Abstract
Background
Plasma tumour DNA (ptDNA) levels on treatment are associated with response in a variety of cancers. However, the role of ptDNA in prostate cancer monitoring remains largely unexplored. Here we characterised on-treatment ptDNA dynamics and evaluated its potential for early assessment of therapy efficacy for metastatic castration-resistant prostate cancer (mCRPC).
Methods
Between 2011 and 2016, 114 sequential plasma samples from 43 mCRPC abiraterone-treated patients were collected. Targeted next-generation sequencing was performed to determine ptDNA fraction. ptDNA progressive disease was defined as a rise in the fraction compared to the pre-treatment.
Results
A ptDNA rise in the first on-treatment sample (interquartile range (IQR) 2.6–3.7 months) was significantly associated with increased risk of early radiographic or any prostate-specific antigen (PSA) rise (odds ratio (OR) = 15.8, 95% confidence interval (CI) 3.5–60.2, p = 0.0002 and OR = 6.0, 95% CI 1.6–20.0, p = 0.01, respectively). We also identified exemplar cases that had a rise in PSA or pseudoprogression secondary to bone flare but no rise in ptDNA. In an exploratory analysis, initial ptDNA change was found to associate with the duration of response to prior androgen deprivation therapy (p < 0.0001) but not to prior taxanes (p = 0.32).
Conclusions
We found that ptDNA assessment for therapy monitoring in mCRPC is feasible and provides data relevant to the clinical setting. Prospective evaluation of these findings is now merited.
Subject terms: Predictive markers, Prostate cancer
BACKGROUND
Prostate cancer is one of the most frequently diagnosed cancers in men and the second-leading cause of cancer-related death worldwide.1 In recent years, treatment of metastatic castration-resistant prostate cancer (mCRPC) has changed with the introduction of different therapeutic options and optimising clinical benefit has become progressively challenging.2 Treating patients with ineffective therapies leads to unnecessary toxicity while also allowing symptomatic cancer progression. However, early discontinuation of a drug that is providing some degree of clinical benefit relative to other available modalities can also be disadvantageous. Currently, standard disease evaluation recommended by the Prostate Cancer Clinical Trials Working Group 3 (PCWG3) guidelines3 specifies imaging tests and prostate-specific antigen (PSA) assessments: these guidelines now also highlights the need for the evaluation of progressive disease (PD) at established timepoints using liquid biopsies to better characterise disease biology and identify potential predictive molecular biomarkers.
Circulating, cell-free DNA, often termed plasma tumour DNA (ptDNA) in the oncology setting, is rapidly becoming a quantitative and qualitative analyte to measure treatment efficacy.4 Its promise as a minimally invasive biomarker is supported by the high concordance of detected genomic alterations with matched metastatic biopsies.5,6
The amount of tumour DNA in the pool of cell-free DNA is called the tumour fraction or tumour content and ranges greatly from almost undetectable up to 90%.7 The presence of a low tumour fraction could be a limitation for plasma DNA analyses. However, the introduction of new genomic technologies with high sensitivity and specificity, including next-generation sequencing (NGS),8 has greatly contributed to the study of ptDNA.9,10
In prostate cancer, we and others have shown an association between pre-treatment ptDNA fraction assessed by NGS and clinical outcome.7,11–14 A recent randomised phase 2 study (NCT02125357)15 combining whole-exome and/or deep targeted sequencing on ptDNA samples from 202 patients with first-line mCRPC treated with abiraterone or enzalutamide showed that genomic alterations in BRCA2, ATM, TP53 and AR were associated with treatment outcome and low pre-treatment ptDNA fraction was correlated with a good prognosis. However, the role of ptDNA change in response to treatment and its relationship with PSA and imaging assessment in prostate cancer monitoring is largely unexplored. Here we aimed to determine if plasma DNA dynamics could be used as an early assessment of therapy efficacy for mCRPC.
METHODS
Study design and patient cohort
Plasma samples were prospectively collected with the primary objective of biomarker evaluation approved by the Institutional Review Board of Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST) IRCCS, Meldola, Italy (REC 2192/2013). Participants had histologically confirmed prostate adenocarcinoma without neuroendocrine differentiation, PD despite “castration levels” of serum testosterone (<50 ng/dL), ongoing luteinizing hormone-releasing hormone (LHRH)-analogue treatment or prior surgical castration. Patients received treatment with abiraterone 1 g once a day and prednisone 5 mg twice daily as first- or second-line therapy. Abiraterone was administered continuously until evidence of PD or unacceptable toxicity. Serum PSA was evaluated within 3 days of beginning therapy and monthly thereafter. Radiographic disease was assessed with the use of computed tomography and bone scan at the time of screening and every 12 weeks on treatment. The study was conducted in accordance with the Declaration of Helsinki and the Good Clinical Practice guidelines of the International Conference of Harmonisation. Written informed consent was obtained from all patients.
Circulating tumour DNA fraction analysis
Serial blood samples were collected pre-treatment and, when possible, on treatment and at progression. Circulating DNA was extracted from 1 to 2 mL of plasma from each patient using the QIAamp Circulating Nucleic Acid Kit (Qiagen) and quantified using the high-sensitivity Quant-iT PicoGreen double-stranded DNA Assay Kit (Invitrogen) as previously described.7,12
Targeted NGS was performed on the PGM Ion Torrent using a 316 or 318 Chip for a target of 1000× coverage for AR, TP53, FOXA1, CYP17A1, SPOP and regions of loss in prostate cancer. The ptDNA fraction for each plasma sample was estimated using the CLONET computational tool estimating genomic deletions.7 This method to determine ptDNA fraction was recently shown to significantly correlate with an orthogonal method of ptDNA assessment involving targeted methylome NGS.11
Statistical analysis
The primary objective of the study was to compare ptDNA fraction changes with radiographic/biochemical response after 3-month therapy. The secondary objective was to evaluate the role of ptDNA in tumour monitoring from starting abiraterone treatment to PD defined as biochemical and radiographic according to the PCWG3 criteria.3 PtDNA progression was considered as any increase of ptDNA from baseline value. Radiographic progression-free survival (PFS) was calculated from the first day of therapy to the date of PD or death, whichever occurs first, or last tumour evaluation. Waterfall plots showed the magnitude of ptDNA fraction or PSA decline in patients with different tumour responses at 3-month therapy (response, stable disease, progression). The odds ratios (ORs) and 95% confidence interval (CI) were calculated using logistic regression analysis. All p values were two sided and a p < 0.05 was considered as statistically significant. Statistical analyses were performed with the SAS 9.4 software (SAS Institute, Cary, NC, USA).
RESULTS
Patient and plasma sample characteristics
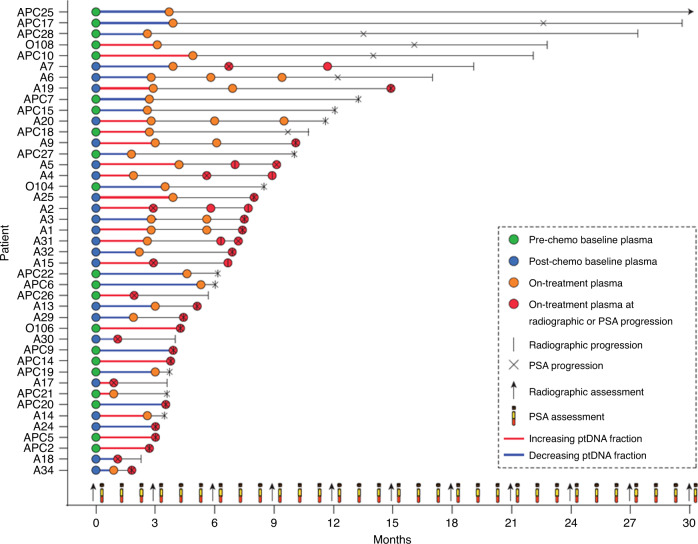
Between January 2011 and June 2016, we collected 114 plasma samples from 43 mCRPC patients treated with abiraterone (21 chemotherapy naive and 22 post docetaxel) at Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST) IRCCS, Meldola, Italy (Fig. 1). Overall, median age was 74 years (interquartile range (IQR) 70–78). Nine (20.9%) patients had visceral metastasis. Chemotherapy-naive patients had a significantly lower incidence of Gleason score ≥8 at diagnosis (p = 0.005) and higher levels of baseline haemoglobin (p = 0.0003) levels compared to post-docetaxel-treated group (Table 1). The first on-treatment plasma samples were taken at a median of 3 months after treatment (IQR = 2.6–3.7, Supplementary Fig. 1).
Fig. 1. Overview of study samples and data points.
Graph showing the time after start of treatment to plasma sample collection and progression by PSA or radiographic assessment for 21 chemotherapy-naive (Pre-chemo) and 22 post docetaxel (Post-chemo) patients.
Table 1.
Patient characteristics.
| Total (n = 43) | Chemotherapy naive (n = 21) | Post docetaxel (n = 22) | p Value | |
|---|---|---|---|---|
| Age (years) | 74 | 76 | 73 | |
| Median (IQR) | (70–80) | (69–81) | (72–77) | 0.532 |
| Gleason score, n (%) | ||||
| <8 | 15 (38.5) | 12 (60.0) | 3 (15.8) | |
| ≥8 | 24 (61.5) | 8 (40.0) | 16 (84.2) | 0.005 |
| Unknown/missing | 4 | 1 | 3 | |
| Bone mts, n (%) | ||||
| No | 8 (18.6) | 5 (23.8) | 3 (13.6) | |
| Yes | 35 (81.4) | 16 (76.2) | 19 (86.4) | |
| <6 | 14 | 3 | 11 | |
| >6 | 21 | 13 | 8 | 0.514 |
| Visceral mts, n (%) | 9 (20.9) | 2 (9.5) | 7 (31.8) | 0.076 |
| Liver mts, n (%) | 2 (4.6) | 1 (4.7) | 1 (4.5) | 0.973 |
| Lymph node mts, n (%) | 20 (46.5) | 11 (52.4) | 9 (40.9) | 0.456 |
| Baseline PSA (ng/mL) | 40.54 | 32.76 | 54.05 | |
| Median (IQR) | (10.17–81.39) | (1.34–403.50) | (11.44–123.40) | 0.174 |
| Baseline LDH (U/L) | 168 | 167 | 177 | 0.449 |
| Median (IQR) | (151–201) | (145–198) | (157–208) | |
| Baseline ALP (IU/L) | 98 | 96 | 103 | 0.449 |
| Median (IQR) | (85–133) | (87–121) | (71–133) | |
| Baseline haemoglobin (g/dL) | 13.9 | 14.0 | 13.1 | 0.0003 |
| Median (IQR) | (13.2–14.2) | (14.0–14.6) | (12.2–13.9) | |
| Baseline albumin (g/dL) | 4.1 | 4.1 | 3.9 | 0.220 |
| Median (IQR) | (3.8–4.2) | (3.9–4.3) | (3.5–4.1) | |
| Baseline NLR (n (%)) | ||||
| <3 | 23 (53.5) | 13 (61.9) | 10 (45.5) | |
| ≥3 | 20 (46.5) | 8 (38.1) | 12 (54.5) | 0.285 |
ALP alkaline phosphatase, IQR interquartile range, LDH lactate dehydrogenase, mts metastasis, n number, NLR neutrophil-to-lymphocyte ratio, PSA prostate-specific antigen.
Early changes in ptDNA and radiographic and biochemical response
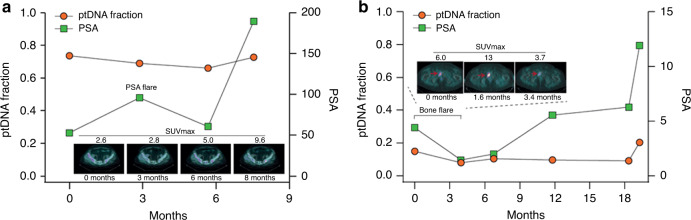
We examined the correlation between ptDNA fraction and disease status at first restaging in patients who completed at least 3 months of abiraterone. Supplementary Fig. 2 shows an overview of changes between the baseline and on treatment ptDNA fractions. We observed that patients with an increase in ptDNA fraction had a significantly increased risk of having PD, based on 3-month early radiographic assessment (OR = 15.8, 95% CI 3.5–60.2, p = 0.0002, Fig. 2a). Conversely, patients with a decrease in ptDNA fraction had significantly higher chance of having a response (OR = 16.7, 95% CI 2.4–188.5, p = 0.0039, Fig. 2a). Moreover, patients with an increase in ptDNA fraction had an increased risk of having a PSA rise (OR = 6.0, 95% CI = 1.6–20, p = 0.012, Fig. 2b). We did not observe an association between on-treatment ptDNA fraction increase and the absence of a ≥50% PSA decline (OR = 2.0, 95% CI = 0.6–6.6, p = 0.32). Interestingly, we identified examples of cases with a PSA flare or pseudoprogression on imaging secondary to a bone flare, but without a rise in ptDNA fraction (Fig. 3). We performed a supplementary analysis to evaluate the association between baseline ptDNA and clinical outcome. Patients were dichotomised into two groups according to the ptDNA fraction cut-off of 0.18 assessed by receiver-operating characteristic curve analysis. Patients with a ptDNA fraction >0.18 had significantly shorter PFS and a trend towards shorter OS (p = 0.02 and p = 0.10, respectively, Supplementary Fig. 3).
Fig. 2. Early changes in ptDNA and radiographic and biochemical response.
Waterfall plot of ptDNA change and a radiographic response and b PSA change in on-treatment vs. baseline samples. PSA change was capped at 100%; dashed line indicates PSA change of −50%. Odds ratio was calculated using Baptista–Pike and p value by Fisher’s exact test.
Fig. 3. Association of ptDNA dynamics with treatment response.
a ptDNA change and PSA flare in patient A3. b ptDNA change and bone pseudoprogression in patient A7. Patient A3 is characterised by an initial PSA elevation followed by a PSA decline and no radiographic progression on 18F-fluorocholine positron emission tomography/computed tomography (FCH-PET/CT) by using semiquantitative criteria based on the maximum standardised radiotracer uptake value (SUVmax). Patient A7 showed a bone flare on abiraterone characterised by an early increase in the intensity of bone lesions in the context of treatment response. In both these cases, ptDNA did not follow PSA and radiographic changes, but reduced in the first ~3 months from starting abiraterone.
Impact of previous treatment on ptDNA fraction change
All patients underwent androgen deprivation treatment (ADT) with an LHRH agonist and antiandrogens before abiraterone. We found that the ptDNA change with abiraterone was significantly associated with previous duration of ADT (Fig. 4a). This was observed both for chemotherapy-naive and post-docetaxel patients (Supplementary Fig. 4). However, no association was seen with previous docetaxel response in the post-docetaxel patients (Fig. 4b).
Fig. 4. Previous response to different treatments according to ptDNA increase or decrease upon abiraterone treatment.
a Previous response to ADT at hormone-sensitive prostate cancer (HSPC) setting in patients with ptDNA increase or decrease upon abiraterone treatment. b Previous response to docetaxel at castration-resistant prostate cancer (CRPC) setting in patients with ptDNA increase or decrease upon abiraterone treatment.
DISCUSSION
One key aim of treating CRPC is to prolong survival while maintaining quality of life and preventing needless toxic effects of ineffective treatments. Currently, tumour response and treatment benefit are determined using radiographic imaging and PSA assessment, but this may result in patient over-treatment and can be confounded by an apparent initial worsening that is in fact due to a flare secondary to bone remodelling at responding metastatic sites. Our study shows the potential of adding ptDNA assessment to routine monitoring of mCRPC patients.
Of note, a rise in ptDNA was strongly associated with radiological progression or any PSA rise, but not with PSA or radiological flares, indicating potential added value.16,17 Future prostate cancer studies utilising more frequent plasma sampling should enable the identification of any lead time benefit of ptDNA assessment for progression, as has been observed in other cancers.18,19
In our study, there was no association with ptDNA change and ≥50% PSA decline. Although an exploratory study, the association of ptDNA change on abiraterone with previous response to ADT at castration-sensitive prostate cancer (CSPC) provides an additional level of support to the relevance of early ptDNA assessment and suggests that patients with rapid progression on ADT are less likely to derive benefit from subsequent androgen signalling targeting even when separated by one line of treatment.
The main limitations of the study are the small cohort of patients, including both taxane-naive and post-docetaxel patients, the retrospective nature of the analysis and the limited number of multiple sequential plasma samples. Another general limitation for ptDNA studies is how to define progression. As we had a baseline and an on-treatment sample for all patients, we took a pragmatic approach and considered any increase of ptDNA fraction as progression. For future studies, we would envision a combination of a cut-off for what is considered a ptDNA change as well as having two subsequent samples showing an increased ptDNA fraction to confirm ptDNA rise. Despite these limitations, the present study represents a first step towards achieving the introduction of longitudinal ptDNA fraction assessment with standard tests utilised in clinical practice. This work underlines the potential of minimally invasive liquid biopsies to better inform future decision making that have suggested circulating tumour cell detection. Serial ptDNA analysis of prostate cancer patients is now being incorporated into larger prospective clinical studies.
CONCLUSION
In this report, we looked at the utility of ptDNA monitoring during abiraterone treatment. We found that ptDNA assessment associated with radiological progression or response and any decline in PSA, but also provided independent information. Future studies could support the integration of ptDNA into composite biomarker tests for early assessment of treatment benefit.
Supplementary information
Acknowledgements
We thank the participating men and their families who suffered from metastatic prostate cancer and nonetheless gave the gift of participation so that others might benefit.
Author contributions
Conception and design: V.C., D.W., G.A. Development of methodology: V.C., D.W., A.R., G.G., A.J., D.G.T., A.W. Patient samples collection: V.C., A.J., C.L., G.S., C.C., U.D.G. Analysis and interpretation of data: V.C., D.W., E.S., A.R., D.G.T., F.M., G.P., E.G.-B., F.D., U.D.G., G.A. Writing, review and/or revision of the manuscript: all authors. Study supervision: F.D., U.D.G., G.A.
Ethics approval and consent to participate
Written informed consent was obtained from each patient prior to entry into the study. The study was conducted in compliance with the principles of the Declaration of Helsinki and local ethical and legal requirements. The protocol and informed consent were approved by the Institutional Review Board of Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST), Meldola, Italy (REC 2192/2013).
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary information data
Competing interests
G.A. certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (e.g. employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received or pending), are the following: G.A. reports receiving commercial research grants from Janssen, Arno Therapeutics and Innocrin Pharma; has received honoraria and/or travel support from the speakers’ bureaus of Janssen, Astellas, Sanofi-Aventis and Roche/Ventana; has received travel support from Pfizer, Abbott Laboratories, Bayer Healthcare and Essa Pharmaceuticals; has ownership interest (including patents) in The Institute of Cancer Research Rewards to Inventors; and is a consultant for/advisory board member of Janssen-Cilag, Veridex, Bayer Healthcare, Roche/Ventana, Astellas, Medivation, Pfizer, Novartis, Millennium Pharma, Abbott Laboratories and Essa Pharma. V.C. has received speaker honoraria or travel support from Astellas, Janssen-Cilag and Sanofi-Aventis, and has received consulting fee from Bayer. E.G.-B. received speaker honoraria or travel support from Astellas, Janssen-Cilag and Sanofi-Aventis. U.D.G. has served as consultant/advisory board member for Astellas, Bayer, BMS, Ipsen, Janssen, Merck, Pfizer and Sanofi; has received travel support from BMS, Ipsen, Janssen and Pfizer; and has received research funding from AstraZeneca, Roche and Sanofi (Inst). No potential conflicts of interest were disclosed by the other authors
Funding information
V.C. was funded by a European Society of Medical Oncology Translational Clinical Research Fellowship. A.J. was supported by a grant from the Medical Research Council (MR/P002072/1). G.A. was supported by a Cancer Research UK Advanced Clinician Scientist Grant (A22744) and Cancer Research UK Accelerator Award (C416/A26822). E.G.-B. was funded by a grant from the “Instituto de Salud Carlos III” (ISCIII) PI15/01499. F.D. was funded by a grant from Fondazione Cassa di Risparmio Trento e Rovereto (CARITRO). The funders of the study had no role in study design, data collection, data analysis, data interpretation or writing of the report. The corresponding authors had full access to all data and had the final responsibility for the decision to submit for publication.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Vincenza Conteduca, Daniel Wetterskog
These authors jointly supervised this work: Francesca Demichelis, Ugo De Giorgi, Gerhardt Attard
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-020-0969-5.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Davies A, Conteduca V, Zoubeidi A, Beltran H. Biological evolution of castration-resistant prostate cancer. Eur. Urol. Focus. 2019;5:147–154. doi: 10.1016/j.euf.2019.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Scher HI, Morris MJ, Stadler WM, Higano C, Basch E, Fizazi K, et al. Trial design and objectives for castration-resistant prostate cancer: updated recommendations from the Prostate Cancer Clinical Trials Working Group 3. J. Clin. Oncol. 2016;34:1402–1418. doi: 10.1200/JCO.2015.64.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.González-Billalabeitia E, Conteduca V, Wetterskog D, Jayaram A, Attard G. Circulating tumor DNA in advanced prostate cancer: transitioning from discovery to a clinically implemented test. Prostate Cancer Prostatic. Dis. 2019;22:195–205. doi: 10.1038/s41391-018-0098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyatt AW, Romanel A, Conteduca V, Casiraghi N, Sigouros M, Franceschini GM, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J. Natl Cancer Inst. 2018;110:78–86. [Google Scholar]
- 6.Vandekerkhove G, Struss WJ, Annala M, Kallio HML, Khalaf D, Warner EW, et al. Circulating tumor DNA abundance and potential utility in de novo metastatic prostate cancer. Eur. Urol. 2019;75:667–675. doi: 10.1016/j.eururo.2018.12.042. [DOI] [PubMed] [Google Scholar]
- 7.Romanel A, Gasi Tandefelt D, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med. 2015;7:312re10. doi: 10.1126/scitranslmed.aac9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 9.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci. Transl. Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu A, Cremaschi P, Wetterskog. D, Conteduca. V, Franceschini. GM, Kleftogiannis. D, et al. The plasma methylome of metastatic prostate cancer. J. Clin. Invest. 2020;130:1991–2000. doi: 10.1172/JCI130887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conteduca V, Wetterskog D, Sharabiani MTA, Grande E, Fernandez-Perez MP, Jayaram A, et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann. Oncol. 2017;28:1508–1516. doi: 10.1093/annonc/mdx155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyatt AW, Azad AA, Volik SV, Annala M, Beja K, McConeghy B, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1598–1606. doi: 10.1001/jamaoncol.2016.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carreira S, Romanel A, Goodall J, Grist E, Ferraldeschi R, Miranda S, et al. Tumor clone dynamics in lethal prostate cancer. Sci. Transl. Med. 2014;6:254ra125. doi: 10.1126/scitranslmed.3009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8:444–457. doi: 10.1158/2159-8290.CD-17-0937. [DOI] [PubMed] [Google Scholar]
- 16.Conteduca V, Caffo O, Lolli C, Aieta M, Scarpi E, Bianchi E, et al. Long-term clinical impact of PSA surge in castration-resistant prostate cancer patients treated with abiraterone. Prostate. 2017;77:1012–1019. doi: 10.1002/pros.23357. [DOI] [PubMed] [Google Scholar]
- 17.Burgio SL, Conteduca V, Rudnas B, Carrozza F, Campadelli E, Bianchi E, et al. PSA flare with abiraterone in patients with metastatic castration-resistant prostate cancer. Clin. Genitourin. Cancer. 2015;13:39–43. doi: 10.1016/j.clgc.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Christensen E, Birkenkamp-Demtröder K, Sethi H, Shchegrova S, Salari R, Nordentoft I, et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J. Clin. Oncol. 2019;37:1547–1557. doi: 10.1200/JCO.18.02052. [DOI] [PubMed] [Google Scholar]
- 19.Hrebien S, Citi V, Garcia-Murillas I, Cutts R, Fenwick K, Kozarewa I, et al. Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial. Ann. Oncol. 2019;30:945–952. doi: 10.1093/annonc/mdz085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Supplementary information data