Abstract
Plants have evolved both cell surface-resident receptor-like kinases (RLKs) and intracellular nucleotide-binding leucine-rich repeat (NLR) proteins as immune receptors to detect infections. It remains enigmatic how RLK- and NLR-mediated signaling is connected. Disruption of an immune-activated MEKK1-MKK1/2-MPK4 MAPK cascade activates the NLR SUMM2 via the MAPK kinase kinase MEKK2, leading to autoimmunity. To gain insights into the mechanisms underlying SUMM2 activation, we deployed an RNAi-based genetic screen for mekk1 autoimmune suppressors, and identified an uncharacterized malectin-like RLK, named LETUM1 (LET1), as a specific regulator of mekk1-mkk1/2-mpk4 autoimmunity via complexing with both SUMM2 and MEKK2. MEKK2 scaffolds LET1 and SUMM2 for protein stability and association, and counter-regulates the F-box protein CPR1-mediated SUMM2 ubiquitination and degradation; thereby, regulating SUMM2 accumulation and activation. Our study indicates that malectin-like RLK LET1 senses the perturbance of cellular homeostasis caused by the deficiency in immune-activated signaling, and activates the NLR SUMM2-mediated autoimmunity via MEKK2 scaffolding.
The innate immune system detects pathogen-derived molecules via specialized immune receptors to prevent infections. Plant immune receptors include cell surface-resident pattern recognition receptors (PRRs) and intracellular nucleotide-binding domain leucine-rich repeat proteins (NLRs)1–3. In plants, PRRs are often receptor-like kinases (RLKs) and receptor-like proteins (RLPs) that sense microbe/damage-associated molecular patterns (MAMPs/DAMPs) and collectively contribute to host immunity against a broad-spectrum of pathogens4–6. Plant NLRs detect pathogen-specific effector proteins that are translocated into host cells and trigger effector-specific immunity, often accompanied by programmed cell death7–9. Mitogen-activated protein kinase (MAPK) cascades, canonically consisting of three-tiered kinases, the MAP kinase kinase kinase (MAPKKK/MKKK/MEKK), the MAP kinase kinase (MAPKK/MKK) and the MAPK (MPK), are shared signaling modules in plant PRR- and NLR-mediated immunity10–12. Perception of MAMPs by PRRs activates two parallel MAPK cascades: MKKK3/5-MKK4/5-MPK3/6 and MEKK1-MKK1/2-MPK4 in Arabidopsis13–15. Interestingly, deficiency in PRR-activated MEKK1-MKK1/2-MPK4 cascade activates the NLR protein suppressor of mkk1 mkk2 2 (SUMM2)-mediated autoimmunity and cell death16–21.
It remains enigmatic how NLR- and PRR-mediated signaling pathways are mechanistically interconnected as the distinct and overlapping responses are activated in the two-tiered plant immune system. The observation that PRR-activated MAPK cascade (MEKK1-MKK1/2-MPK4) suppresses NLR SUMM2 activation presents a unique case to dissect the intertwined regulation of RLK and NLR activation. With a virus-induced gene silencing (VIGS)-based transient RNAi approach, we have established an innovative and high-throughput cell death suppressor screen towards Arabidopsis T-DNA sequence-indexed mutant collections to understand the mechanism underlying PRR co-receptor BAK1/SERK4-mediated cell death22,23. To gain insights into the mechanisms underlying NLR SUMM2 activation, which is otherwise suppressed by a PRR-activated MAPK cascade, we deployed the VIGS-based RNAi screen for the suppressors of mekk1 cell death. The mekk1 null mutant is postembryonic lethal associated with spontaneous cell death and autoimmunity, and is unable to produce enough seeds for classical suppressor screens17–20. Silencing MEKK1 via Agrobacterium-mediated RNAi in wild-type (WT) Col-0 plants triggers plant dwarfism and leaf chlorosis, spontaneous cell death and elevated H2O2 accumulation and defense gene accumulation as observed in the mekk1 mutants (Fig. 1A–C). From a screen of ~12,000 homozygous Arabidopsis T-DNA insertion mutants from the Arabidopsis Biological Resource Center (ABRC), we identified several mutants named lethality suppressor of mekk1 (letum or let) that suppressed the cell death and autoimmunity caused by silencing MEKK124. T-DNA insertional analysis of some mutants instantaneously revealed the identities of the corresponding genes, including SUMM2, MAPK KINASE KINASE 2 (MEKK2), and CALMODULIN-BINDING RECEPTOR-LIKE CYTOPLASMIC KINASE 3 (CRCK3), which are essential players in regulating the mekk1-mkk1/2-mpk4 cell death16,25–27, demonstrating the robustness of the VIGS screen to uncover the components regulating plant cell death24.
Fig. 1. The let1 mutants suppress autoimmunity triggered by silencing MEKK1.
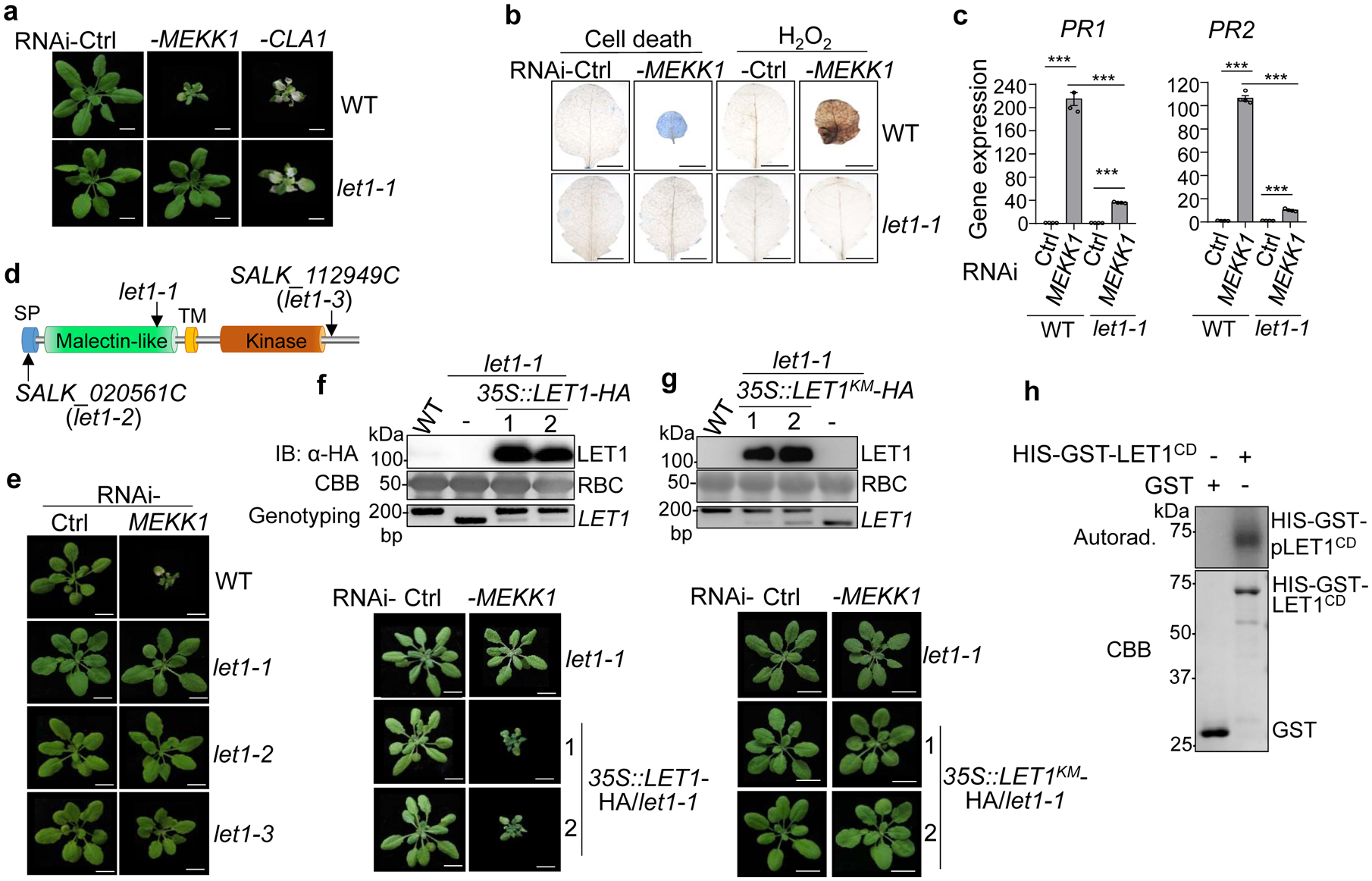
a, The let1–1 mutant suppresses growth defects triggered by RNAi-MEKK1. Plants of WT and let1–1 are shown three weeks after the inoculation of Agrobacterium carrying the VIGS vector control (Ctrl), VIGS-MEKK1, or VIGS-CLA1. Plant dwarfism and leaf chlorosis after silencing MEKK1 were observed in WT but not in let1–1. The albino phenotype of WT and let1–1 after silencing CLA1 (Cloroplastos alterados 1) was used as a visible indicator for VIGS efficiency. Scale bar, 1 cm.
b, The let1–1 mutant suppresses cell death and H2O2 production triggered by RNAi-MEKK1. True leaves after VIGS of MEKK1 were stained with trypan blue for cell death and DAB for H2O2 accumulation. Scale bar, 0.5 cm.
c, The let1–1 mutant suppresses PR1 and PR2 expression triggered by RNAi-MEKK1. The expression of PR1 and PR2 was normalized by the expression of UBQ10. P = 1.42 × 10−6 (PR1, column 1 and 2), P = 6.06 × 10−10 (PR1, column 3 and 4), P = 4.09 × 10−6 (PR1, column 2 and 4), P = 7.28 × 10−9 (PR2, column 1 and 2), P = 3.73 × 10−6 (PR2, column 3 and 4), P = 1.48 × 10−8 (PR2, column 2 and 4). Data are shown as mean ± SE from four independent repeats. The asterisks indicate statistical significance by using two-sided two-tailed Student’s t-test (***, P < 0.001).
d, The schematic diagram of LET1 protein motifs consisting of the signal peptide (SP), malectin-like domain, transmembrane domain (TM) and kinase domain. Arrows indicate T-DNA insertion sites or deletion sites of the different mutant alleles of let1.
e, The let1–2 and let1–3 mutants suppress growth defects triggered by RNAi-MEKK1. Experiments were done similarly as in a.
f, Complementation of the let1–1 mutant with LET1 restores the growth defects by RNAi-MEKK1. #1 and #2 are two homozygous complementation lines carrying 35S::LET1-HA in the let1–1 mutant background. Scale bar, 1 cm. The immunoblot by an α-HA antibody shows LET1-HA proteins (top panel); Coomassie brilliant blue (CBB) staining of Rubisco proteins (RBC) serves as a loading control (middle panel), and genotyping PCR using primer pairs spanning the deletion site in let1–1 was used to confirm the deletion in let1–1 (bottom panel).
g, LET1KM cannot complement the let1–1 mutant for the growth defects by RNAi-MEKK1. #1 and #2 are two homozygous complementation lines carrying 35S::LET1KM-HA in the let1–1 mutant background. Experiments were done similarly as in f.
h, LET1 bears kinase activity. The LET1 cytosolic kinase domain fused with HIS-GST (HIS-GST-LET1CD) was expressed in insect cells. The kinase assay was performed using purified GST or HIS-GST-LET1CD proteins with [γ−32P] ATP (top panel). The CBB staining of input proteins is shown on the bottom panel.
The experiments in a-g were repeated at least three times, and h was repeated twice with similar results.
Here, we report the characterization of let1, which substantially reduced growth defects (Fig. 1a), cell death detected by trypan blue staining (Fig. 1b), elevated H2O2 accumulation by 3,3’-diaminobenzidine (DAB) staining (Fig. 1b), and constitutive expression of pathogenesis-related genes PR1 and PR2 (Fig. 1c), caused by silencing MEKK1. The let1–1 mutant did not affect cell death caused by silencing BAK1/SERK4 or BAK1-interacting RLK BIR1 (Supplementary Fig. 1a), suggesting the specific involvement of LET1 in mekk1 cell death. The let1–1 mutant is SALK_018793C, which bears a T-DNA insertion annotated in the promoter of PIRL1 (AT5G05850), encoding for a member of the Plant Intracellular Ras-group-related Leucine-rich repeat proteins (PIRLs) (Supplementary Fig. 1b & c). However, the transcripts of PIRL1 were not reduced in let1–1, likely due to the insertion in the promoter region (Supplementary Fig. 1d). In addition, another allele of T-DNA insertion mutant of PIRL1, SALK_072332C, did not suppress RNAi-MEKK1-triggered cell death (Supplementary Fig. 1e), implicating that pirl1 might not be the causal mutation in let1–1 suppressing mekk1 cell death. To identify the causal mutation in let1–1, we crossed the let1–1mekk1 double mutant in the Col-0 background with the Ler-0 ecotype and genotyped plants with growth defects from an F2 segregating population. All the plants with growth defects are mekk1 homozygous. Using 157 plants resembling let1–1mekk1, which partially suppressed mekk1 cell death when grown on soil (Supplementary Fig. 2a), the mutation was mapped in a region around 1M base pairs (bp) between markers F5H14 and T20D16 (Supplementary Fig. 2b). The whole-genome resequencing of let1–1 revealed a 17-bp deletion in AT2G23200 in the mapped region (Supplementary Fig. 2b), which leads to an early stop codon (Supplementary Fig. 2c). The deletion was further confirmed by Polymerase Chain Reaction (PCR) analysis and targeted Sanger sequencing (Supplementary Fig. 2d).
LET1 encodes an uncharacterized malectin-like RLK (Fig. 1d). Malectin-like RLKs, also called Catharanthus roseus RLK1-like kinases (CrRLK1Ls), contain one or two extracellular malectin domains with similarity to the animal carbohydrate-binding malectin proteins28–31. There are 17 members in Arabidopsis, some of which, such as FERONIA (FER)32, ANXUR1 (ANX1)/ANX233,34, HERKULES1 (HERK1)35, and BUDDHA’S PAPER SEAL 1 (BUPS1)/BUPS236, are key regulators in various developmental processes including fertilization, cell elongation, and polarized growth. In addition, both FER and ANXs have been shown to be involved in plant immunity37–40. LETUM, also known as Mors, is the personification of death in Roman mythology, which led us to name our mutants as lethality suppressor of mekk1 (letum or let). Two additional T-DNA insertion alleles, let1–2 and let1–3, also suppressed growth defects triggered by RNAi-MEKK1 (Fig. 1e). In addition, expressing LET1, but not its kinase mutant variant bearing a mutation in a conserved ATP binding site (LET1K516E, LET1KM), restored cell death in let1–1 (Fig. 1f & g). Thus, mutations in LET1 suppressed RNAi-MEKK1 cell death and the kinase activity of LET1 is likely required for this function. Consistently, when purified from insect cells, the LET1 cytosolic domain (CD) consisting of the juxtamembrane and kinase domains fused with HIS-tagged glutathione S-transferase (HIS-GST-LET1CD) displayed strong autophosphorylation activity in an in vitro kinase assay (Fig. 1h).
MAPKK MKK1/2 and MAPK MPK4 function downstream of MEKK1 in SUMM2-mediated cell death control17. Similar to mekk1, the mkk1/2 and mpk4 mutants are seedling lethal17,21. To delineate the genetic position of let1 in suppressing mekk1-mkk1/2-mpk4 cell death, we generated the let1–1/mekk1, let1–1/mkk1/2, and let1–1/mpk4 mutants by genetic crosses. The let1–1 mutant suppressed the seedling lethality (Fig. 2a), cell death (Fig. 2b), elevated H2O2 accumulation (Fig. 2c), and defense gene PR1 and PR2 accumulation (Fig. 2d) in the let1–1/mekk1, let1–1/mkk1/2 and let1–1/mpk4 mutants, suggesting that LET1 functions genetically downstream of the MEKK1-MKK1/2-MPK4 cascade. It is known that the MEKK1-MKK1/2-MPK4 cascade negatively regulates the transcript level of MEKK2/SUMM1, encoding another MAPKKK, in SUMM2-mediated autoimmunity25,26. Overexpression of MEKK2 induced stunted growth, cell death, elevated H2O2 accumulation and PR gene expression in WT plants25,26 (Fig. 2e, Supplementary Fig. 3a & b). However, the elevated defense responses triggered by MEKK2 overexpression was largely reduced in the let1–1 mutant (Fig. 2e & f, Supplementary Fig. 3a & b), indicating that LET1 functions genetically downstream of MEKK2. Overexpression of an active form of SUMM2 (SUMM2ac), which bears an asparagine-to-valine mutation at the 478 amino acid residue in the MHD motif, triggers cell death in Nicotiana benthamiana16. Overexpression of SUMM2ac also triggered growth defects and autoimmunity in Arabidopsis WT plants (Fig. 2g & h, Supplementary Fig. 4a & b). However, the let1–1 mutant did not interfere with cell death, H2O2 accumulation, stunted growth, and defense gene activation triggered by overexpression of SUMM2ac (Fig. 2g & h, Supplementary Fig. 4a & b), indicating that LET1 functions genetically upstream or independent of SUMM2. The protein level of MEKK2 and SUMM2ac was similar in WT and let1–1 (Fig. 2f & h), indicating that LET1 might not affect MEKK2 and SUMM2 protein stability. Taken together, the above epistasis analysis demonstrates that LET1 functions genetically downstream of MEKK2 and upstream of SUMM2 in the mekk1 cell death pathway (Fig. 2i).
Fig. 2. LET1 functions genetically downstream of MEKK2 and upstream of SUMM2 in cell death control.
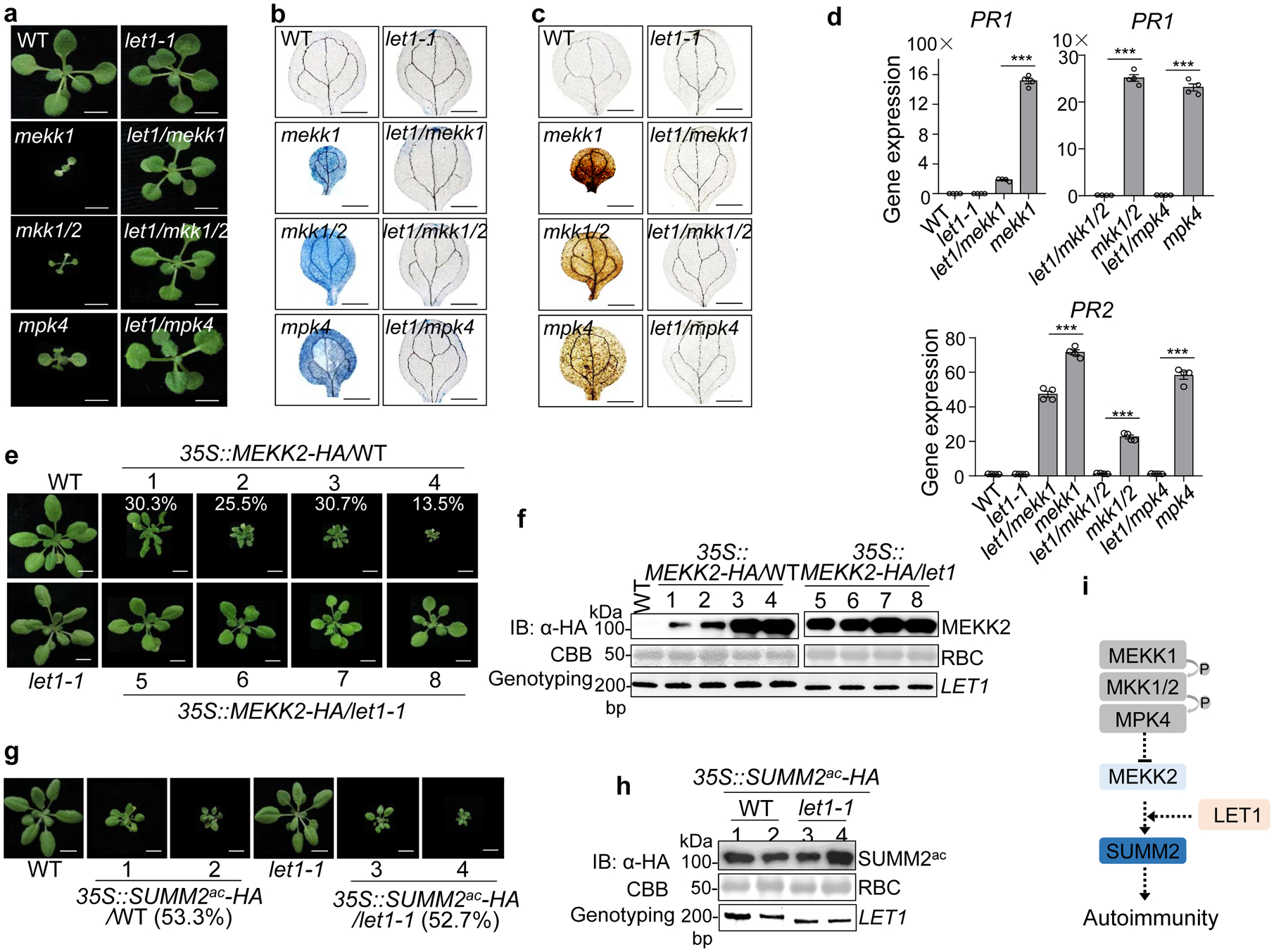
a, The let1–1 mutant suppresses growth defects of mekk1, mkk1/2, and mpk4 mutants. Three-week-old plants grown on ½ MS medium plates with indicated genotypes are shown. Scale bar, 0.5 cm.
b and c The let1–1 mutant suppresses cell death (b) and H2O2 accumulation (c) in mekk1, mkk1/2, and mpk4. Cell death and H2O2 were stained by trypan blue and DAB respectively, with cotyledons of two-week-old plants grown on ½ MS medium plates. Scale bar, 0.1 cm.
d, The let1–1 mutant suppresses the elevated expression levels of PR1 and PR2 in mekk1, mkk1/2, and mpk4. The expression of PR1 and PR2 was normalized by the expression of UBQ10. P = 3.37 × 10−8 (PR1, left, column 3 and 4), P = 3.81 × 10−8 (PR1, right, column 1 and 2), P = 8.32 × 10−8 (PR1, right, column 3 and 4), P = 4.21 × 10−5 (PR2, column 3 and 4), P = 1.21 × 10−6 (PR2, column 5 and 6), P = 2.22 × 10−7 (PR2, column 7 and 8). Data are shown as mean ± SE from four independent repeats. The asterisks indicate statistical significance by using two-sided two-tailed Student’s t-test (***, P < 0.001).
e, The let1–1 mutant suppresses growth defects triggered by overexpressing MEKK2. The 35S::MEKK2-HA transgenic plants in the WT background were grouped into four categories with representative plants showing dwarfism. The numbers are the percentages of plants displaying the indicated phenotype among a total of 251 transgenic plants analyzed for 35S::MEKK2-HA/WT (n=251). The 35S::MEKK2-HA transgenic plants in the let1–1 mutant background do not show severe growth defects and cell death (n=56). Scale bar, 1 cm.
f, MEKK2 protein expression and genotyping of LET1 in transgenic plants from e. Total proteins were subjected to immunoblotting using an α-HA antibody (top panel), and CBB staining serves as a loading control (middle panel). PCR genotyping of LET1 with primers spanning the deletion site in let1–1 is shown (bottom panel).
g, The let1–1 mutant cannot suppress the growth defects triggered by overexpressing the active SUMM2 (SUMM2ac), D478V variant. The 35S::SUMM2ac-HA transgenic plants show growth defects with a similar ratio in WT (53.3%, n=75) and let1–1 (52.7%, n=55) backgrounds. Scale bar, 1 cm.
h, SUMM2ac protein expression and genotyping of LET1 in transgenic plants from g. Assays were done similarly as in f.
i, The schematic diagram of LET1 in the SUMM2-mediated cell death pathway.
All the above experiments were repeated at least three times with similar results obtained.
Since LET1 genetically functions between MEKK2 and SUMM2, we tested whether LET1 complexes with MEKK2 and/or SUMM2. A co-immunoprecipitation (Co-IP) assay with HA epitope-tagged LET1 and FLAG epitope-tagged MEKK2, SUMM2 or MPK4 indicated that LET1 associated with MEKK2 and SUMM2 but not with MPK4, in Arabidopsis protoplasts (Fig. 3a). The associations of LET1 with MEKK2 and SUMM2 were confirmed using reciprocally switched epitope tags in Arabidopsis protoplasts (Supplementary Fig. 5a & b), and in stable transgenic plants expressing LET1 under its native promoter (pLET1::LET1-FLAG) (Supplementary Fig. 5c & d). We also tested the association of LET1 with SUMM2 and MEKK2 with split-luciferase (Split-LUC) assays, in which LET1 was fused with the C-terminal luciferase (LET1-Cluc) and SUMM2 or MEKK2 was fused with the N-terminal luciferase (SUMM2-Nluc or MEKK2-Nluc). Co-expression of LET1-Cluc with SUMM2-Nluc or MEKK2-Nluc significantly increased luciferase signals compared to GFP controls (Fig. 3b, and Supplementary Fig. 5e). Furthermore, Förster resonance energy transfer (FRET)-fluorescence lifetime imaging (FLIM) measurements revealed that LET1-GFP proteins were in the close proximity to MEKK2-mCherry, but not BIR2-mCherry, when co-expressed in Arabidopsis protoplasts (Fig. 3c & d). The FRET efficiency calculated based on GFP fluorescence lifetime of LET1-GFP and MEKK2-mCherry (13.05 ± 1.76%) was similar to that of BAK1-GFP and BIR2-mCherry (13.46% ± 0.17%) (Fig. 3c & d). MEKK2 is a modular protein with an N-terminal regulatory domain (MEKK2N) and a C-terminal kinase domain. MEKK2N directly interacted with the LET1 cytosolic domain (LET1CD) in an in vitro pull-down assay with purified proteins (Fig. 3e) or in a Co-IP assay when expressed in protoplasts (Supplementary Fig. 5f). Notably, MEKK2 also associated with SUMM2 with Co-IP assays (Supplementary Fig. 5a & b). The interaction between MEKK2 and SUMM2 was further confirmed with Split-LUC (Supplementary Fig. 5g) and FRET-FLIM (Supplementary Fig. 6a & b) assays. The FRET efficiency of MEKK2-mCherry and SUMM2-GFP was 7.99 ± 0.19%. Together, the data reveal that LET1, MEKK2 and SUMM2 exist in a protein complex in plant cells.
Fig. 3. MEKK2 stabilizes LET1 and SUMM2 and promotes cell death.
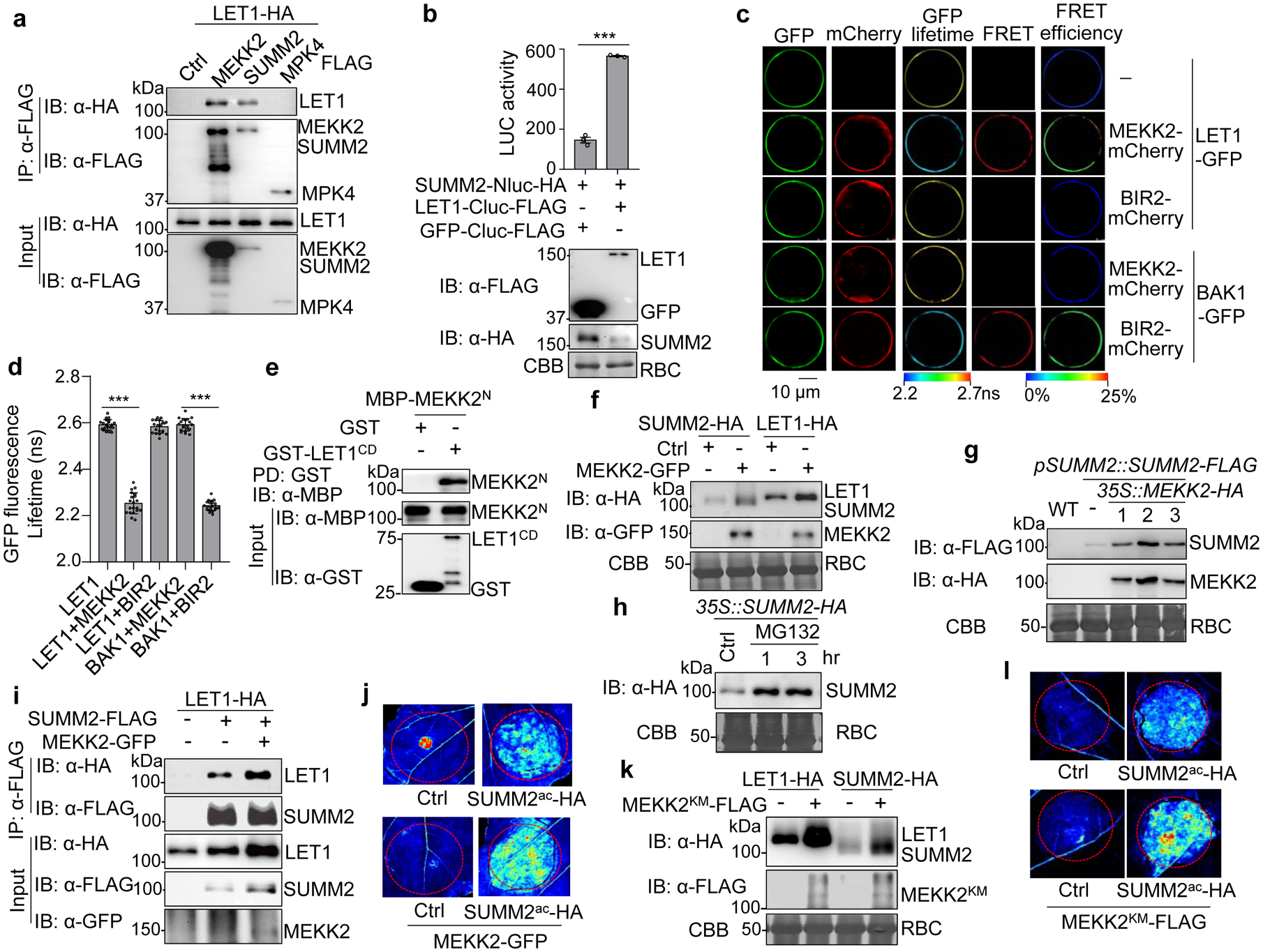
a, LET1 associates with MEKK2 and SUMM2, but not MPK4, with a Co-IP assay. LET1-HA was co-expressed with a vector control (Ctrl), MEKK2-FLAG, SUMM2-FLAG, or MPK4-FLAG, in WT Col-0 protoplasts. Total proteins were immunoprecipitated with α-FLAG affinity beads and then immunoblotted by an α-HA or α-FLAG antibody (top two panels). Proteins before immunoprecipitation are shown as input controls (bottom two panels).
b, LET1 associates with SUMM2 with a split-luciferase assay. The LET1-Cluc-FLAG or GFP-Cluc-FLAG (Ctrl) was co-expressed with SUMM2-Nluc-HA in protoplasts, and luciferase activity was detected 12 hr after transfection, P = 9.00 × 10−6. Protein expression is shown on the bottom with immunoblots. Data are shown as mean ± SE from three independent repeats. The asterisk indicates statistical significance by using two-sided two-tailed Student’s t-test (***, P<0.001).
c and d, FRET-FLIM analysis of LET1 and MEKK2 interaction in Arabidopsis protoplasts. The indicated proteins were transiently expressed in protoplasts for 16 hr, and FRET-FLIM was visualized using a confocal laser scanning microscopy (c). Localization of the LET1-GFP/BAK1-GFP and MEKK2-mCherry/BIR2-mCherry is shown with the first (Green) and second column (Red), respectively. The lifetime (τ) distribution (third column), FRET (fourth column), and apparent FRET efficiency (fifth column) are presented as pseudo-color images according to the scale. The GFP mean fluorescence lifetime (τ) values, ranging from 2.2 to 2.7 nanoseconds (ns), were statistically analyzed and data are shown as mean ± SE from twenty independent cells (d), P <1.00 × 10−15 (column 1 and 2), P <1.00 × 10−15 (column 4 and 5). The pairs of BAK1-GFP/MEKK2-mCherry and LET1-GFP/BIR2-mCherry were used as a negative control, and BAK1-GFP and BIR2-mCherry as a positive control53. Asterisks represent significant differences by using two-sided two-tailed Student’s t-test (***, P < 0.001). Scale bar, 10 μm.
e, LET1CD interacts direcstly with the N-terminal MEKK2 (MEKK2N). LET1CD consists of the juxtamembrane and kinase domains (425–834aa). MBP-MEKK2N and GST-LET1CD proteins purified from E. coli were used in an in vitro pull-down assay using glutathione agarose beads followed by immunoblotting using an α-MBP antibody (top panel). Input proteins were immunoblotted by an α-MBP or α-GST antibody before pull-down (middle and bottom panels).
f, MEKK2 enhances the LET1 and SUMM2 protein levels. LET1-HA or SUMM2-HA was co-expressed with a vector control (Ctrl) or MEKK2-GFP in N. benthamiana. Total proteins were isolated and immunoblotted by an α-HA or α-GFP antibody two days after infiltration. CBB was used for a loading control.
g, MEKK2 enhances SUMM2 protein accumulation in transgenic plants. The pMDC-35S::MEKK2-HA construct was transformed into pSUMM2::SUMM2-FLAG transgenic plants. SUMM2 and MEKK2 protein levels were examined by α-FLAG and α-HA antibodies in T1 generation of pSUMM2::SUMM2-FLAG/35S::MEKK2-HA transgenic plants (Line #1, #2, and #3). The SUMM2 protein level in the parental pSUMM2::SUMM2-FLAG line was used as a control (lane 2). The protein loading is shown by CBB staining.
h, MG132 increases SUMM2 protein level. Transgenic plants of 35S::SUMM2-HA in WT were treated with DMSO (Ctrl) or 2 μM MG132 for 1 or 3 hr. Proteins were immunoblotted using an α-HA antibody (top panel), and CBB was used for a loading control (bottom panel).
i, MEKK2 enhances the association of LET1 and SUMM2. LET1-HA was co-expressed with a vector (−), or SUMM2-FLAG with or without MEKK2-GFP in N. benthamiana and followed by a Co-IP assay.
j, MEKK2 enhances the cell death triggered by SUMM2ac. SUMM2ac-HA was co-expressed with or without MEKK2-GFP in N. benthamiana. The GFP construct was used as a Ctrl. The images were taken at two days after infiltration under the UV light.
k, The MEKK2 kinase mutant (MEKK2KM) enhances the LET1 and SUMM2 protein levels. The experiment was performed similarly as in f.
l, MEKK2KM enhances the cell death triggered by SUMM2ac. The experiment was performed similarly as in j.
All the above experiments were repeated at least three times with similar results obtained.
Interestingly, we consistently observed that the protein levels of LET1-HA and SUMM2-HA were increased whenever co-expressed with MEKK2-GFP in N. benthamiana (Fig. 3f). Ectopic expression of MEKK2-GFP in protoplasts of 35S::SUMM2-HA transgenic plants also increased the protein levels of SUMM2-HA (Supplementary Fig. 7a). When 35S::MEKK2-HA was transformed into pSUMM2::SUMM2-FLAG or pLET1::LET1-FLAG transgenic plants, the protein levels of SUMM2 or LET1 were considerably increased in the double transgenic lines compared to the parental transgenic lines (Fig. 3g, and Supplementary Fig. 7b). In addition, the SUMM2-GFP signal intensity was enhanced in the presence of MEKK2-FLAG when transiently expressed in N. benthamiana (Supplementary Fig. 7c). The data suggest that SUMM2 and LET1 may undergo protein degradation, and MEKK2 likely stabilizes SUMM2 and LET1. Treatment of MG132, a proteasome-dependent protein degradation inhibitor, substantially stabilized SUMM2 and LET1 (Fig. 3h, and Supplementary Fig. 7d). Notably, in the presence of MEKK2-GFP, MG132 exhibited a less pronounced effect on stabilizing LET1-HA (Supplementary Fig. 7d). Collectively, the data suggest that MEKK2 might regulate the protein homeostasis of SUMM2 and LET1, by protecting them from protein degradation. In line with this observation, MEKK2 promoted the association between LET1 and SUMM2 in N. benthamiana (Fig. 3i). In addition, expression of MEKK2 further aggravated cell death triggered by SUMM2ac in N. benthamiana, likely due to the MEKK2-mediated stabilization of SUMM2ac (Fig. 3j). Interestingly, the MEKK2 kinase mutant (MEKK2KM) could also stabilize SUMM2 and LET1 (Fig. 3k), and enhance SUMM2ac-triggered cell death (Fig. 3l). This echoes the observations that the kinase activity of MEKK2 is not required for triggering cell death, and MEKK2 may act as a structural protein, rather than a functional kinase, in the activation of SUMM224,41. Taken together, the data indicate that MEKK2 likely scaffolds and stabilizes the SUMM2 and LET1 protein complex to activate autoimmunity.
The stabilization of SUMM2 by MG132 treatment prompted us to test whether any E3 ubiquitin ligase could mediate SUMM2 ubiquitination and degradation. The stability of several NLR proteins, including RPS2 and SNC1, is regulated by the SKP1-CULLIN1-F-box (SCF) complex-mediated proteasome degradation pathway42,43. The F-box protein CPR1 interacts with RPS2 and SNC1 in vivo42,43. We tested whether CPR1 could affect SUMM2 protein stability. When co-expressing with CPR1 in N. benthamiana, the protein level of SUMM2, but not a GFP control, was considerably reduced (Fig. 4a). Similarly, expression of CPR1 reduced the SUMM2 protein level in 35S::SUMM2-HA transgenic plants, which could be suppressed by MG132 treatment (Supplementary Fig. 8a), suggesting that CPR1 may mediate proteasome degradation of SUMM2. Importantly, CPR1 did not affect the protein level of LET1 nor MEKK2, indicating a specific regulation of CPR1 towards NLRs (Supplementary Fig. 8b). Further, CPR1 associates with SUMM2 in both Arabidopsis protoplasts (Fig. 4b) and N. benthamiana (Supplementary Fig. 8c), similar to its association with RPS2 (Supplementary Fig. 8d), in Co-IP assays. We next determined whether SUMM2 is ubiquitinated and the effect of CPR1 on its ubiquitination. We have previously established an in vivo ubiquitination assay in which HA-tagged ubiquitin (HA-UBQ) is co-expressed with FLAG-tagged target proteins in planta to detect the ubiquitinated proteins with an α-HA immunoblot upon extensive washing to remove associated proteins and immunoprecipitation with an α-FLAG antibody for target proteins44. As shown in Fig. 4c, in the presence of HA-UBQ, immunoprecipitated SUMM2-FLAG proteins were detected as a ladder-like smear migrating above its predicted molecular weight (MW, ~105 kDa with 2 x FLAG epitopes), suggesting that SUMM2 was polyubiquitinated in planta. The smear bands of the ubiquitinated SUMM2 were enhanced when co-expressing CPR1 (Fig. 4c, compare lane 2 and 3) and reduced in the cpr1–2 mutant (Fig. 4d, compare lane 2 and 4). The data suggest that SCFCPR1 ubiquitinates SUMM2, contributing to its degradation.
Fig. 4. MEKK2 counter-regulates CPR1-mediated SUMM2 ubiquitination and degradation.
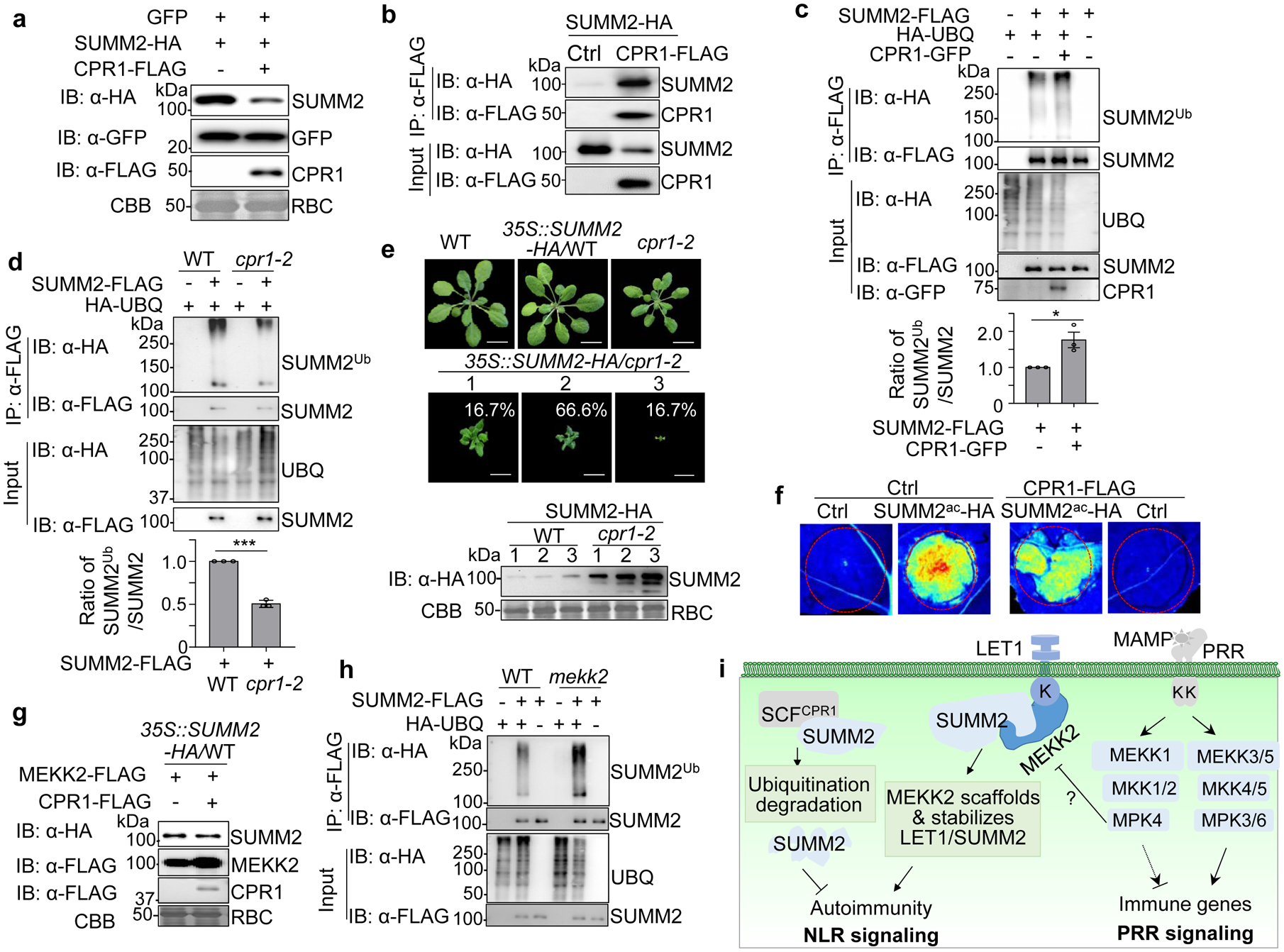
a, CPR1 promotes SUMM2 protein degradation. SUMM2-HA was co-expressed with a vector or CPR1-FLAG in N. benthamiana. GFP was included as a control. Total proteins were immunoblotted by an α-HA, α-FLAG or α-GFP antibody (top three panels). CBB is a loading control.
b, SUMM2 associates with CPR1. SUMM2-HA was co-expressed with a vector (Ctrl) or CPR1-FLAG in WT protoplasts. Total proteins were immunoprecipitated with α-FLAG affinity beads and then immunoblotted by an α-HA or α-FLAG antibody (top two panels). Immunoblots using total proteins before immunoprecipitation are shown as protein inputs (bottom two panels).
c, CPR1 enhances SUMM2 ubiquitination. SUMM2-FLAG was co-expressed with HA-UBQ with or without CPR1-GFP in WT protoplasts. Total proteins were immunoprecipitated with α-FLAG affinity beads and then immunoblotted by an α-HA or α-FLAG antibody (top two panels). Immunoblots using total protein before immunoprecipitation are shown as protein inputs (bottom three panels). The input for SUMM2 was adjusted to the similar level for Co-IP and IB. The quantification of SUMM2 ubiquitination with and without CPR1-GFP is shown on the bottom, P = 2.41 × 10−2. Data are shown as mean ± SE from three independent repeats. The asterisk indicates statistical significance by using two-sided two-tailed Student’s t-test (*, P < 0.05).
d, The cpr1–2 mutant reduces SUMM2 ubiquitination. Ubiquitination assay was performed as in c with protoplasts from WT and cpr1–2. The input for SUMM2 in WT and cpr1–2 was adjusted to a similar level for Co-IP and IB. The quantification of SUMM2 ubiquitination is shown on the bottom, P = 3.36 × 10−5. Data are shown as mean ± SE from three independent repeats. The asterisks indicate statistical significance by using two-sided two-tailed Student’s t-test (***, P < 0.001).
e, Overexpressing SUMM2 aggravates growth defects in cpr1–2. The 35S::SUMM2-HA construct was transformed into WT and cpr1–2. Plants were grown at 26 °C to reduce the growth defects of cpr1–2. All transgenic plants of 35S::SUMM2-HA/WT were phenotypically similar to WT (n>200). Total 120 transgenic plants of 35S::SUMM2-HA/cpr1–2 were grouped into three categories (the ratio of each category is indicated) based on the severity of growth defects. Pictures were taken four weeks after germination. Immunoblots with an α-HA antibody show SUMM2-HA protein expression, and CBB was used as a loading control (bottom). Scale bar, 1 cm.
f, CPR1 attenuates SUMM2ac-triggered cell death. SUMM2ac-HA was co-expressed with or without CPR1-FLAG in N. benthamiana. The GFP construct was used as a Ctrl. The images were taken two days after infiltration under the UV light.
g, MEKK2 antagonizes CPR1-mediated SUMM2 protein degradation. MEKK2-FLAG was co-expressed with or without CPR1-FLAG in protoplasts of 35S::SUMM2-HA transgenic plants.
h, Elevated SUMM2 ubiquitination in mekk2 compared to WT plants. SUMM2-FLAG was co-expressed with or without HA-UBQ in protoplasts of WT and mekk2. IP and IB were performed similarly as in c. The input for SUMM2 in WT and mekk2 was adjusted to a similar level for Co-IP and IB.
i, A model for MEKK2 scaffolding LET1-SUMM2 complex and protecting SUMM2 from CPR1-mediated degradation in cell death control. MAMP-activated MEKK1-MKK1/2-MPK4 cascade regulates PRR-mediated immune signaling and suppresses SUMM2-mediated autoimmunity via modulating MEKK2 protein. MEKK2 scaffolds and stabilizes LET1-SUMM2 complex, which blocks SUMM2 ubiquitination and degradation mediated by SCFCPR1 complex.
All the experiments were repeated at least three times with similar results.
To determine the biological significance of CPR1-mediated ubiquitination and degradation of SUMM2 in cell death regulation, we generated 35S::SUMM2-HA transgenic plants in WT and cpr1–2 backgrounds. Significantly, 35S::SUMM2-HA caused severe dwarfism and growth defects in cpr1–2 but not in WT plants (Fig. 4e). The severity of transgenic plant dwarfism was positively correlated with the SUMM2-HA protein accumulation, and SUMM2 proteins in cpr1–2 were much higher than those in WT (Fig. 4e). Similarly, SUMM2ac-HA accumulated more and caused much more pronounced growth defects in cpr1–2 than in WT (Supplementary Fig. 9a & b). Furthermore, expression of CPR1 ameliorated SUMM2ac-triggered cell death (Fig. 4f) and reduced protein accumulation of SUMM2ac in N. benthamiana (Supplementary Fig. 9c). Taken together, our data indicate that CPR1 interacts with and ubiquitinates SUMM2, and negatively regulates the homeostasis of SUMM2. The observation that MEKK2 stabilized SUMM2 (Fig. 3f & g) prompted us to test whether MEKK2 counter-regulates SCFCPR1-mediated SUMM2 ubiquitination and turnover. Indeed, co-expression of MEKK2 blocked the CPR1-mediated SUMM2 degradation (Fig. 4g). Furthermore, the ubiquitinated SUMM2 appeared to be increased in the mekk2 mutant compared to WT (Fig. 4h). Conversely, SUMM2 ubiquitination was decreased when co-expressing MEKK2 (Supplementary Fig. 9d). Thus, the data implicate that MEKK2 suppresses SUMM2 ubiquitination, which is likely mediated by SCFCPR1, thereby stabilizing SUMM2.
It has been reported that Pseudomonas syringae effector HopAI1 associated with MPK4 and inhibited MAMP-induced MPK4 activation in vivo16. HopAI1 functions a phosphothreonine lyase that targets multiple MAPKs, including MPK3 and MPK645. It has been hypothesized that HopAI1-mediated inactivation of MPK4 results in the activation of SUMM2-mediated defense16. We tested whether HopAI1 affected the integrity of MEKK2-LET1-SUMM2 complex. Co-expression of HopAI1 with MEKK2, LET1, and SUMM2 in Arabidopsis protoplasts indicated that HopAI1 did not affect the association of LET1 with MEKK2 or SUMM2 (Supplementary Fig. 10a & b), but reduced the association of MEKK2 and SUMM2 (Supplementary Fig. 10c). The data suggest that HopAI1 might induce SUMM2 dissociation from MEKK2. HopAI1 also reduced the association of CPR1 and SUMM2 (Supplementary Fig. 10d), consistent with the reduced ubiquitination of SUMM2 in the presence of HopAI1 (Supplementary Fig. 10e). The data suggest that HopAI1 might induce SUMM2 dissociation from CPR1 for its abundance and activation.
Plant RLKs not only sense MAMPs but also recognize plant-derived DAMPs to regulate immunity6. Deficiency in the MAMP-activated MEKK1-MKK1/2-MPK4 cascade triggers the NLR SUMM2-mediated autoimmunity16–21, suggesting that NLRs also sense plant-derived molecules due to the disturbance of host cellular homeostasis. Here we show that a malectin-like RLK LET1 complexes with SUMM2 and is required for the NLR SUMM2 activation, linking RLKs directly to plant NLR-mediated signaling (Fig. 4i). In addition, the related malectin-like RLK, ANX1, complexes with NLR RPS2 and negatively modulates RPS2 immunity38. Furthermore, FER and ANX1/2 regulate plant MAMP-triggered immunity through association with PRRs37–40. Apparently, malectin-like RLKs regulate two-tiered plant immunity through modulation of both PRR and NLR immune receptors. Emerging evidence indicates the extracellular peptides of the RALF family act as the ligands of CrRLK1Ls36,37,46,47. RALFs or another type of ligands could be the potential ligands of LET1 in regulating SUMM2 activation.
Although the exact mechanism is not clear, the abundance of MEKK2 transcripts and proteins is positively correlated with the autoimmunity observed in the mekk1, mkk1/2, and mpk4 mutants25,26. Interestingly, MEKK2 has no detectable kinase activity and overexpression of WT MEKK2 and its kinase catalytic site mutant triggered a similar level of autoimmunity24,41. Thus, MEKK2 may play a structural role, rather than function as a kinase, in regulating SUMM2 accumulation. A recent study shows that MEKK2 might inhibit MPK4 activation by upstream MKKs41. We found here that MEKK2 interacts with both LET1 and SUMM2, and stabilizes LET1 and SUMM2 in a kinase activity-independent manner. The data hint that MEKK2 acts as a scaffold that stabilizes RLK LET1 and NLR SUMM2 for immune activation. The stabilization may be achieved by preventing proteasome-mediated SUMM2 degradation by SCFCPR1 complex. In WT plants, MEKK2 proteins remain at a basal level, resulting in a low abundance of LET1-SUMM2 complex. In the mekk1, mkk1/2, and mpk4 mutants, MEKK2 protein abundance is increased and sufficient to stabilize LET1-SUMM2 complex for autoimmune activation.
Materials and Methods
Plant materials and growth conditions
The Arabidopsis thaliana ecotype Col-0 was used as wild-type (WT) in this study. The individual T-DNA insertion mutant lines (SALK_052557, mekk1–1; SALK_018793C, let1–1; SALK_020561C, let1–2; SALK_112949C, let1–3; SALK_072332C, pirl1) and confirmed T-DNA insertion libraries (CS27941, 6,866 lines; CS27942, 3,980 lines; CS27943, 3,739 lines; CS27944, 3,263 lines; CS27945, 3,705 lines; CS27946, 2,372 lines) were obtained from the Arabidopsis Biological Resource Center (ABRC). The mutants of mkk1/2 from Dr. Patrick Krysan, mpk4 and summ2 (summ2–8) from Dr. Yuelin Zhang, and cpr1–2 from Dr. Jian Hua were reported previously16,26,43. The genotypes of the mutants were confirmed by PCR using the primers listed in Supplementary Table 1. The Arabidopsis and N. benthamiana plants used in this study were grown in soil (Metro Mix 366 for Arabidopsis and LP5 for N. benthamiana) in a growth room at 23 °C (except where indicated), 45% relative humidity, 100 μE m−2 s−1 light with a photoperiod of 12-hr light/12- hr dark. Seedlings were grown on plates containing half-strength Murashige and Skoog medium (½MS) with 0.5% sucrose, 0.8% agar and 2.5 mM MES at pH 5.7, in a growth room with the same condition as the above.
Quantification and statistical analysis
Data for quantification analyses are presented as mean ± standard error (SE) or standard deviation (SD). The statistical analyses were performed by using two-sided two-tailed Student’s t-test or one-way analysis of variance (ANOVA) test (* P < 0.05, ** P < 0.01, *** P < 0.001). Number of replicates is shown in the figure legends. The Microsoft Excel 2016 and Graphpad Prism 8 (Graphpad Prism software, version 8.0.1) were used for statistics and bar graphs overlaid with dot plots.
Plasmid constructs and generation of transgenic plants
The VIGS constructs of BAK1SERK4, BIR1 and CLA1 were reported previously22. The fragments of MEKK1 (261 bp) were amplified from Col-0 cDNA by PCR using the primers containing EcoRI at the 5’ end and KpnI at the 3’ end and cloned into the pYL156 (pTRV–RNA2) vector. The LET1 (AT2G23200) gene (2502 bp) was amplified by PCR from Col-0 cDNA using the primers containing BamHI at the 5’ end and StuI at the 3’ end. PCR products were digested by BamHI and StuI into two fragments, LET1N (BamHI-LET11–1969 bp-BamHI) and LET1C (BamHI-LET11970–2502 bp-StuI), due to an internal BamHI site in LET1. LET1C was ligated into a plant protoplast expression vector pHBT under the control of a CaMV 35S promoter with an HA tag at C-terminus by BamHI and StuI, and subsequently, LET1N was introduced by BamHI digestion and ligation to obtain pHBT-p35S::LET1-HA. To sub-clone LET1 into other vectors by BamHI and StuI, site-directed mutagenesis by Platinum Pfx DNA polymerase-mediated PCR was used to mutate the internal BamHI site without changing its codons in LET1 and generate pHBT-p35S::LET1mBamHI-HA. The fragments of MPK4 (AT4G01370, 1128 bp), MEKK2 (AT4G08480, 2319 bp), MEKK2N (1–1389 bp), SUMM2 (AT1G12280, 2682 bp), and CPR1 (AT4G12560, 1239 bp) were amplified from Col-0 cDNA using the primers containing BamHI at the 5’ end and StuI at the 3’ end, and ligated into a pHBT vector with a HA, FLAG or GFP tag at C-terminus. RPS2 was reported previously38. LET1KM, MEKK2KM and SUMM2ac were generated by site-directed mutagenesis using Platinum Pfx DNA polymerase-mediated PCR. LET1, MEKK2, SUMM2, LET1KM, MEKK2KM and SUMM2ac were sub-cloned into the binary vectors pCB302–35S::HA, pMDC32–2×35S::HA, pMDC32–2×35S::FLAG or pMDC32–2×35S::GFP by BamHI and StuI digestions. The promotor of LET1 (−1‒ −1543 bp upstream of ATG) was amplified by PCR from Col-0 genomic DNA and sub-cloned into the binary vector pCB302::FLAG by XhoI and BamHI digestions to generate pCB302-pLET1::FLAG binary vector. LET1 was sub-cloned into the binary vector pCB302-pLET1::FLAG by BamHI and StuI digestions, generating binary vector pCB302-pLET1::LET1-FLAG. To generate binary vector pCB302-pSUMM2::SUMM2-FLAG, the promotor of SUMM2 (−1‒ −2685 bp upstream of ATG) was subcloned into the binary vector pCB302::FLAG by XhoI and BamHI digestions and SUMM2 was inserted into pCB302-pSUMM2::FLAG by BamHI and StuI digestions. To construct the E. coli expression vectors, LET1CD (1267–2502 bp) was amplified from pHBT-p35S::LET1-HA using the primers containing BamHI at the 5’ end and StuI at the 3’ end and ligated into a modified GST fusion protein expression vector pGEX4T-1 (Pharmacia, USA). MEKK2N was sub-cloned from pHBT-p35S::MEKK2N-HA into a modified MBP fusion protein expression vector pMAL-1 (NEB, USA) by BamHI and StuI digestions to generate pGST-LET1CD and pMBP-MEKK2N. LET1CD (1267–2502 bp) was amplified by PCR from pHBT-p35S::LET1-HA using the primers containing StuI at the 5’ end and KpnI at the 3’ end and cloned into an insect cell expression vector pAcGHLT-C to generate pAcGHLT-LET1CD. To generate pHBT-p35S::LET1-Cluc-FLAG and pHBT-p35S::SUMM2-Cluc-FLAG plasmids, Cluc (1–456bp) was amplified by PCR from pHBT-pFRK::LUC plasmids48 and sub-cloned into pHBT-p35S::LET1-FLAG and pHBT-p35S::SUMM2-FLAG using a one-step cloning kit (Vazyme Biotech, USA) by StuI digestion. pHBT-p35S::GFP-Cluc-FLAG was generated by PCR amplifying GFP from pHBT-p35S::GFP and sub-cloned into pHBT-p35S::Cluc-FLAG by BamHI and SpeI digestion. pHBT-p35S::MEKK2-Nluc-HA and pHBT-p35S::SUMM2--Nluc-HA were generated by PCR amplifying Nluc (457–1746 bp) from pHBT-pFRK::LUC and sub-cloned into pHBT-p35S::MEKK-HA and pHBT-p35S::SUMM2-HA using a one-step cloning kit by StuI digestion. The fragment of UBQ10 (228 bp) was digested from pHBT-p35::FLAG-UBQ1044 with BamHI at the 5’ end and StuI at the 3’ end, and inserted into pHBT vector which has a HA tag at N-terminus. The HopAI1 (NC_004578) gene (783 bp) was amplified by PCR from Pseudomonas syringae genomic DNA using the primers containing BamHI at the 5’ end and StuI at the 3’ end, and sub-cloned into pHBT-HA vector.
For FRET-FLIM constructs, genes-of-interest tagged by the fluorescence protein EGFP or mCherry were developed according to previously reported49. Briefly, the LET1, SUMM2, and BAK1 fragments were released from pHBT-p35S::LET1-, SUMM2- or BAK1-FLAG by BamHI and StuI digestion, and ligated into the pHBT vector with an EGFP epitope-tag at C-terminus. Similarly, the MEKK2 and BIR2 fragments were released from pHBT-p35S::MEKK2 or BIR2-FLAG by BamHI and StuI digestion, and ligated into the pHBT vector with a mCherry epitope-tag at C-terminus.
All the primer sequences are listed in Supplemental Table 1. The sequences of all genes and mutation were verified by the Sanger-sequencing. The binary plasmids were transformed into Agrobacterium tumefaciens strain GV3101 and introduced into Arabidopsis using the floral dipping method. Transgenic plants were selected by Glufosinate-ammonium (Basta, 50 μg ml−1) for the pCB302 vector and hygromycin (50 μg ml−1) for the pMDC vector. Multiple transgenic lines were analyzed by immunoblot (IB) for protein expression.
Agrobacterium-mediated virus-induced gene silencing (VIGS) assay
The binary TRV vector pTRV-RNA1 and pTRV-RNA2 derivatives, pTRV-MEKK1, pTRV-CLA1 and pTRV-GFP (the vector control), were transferred into A. tumefaciens strain GV3010 by electroporation. Positive transformants were selected on LB plates containing 50 μg ml−1 kanamycin and 25 μg ml−1 gentamicin by incubating at 28°C for 36 hr. An individual transformant was transferred into 2 ml LB liquid medium containing 50 μg ml−1 kanamycin and 25 μg ml−1 gentamicin in 20 ml-glass culture tubes for overnight at 28°C in a roller drum, and sub-cultured in 100 times of volume of fresh LB liquid medium containing 50 μg ml−1 kanamycin, 25 μg ml−1 gentamicin, 10 mM MES and 20 μM acetosyringone for overnight at 28°C with 200 rpm shaking. Cells were pelleted by 1,300 g centrifugation, re-suspended in buffer containing 10 mM MgCl2, 10 mM MES and 200 μM acetosyringone, adjusted to OD600 of 1.5 and incubated at 25°C for at least 3 hr. Bacterial cultures containing pTRV-RNA1 and pTRV-RNA2 derivatives were mixed at a 1:1 ratio and inoculated into the first pair of true leaves of two-week-old soil-grown plants using a needleless syringe.
Map-based cloning of the LET1 gene
The let1–1 mutant (SALK_018793) was crossed to the heterozygous mekk1–1 mutant to obtain the let11–1mekk1 double homozygous mutant, which suppressed the mekk1 seedling lethality but still showed growth defects distinguishable from WT plants. The let11–1mekk1 mutant was crossed to the ecotype Landsberg (Ler-0). Two pools of plants, including 20 WT-looking plants and 20 let1–1mekk1-looking plants from the F2 population, were selected for genomic DNA isolation and mapping-PCR analysis. The genetic mapping, next-generation sequencing (NGS), and data analyses followed a previous report48. Briefly, twenty-two pairs of simple sequence length polymorphism (SSLP) markers evenly distributed on five chromosomes of Arabidopsis were used to screen the length polymorphisms between two mixed DNA pools consisting of an equal amount of DNAs from 20 individual plants in each pool. The initial mapping placed LET1 on Chromosome 2 and MEKK1 (AT4G08500) on chromosome 4 based on the bulked segregation analysis. Additional SSLP makers from Chromosome 2 with 157 let1–1mekk1-looking plants from F2 population further mapped LET1 in a ~1 M bp region between makers of F5H14 and T20D16. We then performed NGS of let1–1 with 100 nt paired-end sequencing on an Illumina HiSeq 2000 platform at Texas AgriLife Genomics and Bioinformatics Service (TAGS) (College Station, TX, USA). Forty-fold genome coverage was obtained. Illumina reads were mapped to the TAIR10 release of the Col-0 genome using CLC Genomics Workbench 6.0.1 software (http://www.clcbio.com). The candidate variants between F5H14 and T20D16 were selected, and 17-bp deletion in AT2G23200 (LET1) was identified. The mutation was further confirmed with Sanger sequencing using genomic DNA.
Transient expression assay in Arabidopsis protoplasts and N. benthamiana
The indicated pHBT constructs were used for protoplast transfection following the protocol50. Briefly, for Co-IP assay, 100 μl of plasmid DNA (2 μg μl−1) was mixed with 1 ml of protoplasts (2 × 105 cells ml−1), and for split-luciferase assays, 50 μl of plasmid DNA (2 μg μl−1) were mixed with 500 ul cells, for the PEG-mediated transfection.
For transient assays in N. benthamiana, the indicated constructs were transferred into A. tumefaciens strain GV3010 by electroporation. A single transformant was transferred into 2 ml LB liquid medium containing 50 μg ml−1 kanamycin and 25 μg ml−1 gentamicin for overnight incubation at 28 °C. Bacteria were harvested by centrifugation at 1,200 g and resuspended in the buffer of 10 mM MgCl2 at OD600=1. The Agrobacterial cultures were infiltrated into the leaves of four-week-old N. benthamiana. Proteins were isolated 2–3 days after inoculation from the infiltrated area and subjected for immunoblot analysis. Cell death was observed 3 days after infiltration, and pictures were taken under the UV light with the ChemiDoc system.
Trypan blue and DAB staining
Detached leaves were soaked in trypan blue staining solution (2.5 mg ml−1 trypan blue dissolved in lactophenol containing an equal volume of lactic acid, glycerol, liquid phenol, and ddH2O or 3,3’-diaminobenzidine (DAB) solution (1 mg ml−1 DAB dissolved in ddH2O, pH 3.8) for overnight incubation. Samples were transferred into trypan blue destaining solution containing lactophenol and ethanol in a ratio of 1:2 or DAB destaining solution containing glycerol, acetic acid, and ethanol in a ratio of 1:1:3 and incubated at room temperature with gentle shaking until entirely destained. Samples were observed and recorded under a dissecting microscope.
Co-IP and transient expression assays
Proteins were expressed overnight in Arabidopsis protoplasts or N. benthamiana leaves for three days. Protoplasts were lysed by vortexing and leaves were grounded in the extraction buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 7.5, 2 mM NaF, 2 mM Na3VO4, 1 mM DTT, 0.5% Triton X-100, 10% glycerol, and 1 x protease inhibitor). After centrifugation at 12,500 g at 4 °C for 15 min, 250 μl of extraction buffer were added to dissolve pellets, and 20 μl of supernatant were collected for input controls, and the remaining was incubated with α-FLAG affinity beads (Sigma, USA) at 4 °C for 2 hr with gentle shaking. Beads were collected and washed three times with washing buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100), and once with 50 mM Tris-HCl, pH7.5. Proteins were eluted by 2 × SDS-PAGE loading buffer and boiled at 94 °C for 5 min. Immunoprecipitated and input proteins were analyzed by immunoblot with indicated antibodies. For Co-IP assay with samples from transgenic plants, total proteins were extracted from 2 g leaves of four-week-old plants with 2 ml IP buffer containing 1% Triton X-100. The supernatant was collected after centrifugation at 12,500 g for 10 min at 4 °C, followed by another centrifugation at 12,500 g for 30 min at 4 °C. The FLAG-tagged proteins and the associated proteins were purified from the cell lysates with α-FLAG affinity beads by immunoprecipitation. The beads were boiled with 2 × SDS-PAGE loading buffer at 94 °C for 5 min to elute the proteins. Immunoprecipitated and input proteins were analyzed by immunoblots with indicated antibodies.
Split-luciferase assay
The Cluc-FLAG and Nluc-HA tagged proteins were co-expressed in protoplasts for 12 hr. The cells were lysed by extraction buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 7.5, 2 mM NaF, 2 mM Na3VO4, 1 mM DTT, 0.5% Triton X-100, 10% glycerol, and 1 x protease inhibitor). The cell lysis was centrifuged at 12,500 g at 4 °C for 15 min. The luciferase activity of the supernatant was measured with 0.2 mM luciferin by a luminometer (Perkin Elmer, USA).
In vivo protein ubiquitination assay
FLAG-tagged proteins (40 μg DNA) were co-expressed with HA-UBQ or vector control (40 μg DNA) in 400 μl protoplasts at the density of 2 × 105 ml−1 for 12 hr. Cells were collected for the above-mentioned Co-IP assays in a Co-IP buffer containing 1% Triton X-100.
RNA isolation, qRT-PCR analysis, and genotyping PCR
Plant total RNAs were extracted by TRIzol reagent (Sigma/Invitrogen, USA). Genomic DNA was degraded by treatment with RNase-free DNase I (NEB, USA). Complementary DNAs (cDNAs) were synthesized with M-MuLV Reverse Transcriptase (NEB, USA) and oligo(dT) primers. Quantitative RT-PCR analysis was performed by iTaq Universal SYBR green Supermix (Bio-Rad, USA) with a Bio-Rad CFX384 Real-Time PCR System (Bio-Rad, USA). UBQ10 was used as an internal reference.
Recombinant protein isolation and in vitro pull-down assay
Fusion proteins were produced from E. coli BL21 at 16 °C using LB medium with 0.25 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Glutathione S-transferase (GST) fusion proteins were purified with Pierce glutathione agarose (Thermo Scientific, USA), and maltose-binding protein (MBP) fusion proteins were purified using amylose resin (New England Biolabs, USA) according to the standard protocol from companies. MBP-MEKK2N proteins were incubated with GST or GST-LET1CD in the pull-down buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 0.2% Triton X-100) for 1 hr with gentle shaking, subsequently incubated with 20 μl of glutathione agarose beads at 4 °C for another 2 hr with gentle shaking. Beads were washed five times with pull-down buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 0.2% Triton X-100). Beads were boiled in 50 μl of 2 x SDS protein loading buffer for 10 min and detected by immunoblotting with an α-MBP antibody.
Protein isolation from insect cells and in vitro kinase assay
The fragment of the cytoplasmic domain of LET1 (LET1CD) was inserted into pAcGHLT-C vector that is used to express HIS-GST tag at the N-terminus of expressed protein51. About 2 μg of pAcGHLT-C-LET1CD and 2.5 μl of Baculovirus DNA (AB Vector, USA) were mixed and transfected into 3.5 × 106 Spodoptera frugiperda sf9 insect cells. The insect cells were incubated with 1 L ESF921 medium (Expression systems, USA) at 37 °C with 8% CO2 and gentle shaking (130 rpm) for 2 days. The cells were collected at 2,000 rpm for 5 min. The pellets were dissolved by 50 ml lysis buffer (20 mM Tris, pH 8.8, 150 mM NaCl, 1 mM PMSF, 1% NP-40) for 2 hr at 4°C with gentle shaking. After centrifugation at 16,000 rpm at 4 °C for 30 min, the supernatant of cell lysis was incubated with 5 ml Ni-NTA agarose (Qiagen, USA) for 2 hr with gentle shaking. The beads were washed with washing buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 10 mM imidazol) for four times. The protein was eluted by 20 ml elution buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 250 mM imidazole, 5 mM DTT and 1 mM PMSF). The in vitro kinase assays were performed with 0.5 μg HIS-GST-LET1CD in 20 μl kinase reaction buffer (20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 5 mM EGTA, 100 mM NaCl, 1 mM DTT and 1 μl [γ−32P] ATP). After gentle shaking at room temperature for 2 hr, samples were denatured with 4 x SDS loading buffer and separated by 10% SDS-PAGE gel. Phosphorylation was analyzed by autoradiography.
Confocal microscopy and FRET-FLIM assays
The GFP and mCherry fusion proteins were detected using a Leica TCS SP8 confocal laser scanning microscope (Germany). The GFP fluorescence was excited at 488 nm, and emissions were detected between 490 and 530 nm. The mCherry fluorescence was excited at 587 nm, and emissions were detected between 590 and 620 nm. The pinhole was set at 1 Airy unit. Images and FLIM/FRET analyses were performed by using Leica Application Suite X (LAS X) software as described52. Briefly, FRET measurements were done with a pair of GFP/mCherry fusion proteins. The image of GFP donor fluorescence was analyzed and scanned at 488 nm and detected between 490 and 530 nm. The fluorescence lifetime (τ) was calculated as the average of 20 τ values randomly measured in the protoplast cells. The values obtained for 20 protoplasts were used to determine the average value of τ for each pair of proteins analyzed. The relative fluorescence intensity (I) in a certain region of interest (ROI), lifetime (τ) and FRET efficiency were measured by the Leica LAS X software. FRET efficiency (E) was calculated by using the formula E = 1-(τDA/τD), τDA is the lifetimes of the donor in the presence of acceptor and τD is fluorescence lifetime of the donor alone. The statistical analysis was performed by using two-sided two-tailed Student’s t-test.
Data Availability
Original data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
We thank the Arabidopsis Biological Resource Center (ABRC) for Arabidopsis T-DNA insertion library and various mutant seeds, Drs. Patrick Krysan (University of Wisconsin, USA), Yuelin Zhang (University of British Columbia, Canada), and Jia Hua (Cornell University, USA) for Arabidopsis seeds, Drs. Christina Franck and Cyril Zipfel for the critical reading of the manuscript, and members of the laboratories of L.S. and P.H. for discussions and comments of the experiments. The work was supported by National Institutes of Health (NIH) (R01GM092893) and National Science Foundation (NSF) (MCB-1906060) to P.H, and NIH (R01GM097247) and the Robert A. Welch Foundation (A-1795) to L.S. Y.H. and D.G. were partially supported by China Scholarship Council (CSC), and G.C.M was partially supported by INCT/CNPq Fellowship, Brazil.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Reference
- 1.Jones JDG & Dangl JL The plant immune system. nature 444, 323–329, (2006). [DOI] [PubMed] [Google Scholar]
- 2.Chisholm ST, Coaker G, Day B & Staskawicz BJ Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814, (2006). [DOI] [PubMed] [Google Scholar]
- 3.Spoel SH & Dong X How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12, 89–100, (2012). [DOI] [PubMed] [Google Scholar]
- 4.Couto D & Zipfel C Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16, 537–552, (2016). [DOI] [PubMed] [Google Scholar]
- 5.Yu X, Feng B, He P & Shan L From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu Rev Phytopathol 55, 109–137, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gust AA, Pruitt R & Nurnberger T Sensing Danger: Key to Activating Plant Immunity. Trends Plant Sci 22, 779–791, (2017). [DOI] [PubMed] [Google Scholar]
- 7.Cui H, Tsuda K & Parker JE Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66, 487–511, (2015). [DOI] [PubMed] [Google Scholar]
- 8.Elmore JM, Lin ZJ & Coaker G Plant NB-LRR signaling: upstreams and downstreams. Curr Opin Plant Biol 14, 365–371, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeYoung BJ & Innes RW Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol 7, 1243–1249, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez MC, Petersen M & Mundy J Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61, 621–649, (2010). [DOI] [PubMed] [Google Scholar]
- 11.Meng X & Zhang S MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51, 245–266, (2013). [DOI] [PubMed] [Google Scholar]
- 12.Tena G, Boudsocq M & Sheen J Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14, 519–529, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun T et al. Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep 19, e45324, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi G et al. Receptor-Like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-Activated Protein Kinase Cascades in Arabidopsis. Plant Cell 30, 1543–1561, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asai T et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983, (2002). [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z et al. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11, 253–263, (2012). [DOI] [PubMed] [Google Scholar]
- 17.Gao M et al. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell research 18, 1190, (2008). [DOI] [PubMed] [Google Scholar]
- 18.Ichimura K, Casais C, Peck SC, Shinozaki K & Shirasu K MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem 281, 36969–36976, (2006). [DOI] [PubMed] [Google Scholar]
- 19.Suarez-Rodriguez MC et al. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol 143, 661–669, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagami H, Soukupova H, Schikora A, Zarsky V & Hirt H A Mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem 281, 38697–38704, (2006). [DOI] [PubMed] [Google Scholar]
- 21.Petersen M et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120, (2000). [DOI] [PubMed] [Google Scholar]
- 22.de Oliveira MVV et al. Specific control of Arabidopsis BAK1/SERK4-regulated cell death by protein glycosylation. Nature Plants 2, 15218, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X et al. The Receptor Kinases BAK1/SERK4 Regulate Ca(2+) Channel-Mediated Cellular Homeostasis for Cell Death Containment. Curr Biol 29, 3778–3790 e3778, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y et al. RNAi-based screen reveals concerted functions of MEKK2 and CRCK3 in plant cell death regulation. Plant Physiol DOI: 10.1104/pp.1119.01555, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong Q et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24, 2225–2236, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su SH et al. Deletion of a tandem gene family in Arabidopsis: increased MEKK2 abundance triggers autoimmunity when the MEKK1-MKK1/2-MPK4 signaling cascade is disrupted. Plant Cell 25, 1895–1910, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z et al. The NLR protein SUMM2 senses the disruption of an immune signaling MAP kinase cascade via CRCK3. EMBO Rep 18, 292–302, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nissen KS, Willats WG & Malinovsky FG Understanding CrRLK1L Function: Cell Walls and Growth Control. Trends Plant Sci 21, 516–527, (2016). [DOI] [PubMed] [Google Scholar]
- 29.Li C, Wu HM & Cheung AY FERONIA and Her Pals: Functions and Mechanisms. Plant Physiol 171, 2379–2392, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindner H, Muller LM, Boisson-Dernier A & Grossniklaus U CrRLK1L receptor-like kinases: not just another brick in the wall. Curr Opin Plant Biol 15, 659–669, (2012). [DOI] [PubMed] [Google Scholar]
- 31.Franck CM, Westermann J & Boisson-Dernier A Plant Malectin-Like Receptor Kinases: From Cell Wall Integrity to Immunity and Beyond. Annu Rev Plant Biol 69, 301–328, (2018). [DOI] [PubMed] [Google Scholar]
- 32.Huck N, Moore JM, Federer M & Grossniklaus U The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130, 2149–2159, (2003). [DOI] [PubMed] [Google Scholar]
- 33.Boisson-Dernier A et al. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazaki S et al. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol 19, 1327–1331, (2009). [DOI] [PubMed] [Google Scholar]
- 35.Guo H et al. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci U S A 106, 7648–7653, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge Z et al. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358, 1596–1600, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stegmann M et al. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–+, (2017). [DOI] [PubMed] [Google Scholar]
- 38.Mang H et al. Differential Regulation of Two-Tiered Plant Immunity and Sexual Reproduction by ANXUR Receptor-Like Kinases. Plant Cell 29, 3140–3156, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H et al. FERONIA Receptor Kinase Contributes to Plant Immunity by Suppressing Jasmonic Acid Signaling in Arabidopsis thaliana. Curr Biol 28, 3316–3324 e3316, (2018). [DOI] [PubMed] [Google Scholar]
- 40.Kessler SA et al. Conserved molecular components for pollen tube reception and fungal invasion. Science 330, 968–971, (2010). [DOI] [PubMed] [Google Scholar]
- 41.Nitta Y et al. MEKK2 inhibits activation of MAP kinases in Arabidopsis. Plant J, doi: 10.1111/tpj.14763, (2020). [DOI] [PubMed] [Google Scholar]
- 42.Cheng YT et al. Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. P Natl Acad Sci USA 108, 14694–14699, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gou M et al. The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J 69, 411–420, (2012). [DOI] [PubMed] [Google Scholar]
- 44.Lu D et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1, 175–185, (2007). [DOI] [PubMed] [Google Scholar]
- 46.Haruta M, Sabat G, Stecker K, Minkoff BB & Sussman MR A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao Y et al. Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572, 270–274, (2019). [DOI] [PubMed] [Google Scholar]
- 48.Li F et al. Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16, 748–758, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bajar BT, Wang ES, Zhang S, Lin MZ & Chu J A Guide to Fluorescent Protein FRET Pairs. Sensors-Basel 16, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He P, Shan L & Sheen J in Plant-Pathogen Interactions 1–9 (Springer, 2007). [Google Scholar]
- 51.Shu C et al. Structural Insights into the Functions of TBK1 in Innate Antimicrobial Immunity. Structure 21, 1137–1148, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bucherl C, Aker J, de Vries S & Borst JW Probing protein-protein Interactions with FRET-FLIM. Methods Mol Biol 655, 389–399, (2010). [DOI] [PubMed] [Google Scholar]
- 53.Halter T et al. The Leucine-Rich Repeat Receptor Kinase BIR2 Is a Negative Regulator of BAK1 in Plant Immunity. Current Biology 24, 134–143, (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data that support the findings of this study are available from the corresponding author upon request.


