Abstract
Analysis of metal-organic framework (MOF) structure by electron microscopy and electron diffraction offers an alternative to growing large single crystals for high-resolution X-ray diffraction. However, many MOFs are electron beam-sensitive, which can make structural analysis using high-resolution electron microscopy difficult. In this work we use the microcrystal electron diffraction (MicroED) method to collect high-resolution electron diffraction data from a model beam-sensitive MOF, ZIF-8. The diffraction data could be used to determine the structure of ZIF-8 to 0.87 Å from a single ZIF-8 nanocrystal, and this refined structure compares well with previously published structures of ZIF-8 determined by X-ray crystallography. This demonstrates that MicroED can be a valuable tool for the analysis of beam-sensitive MOF structures directly from nano and microcrystalline material.
Keywords: Metal-organic framework, microcrystal electron diffraction, crystallography, cryoEM, MicroED, Electron diffraction
1. Introduction
Metal-organic frameworks represent a new class of microporous materials. Zeolitic imidazolate frameworks (ZIFs) are a subcategory of highly crystalline metal-organic framework materials bearing zeolitic topologies finding wide variety of applications in adsoption, separation, gas storage, and heterogeneous catalysis. ZIF-8 with sodalite (SOD) topology has been among the most extensively studied ZIF material [1, 2]. Many MOF structures are determined by single crystal X-ray diffraction [3], however this requires the growth of large crystals to withstand the effects of radiation damage and allow the collection of sufficient diffraction data. In some cases, powder diffraction data alone can be used for structure determination; however, this can be a more difficult approach. Because MOFs are often synthesized as micro and nanocrystals, the use of transmission electron microscopy offers a way to study the MOF structure directly from the synthesized material. Collecting data at cryogenic temperatures improves data collection from beam-sensitivity MOFs, and this can be further improved by using low dose imaging conditions (in the range of 4 – 12 e−/Å2 total exposure) and direct electron detection [4–7]. Imaging thin sections of MOF fragments provides an excellent approach for directly visualizing interfacial, surface, and local structure of these beam sensitive materials.
As an alternative to imaging based TEM structure determination methods for overall structure determination, electron diffraction can also be employed for high-resolution structure determination from crystalline materials, including MOFs [8–11]. In recent years, electron microscopy methods, such as microcrystal electron diffraction (MicroED) [12, 13], have been employed to determine the structures of a variety of samples from various fields including materials science, organic chemistry, and structural biology [12–18]. A key benefit of electron diffraction is that crystals several orders of magnitude smaller than those used for single crystal X-ray methods can be used for high-resolution structure determination, which bypasses the often times difficult and time-consuming steps of growing large single crystals for structural analysis.
As is the case with imaging, very low electron dose must be used when beam-sensitive materials are being analyzed by electron diffraction. MicroED is a technique that uses ultra-low dose to collect electron diffraction data at cryogenic temperature from very small microcrystals as the stage continually rotates [12, 13, 19]. By using sufficiently low dose and continuous rotation of the sample, large volumes of reciprocal space can be sampled before the crystal is overcome by radiation damage [20, 21], which allows for the indexing and integration of the diffraction data set. MicroED has been successfully applied to many samples including proteins, peptides, small molecules, and natural products [22–32].
In this work, we make use of ZIF-8 as a model beam-sensitive MOF [5] and demonstrate that MicroED data collection procedures are suitable for rapid structure determination of these materials at very high-resolution. The ZIF-8 MicroED data were collected and processed using a cryo-TEM equipped with a high-speed camera for diffraction data collection, and did not require any other specialized hardware. The ZIF-8 data from a single crystal were processed to 0.87 Å and an ab initio structure was determined, which compares well to previously solved structures of ZIF-8 determined by single crystal X-ray crystallography.
2. Methods
2.1. ZIF-8 synthesis
ZIF-8 crystals were synthesized by modifying the procedure reported by Tanaka and coworkers [33]. All chemicals were purchased from Sigma-Aldrich (U.S.A) unless otherwise stated. Zinc nitrate hexahydrate [Zn(NO3).6H2O] (98%, 0.297g) was dissolved in 20 ml of deionized water and 3.28 g of 2-methylimidazole (MeIM) was dissolved separately in 20 ml of deionized water. The separate dissolved solutions were stirred for 10 min. The imidazole solution was poured into the zinc solution and stirred for 1h. The solution quickly became cloudy and a suspension was obtained. The final molar composition of the synthesis solution was Zn+: MeIM: Water = 1: 40: 2,227. After mixing, the resultant white solution was centrifuged at 5,000 g for 10 min and washed with methanol (VWR, 99.8%) 3 times. The Products were then dried overnight in vacuum oven at room temperature. The morphology and crystallographic properties of ZIF-8 crystals was examined using scanning electron microscope (SEM, Amray 1910) and X-ray powder diffraction (XRD, AXS-D8, Bruker) with a scan step of 0.015° using Cu Kα radiation (λ = 1.543 Å), respectively.
2.2. MicroED ZIF-8 sample preparation
Dried, lightly ground ZIF-8 crystals (0.004 g) were placed in a glass vial containing 100 ml of methanol to form 0.005 wt. % colloidal ZIF-8 solution. 2 μL of prepeared solution were applied onto the surface of glow-discharged Quantifoil 300-mesh 2/2 grids, and dried by evaporation for a few minutes. Grids were subsequently placed into autoloader cartridges under liquid nitrogen and loaded into a Titan Krios cryo-TEM (Thermo Fisher Scientific) operated at 300 kV and equipped with a CETA D detector (Thermo Fisher Scientific), which was used for diffraction data collection.
2.3. MicroED data collection
MicroED data were collected using similar protocols described previously [19, 34]. Low-dose settings for the Titan Krios cryo-TEM were employed throughout screening and data collection steps. Crystals that were well-separated from other crystals on the grid were identified using low magnification search mode. Upon the identification of single microcrystals suitable for data collection, initial diffraction patterns were collected by switching the microscope into diffraction mode using the pre-calibrated and aligned “exposure mode” within the low dose settings. If a high quality diffraction pattern was obtained, the microscope was put back into search mode and the maximal tilt range of for data collection (i.e. the range where crystal remained centered and no other crystals or grid bars moved into the path of the beam) was determined. The stage was tilted to the maximum angle and the selected area aperture and beam stop were inserted. The selected area aperture size was selected such that the entire crystal was within the aperture while also minimizing the area surrounding the crystal. MicroED data sets were collected by continuously rotating the crystal (0.7 degrees per second) in the beam as the CETA D camera was continuously acquiring diffraction frames at a rate of 2 seconds per frame. The exposure rate was approximately 0.02 e−/Å2 per frame.
2.4. Data processing and structure determination
Diffraction data sets were converted to SMV format [35] and indexed, integrated and scaled using XDS [36]. The structure of ZIF-8 was ultimately determined with data from a single crystal by direct methods using SHELXT [37] followed by refinement using the the ShelXle interface [38].
3. Results and Discussion
The morphology of the synthesized ZIF-8 crystals were observed by SEM as shown in Fig. 1A. ZIF-8 crystals shows an approximate isotropic hexagon envelope with particle size range of 200 to 800 nm. The XRD powder diffraction pattern from prepared ZIF-8, is consistent with that of standard ZIF-8 samples reported in literature [39–41] (Fig. 1B), thus revealing the successful synthesis of ZIF-8. The as-synthesized ZIF-8 particles were suspended in methanol, and this solution was directly deposited on a holey carbon coated EM grid. Following the evaporation of the methanol from the surface of the grid, the EM grid was loaded into the Titan Krios cryo-TEM and the ZIF-8 particle distribution was assessed. The distribution of particles is important for MicroED because individual crystals need to be isolated for data collection to ensure that data from multiple crystals is not collected. When viewed at lower magnification, it can be seen that while many of the ZIF-8 nanoparticles preferred to clump together, there were a significant fraction of particles that were well-dispersed and amenable to data collection (Fig. 2A).
Figure 1.
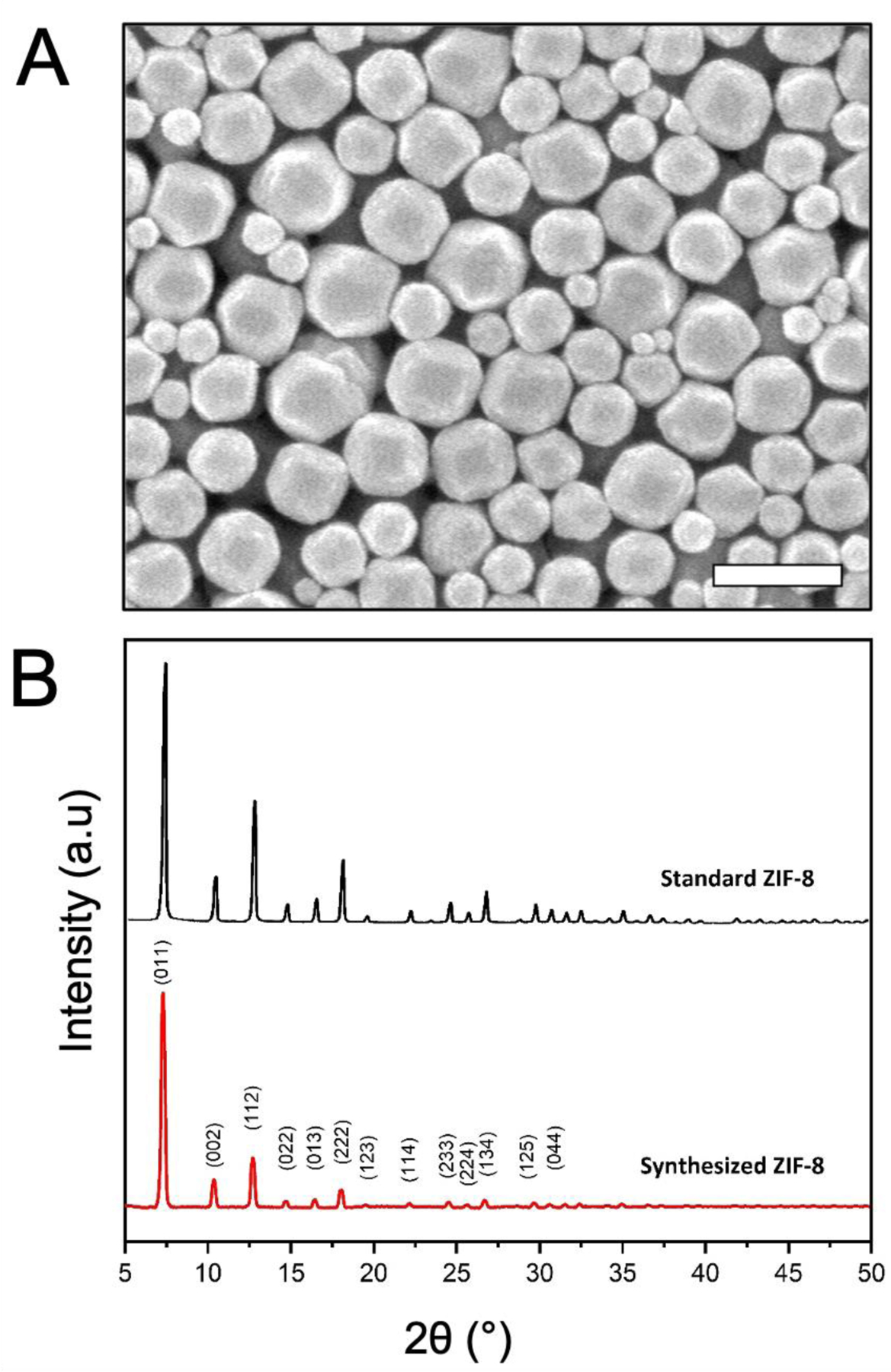
(A) SEM micrograph of synthesized ZIF-8 and (B) XRD powder diffraction data of standard [41] and synthesized ZIF-8 crystals. Scale bar in A represents 1 μm.
Figure 2.
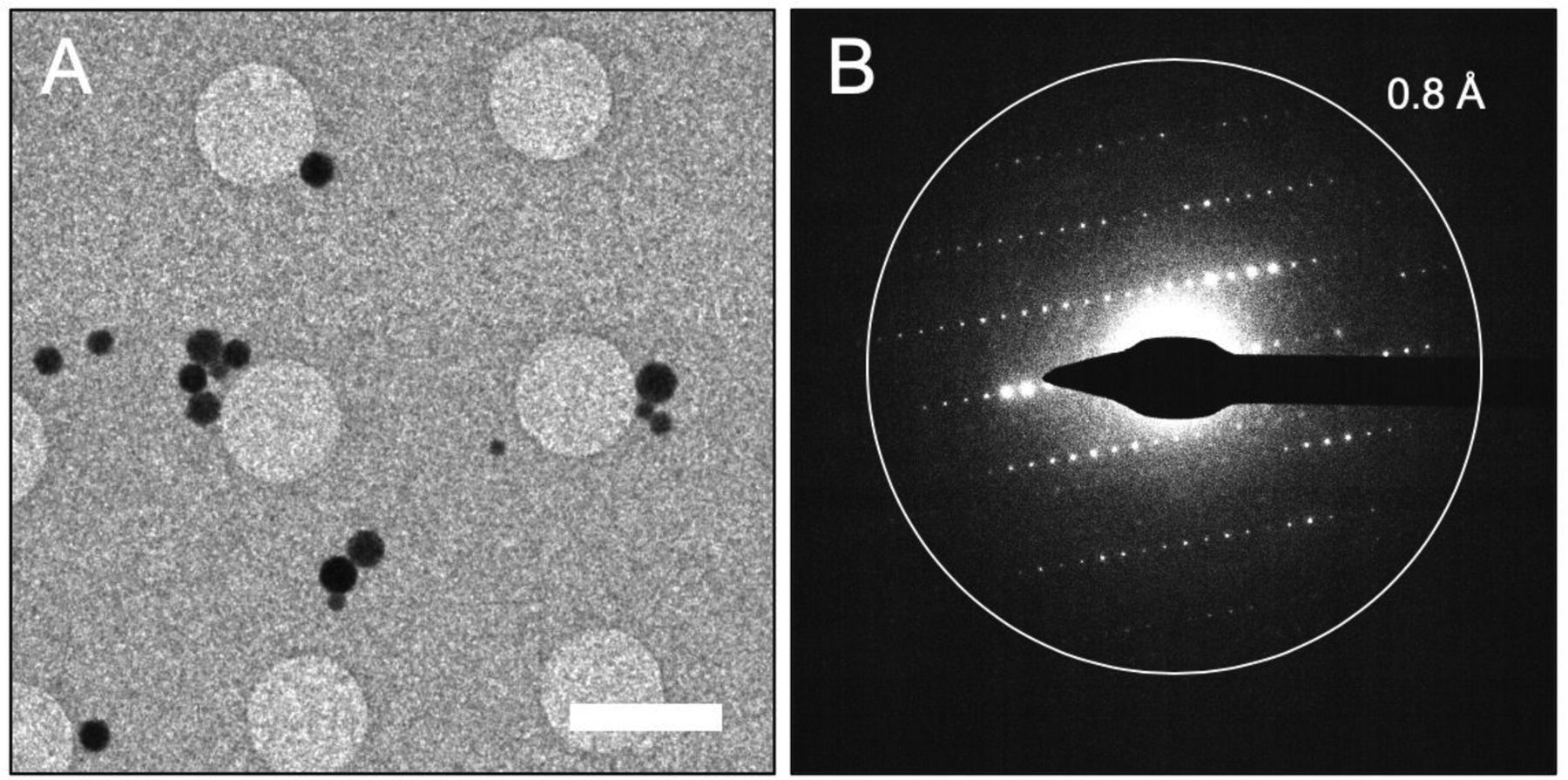
ZIF-8 crystals yield high resolution diffraction patterns. (A) A low magnification search of the EM grid shows well-dispersed ZIF-8 crystals on the surface of the holey-carbon. (B) Electron diffraction pattern collected from a high-quality ZIF-8 nanocrystal shows sharp and well-separated spots extending to high-resolution. This diffraction pattern represents what was typically seen for crystals which were used for MicroED data collection. Scale bar in A represents 3 μm.
ZIF-8 crystals ranging in diameter from approximately 200 nm to 800 nm could be seen on the grids. While screening crystal quality by taking initial diffraction patterns from each crystal, it was found that the diffraction data became qualitatively worse (i.e. lower resolution and more background noise from the increased diffuse scattering) as the size of the ZIF-8 nanoparticles approached the larger end of the size range (approximately 400 nm to 800 nm in diameter). The crystals which were on the smaller end of the size range (approximately 200 nm to 400 nm in diameter), generally produced better diffraction, and crystals that produced the data with the highest resolution and sharpest diffraction spots were used for data collection (Fig 2B). Ultimately, the crystal that produced the data used for structure determination was approximately 200 nm across and 200 nm thick. This is significantly smaller than the size of crystals used for single crystal X-ray methods which have dimensions on the order of tens to hundreds of micrometers.
In a previous study, it was demonstrated that ZIF-8 samples rapidly lose their crystallinity in the electron beam [5]. Therefore, the total exposure for each crystal during the course of MicroED data collection was kept to ∼1 e−/Å2, which is much lower than the what was used the recent high-resolution imaging studies of ZIF-8 particles [4, 5, 7]. The time to collect a MicroED data set is relatively short (in the range of 1 to 2 minutes for the ZIF-8 samples), and because many of the ZIF-8 nanocrystals in the 200 to 400 nm size range diffracted to beyond 1Å, we could specifically focus on crystals of this size. This allowed the collection of many high-resolution data sets in a relatively short time (about 1–2 minutes per crystal). With sample loading, microscope setup, crystal screening for only the best crystals, and data collection are all taken into account, we collected 12 high-quality data sets in approximately 2 hours. The ability to rapidly collect a large number of data sets is a great advantage for the use of MicroED in the study of MOFs and other organic, biological, or beam-sensitive crystalline samples.
While data from many crystals were collected, because of the high symmetry of the ZIF-8 nanocrystals, data from a single crystal could ultimately be used for structure determination to 0.87 Å (Table 1). When processed, data from one single crystal resulted in a data set that was 99.8% complete. It should be noted that many other crystals in the data sets collected could also be used for structure determination as well. The direct methods solution found was in space group I-43m and 5 atoms were placed within the asymmetric unit, which is what would be expected for ZIF-8. Following structure refinement (R1 = 0.1779, wR2 = 0.3411, and GooF = 1.011) and expanding the asymmetric unit the structure of ZIF-8 can be seen, with each zinc being coordinated by 4 MeIM ligands and each ligand bridges 2 zinc atoms (Fig 3). This produces the typical SOD zeolite type structure with hex-zinc-atomic open ring channels of 3.4 Å in diameter. Both the unit cell parameters and atom positions of the MicroED structure compare well with previous ZIF-8 structures determined by single crystal X-ray diffraction [39, 42, 43], with differences in bond lengths between the MicroED structures and the X-ray structures being in the range of 0.03 to 0.07 Å.
Table 1.
Data collection and refinement statistics
| ZIF-8 | |
| Data collection | |
| Excitation Voltage | 300 kV |
| Wavelength (Å) | 0.019687 |
| Number of crystals | 1 |
| Data Processing | |
| Space group | l-43m |
| Unit cell length a = b = c (Å) | 16.880 |
| Angles α = β = γ (°) | 90.00 |
| Resolution (Å) | 8.45 – 0.87 |
| Number of reflections | 7,466 |
| Unique reflections | 407 |
| Robs(%) | 34.3 (159.0) |
| Rmeas(%) | 35.8 (163.3) |
| I/σI | 4.37 (1.11) |
| CC1/2 (%) | 97.2 (57.0) |
| Completeness (%) | 99.8 (100.0) |
| Structure Refinement | |
| R1 | 0.1779 |
| wR2 | 0.3411 |
| GooF | 1.011 |
Values in parentheses represent numbers in the highest resolution shell (1.02 – 0.97 Å)
Figure 3.
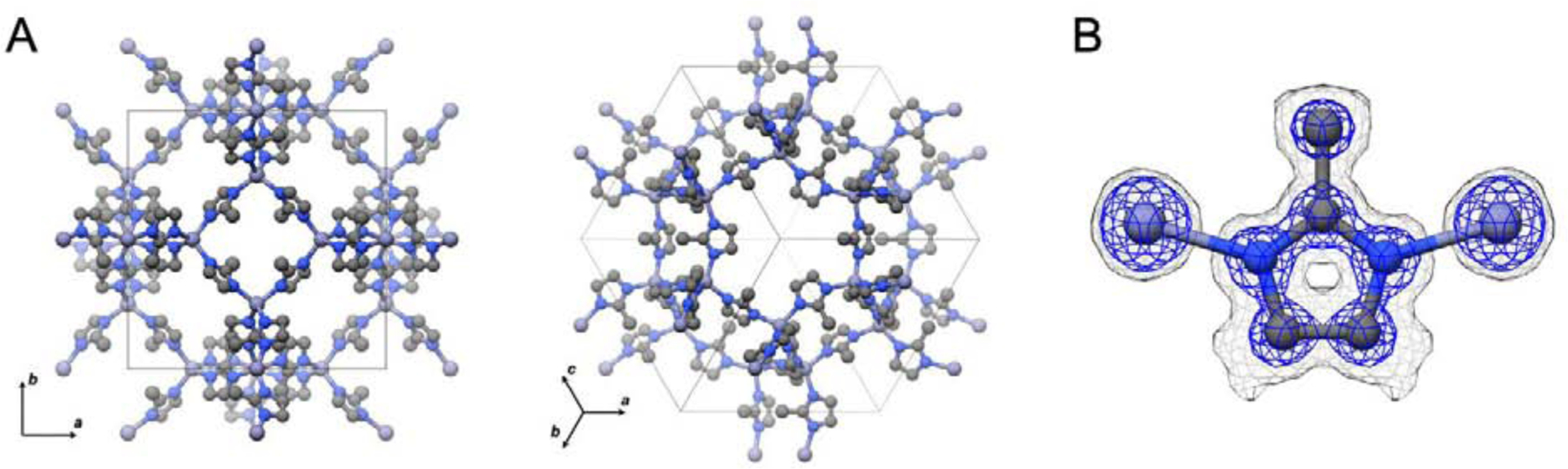
MicroED structure and unit cell of ZIF-8 veiwed in different orientations (A) and the surrounding density (B). The maps in panel B are contoured at 0.8σ (gray) and 2.4σ (blue). Hydrogens are omitted from the figures for clarity.
4. Conclusion
The accurate determination MOF structure is critical to understanding and engineering the properties of these materials, and it is important to make use of methods capable of facilitating the rapid determination of beam-sensitive MOFs. The cryo-EM method of MicroED is a fast and reliable method for structure determination from micro and nanocrystals, and we have shown here that this procedure can be easily extended to the ab initio structure determination and analysis of beam sensitive MOFs. A benefit of the MicroED method is that it can be performed on many of the same cryo-TEM instruments used for high-resolution imaging. Future combinations of MicroED structure determination with low-dose high-resolution imaging could allow multi-leveled analysis of beam-sensitive materials from a single sample, where diffraction is used for high-resolution structure determination and the images can analyze surface and interfacial structure. This would further cement low-dose cryo-TEM as an invaluable tool for understanding MOF structure and function.
Highlights.
Microcrystal electron diffraction (MicroED) procedures can be used to determine the structure of beam-sensitive metal-organic frameworks (MOFs)
The diffraction data were collected at ultralow dose (∼1 e−/Å2 per data set) to reduce the damaging effects of the electron beam
MicroED data sets could be collected from ZIF-8 crystals quickly, and data were processed rapidly using common crystallographic programs.
Data from a single ZIF-8 crystal was sufficient for structure determination
The structure of the model MOF ZIF-8 was determined to 0.87 Å by MicroED
Acknowledgements
FB and JYSL would like to acknowledge support from the National Science Foundation (CBET-1511005). BLN would like acknowledge the support from the National Science Foundation DMR-1942084) and the National Institutes of Health (R21GM135784). We would like to acknowledge the use of the Titan Krios at the Eyring Materials Center at Arizona State University and the funding of this instrument by NSF MRI 1531991.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Coordinates for ZIF-8 have been deposited in the Cambridge Structural Database (CCDC 1997958)
References
- [1].Zhang C, Lively RP, Zhang K, Johnson JR, Karvan O, Koros WJ, Unexpected Molecular Sieving Properties of Zeolitic Imidazolate Framework-8, The Journal of Physical Chemistry Letters, 3 (2012) 2130–2134. [DOI] [PubMed] [Google Scholar]
- [2].Zhao Z, Li Z, Lin YS, Adsorption and Diffusion of Carbon Dioxide on Metal−Organic Framework (MOF-5), Industrial & Engineering Chemistry Research, 48 (2009) 10015–10020. [Google Scholar]
- [3].Gandara F, Bennett TD, Crystallography of metal-organic frameworks, Iucrj, 1 (2014) 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li Y, Wang K, Zhou W, Li Y, Vila R, Huang W, Wang H, Chen G, Wu G-H, Tsao Y, Wang H, Sinclair R, Chiu W, Cui Y, Cryo-EM Structures of Atomic Surfaces and Host-Guest Chemistry in Metal-Organic Frameworks, Matter, 1 (2019) 428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhu YH, Ciston J, Zheng B, Miao XH, Czarnik C, Pan YC, Sougrat R, Lai ZP, Hsiung CE, Yao KX, Pinnau I, Pan M, Han Y, Unravelling surface and interfacial structures of a metal-organic framework by transmission electron microscopy, Nat Mater, 16 (2017) 532−+. [DOI] [PubMed] [Google Scholar]
- [6].Li X, Wang J, Liu X, Liu L, Cha D, Zheng X, Yousef AA, Song K, Zhu Y, Zhang D, Han Y, Direct Imaging of Tunable Crystal Surface Structures of MOF MIL-101 Using High-Resolution Electron Microscopy, Journal of the American Chemical Society, 141 (2019) 12021–12028. [DOI] [PubMed] [Google Scholar]
- [7].Zhang D, Zhu Y, Liu L, Ying X, Hsiung C-E, Sougrat R, Li K, Han Y, Atomic-resolution transmission electron microscopy of electron beam–sensitive crystalline materials, Science, (2018) eaao0865. [DOI] [PubMed] [Google Scholar]
- [8].Zhu L, Zhang D, Xue M, Li H, Qiu S, Direct observations of the MOF (UiO-66) structure by transmission electron microscopy, CrystEngComm, 15 (2013) 9356–9359. [Google Scholar]
- [9].Feyand M, Mugnaioli E, Vermoortele F, Bueken B, Dieterich JM, Reimer T, Kolb U, de Vos D, Stock N, Automated Diffraction Tomography for the Structure Elucidation of Twinned, Submicrometer Crystals of a Highly Porous, Catalytically Active Bismuth Metal–Organic Framework, Angewandte Chemie International Edition, 51 (2012) 10373–10376. [DOI] [PubMed] [Google Scholar]
- [10].Yuan S, Qin J-S, Xu H-Q, Su J, Rossi D, Chen Y, Zhang L, Lollar C, Wang Q, Jiang H-L, Son DH, Xu H, Huang Z, Zou X, Zhou H-C, [Ti8Zr2O12(COO)16] Cluster: An Ideal Inorganic Building Unit for Photoactive Metal–Organic Frameworks, ACS Central Science, 4 (2018) 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Denysenko D, Grzywa M, Tonigold M, Streppel B, Krkljus I, Hirscher M, Mugnaioli E, Kolb U, Hanss J, Volkmer D, Elucidating gating effects for hydrogen sorption in MFU-4-type triazolate-based metal-organic frameworks featuring different pore sizes, Chemistry, 17 (2011) 1837–1848. [DOI] [PubMed] [Google Scholar]
- [12].Nannenga BL, MicroED methodology and development, Struct Dyn, 7 (2020) 014304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nannenga BL, Gonen T, The cryo-EM method microcrystal electron diffraction (MicroED), Nature Methods, 16 (2019) 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gemmi M, Mugnaioli E, Gorelik TE, Kolb U, Palatinus L, Boullay P, Hovmöller S, Abrahams JP, 3D Electron Diffraction: The Nanocrystallography Revolution, ACS Central Science, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gruene T, Wennmacher JTC, Zaubitzer C, Holstein JJ, Heidler J, Fecteau-Lefebvre A, De Carlo S, Muller E, Goldie KN, Regeni I, Li T, Santiso-Quinones G, Steinfeld G, Handschin S, van Genderen E, van Bokhoven JA, Clever GH, Pantelic R, Rapid Structure Determination of Microcrystalline Molecular Compounds Using Electron Diffraction, Angew Chem Int Ed Engl, 57 (2018) 16313–16317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jones CG, Martynowycz MW, Hattne J, Fulton TJ, Stoltz BM, Rodriguez JA, Nelson HM, Gonen T, The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination, ACS Central Science, 4 (2018) 1587–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Brázda P, Palatinus L, Babor M, Electron diffraction determines molecular absolute configuration in a pharmaceutical nanocrystal, Science, 364 (2019) 667. [DOI] [PubMed] [Google Scholar]
- [18].Palatinus L, Brazda P, Boullay P, Perez O, Klementova M, Petit S, Eigner V, Zaarour M, Mintova S, Hydrogen positions in single nanocrystals revealed by electron diffraction, Science, 355 (2017) 166–169. [DOI] [PubMed] [Google Scholar]
- [19].Nannenga BL, Shi D, Leslie AG, Gonen T, High-resolution structure determination by continuous-rotation data collection in MicroED, Nat Methods, 11 (2014) 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hattne J, Shi D, Glynn C, Zee CT, Gallagher-Jones M, Martynowycz MW, Rodriguez JA, Gonen T, Analysis of Global and Site-Specific Radiation Damage in Cryo-EM, Structure, 26 (2018) 759–766 e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shi D, Nannenga BL, Iadanza MG, Gonen T, Three-dimensional electron crystallography of protein microcrystals, Elife, 2 (2013) e01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Warmack RA, Boyer DR, Zee C-T, Richards LS, Sawaya MR, Cascio D, Gonen T, Eisenberg DS, Clarke SG, Structure of amyloid-β (20–34) with Alzheimer’s-associated isomerization at Asp23 reveals a distinct protofilament interface, Nature Communications, 10 (2019) 3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ting CP, Funk MA, Halaby SL, Zhang Z, Gonen T, van der Donk WA, Use of a scaffold peptide in the biosynthesis of amino acid–derived natural products, Science, 365 (2019) 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Purdy MD, Shi D, Chrustowicz J, Hattne J, Gonen T, Yeager M, MicroED structures of HIV-1 Gag CTD-SP1 reveal binding interactions with the maturation inhibitor bevirimat, Proceedings of the National Academy of Sciences, (2018) 201806806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Krotee P, Griner SL, Sawaya MR, Cascio D, Rodriguez JA, Shi D, Philipp S, Murray K, Saelices L, Lee J, Seidler P, Glabe CG, Jiang L, Gonen T, Eisenberg DS, Common fibrillar spines of amyloid-beta and human islet amyloid polypeptide revealed by microelectron diffraction and structure-based inhibitors, J Biol Chem, 293 (2018) 2888–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gallagher-Jones M, Glynn C, Boyer DR, Martynowycz MW, Hernandez E, Miao J, Zee CT, Novikova IV, Goldschmidt L, McFarlane HT, Helguera GF, Evans JE, Sawaya MR, Cascio D, Eisenberg DS, Gonen T, Rodriguez JA, Sub-angstrom cryo-EM structure of a prion protofibril reveals a polar clasp, Nat Struct Mol Biol, 25 (2018) 131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sawaya MR, Rodriguez J, Cascio D, Collazo MJ, Shi D, Reyes FE, Hattne J, Gonen T, Eisenberg DS, Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED, Proc Natl Acad Sci U S A, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nannenga BL, Shi D, Hattne J, Reyes FE, Gonen T, Structure of catalase determined by MicroED, Elife, 3 (2014) e03600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dick M, Sarai NS, Martynowycz MW, Gonen T, Arnold FH, Tailoring Tryptophan Synthase TrpB for Selective Quaternary Carbon Bond Formation, Journal of the American Chemical Society, 141 (2019) 19817–19822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rodriguez JA, Ivanova MI, Sawaya MR, Cascio D, Reyes FE, Shi D, Sangwan S, Guenther EL, Johnson LM, Zhang M, Jiang L, Arbing MA, Nannenga BL, Hattne J, Whitelegge J, Brewster AS, Messerschmidt M, Boutet S, Sauter NK, Gonen T, Eisenberg DS, Structure of the toxic core of alpha-synuclein from invisible crystals, Nature, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Xu H, Lebrette H, Clabbers MTB, Zhao J, Griese JJ, Zou X, Högbom M, Solving a new R2lox protein structure by microcrystal electron diffraction, Science Advances, 5 (2019) eaax4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Levine AM, Bu G, Biswas S, Tsai EHR, Braunschweig AB, Nannenga BL, Crystal structure and orientation of organic semiconductor thin films by microcrystal electron diffraction and grazing-incidence wide-angle X-ray scattering, Chemical Communications, 56 (2020) 4204–4207. [DOI] [PubMed] [Google Scholar]
- [33].Kida K, Okita M, Fujita K, Tanaka S, Miyake Y, Formation of high crystalline ZIF-8 in an aqueous solution, CrystEngComm, 15 (2013) 1794–1801. [Google Scholar]
- [34].Shi D, Nannenga BL, de la Cruz MJ, Liu J, Sawtelle S, Calero G, Reyes FE, Hattne J, Gonen T, The collection of MicroED data for macromolecular crystallography, Nat Protoc, 11 (2016) 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hattne J, Reyes FE, Nannenga BL, Shi D, de la Cruz MJ, Leslie AG, Gonen T, MicroED data collection and processing, Acta Crystallogr A Found Adv, 71 (2015) 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kabsch W, Xds, Acta Crystallogr D Biol Crystallogr, 66 (2010) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sheldrick GM, SHELXT - integrated space-group and crystal-structure determination, Acta Crystallogr A Found Adv, 71 (2015) 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hubschle CB, Sheldrick GM, Dittrich B, ShelXle: a Qt graphical user interface for SHELXL, J Appl Crystallogr, 44 (2011) 1281–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Park KS, Ni Z, Côté AP, Choi JY, Huang R, Uribe-Romo FJ, Chae HK, O’Keeffe M, Yaghi OM, Exceptional chemical and thermal stability of zeolitic imidazolate frameworks, Proceedings of the National Academy of Sciences, 103 (2006) 10186–10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liu G, Jiang Z, Cao K, Nair S, Cheng X, Zhao J, Gomaa H, Wu H, Pan F, Pervaporation performance comparison of hybrid membranes filled with two-dimensional ZIF-L nanosheets and zero-dimensional ZIF-8 nanoparticles, Journal of Membrane Science, 523 (2017) 185–196. [Google Scholar]
- [41].Liu G, Xu Y, Han Y, Wu J, Xu J, Meng H, Zhang X, Immobilization of lysozyme proteins on a hierarchical zeolitic imidazolate framework (ZIF-8), Dalton Transactions, 46 (2017) 2114–2121. [DOI] [PubMed] [Google Scholar]
- [42].Morris W, Stevens CJ, Taylor RE, Dybowski C, Yaghi OM, Garcia-Garibay MA, NMR and X-ray Study Revealing the Rigidity of Zeolitic Imidazolate Frameworks, The Journal of Physical Chemistry C, 116 (2012) 13307–13312. [Google Scholar]
- [43].Kwon HT, Jeong H-K, Lee AS, An HS, Lee JS, Heteroepitaxially Grown Zeolitic Imidazolate Framework Membranes with Unprecedented Propylene/Propane Separation Performances, Journal of the American Chemical Society, 137 (2015) 12304–12311. [DOI] [PubMed] [Google Scholar]


