzat MO, Majid AS, Majid AM. The anticancer, antioxidant and antimicrobial properties of the sequiterpene β-caryophyllene from the essential oil of Aquilaria crassna. Molecules. 2015;20:11808–11829. doi: 10.3390/molecules200711808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekanayake S, Skog K, Asp NG. Canavanine content in sword bean (Canavalia gladiata): Analysis and effect of processing. Food Chem. Toxicol. 2007;45:797–803. doi: 10.1016/j.fct.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Fidyt K, Fiedorowicz A, Strządała L, Szumny A. β-Caryophyllene and β-caryophyllene oxide–natural compounds of anticancer and analgesic properties. Cancer Med. 2016;5:3007–3017. doi: 10.1002/cam4.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro RS, Maquilang QMA, Sanjorjo RAS, Tradio MD, Shen CC, Ragasa CY. Secondary methabolites from Cinnamomum cebuense. J. Med. Plants Res. 2012;6:2146–2149. [Google Scholar]
- Gan RY, Kong KW, Li Li HB, Wu K, Ge YY, Chan CL, Shi XM, Corke H. Separation, identification, and bioactivities of the main gallotannins of red sword bean (Canavalia gladiata) coats. Front. Chem. 2018;6:39. doi: 10.3389/fchem.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan RY, Lui WY, Corke H. Sword bean (Canavalia gladiata) as a source of antioxidant phenolics. Int. J. Food. Sci. Technol. 2016;51:156–162. doi: 10.1111/ijfs.12979. [DOI] [Google Scholar]
- Hussain N, Moden MH, Shabbir SG, Zaidi SAH. Antimicrobial principles in Mimosa hamata. J. Nat. Prod. 1979;42:525–527. doi: 10.1021/np50005a014. [DOI] [PubMed] [Google Scholar]
- Jeon KS, Na HJ, Kim YM, Kwon HJ. Antiangiogenic activity of 4-O-methylgallic acid from Canavalia gladiata, dietary legume. Biochem. Biophys. Res. Commum. 2005;33:1268–1274. doi: 10.1016/j.bbrc.2005.03.109. [DOI] [PubMed] [Google Scholar]
- Ji KY, Jang JH, Lee EH, Kim SM, Song HW, Yang WK, Kim HY, Kim KH, Lee YS, Kim DS, Kang HS, Kim SH. Canavalia gladiata and Arctium lappa extracts ameliorate dextran sulphate sodium-induced inflammatory bowel disease by enhancing immune responses. J. Fun. Foods. 2018;45:24–33. doi: 10.1016/j.jff.2018.03.018. [DOI] [Google Scholar]
- Kim JP, Lee HH, Moon JH, Ha DR, Kim ES, Kim JH, Seo KW. Isolation and identification of antioxidants from methanol extract of sword bean (Canavalia gladiata) Korean J. Food Sci. Technol. 2013;45:777–784. doi: 10.9721/KJFST.2013.45.6.777. [DOI] [Google Scholar]
- Kim OK, Nam DE, You Y, Jun W, Lee J. Protective effect of Canavalia gladiata on gastric inflammation induced by alcohol treatment in rats. J. Korean Soc. Food Sci. Nutr. 2013;42:690–696. doi: 10.3746/jkfn.2013.42.5.690. [DOI] [Google Scholar]
- Kim SH, Kim K, Chi GY, Cho IS, Kim HY, Lee YC. Enhancing effect of Canavalia gladiata DC semen on the hematopoietic expansion and function of stem cells. Kor. J. Herbology. 2012;27:9–16. doi: 10.6116/kjh.2012.27.4.9. [DOI] [Google Scholar]
- Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: An overview. Sci. World J. 2013;2013:162750. doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Jeong HS. Isolation and identification of antimicrobial substance from Canavalia gladiata. Food Sci. Biotechnol. 2005;14:268–274. [Google Scholar]
- Megawati AD, Fajriah S. 3′,4′-Dimethoxyquercetin, a flavonol compound isolated from Kalanchoe pinnata. J. Appl. Pharm. Sci. 2013;3:90–99. [Google Scholar]
- Murakami T, Kohno K, Kishi A, Matsuda H, Yoshikawa M. Medicinal foodstuffs XIX 1) Absolute stereostructures of canavalioside. Chem. Pharm. Bull. 2000;48:1673–1680. doi: 10.1248/cpb.48.1673. [DOI] [PubMed] [Google Scholar]
- Okoye FBC, Esimone CO, Proksch P. Novel kaempferol triglycosides from Olax manni leaves: Structures and biological activities. Planta Med. 2012;78:139. doi: 10.1055/s-0032-1320826. [DOI] [Google Scholar]
- Raffauf RF, Pastore MP, Kelley CJ, Le Quesne PW, Miura I, Nakanishi K, Finer J, Clardy J. Lychnopholic acid, a novel trioxygenated caryophyllene derivative from Lychnophora affinis Gardn. J. Am. Chem. Soc. 1987;100:7437–7439. doi: 10.1021/ja00491a064. [DOI] [Google Scholar]
- Rho T, Yoon KD. Chemical constituents of Nelumbo nucifera seeds. Nat. Prod. Sci. 2017;23:253–257. doi: 10.20307/nps.2017.23.4.253. [DOI] [Google Scholar]
- Tanase C, Coşarcă S, Muntean DL. A critical review of phenolic compounds extracted from the bark of woody vascular plants and their potential biological activity. Molecules. 2019;24:1182. doi: 10.3390/molecules24061182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichewski W, Lins AP, Herz W, Murari R. Lychnopholic acid and its acetate from Lychnophora martiana. Phytochemistry. 1980;19:685–686. doi: 10.1016/0031-9422(80)87040-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.