Abstract
Liquid biopsy, screening cancer non-invasively and frequently by detecting and quantifying molecular markers in physiological fluids, would significantly improve cancer survival rate but it remains a distant goal. The key obstacles presented by the highly heterogeneous samples are rapid/high-yield purification and precise/selective marker capture by their antibody and oligo probes. As irregular expressions of these molecular biomarkers are the key signals, quantifying only those from the cancer cells would greatly enhance the performance of the screening tests. The recent discovery that the biomarkers are carried by nanocarriers, such as exosomes, with cell-specific membrane proteins suggests that such selection may be possible, although a new suite of fractionation and quantification technologies would need to be developed. Although under-appreciated, membrane microfluidics has made considerable contributions to resolving these issues. We review the progress made so far, based on ion-selective, track-etched, and gel membranes and advanced electrophoretic and nano-filtration designs, in this perspective and suggest future directions.
Keywords: cancer, extracellular vesicles, high-throughput, liquid biopsy, nanopore
1. Challenges of liquid biopsy
The ability to non-invasively and frequently screen cancer by identifying and quantifying cancer biomarkers in a small sample of physiological fluids (blood, saliva, urine), ideally before the tumors have grown or metastasized sufficiently to be detected by imaging or tissue biopsy, has been a dream for many decades. Mortality rate decreases and life expectancy increases dramatically with early cancer detection. This is particularly true with the advent of immunotherapy and other very promising cancer therapies currently being developed—curing cancer is becoming a reality and early screening would further magnify the curing efficacy of new therapeutics. In fact, with the plethora of next-generation therapeutics, selecting and optimizing the proper therapy for individual patients (personalized medicine) would also require frequent biomarker monitoring with the same liquid biopsy technologies.
Paradoxically, even though hundreds of protein cancer biomarkers have been discovered in the last few decades, only a handful have been FDA approved and only one of them (PSA) is used in liquid biopsy for early screening [1,2]. The others are used for phenotype identification in tissue biopsies through imaging techniques like fluorescence in situ hybridization (FISH). The same is true of potential microRNA biomarkers that have been discovered in the last decade [3]. None of the microRNA diagnostics technologies have been FDA cleared. However, ThyraMIR® by Interpace Diagnostics for thyroid cancer screening is now available to clinicians and is covered by medical insurance [4]. The scarcity of approved marker-based screening test is quite surprising, given that there are numerous extremely sensitive molecular detection technologies. For example, single-exosome markerprofiling assays by digital PCR in droplets and micro-wells with barcoded miRNA primers and with antibodies conjugated with barcoded nucleic acids, followed by rapid sequencing, can theoretically achieve single-molecule resolution of an entire library of markers. It would, however, be prohibitively expensive and slow. Other than the obvious issues of cost and throughput, early cancer screening tests are only effective if they are conducted as part of the annual checkup or even more frequently for high-risk individuals and cancer survivors, an often-overlooked issue is robustness (or specificity) in its quantification of irregularly expressed biomarkers in a heterogeneous physiological sample.
Lack of robustness can be because the markers are extremely small in number. For example, there are too few circulating tumor cells (CTCs) to enable a viable CTC liquid biopsy platform. It would need to detect and quantify 5–10 CTCs per milliliter of whole blood in early-stage relapse patients. It may still be useful for phenotype identification or therapy management for late-stage patients, but reproducible detection and quantification of such a small number (out of billions of blood cells) is not likely even with the most sensitive technologies.
Lack of specificity is also because the probes we use to detect and quantify the molecular markers are not sufficiently selective. Hence, even if the technology can detect the probetarget complex at a single-molecule resolution, the fact that the captured molecule may not be the correct target would produce too high a false positive rate to earn FDA approval. In a recent mass spectrometry and Western Blot study [5], 84% of 70 viable antibodies that are able to capture their targets in plasma are found to also capture nontarget proteins in ELISA assays. Importantly, the dissociation constants of the targets and non-targets (most likely isoforms) with the antibodies are often within one order of magnitude. One order of magnitude is also roughly the overexpression level of most markers in cancer patients. There are two main reasons for such high rates of false positives. False positive is usually not an issue with natural immune response within the body, as erroneous attacks on nontarget cells usually do not lead to serious auto-immune diseases. Hence, natural antibodies from evolution may not be sufficiently selective innately. Moreover, recent studies have shown that immune cells, particularly lymphocytes, utilize both normal and shear mechanical forces exerted by their intracellular myosin/actin polymers to enhance the selectivity of immune-response to antigen-presenting cells [6, 7]. Such force-enhanced selectivity for antigens with very similar affinities for the antibodies is often absent in our immunoassays, other than a simple wash to remove unbound molecules. Hence, even though highly sensitive technologies exist for single-protein detection, for example, by droplet microfluidics [8], none of them are sufficiently selective because of these issues.
For the lone FDA-approved liquid biopsy screening test for prostate cancer, the PSA target is detected and quantified after several laboratory-bound pretreatment steps to purify the target. This remains the only solution to the selectivity issue—high-yield purification of relatively abundant (>nanograms per milliliter) markers. However, this is not an option for the remaining protein markers with typical concentrations at or below picograms per milliliter. Also, PSA purification still requires large laboratory-bound in strumentation and hence cannot be used for more extensive large-population screening.
The selectivity issue becomes even more acute with the most recent cancer biomarkers—extracellular RNAs (exRNAs) that are mostly microRNAs and messenger mRNAs. Quantification of the microRNA is particularly problematic. As it is short with about 20–25 nucleotides in length, a ligation step is required to convert it into longer cDNAs that can be amplified by PCR primers of roughly the same length as the original target. The ligation step is done with hairpin or toe-hold adaptor probes that form a pairing sequence of no longer than 10 bases with the target. Such a short pairing sequence is prone to interference by nontarget microRNA with similar sequences. Due also to the short pairing sequence, the yield of the cDNA reverse transcription step can also vary by over three decades for different sequences [9]. Hence, either careful normalization is done with each microRNA in each sample for PCR-based quantification or one needs to resort to digital PCR or PCR-free quantification technologies. As with all PCR-based assay technologies, PCR inhibitors in the blood can suppress PCR amplification completely [10], thus compromising even the most precise PCR-related technologies like rapid sequencing and digital PCR. All these issues render PCR-based quantification of the specific exRNA noise sensitive and not sufficiently robust.
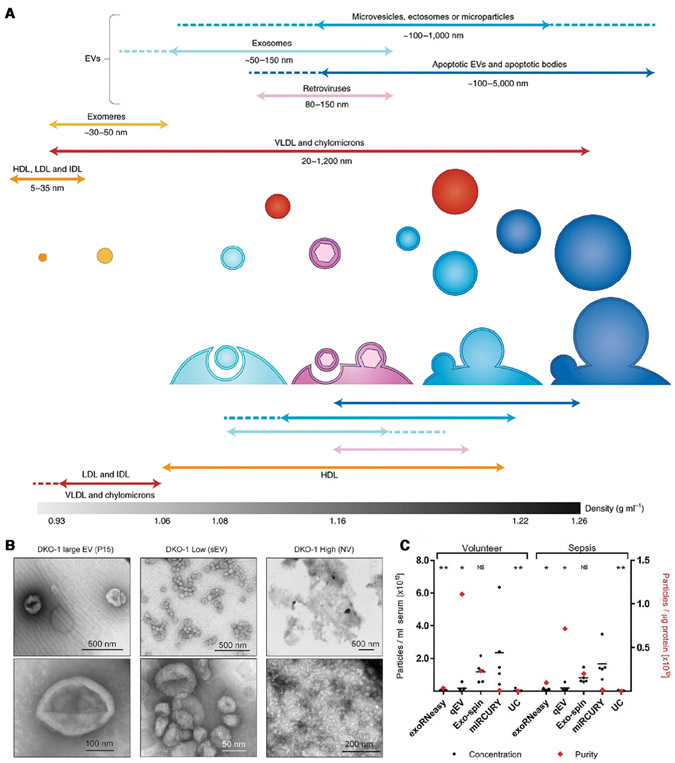
There are, however, even more complex selectivity issues with exRNA-based liquid biopsies. It has now been established that the exRNA biomarkers, instrumental in metastasis and immune suppression, are trafficked by extracellular vesicles (EVs) and other nanocarriers in eukaryotic cells [11]. These nanocarriers range from 10 nm to 1 micron in size and are quite abundant in the blood (about 1 billion to 1 trillion particles per milliliter of blood). These carriers are generally divided into several subclasses: microvesicles, exosomes, exomeres, ribonucleoproteins, and high-density lipoproteins (HDLs), in the order of decreasing size (see Fig. 1A). Their size ranges overlap but the most abundant vesicles, the exosomes, are almost entirely within the 50–200 nm range. Nevertheless, a large percentage of the nanocarriers within this range are not exosomes (see TEM image, Fig. 1B). It is now known that different EVs have different functionalities and hence carry different exRNA markers [12]. It is hence important to isolate the EVs that originate from cancer cells and carry true cancer biomarkers. Some nanocarriers like exomeres [13] and ribonucleoproteins are not vesicles at all—they do not consist of lipid bilayers. Yet, they also carry exRNAs that may or may not be markers. Fractionation of different vesicles and nanocarriers is hence necessary for very precise quantification of the cancer markers.
Figure 1.
(A) The different kinds of nanocarriers for cancer biomarkers and their characteristics (Reproduced from [17] with permission). (B) Negative stain transmission electron microscopy (TEM) images of EVs (Reproduced from [12] with permission). (C) Comparison of different EV and microRNA isolation methods. They include ultra-centrifugation (UC), size exclusion chromatography (SEC, qEV), precipitation (miRCURY, Exospin) and affinity column (exRNA easy) techniques. The yield (concentration) varies by a factor of 10, suggesting the lower-yield techniques capture less than 10% of the EVs and microRNAs (Reproduced from the open access resource [14], https://www.tandfonline.com/doi/pdf/10.1080/20013078.2018.1481321?needAccess=true).
There are commercial precipitation-based EV isolation technologies to indiscriminately isolate EVs in organic or polymer solutions, such as ExoQuick™ and Total Exosome Isolation™ technologies. They are attractive because of their simplicity and inexpensive equipment. However, these kits are still time consuming as they require overnight incubation. They are also known to produce very low yields that are EV concentration dependent (see Fig. 1C) [14-16]. Size- or mass-based EV isolation by ultracentrifugation (UC) is commonly used in medical research but the yield of UC isolation is among the lowest (<10%) of all techniques. More precise separation can be achieved with gradient ultracentrifugation (UC) and size-exclusion chromatography. Because of the size of the nanocarriers and their comparable specific density [17], UC fractionation, and size-exclusion chromatography have very low yield (<10%) and very high bias (see Fig. 1C) [15,18,19]. For example, van Deun et al. found that using an iodixanol gradient reduced protein contamination, but EV isolation yield got decreased by twofold and the UC times increased by 20 h [15].
There are now more advanced size-based EV isolation technologies like AF4 (asymmetric-flow field-flow fractionation) [20] and NLDA (nanoscale lateral displacement array) [21] technologies. AF4 can accommodate only small amounts of sample (e.g., 40–100 μg), which is often not efficient for large-scale preparations in more detailed assessments of EVs. NLDA, while not yet on the market, would require highly expensive nanofabrication and utilize very high pressures (≈10 atm) that lead to nanocarrier lysing and low yield. A viable liquid biopsy platform hence requires a high-throughput, high yield, and low-cost nanocarrier fractionation technology.
More importantly, there are many kinds of exRNA carriers and they seem to pack different exRNA cargoes [20]. Both healthy cells and cancer tumor cells release the same carriers, but the tumor cells tend to release more of the EVs with specific cancer protein biomarkers that have inherited from the tumor cell membranes (CD9, CD63, CD81 of the tetraspanin family, annexin or EpCAM, see, for example, Jeppesen et al. [12]). These EVs can also be captured with antibody-based technologies like fluorescence activated flow cytometry, immune-magnetic beads [22,23], and immunoprecipitation. There are several issues here. The nanocarriers’ small dimension also means there are not many labeled fluorophores (no more than one antibody for sub-50 nm EVs) for flow cytometry. Each antibody can be as large as 20 nm and hence there is not much room for multiple fluorescently labeled antibodies on a sub-50 nm EV. The selectivity and yield of the capture antibodies are often significantly compromised by interference from other nanocarriers and other antigens. Like molecular immunocapture, the selectivity of EV immunocapture can be enhanced by purification but it must also be done with high yield and without bias.
Even if nanocarrier fractionation is possible such that only specific nanocarriers from cancer cells are isolated, their molecular cargo still represents a large heterogeneous population of proteins and RNAs, only a small fraction of which are the desired biomarkers. Rapid and high-yield pretreatment technologies to enhance detection selectivity of lowabundance markers in a heterogeneous molecule population remains a main technical challenge to liquid biopsy. Falsepositives are minimized in a laboratory-bound assay with laborious pretreatment, such that the targets are purified or are at least the majority molecule in a far-less heterogeneous population. Molecular separation by HPLC, affinity chromatography, column capture/elution, mobility-based gel electrophoresis, isoelectric separation, SDS-PAGE, etc. is used to remove interfering nontargets for more selective quantification downstream. However, the yield of these purification steps is low and hence statistically significant quantification can only be done for abundant markers, like PSA.
Another potential solution to the selectivity issue, for both nanocarriers and molecules, is to mimic immune cells and use force to achieve more selective probe capture of the targets. As is practiced by the immune cells, this force-enhanced selectivity is best done by selective removal of the nontargets after nonselective capture. It would be a scientifically designed “washing” step. This direction has not been explored in the community but we believe it is a very promising one.
Microfluidics can offer such a high-throughput liquid biopsy platform, with integrated high-yield purification pretreatment for nanocarriers and force-enhanced highly selective sensor modules for their molecular cargo. Its miniature dimensions allow the integration of pretreatment modules without sacrificing throughput or producing analyte loss. It permits precisely controlled flow rate and electric field for selectivity enhancement—so the “wash” step in any assay can be optimized for selectivity. Microfluidics also offers several technologies that can overcome a key obstacle in purification: hydrodynamic dispersion. With the high shear rates and the long channels, dispersion determines the plate height of the isolated analyte and hence the quality and yield of the pretreatment, as the dispersion coefficient is related to the diffusivity by the square of the Peclet number UR/D [24]. Even with a typical (high-throughput) flow velocity of U = 0.1 mm/s in a typical micro-channel width R = 100 microns, the small diffusivities of biomolecules and EVs (D~10−6–10−8 cm2/s) still produce a large Peclet number between 100 and 104 or a dispersion coefficient that is four to eight orders higher than the diffusivity!!! The plate height based on the hydrodynamic dispersion coefficient would hence be roughly a few centimeters, comparable to the slug length of a small-volume (~1 mL) sample that is flowing through the device. With such a large plate height, purification of the target is extremely difficult. The new concentration technologies hence enable high-yield purification modules that can be integrated with the sensor technologies to allow both high throughput and high selectivity. We would like to encourage the microfluidics community to focus on such multiplexed designs with high-yield pretreatment modules and with high-selectivity force-enhanced sensor modules. We will point out a few remaining obstacles in this review but we believe the first FDA approved comprehensive liquid biopsy technology will be a turn-key integrated microfluidic platform that allows robust and highly accurate quantification of a panel of biomarkers in blood and other physiological fluids. In fact, such a microfluidic platform can also be used to screen for more selective antibodies. We will make the case that membrane microfluidics, with integrated ion-selective, track-etched and gel membranes, is the microfluidic platform of choice in this perspective.
2. Membrane microfluidics and analyte concentration/purification by localized ion-depleted regions
We have developed an array of membrane microfluidics for microfluidics molecular sensing and pretreatment applications [25]. We shall focus on its application to molecular and EV isolation here. It has been known for decades that, due to the conductivity difference between an ion-selective membrane and the bulk electrolyte, an external ion depletion front can develop on one side of the membrane (see our recent reviews [3,25]). Depending on the geometry and the size of the membrane relative to the electrode, this depletion front can propagate indefinitely (to the electrode) or stop at a distance from the membrane that is roughly the membrane width [26]. Quite recently, we used a gated design to control the length of this depletion region in a long channel [27]. The analyte concentration features of the ion depletion front have recently been used extensively for biosensing applications [10,28-32]).
The ion-depleted region sustains a high field (as high as 1000 V/cm) that can be utilized for several local electrophoretic separation tasks. It would be difficult to apply such a high field over the entire device that is filled with high-ionic strength physiological fluids. It is, however, possible in a localized ion-depleted region without significant Ohmic heating. A good common example is the depletion zone on one side of an ion-selective membrane under limitingcurrent conditions. Over a region no more than 100 nm (several Debye lengths), the ionic strength is essentially at DI water level and the electric field exceeds million V/cm and yet there is an insignificant change in temperature due to Ohmic heating [33]. We will need to extend this ion depleted region to millimeter range but the field remains high at 1000 V/cm and Ohmic heating is still negligible, particularly in the presence of a net flow with large thermal Peclet number.
As this ion-depleted region is surrounded by the high-ionic strength buffer with a low field, any analyte (molecule or EV) that is electrophoretically driven from the depleted region to the high-ionic strength region would be concentrated at the boundary. The width (plate height) of the concentrated band is D/U, where U is the high free-space electrophoretic velocity (mm/s) in the depleted region, as small as 10–100 microns, compared to the centimeter-long plate height due to hydrodynamic dispersion [25]. Very effective (>1000×) and very rapid (<minutes) analyte concentration can hence be achieved both in free flow and in gel [34]. Such a high analyte concentration factor enhances the sensitivity of the sensors by orders of magnitude. For example, we can extend the limit of detection of our membrane sensor for microRNA from nM to pM [35,36] and for our nanoparticle sensors from 40 nM to 500 pM [37]. We shall, however, focus on how they can improve the pretreatment portion of the liquid biopsy platform here. Our recent study demonstrates the effect of this depletion front in trapping all molecules in a flowing stream and concentrating a 5 mm band of fluorescently labeled short DNAs in gel into a single sub-millimeter band [37].
We have developed numerous microfluidic molecular purification designs, based on the high-field and analyte concentration features of on-chip ion depleted regions activated by ion-selective membranes. The concentrated band can be pushed through hydrogel and through a micro-channel against a pressure-driven flow.
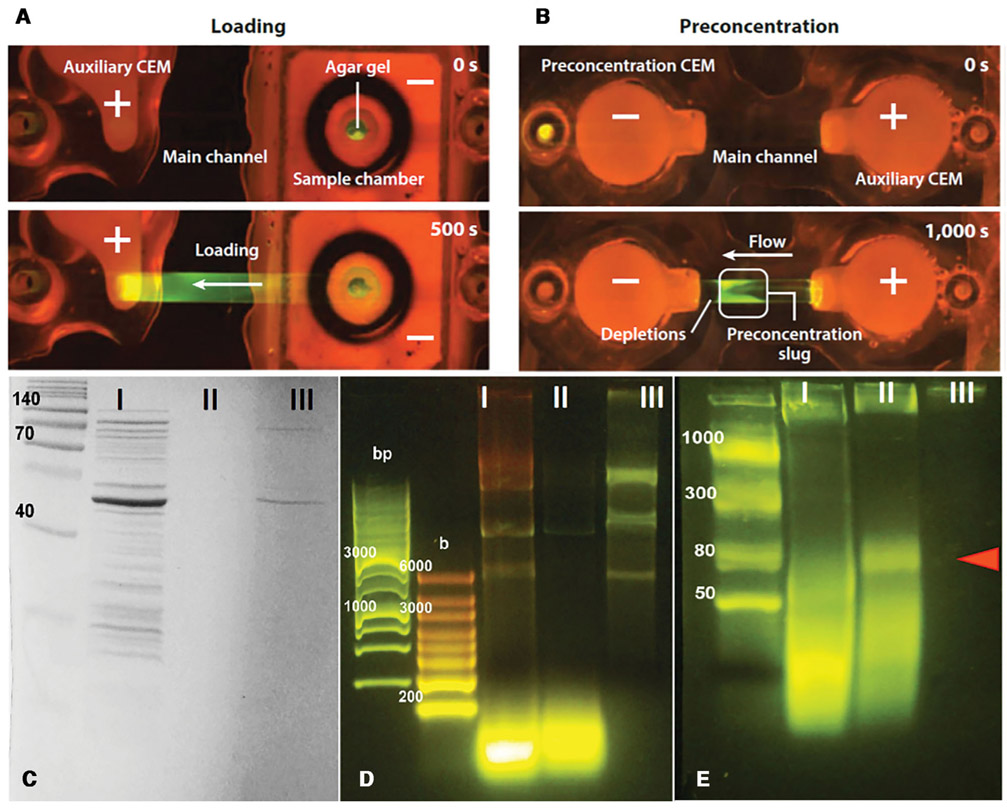
If the depletion front propagates against a counter flow in a free-flow electrophoresis format but with a localized ion depletion zone to enhance the field and to minimize hydrodynamic dispersion, the electric field can be tuned such that the target molecules are trapped in midstream because its hydrodynamic drag is exactly equal to the electrophoretic force applied by the electric field in the depletion front. Molecules of opposite charge would be rapidly removed from this stationary slug, as the electrophoretic force and the hydrodynamic drag they experience are in the same direction. Even molecules of the same charge can be removed if their electrophoretic mobility is higher or lower than that of the target. It is typically easier to trap targets if they have the highest electrophoretic mobility, as all non-targets would be eluted by the flow and high-mobility targets may still be trapped at the boundary of the depletion front. This is particularly convenient for microRNA, as it has the highest mobility of all negatively charged nucleic acids and vesicles in the blood. In Fig. 2, we show the effect of pushing a cell lysate slug by the depletion front against a flow [3, 25]. All proteins are removed, whether they are of opposite charge from the nucleic acids or have the same charge but lower mobility. Moreover, all DNAs and RNAs longer than the microRNA population are also removed.
Figure 2.
After loading cell lysate (frames A), the polarity is reversed to advance a depletion front to the right against a flow to the left (frames B). The depletion front advancement is arrested by the flow at a particular position (Reproduced with permission from the Annu. Rev. Anal. Chem., Volume 7 © 2014 by Annual Reviews, http://www.annualreviews.org). Gel electrophoresis (second non-ladder lanes of D and E) and SDS-PAGE (second lane of C) analyses of the trapped lysate at the boundary of the depletion front shows only high-mobility microRNA remains. Longer nucleic acids and all proteins evident in the first unfiltered lysate lane have been removed. Experiment E is to verify the trapped molecules are microRNAs and other short RNAs (Reproduced from [3] with permission).
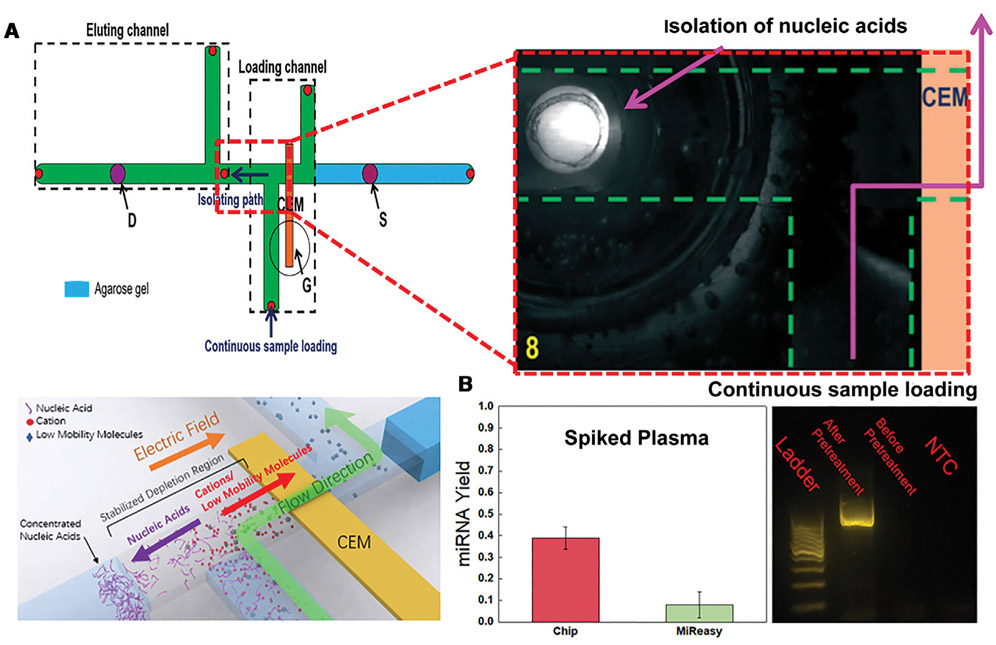
If the same high field of the ion depleted region is applied across a flowing high-ionic strength buffer, the high field can be used to extract charged molecules from the flowing stream and insert them into a different buffer. Such a design is shown in Fig. 3 to extract microRNAs and DNAs from whole blood and insert them into a PCR cocktail for PCR amplification. Without this extraction, PCR amplification is not possible (Fig 3). The reverse-transcription PCR yield for microRNA is shown to be more than four times that of the best commercial microRNA extraction kit. This continuous extraction is not only more rapid than the commercial DNA extraction technology but can be integrated with downstream PCR units and sensor units in a microfluidic chip [10].
Figure 3.
(A) A depletion front generated by the ion-exchange membrane G extends to the junction between the two cross-channels, the bottom vertical channel and the horizontal channel. Spiked DNA serum flows from the bottom vertical channel to the right of the horizontal channel before exiting by another vertical channel, as shown by the blue line. The high field in the junction extracts the DNA and places it in a buffer to the left of the horizontal channel. (B) Spiked microRNA and DNA into serum is tested with the pretreatment chip. The reverse-transcription PCR yield of microRNA is shown to be more than 4 times higher than the best commercial pretreatment unit. There is no PCR amplification of the DNA without pretreatment (Reproduced from [10] with permission from the Royal Society of Chemistry).
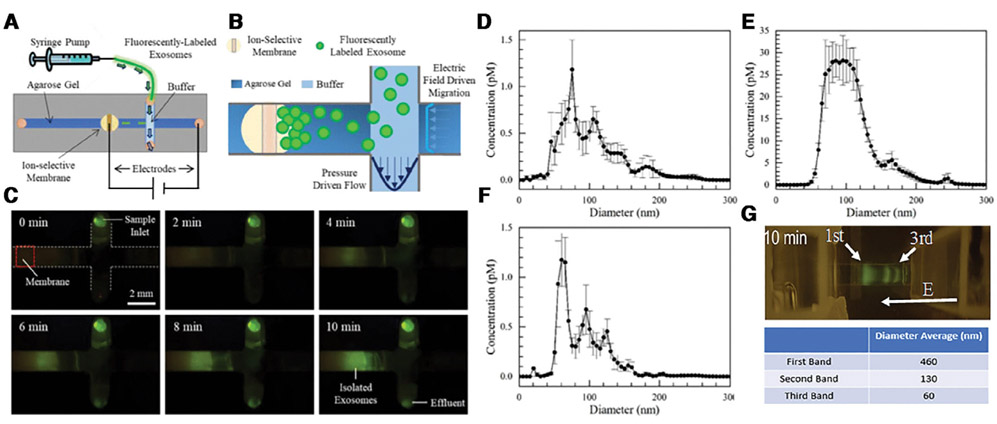
It is known the exosomes and microvesicles are negatively charged and hence can be isolated from the sample by electric field. Davies et al. demonstrated direct isolation of EVs from blood using electrophoresis with field perpendicular to the flow direction [38]. However, the low electric field strength (<10 V/cm) employed in their study allowed them to recover only 2% of EVs [38]. Another electrophoretic technique developed by Cho et al. applied a higher electric field (>10 V/cm) across a dialysis membrane to achieve a 65% recovery rate in 30 min [39]. The need for higher field to electrophoretically isolate EVs suggests the use of depletion front. Correspondingly, Marczak et al. used a depletion front to laterally extract EVs from a fast flowing (~3 μL/min) of cell culture media and serum in less than 30 min, with more than 15× concentration factor and a yield of about 70% (Fig. 4) [40]. The depletion front invades into a gel reservoir where it deposits the EVs. The gel medium is also used to filter out cell debris. We have also used the same depletion action in a gel to concentrate and separate nanoparticles for very precise and very selective quantification of molecular targets [37].
Figure 4.
Schematic of the EV isolation chip (A). A zoom view of the isolated region (B). The pictures of the exosome isolation process with fluorescently labeled exosomes (C). Particle-tracking analysis shows that an optimized 1-step extraction protocol can achieve 70% extraction yield in 10 minutes, based on the particle-tracking analysis of the original suspension (D), the trapped EVs (E), and the flow through (F) (Reproduced from [40] with permission). (G) Preliminary data on chip electrophoretic separation of EVs. Three distinct bands appear, with sizes corresponding to microvesicles, exosomes and lipoproteins (see Fig. 1A). Curiously the mobility is proportional to the size, indicating a significant increase in charge density for larger particles.
3. High-throughput and high-yield EV purification by membrane microfluidics
3.1. Size-based separation
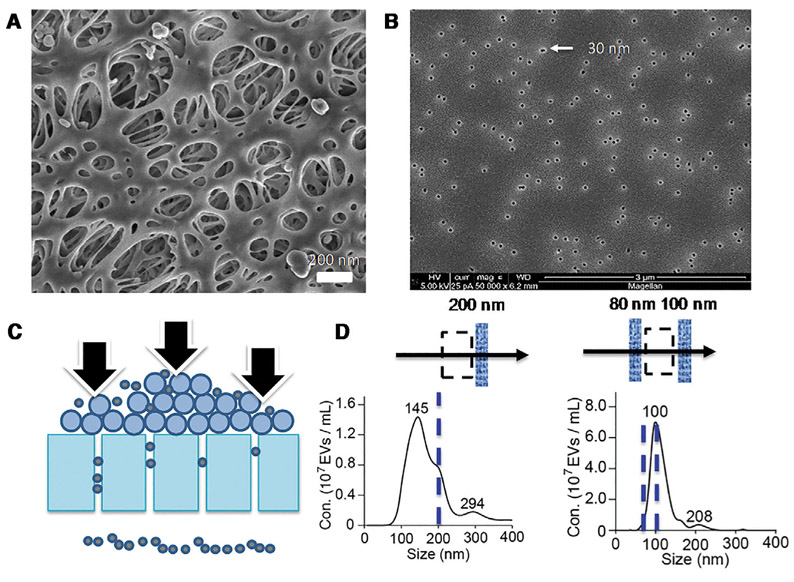
Once the EVs are isolated, they need to be further fractionated to get better purity and better statistics for the molecular biomarkers they carry. Because of undesirable yield and selectivity for flow cytometry and ultra-centrifugation, other separation technologies should be employed. As the exosomes have a size range (50–200 nm) that is reasonably separated from microvesicles and exomeres. Exosome separation by size has attracted considerable attention recently. The most established size-based fractionation technology is nanofiltration. They are either membrane-based or micropore-based. Lysing is a typical concern with these size-based filtration technologies, as its yield is known to be low [38]. Low-yield is most likely due to dead-end pores of varying sizes that can permanently trap the EVs. Contamination by other filtrates and adsorbed proteins is also a concern. For example, Davies et al. used a porous polymer monolith membrane to filter EVs directly from blood yet experienced significant contamination by proteins [38]. Size-exclusion chromatography can remove the proteins with small dead-end pores but, because it is hard to control the size of the dead-end pores, the yield goes down correspondingly (see Fig. 1). Recently, Liu et al. used track-etched membranes with straight pores of uniform larger radius to prevent protein fouling and to increase the yield. However, such ultrafiltration membranes have low throughputs even at high pressures (~2 atm), due to concentration polarization [41] and filter cake formation. Filter-cake formation and high pressure lead to nanocarrier lysing and coalescence, which are the main issues that plague track-etched filters [42] (see Fig. 5). The coalesced particles are twice the mean EV radius and appear in the flowthrough despite the fact they are larger than the pore size. Consequently, the coalescence most likely occurred within the pores due to filter-cake formation. Such coalescence events will release the molecular cargo and compromise the performance of EV fractionation. Filter cake formation often restricts the transit of small particles that are not supposed to be retained, thus giving rise to very poor size cutoff. Concentration polarization and filter-cake formation are commonly minimized by a cross-flow in membrane science [43] but the optimum cross-flow conditions for EV nanofiltration by ion-track membranes have not been reported.
Figure 5.
Representative EM images of (A) an ultrafiltration polyethersulfone (PES) membrane showing irregular pores with different radii and (B) our track-etched membranes with uniform pore radius. Fractionation of cell culture media using track-etched membranes show significant filter cake formation and EV coalescence as schematically shown in (C) that results in a poor size fractionation and leads to particles about twice the average EV size in the flow through (D) (Reproduced from Supplementary Materials of [42] with permission). These large particles are larger than the pore size by 100 nm and hence are believed to be created by coalescence events in the pore.
Asymmetric-flow field-flow fractionation (AF4) is yet another size-based EV separation technology. A most recent paper [20] used state-of-the-art asymmetric (Cross-Flow) Field-Flow Fractionation to identify three bands of exosomes with very distinct hydrodynamic radii (44, 59, and 140 nm). Downstream proteomic analyses suggest the middle-sized are of intraluminal origin and the largest exosomes may have come from membrane budding. AF4 has high throughput but analyte loss (recovery rate) is always an issue, as the exosomes are driven toward one side of the channel and across it. A recent study with nanoparticles [44] shows a loss as much as 74%. Yet another size-based technology is the Nanoscale Deterministic Lateral Displacement (NDLD) technology [21] that uses nanopillars to laterally deflect EVs in a flowing stream. As the larger particles are deflected more and hence can cross the separatrix streamlines of the nanopillars, they exit on the side of the less-displaced smaller EVs. It offers a higher yield than AF4 but, as 10 atm of pressure is required to pump the suspension across the long channel with nanopillars, it is an expensive equipment with a long assay time (~hours). An acoustic nanofilter in which ultrasound standing waves were applied orthogonally across a continuous sample flow resulted in EV recovery rates of up to 80%, which is one of the highest yield reported [45]. However, like NDLD, acoustophoresis suffer from challenges in fabrication and robust operation.
We believe that nanofiltration, with its high throughput and small membrane that can be integrated into microfluidic chips, is the most promising EV isolation technology. Straight and uniform sized nanopores offered by track-etched membranes should offer the highest yield if filter-cake formation and clogging issues can be resolved. In this direction, the use of cross-flow in classical filtration design should be able to minimize both phenomena. The same applies to asymmetric nanopore membranes (ANM) with conic geometries that can be fabricated from track-etched membranes [46]. Ghosal et al. have recently shown that conic nanopores have an electrical and hydrodynamic resistance that is lower than the same cylindrical nanopores, with the same radius as the conic tip, by a factor equal to the ratio ofthe base to tip radii, or as high as a factor of 10 [47]. This means that instead of 2 atm used in Fig. 5, less than 0.2 atm is required for the filtration of 50–200 nm exosomes. This is compared to 10 atm for NDLD and 500 atm for the centrifugal Bernoulli pressure of UC.
3.2. Separation by charge and electrophoretic mobility
It is known that exomeres, exosomes, and microvesicles are negatively charged with a −10 mV Zeta potential [48]. Lipoproteins are neutrally charged [49] and RBPs can even be positively charged with their relative abundance of proteins [50]. The latter carriers are probably removed during fieldbased isolation such as the depletion front. The larger EVs with high hydrodynamic drag would be difficult to isolate with the depletion based free-flow electrophoresis design of Fig. 5. However, the size difference in microvesicles and exosomes suggests they can be separated by gel electrophoresis. The advantage of gel electrophoretic separation lies mostly in assay time, reducing it from days to hours with the high fields we can achieve with ion depletion. In Fig. 4G, we show preliminary EV gel electrophoresis data with three bands. Particle tracking analysis of the three bands suggests they correspond to sizes corresponding to microvesicles, exosomes, and exomeres/HDLs. Lateral ion depletion can also be utilized to extract/concentrate the separated bands from the gel.
We are also optimistic about affinity-based and size-based chromatographic separation based on electrophoresis in gel, polymer membranes or even fiber/particle assemblies. Antibodies can be functionalized onto the medium to selectively separate specific EVs [51]. We envision different stages, each with a different pore size and embedded antibodies to trap specific EVs. Electrophoretic separation based on the high field of membrane-induced ion depletion in the chromatographic medium is much preferred over pressure-driven flow to achieve high throughput. Depletion front into such affinity media would also allow the extraction of the separated bands.
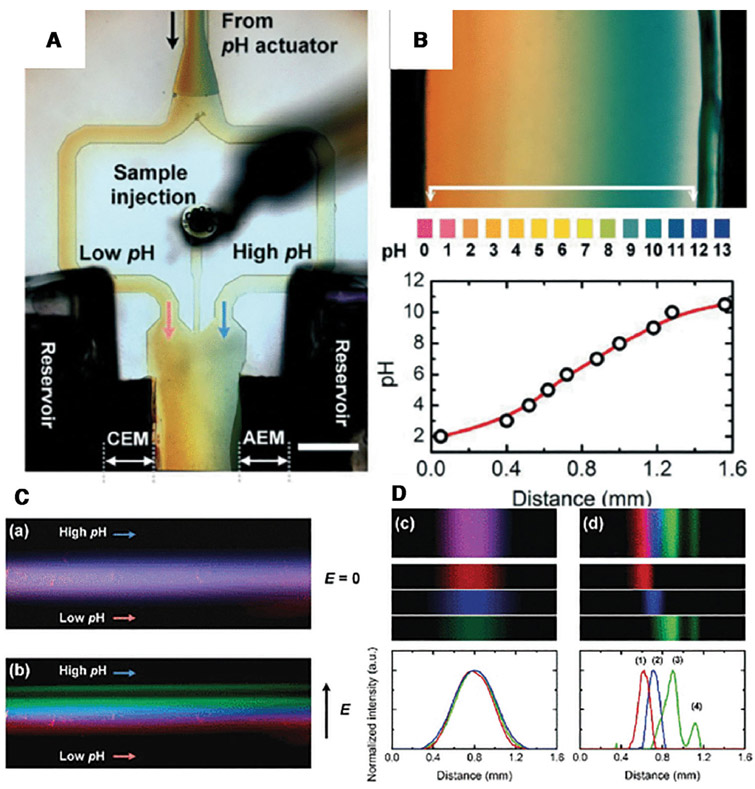
Yet another related electrophoretic separation technology is continuous isoelectric fractionation (CIF). Cheng and Chang [52,53] have shown that when two oppositely charged ion-selective membranes are assembled together, such that the depletion action by both is at the junction, the entire voltage drop of the system occurs at a junction region with a 10 nm Debye thickness. The result is a field in excess of 10 million volts/cm with just a 70 V bias, enough to split water. We are hence able to produce highly acidic and basic solutions from two different sides of the bipolar membrane. This then removes the need of adding acid and base to produce a pH gradient for isoelectric separation. In Fig. 6, we used two bipolar membranes as on-chip sources of acid and base at the two sides of a flow channel to produce a robust pH gradient between 2 and 10 across a 2 mm channel with a strong through flow. It enabled high throughput protein separation with different isoelectric points. The same format can be used for EV fractionation at a throughput that is even higher than gel electrophoretic separation accelerated by ion depletion. There may be a tradeoff between throughput and resolution such that the two technologies are good for different samples and for separating different EVs. A multistage design involving both may also be desirable.
Figure 6.
A picture of the pH chip (A). Two bipolar membranes generate a pH gradient between 2 and 10 in a 2 mm channel (B). A flowing mixture of four fluorescent proteins (C) with different isoelectric points are separated into four different channels (D) (Reproduced from [53] with permission from the Royal Society of Chemistry).
3.3. Separation by nanomagnetic immunobeads
Immunocapture technologies, like precipitation and antibody functionalized pillars, can capture EVs with specific membrane protein [54], but they have a rather low yield, throughput, and, like other antibody-based technologies, low selectivity. The low yield is often a result of probe saturation and the low throughput the long transport time (~1 day) of the EVs and antibodies. The same can be said of micron-sized or larger magnetic beads [22]. Their surface area per unit volume may be too low for more abundant EVs and hence probe saturation can reduce its yield. One solution to enhance its throughput and yield is to use immune-nanomagnetic beads. They are smaller than exosomes and other larger EVs and hence can be inserted in large numbers to achieve high-yield capture. In fact, with nanomagnetic beads outnumbering the EVs, the assay time is determined by the diffusion of the nanobeads to the EVs, which is now much shorter because of the reduced diffusion length and the larger diffusivity of the nanobeads. Consequently, EV capture by nanomagnetic beads is an attractive way of sorting EVs by surface protein [23,55].
However, the nanomagnetic bead has a major disadvantage—they are difficult to trap. Unlike larger magnetic beads, they are paramagnetic rather than ferromagnetic. Their single magnetic domain is too small to sustain any collective magnetic dipole against thermal noise. As such, their magnetic dipole is induced and, like dielectrophoresis with an induced electric dipole, the magnetic force on them is proportional to the gradient of field squared rather than to the field [23]. This presents an issue, as magnetic fields with high gradients are typically produced by small magnets with very low field penetration. Hence, they become very short-range traps that can only trap nanomagnetic beads within one radius of the curvature of the sharp tip at the magnet. One hence can trap them by flowing the nanomagnetic beads in a very short channel or use another mechanism to bring the nanobeads to the domain of attraction of the region with a high-field gradient.
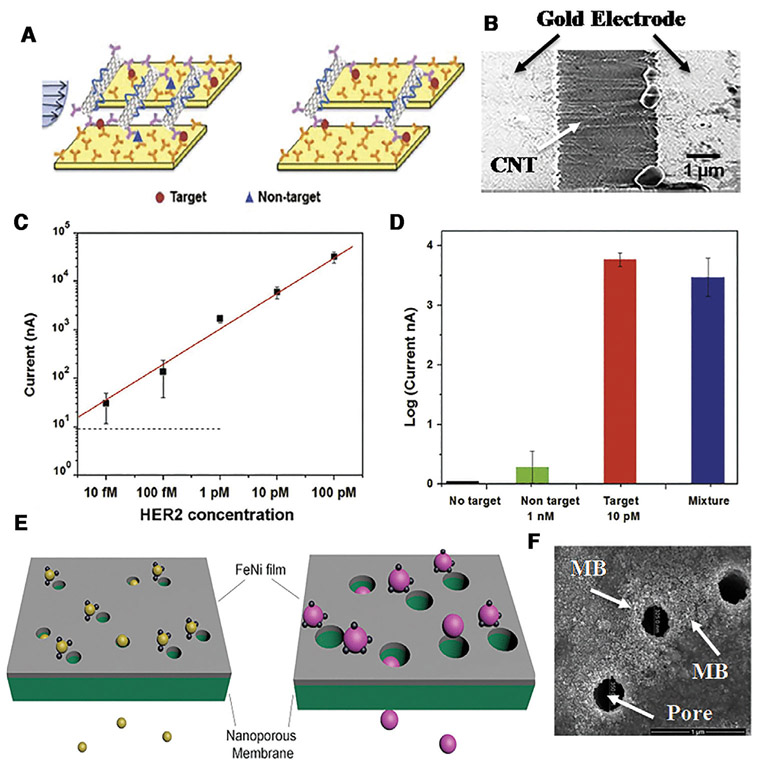
We faced a similar design challenge in our design of dielectrophoretic trapping CNTs at two planar electrode pairs. Our first solution was to use a small microfluidic channel with interdigitated electrodes at the top and bottom channel urface to ensure the field gradient persists over the entire height of the channel [56]. Our second solution for a larger channel is a combined DC and AC electric field [57]. The DC field is applied by a far electrode, with both planar electrodes acting as the counter electrode, such that there is a long-range DC field that can drive the charged CNTs to the vicinity of the planar electrodes. An AC field is then applied across the planar electrode pair to trap the CNTs dielectrophoretically [57]. The CNTs are transported from more than 1 mm away and then placed precisely across the micron-gap of the electrode pair by this hybrid DC electrophoresis and AC dielectrophoresis design, as seen in Fig. 7B.
Figure 7.
(A) Schematic of irreversible hydrodynamic shearing of non-specifically bounded CNTs. (B) Trapping of CNTs across a 3-micron gap by a DC and AC protocol. (C) Using an optimized shear protocol, we can detect HER2 marker at concentrations (10 fM) orders of magnitude lower than its dissociation constant at nM and (D) selective detection of HER2 in the presence of a 100 times more abundant isoform in serum. (Reproduced from [57] with permission. (E) Schematic of nanomagnetic bead trapping. (F) The high magnetic field gradient at the pore tip of a track-etched membrane coated with a Fe/Ni film can also trap magnetic bead (MB) particles.
The same long-range DC electrophoretic transport can be used to bring the nanomagnetic beads to small magnets with high field gradient, provided electrodes can be fabricated near the magnets or if the magnets themselves can act as electrodes. An opposite alternative was suggested by Issadore’s group [55]. If magnetic films are coated on surfaces with nano-sized pores, such as track-etched membranes, the magnetic field and field gradient would be high at the pore edge. The pores, on the other hand, allow fluid passage and the flow can convect the nanomagnetic beads to these pore traps, thus offering the long-range transport mechanism (see Fig. 7F). However, for this trap to be efficient, the pore radius must be comparable in size to the nanomagnetic bead-EV complex. This means that larger particles would clog the pores and hence upstream size separation must be carried out prior to trapping them with the magnetic nanopore membranes. An integrated platform with both size and nanomagnetic bead separation is hence required. This is the main challenge facing nanomagnetic bead capture technologies for EVs or even molecules.
There is an exciting new EV characterization method based on probe-functionalized nanomagnetic particles—miniature nuclear magnetic resonance [58]. The beads produce a unique NMR signature that can be used to quantify the proteins the antibody probe has docked with. Its sensitivity is orders of magnitude higher than colorimetric ELISA technologies. It would be profitable to integrate it with the above EV capture technologies to increase its throughput. The selectivity of the probe remains an issue.
There are some probe-free exosome characterization technologies based on SERS [59]. However, the Raman spectra of different proteins tend to be different in their amplitude and not in the frequency shift, as they are made of the same amino acids. Unless isolation of individual exosome is possible, it also would be difficult to deconvolute the heterogeneity of exosomes. Consequently, SERS characterization is probably best used downstream of other size-based fractionation and probe-based nanomagnetic bead technologies and perhaps only to differentiate between vesicle nanocarriers and non-vesicle carriers like exomeres and ribonucleoproteins. It would be good for small number of EVs with very distinct Raman signals.
4. Force-enhanced highly selective sensors
Separated, the EVs can be lysed either thermally or by Surface Acoustic Wave [35, 36] to release their molecular cargo. For integrated devices, it is best not to inject any additional reagent, such as is done with chemical/surfactant lysing, as it would necessitate additional extraction steps downstream. With proper tuning, the same purification technologies that we use to fractionate the EVs can also be used on the released molecules to purify the target to improve the selectivity of the molecular sensors. However, it is clear from earlier discussions that, other than isolation and purification of the analyte to remove the interfering agent, selectivity can be enhanced by utilizing force, as the immune cells do with their actin/myosin driven catch and slide bonds [6, 7]. Analytes with just slightly less affinity (larger dissociation constant KD) than the target can be removed with force but not with thermal noise. The trick is to render the dissociation reaction irreversible by reducing the barrier by force [60]. Typical dissociation constants between nM and μM for Ab-Ag and microRNA-oligo binding correspond to a Gibbs energy of about 20–30 RT (40–60 kJ/mole). Hence, isoforms with a dissociation constant of the same order as that of the target is only about 2 or 3 RT difference in their Gibbs energy of association or 2 or 3 kT for a single complex. A 10% difference in the Gibbs energy of association is too small a difference to be selective with thermal noise and equilibrium assays, which would only produce a dissociation constant ratio of 10. However, a small voltage (~1 V) or a sufficiently high shear force can remove the barrier for the nontarget while retaining a barrier of a few kT—enough to retain the targets but irreversibly remove the non-targets. Electrical and hydrodynamic shear forces are hence excellent selectivity-enhancing mechanisms. Optimization of these mechanisms can lead to highly selective immunocapture agents for EVs and their molecular cargoes. However, assuming the barrier is roughly the same height as the Gibbs energy of association of 20 kT, the dissociation force required over the 10 nm length scales of EVs is on the order of pN. This translates to a Stokes drag at a velocity at the range of 1 mm/s to 1 cm/s. Such a high velocity is typically not reached in a microfluidic channel and hence means of enhancing hydrodynamic drag by roughly a factor of 10 to 100 is necessary for shear-enhanced selectivity designs. Another possibility is to increase the viscosity by the same factor.
A good example of selectivity-enhanced sensor is the membrane sensor we have developed is based on shifts in the over-limiting current of the ion-selective membrane due to target capture [25,35,36]. The overlimiting current arises because of the appearance of an extended polarized layer at the membrane surface when all the ions are depleted [33]. The extended polarized layer is unstable to a vortex hydrodynamic instability and the resulting electroconvection brings ions from the bulk to the depleted region to return the latter’s ionic strength to the bulk value, thus producing an overlimiting current that is much higher than the limiting current. The sensor membrane is chosen such that the probe and the target are its counterions. As such, ion depletion is not complete as the target/probe counterions are immobilized by covalent and target/probe bonds by functionalization and target capture. This reduces the dimension of the extended polarized region and hence a shift to lower overlimiting currents. Nevertheless, the electroconvection vortices of the overlimiting current provide hydrodynamic shear to enhance the selectivity of the sensor. The actual selectivity enhancement has not been quantified in detail and we often rely on a controlled wash to remove the non-targets. However, surface patterns can be added to produce high-shear vortices on the membrane sensor surface to achieve autonomous selectivity enhancement [61].
We have developed several microfluidic designs that allow tuning of the hydrodynamic drag to enhance the selectivity of our probes without compromising the sensitivity. In Cheng et al., the cusp geometry of oligo-functionalized microbead assembly was used to enhance the shear rate and the dielectric force at the cusp [62]. We were able to balance the shear force of flow with a trapping dielectric force achieve such a high selectivity that we can discriminate against a onemismatch target (in a 26-bp pairing sequence) and yet retain high sensitivity (pM or 1 million copies with a 106 concentration factor) [62].
Another design is to use the high aspect ratio of CNTs (>100), such that its hydrodynamic drag in a transverse flow field is higher than that of sphere of the same radius by a factor that is the logarithm of the aspect ratio. In Fig. 7D, we are able to selectively remove isoforms with similar affinity as the target (the dissociation constant is off by no more than a factor of 10) to get a selectivity of higher than 103—the signal from the isoform at nM is 10 times lower than that of the target at 10 pM and this signal of the target remains roughly the same with a mixture of both isoform and target at the above disproportional ratios. This difference is only observed after shear. It is hence a dramatic demonstration of how shear can improve selectivity by orders of magnitude over static immunoassays.
The same shear-enhanced selectivity should also be implementable to the nanomagnetic beads trapped at the pore edge of magnetic film covered track-etched membranes in Fig. 7. The flow field, like the magnetic field, varies quite a bit at such corners, thus producing a high shear rate and a large drag force as we have seen at the cusp of the assembled oligo-functionalized nanoparticles [62]. This may allow selective detachment of nontarget EVs that would then be convected through the pores with the EVs without the nanomagnetic beads. Such selectivity optimization has not been reported. The shear rate at corners, such as cusps or edges, is infinitely large in principle and hence can be used to enhance selectivity.
Since the free-flow electrophoretic velocity of EVs and molecules in typical capillary and gel electrophoresis is no larger than 0.1 mm/s, the electric force imparted on them would hence need to be increased by more than 10–100 times to pull them off an immobilized probe. This can be done with the high field in the depletion region. Marczak et al. were able to achieve a selectivity of nearly 1000 for two mismatches in a 35 bp pairing sequence of a 69 base ssDNA target using this enhanced electric force [60]. We are able to quantify the target to a limit of detection of 10 pM even if the twomismatch non-target outnumbers the target by a factor 104. It is a good example of how microfluidics allows optimization of the force, electric in this case, to enhance selectivity without compromising sensitivity. The dissociation constants of the target and the non-target with two mismatches at specific location were estimated by melting curve analysis to be 42 and 400 nM, respectively, with about a 2kT difference in the hybridization Gibbs energy. We have hence increased the sensitivity and selectivity based on equilibrium values by factors of 103 by using the depletion front for both concentrating the analyte for the sensor and enhancing the selectivity of the sensor. The electric field has presumably rendered the dehybridization reaction for the nontarget irreversible while retaining the target within a 2 kT energy minimum.
There are other mechanisms to enhance sensor selectivity. Thermal programming, extreme pH dissociation, and ionic strength shock are all “equilibrium” approaches that do not involve force fields. They may be easier to implement for certain applications and are gentler. However, they are typically too slow and do not produce a sufficiently high selectivity. Force-induced selectivity has a major advantage: it is irreversible. Because of flow and field removal, the reverse hybridization/complexification reaction is eliminated even if it has a low barrier. Once detached, the non-targets have zero probability of returning to the immobilized probe. Force hence renders the process non-equilibrium to allow winnowing of nontargets that would otherwise reattach with finite probability and reduce the selectivity. We have developed a theory to delineate such kinetic from thermodynamic equilibrium selectivity enhancement mechanisms in ion-depleted zones produced by ion-selective membranes [60]. Obviously, ion depletion with a higher field and a lower ionic strength to reduce the screening of target-probe electrostatic repulsion could introduce both thermodynamic and kinetic mechanisms to achieve the highest selectivity. Similarly, a combination of thermophoresis and melting due to controlled Ohmic heating in a depletion zone can also enhance selectivity more than each by itself. This has not been attempted before and is a worthwhile future direction to achieve very selective quantification of cancer markers in a sea of other more abundant molecules with similar affinity for the probes.
Because there are typically a multitude of biomarkers, these sensors must be scaled up to a multitarget platform. Also, the optimum force to apply to each sensor will vary because of the different affinities of the targets and nontargets in each sensor. For example, the location of the single mismatch in the 26 pairing sequence produced a different optimum shear rate for selectivity enhancement [62]. We hence see the development of multitarget sensor arrays with individualized actuators as an important future direction. Obviously, the depletion-front concentration modules of Fig. 2 can also benefit from such multisensor/actuator design with a distributed control architecture.
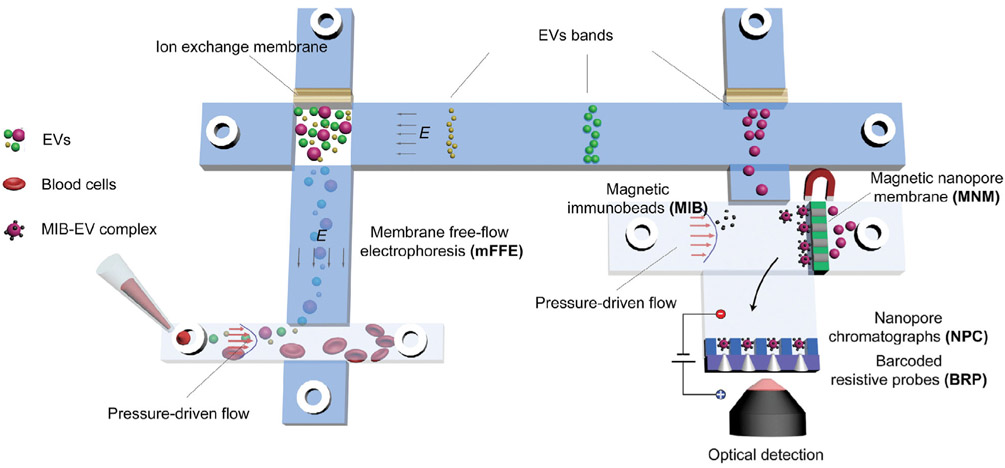
5. Future direction: Integration and multiplexing
We hope the prior sections have outlined the required technologies for liquid biopsy, i.e., pretreatment and sensing technologies for selective fractionation of the nanocarriers and quantification of their molecular cargo. To avoid analyte loss, these pretreatment and sensing modules of the liquid biopsy platform are best integrated with little dead volume on a single microfluidic chip. Such integration will be areas of active research in the future. It would also be preferable that negligible fluid is introduced at midstream to avoid dilution and dispersion. Probes, nanomagnetic beads, and pH/ionic strength changes are best done with little buffer addition. Consequently, electric and magnetic field activation we have reviewed is preferred. The ion depletion feature of ion-selective membranes is particularly advantageous because of its analyte extraction/concentration, high-field actuation, and pH generation features. Several of such membrane and gel modules on the chip essentially render our chip an ionic circuit. One can then use output from such sensors to control the various modules in a massively automated design. The same circuitry can be utilized to optimize the force at each sensor to enhance the selectivity of individual sensor for its specific target. We envision a platform such as the one shown in Fig. 8 with a distributed sensor/actuator system controlled by both a layer of electronics at the bottom and perhaps with wireless connectivity for more precise online control. Such an integrated liquid biopsy system would truly be a Lab Chip. However, integration is not the main difficulty. The key obstacle is the proper design that will enable rapid and selective quantification of multiple markers in a small heterogeneous sample with probe-based technologies. Many of the numerous potential cancer biomarkers may then be targeted with the integrated liquid biopsy platform. The return of our astronomical investment in discovering them could then be realized through membrane microfluidics. Finally, we observe that membrane microfluidics is quite amenable to rapid prototyping and scalable manufacturing, such that the price of the final platform will be much lower than the current technologies—about $0.5 million each for ultra-centrifugation, AF4, and other stand-alone instrumentation. For example, each biochip containing the membrane microfluidics is expected to cost less than $1, compared to >$50 per sample cost of current extraction kits.
Figure 8.
A simple integrated platform with EV fractionation pretreatment, capture of specific EVs by nanomagnetic beads and a multitarget sensor array with probe-based quantification and force-enhanced selectivity enhancement.
Acknowledgments
This work in the Chang lab has been supported in part by NSF-CBET 1065652, USDA 2012-67005-19589, NIH 1R21AI105361-01A1, NIH 1R21CA206904-01, NIH HG009010-01 and the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director, 1UG3CA241684-01.
Abbreviations:
- AC
alternating current
- AF4
asymmetric-flow field-flow fractionation
- ANM
asymmetric nanopore membrane
- cDNA
complementary DNA
- CIF
continuous isoelectric fractionation
- CNT
carbon nanotube
- CTCs
circulating tumor cells
- DC
direct current
- EpCAM
epithelial cell adhesion molecule
- EV
extracellular vesicle
- exRNA
extracellular RNA
- FDA
Food and Drug Administration
- FISH
fluorescence in-situ hybridization
- HDL
high-density lipoprotein
- kT
Boltzmann constant
- mRNA
messenger RNA
- NDLD
nanoscale deterministic lateral displacement
- PSA
prostate specific antigen
- RNA
ribonucleic acid
- SDS
sodium dodecyl sulfate
- SEC
size exclusion chromatography
- SERS
surface-enhanced Raman scattering
- TEM
transmission electron microscope
- UC
ultracentrifugation
- V
volt
6. References
- [1].Füzéry AK, Levin J, Chan MM, Chan DW, Clin. Proteomics 2013, 10, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kirwan A, Utratna M, O’Dwyer ME, Joshi L, Kilcoyne M, BioMed. Res. Int 2015, 2015, 490531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Egatz-Gomez A, Wang C, Klacsmann F, Pan Z, Marczak S, Wang Y, Sun G, Senapati S, Chang H-C, Biomicrofluidics 2016, 10, 032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bonneau E, Neveu B, Kostantin E, Tsongalis G, De Guire V, EJIFCC 2019, 30, 114. [PMC free article] [PubMed] [Google Scholar]
- [5].Fredolini C, Byström S, Sanchez-Rivera L, loannou M, Tamburro D, Ponten F, Branca RM, Nilsson P, Lehtio J, Schwenk JM, Sci. Rep 2019, 9, 8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Huse M, Nat. Rev. Immunol 2017, 17, 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pageon SV, Govendir MA, Kempe D, Biro M, Mol. Biol. Cell 2018, 29, 1919–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yelleswarapu V, Buser JR, Haber M, Baron J, Inapuri E, Issadore D, Proc. Natl. Acad. Sci. USA 2019, 116, 4489–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fuchs RT, Sun Z, Zhuang F, Robb GB, PLoS One 2015, 10, e0126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang C, Sun G, Senapati S, Chang H-C, Lab Chip 2019, 19,3853–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Koga K, Matsumoto K, Akiyoshi T, Kubo M, Yamanaka N, Tasaki A, Nakashima H, Nakamura M, Kuroki S, Tanaka M, Anticancer Res. 2005, 25, 3703–3707. [PubMed] [Google Scholar]
- [12].Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Cell 2019, 177, 428–445, e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Q, Higginbotham JN, Jeppesen DK, Yang Y-P, Li W, McKinley ET, Graves-Deal R, Ping J, Britain CM, Dorsett KA, Cell Rep. 2019, 27, 940–954. e946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Buschmann D, Kirchner B, Hermann S, Marte M, Wurmser C, Brandes F, Kotschote S, Bonin M, Steinlein OK, Pfaffl MW, J. Extracell. Vesicles 2018, 7, 1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, Bracke M, De Wever O, Hendrix A, J. Extracell. Vesicles 2014, 3, 24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yamada T, Inoshima Y, Matsuda T, Ishiguro N, J. Vet. Med. Sci 2012, 12–0032. [DOI] [PubMed] [Google Scholar]
- [17].Mathieu M, Martin-Jaular L, Lavieu G, Thery C, Nat. Cell Biol 2019, 21, 9–17. [DOI] [PubMed] [Google Scholar]
- [18].Momen-Heravi F, Balaj L, Alian S, Mantel P-Y, Halleck AE, Trachtenberg AJ, Soria CE, Oquin S, Bonebreak CM, Saracoglu E, Biol. Chem 2013, 394, 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ, Methods 2012, 56, 293–304. [DOI] [PubMed] [Google Scholar]
- [20].Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Nat. Cell Biol 2018, 20, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Smith JT, Wunsch BH, Dogra N, Ahsen ME, Lee K, Yadav KK, Weil R, Pereira MA, Patel JV, Duch EA, Lab Chip 2018, 18, 3913–3925. [DOI] [PubMed] [Google Scholar]
- [22].Zhao Z, Yang Y, Zeng Y, He M, Lab Chip 2016, 16, 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ha Y, Ko S, Kim I, Huang Y, Mohanty K, Huh C, Maynard JA, ACSAppl. Nano Mater 2018, 1,512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chang H, Yeo L Electrokinetically driven microfluidics and nanofluidics, Cambridge University Press, London: 2009. [Google Scholar]
- [25].Slouka Z, Senapati S, Chang H-C, Annu. Rev. Anal. Chem 2014, 7,317–335. [DOI] [PubMed] [Google Scholar]
- [26].Yossifon G, Mushenheim P, Chang H-C, Europhys. Lett 2010, 90, 64004. [Google Scholar]
- [27].Sun G, Pan Z, Senapati S, Chang H-C, Phys. Rev. Appl 2017, 7, 064024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cheung LS, Wei X, Martins D, Song Y-A, Biomicrofluidics 2018, 12,014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Berzina B, Anand RK, Lab Chip 2019, 19, 2233–2240. [DOI] [PubMed] [Google Scholar]
- [30].Sun G, Wan J, Lu H, Biomicrofluidics 2019, 13, 064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Park S, Abu-Rjal R, Rosentsvit L, Yossifon G, ACS Sens. 2019, 4, 1806–1815. [DOI] [PubMed] [Google Scholar]
- [32].Yin Z, Ramshani Z, Waggoner JJ, Pinsky BA, Senapati S, Chang H-C, Sens. Actuators B 2020, 127854. [Google Scholar]
- [33].Chang H-C, Yossifon G, Demekhin EA, Annu. Rev. Fluid Mech 2012, 44, 401–426. [Google Scholar]
- [34].Sun G, Senapati S, Chang H-C, Lab Chip 2016, 16, 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Taller D, Richards K, Slouka Z, Senapati S, Hill R, Go DB, Chang H-C, Lab Chip 2015, 15,1656–1666. [DOI] [PubMed] [Google Scholar]
- [36].Ramshani Z, Zhang C, Richards K, Chen L, Xu G, Stiles BL, Hill R, Senapati S, Go DB, Chang H-C, Nat. Commun. Biol 2019, 2, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Marczak S, Senapati S, Slouka Z, Chang H-C, Biosens. Bioelectron 2016, 86, 840–848. [DOI] [PubMed] [Google Scholar]
- [38].Davies RT, Kim J, Jang SC, Choi E-J, Gho YS, Park J, Lab Chip 2010, 12, 5202–5210. [DOI] [PubMed] [Google Scholar]
- [39].Cho S, Jo W, Heo Y, Kang JY, Kwak R, Park J, Sens. Actuators B 2016, 233, 289–297. [Google Scholar]
- [40].Marczak S, Richards K, Ramshani Z, Smith E, Senapati S, Hill R, Go DB, Chang HC, Electrophoresis 2018, 39, 2029–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bhattacharjee S, De S, Int. J. Heat Mass Transfer 2018, 118, 116–128. [Google Scholar]
- [42].Liu F, Vermesh O, Mani V, Ge TJ, Madsen SJ, Sabour A, Hsu E-C, Gowrishankar G, Kanada M, Jokerst JV, ACS Nano 2017, 11, 10712–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Horie T, Shiota S, Akagi T, Ohmura N, Wang S, Eze V, Harvey A, Hirata Y, J. Membr. Sci 2018, 554, 134–139. [Google Scholar]
- [44].Jochem A-R, Ankah GN, Meyer L-A, Elsenberg S, Johann C, Kraus T, Anal. Chem 2016, 88,10065–10073. [DOI] [PubMed] [Google Scholar]
- [45].Lee K, Shao H, Weissleder R, Lee H, ACS Nano 2015, 9, 2321–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang C, Fu Q, Wang X, Kong D, Sheng Q, Wang Y, Chen Q, Xue J, Anal. Chem 2015, 87, 8227–8233. [DOI] [PubMed] [Google Scholar]
- [47].Ghosal S, Sherwood JD, Chang H-C, Biomicrofluidics 2019, 13, 011301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Akagi T, Kato K, Kobayashi M, Kosaka N, Ochiya T, Ichiki T, PLoS One 2015, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Camus M, Chapman M, Forgez P, Laplaud P, J. Lipid Res 1983, 24, 1210–1228. [PubMed] [Google Scholar]
- [50].Dreyfuss G, Annu. Rev. Cell Biol 1986, 2, 459–498. [DOI] [PubMed] [Google Scholar]
- [51].Kubota K, Kubo T, Tanigawa T, Naito T, Otsuka K, Sci. Rep 2017, 7, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cheng L-J, Chang H-C, Biomicrofluidics 2011, 5, 046502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cheng L-J, Chang H-C, Lab Chip 2014, 14, 979–987. [DOI] [PubMed] [Google Scholar]
- [54].Clayton A, Court J, Navabi H, Adams M, Mason M, Hobot J, Newman G, Jasani B, J. Immunol. Methods 2001, 247, 163–174. [DOI] [PubMed] [Google Scholar]
- [55].Ko J, Bhagwat N, Yee SS, Ortiz N, Sahmoud A, Black T, Aiello NM, McKenzie L, O’Hara M, Redlinger C, ACS Nano 2017, 11, 11182–11193. [DOI] [PubMed] [Google Scholar]
- [56].Basuray S, Senapati S, Aijian A, Mahon AR, Chang H-C, ACS Nano 2009, 3, 1823–1830. [DOI] [PubMed] [Google Scholar]
- [57].Li D, Wang C, Sun G, Senapati S, Chang H-C, Biosens. Bioelectron 2017, 97, 143–149. [DOI] [PubMed] [Google Scholar]
- [58].Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H, Nat. Med 2012, 18, 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yan Z, Dutta S, Liu Z, Yu X, Mesgarzadeh N, Ji F, Bitan G, Xie Y-H, ACS Sens. 2019, 4, 488–497. [DOI] [PubMed] [Google Scholar]
- [60].Marczak S, Smith E, Senapati S, Chang HC, Electrophoresis 2017, 38, 2592–2602. [DOI] [PubMed] [Google Scholar]
- [61].De Valença J, Jõgi M, Wagterveld RM, Karatay E, Wood JA, Lammertink RG, Langmuir 2018, 34,2455–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cheng I-F, Senapati S, Cheng X, Basuray S, Chang H-C, Chang H-C, Lab Chip 2010, 10, 828–831. [DOI] [PubMed] [Google Scholar]