Abstract
Background
The authors sought to forecast survival and enhance treatment decisions for patients with liver metastatic colorectal cancer by using on-treatment radiomics signature to predict tumor sensitiveness to irinotecan, 5-fluorouracil, and leucovorin (FOLFIRI) alone (F) or in combination with cetuximab (FC).
Methods
We retrospectively analyzed 667 metastatic colorectal cancer patients treated with F or FC. Computed tomography quality was classified as high (HQ) or standard (SD). Four datasets were created using the nomenclature (treatment) – (quality). Patients were randomly assigned (2:1) to training or validation sets: FCHQ: 78:38, FCSD: 124:62, FHQ: 78:51, FSD: 158:78. Four tumor-imaging biomarkers measured quantitative radiomics changes between standard of care computed tomography scans at baseline and 8 weeks. Using machine learning, the performance of the signature to classify tumors as treatment sensitive or treatment insensitive was trained and validated using receiver operating characteristic (ROC) curves. Hazard ratio and Cox regression models evaluated association with overall survival (OS).
Results
The signature (area under the ROC curve [95% confidence interval (CI)]) used temporal decrease in tumor spatial heterogeneity plus boundary infiltration to successfully predict sensitivity to antiepidermal growth factor receptor therapy (FCHQ: 0.80 [95% CI = 0.69 to 0.94], FCSD: 0.72 [95% CI = 0.59 to 0.83]) but failed with chemotherapy (FHQ: 0.59 [95% CI = 0.44 to 0.72], FSD: 0.55 [95% CI = 0.43 to 0.66]). In cetuximab-containing sets, radiomics signature outperformed existing biomarkers (KRAS-mutational status, and tumor shrinkage by RECIST 1.1) for detection of treatment sensitivity and was strongly associated with OS (two-sided P < .005).
Conclusions
Radiomics response signature can serve as an intermediate surrogate marker of OS. The signature outperformed known biomarkers in providing an early prediction of treatment sensitivity and could be used to guide cetuximab treatment continuation decisions.
Colorectal cancer (CRC) is a leading cause of cancer death globally. Liver metastasis affects more than one-half of CRC patients (1). Anti–epidermal growth factor receptor (EGFR) therapies, including tyrosine kinase inhibitors and monoclonal antibodies, demonstrate activity in both CRC and other tumor types. In metastatic CRC (mCRC) patients, the assessment of anti-EGFR monoclonal antibody efficacy relies on computed tomography (CT) scan response endpoints. The on-treatment shrinkage of metastases on CT scans is considered a hallmark of EGFR signaling pathway dependency (2–5) and of treatment sensitivity (6). In unresectable mCRC, tumor shrinkage guides the clinical decision to pursue a curative opportunity (downstage to resection) or palliative treatment to maximize overall survival (OS) and improve quality of life by symptom relief (1). As the decision to continue EGFR-targeted therapy must balance the risks and potential rewards of treatment, there is a pressing need for biomarkers that can estimate the likelihood of clinical benefit in individual patients.
We sought to meet this need for alternative biomarkers by using artificial intelligence (AI). We utilized machine learning to create an AI signature that evaluated a change in tumor phenotype between baseline and 8 weeks on CT-scan images to predict clinical outcome (OS). Radiology is undergoing rapid change because of remarkable advances in the field of AI, particularly algorithms for machine learning and deep learning, which enable automated, high-throughput quantification of medical images as a set of quantitative features. Traditional radiomic features are identified according to a priori definitions (eg, tumor diameter or density), and artificial neural networks adaptively define deep learning features as spatial hierarchies of representations. The dataset of radiomic and deep learning features can be mined by machine learning to identify statistically significant associations between variables of interest, such as clinical outcome or tumor mutational status. This big data approach is applicable to any imaging modality but requires large and consistent datasets, which CT scanning currently best provides because of its use as a standard of care in oncology. In particular, the widespread adoption of CT measurement of tumor diameter (eg, RECIST 1.1) as an endpoint in clinical trials has generated an invaluable resource for AI research. The current study leverages this resource by analyzing data from the multicenter clinical trial NCT00154102.
AI techniques allow objective and reproducible analysis of CT image characteristics, including imaging features not apparent to the human eye. The overall goal of this study is to determine whether AI techniques can offer oncologists additional clinical information regarding response assessment. Our test case is the decision to continue anti-EGFR treatment. We utilized data from mCRC patients treated with cetuximab in combination with FOLFIRI (irinotecan, 5-fluorouracil, and leucovorin) in the randomized multicenter CRYSTAL trial (NCT00154102). Data from this trial were used to obtain regulatory approval for the clinical use of cetuximab in mCRC. Pretreatment selection is currently based on a biopsy sample (primary tumor or metastasis) or on liquid biopsy. In approximately one-half of mCRCs patients (7), a RAS wild-type mutational status (KRAS and NRAS exons 2, 3, 4) guides the clinical use of cetuximab in addition to either FOLFIRI (8–11) or FOLFOX (12). This treatment has been found to prolong median OS by 8 months in RAS wild type (10) and increase overall response rate, but also to increase the rate of adverse events (8). However, several tumor biopsy–driven clinical trials have shown the limitations of predicting clinical benefit from EGFR-targeted therapies in mCRC and other solid tumors based on a single biomarker (13). The exception is the presence of EGFR mutation in non-small cell lung cancer, which does predict a higher response rate and prolonged survival (13).
To determine whether an AI-driven signature could guide clinicians to continue EGFR-targeted therapies on an individualized basis, we aimed to develop and validate an on-treatment signature detecting mCRC patients sensitive to FOLFIRI+cetuximab (FC) using quantitative assessment of tumor changes between baseline and 8-week CT images.
Methods
Study Design
Our primary endpoint was to train and validate an on-treatment signature detecting mCRC patients sensitive to FC. Our secondary endpoint was to generalize the on-treatment signature to other datasets with different quality of imaging settings and treatment types (Figures 1 and 2).
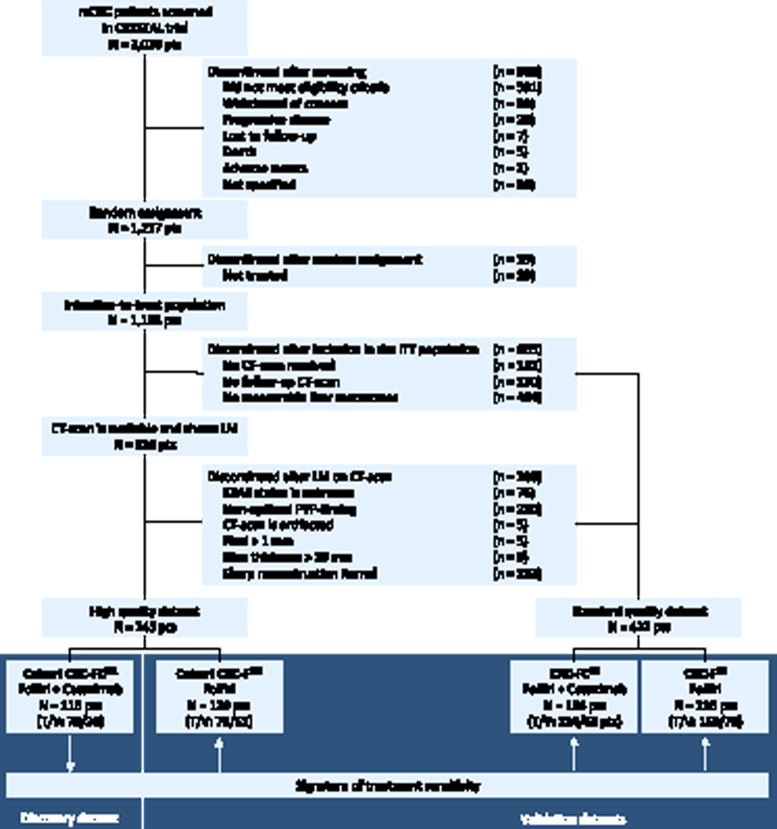
Figure 1.
CONSORT diagram. Patients could be excluded for multiple reasons. The withdrawal boxes show the number of patients excluded at each step. LM = liver metastasis; T = training set; V = validation set. The artificial intelligence (AI) signature was developed (training set) and validated (validation set) in the FOLFIRI+cetuximab discovery cohort with high-quality (HQ) dataset.
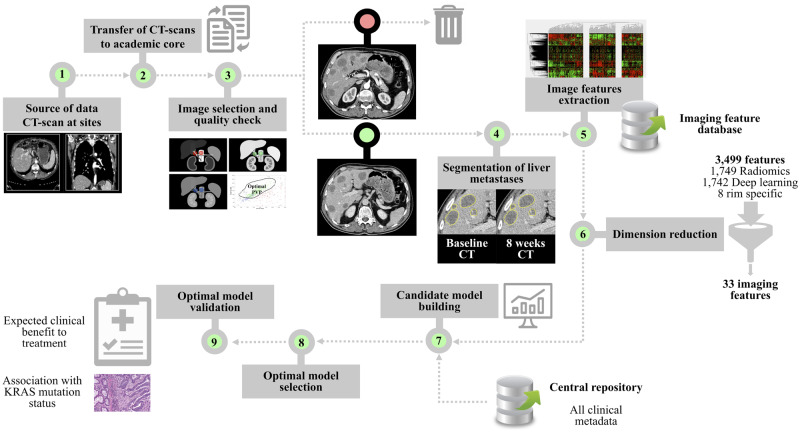
Figure 2.
Artificial intelligence (AI) workflow. Steps 1–2: Computed tomography (CT) scans acquired at study sites are transferred to our academic core. Step 3. Image selection and quality check using a computer-aided algorithm designed by machine-learning. Step 4. Segmentation of liver metastases on CT scan by an expert radiologist at baseline and 8 weeks in each patient. Step 5. Combination of all segmented lesions to compute a tumor imaging phenotype in each patient based on imaging features extraction in each segmented liver metastasis (3499 imaging features characterizing changes between baseline and 8 weeks). Step 6. Dimension reduction using machine learning. Identification of reproducible, nonredundant and informative candidate imaging features for model building. Step 7. Candidate model building to enhance strategic decision-making (training set). Step 8. Optimal model selection in the training set using threefold cross-validation to evaluate the performance of candidate models in terms of area under the receiver operating characteristic curve. Step 9. Signature validation (validation set).
Participants and Datasets
We retrospectively analyzed one clinical trial involving two treatment arms and 667 CRC patients. The two treatment arms involved anti-EGFR treatment regimens (cetuximab: C) and FOLFIRI (F). The CRYSTAL trial (NCT00154102) was a randomized, open-label, multicenter phase III trial comparing FC (cohort FC) with FOLFIRI alone (cohort F) in the first-line treatment of mCRC (Table 2). Data were gathered up to the clinical trials completion date. Patients with missing data were excluded. More information on this clinical trial can be found in the Supplementary Methods (available online).
Table 2.
Patient characteristics in the CRYSTAL trial (NCT00154102)*
| Patient characteristics | LM mCRC patients with liver metastases | SD quality Cohorts CRC-FCSD and CRC-FSD No. (%) |
|||
|---|---|---|---|---|---|
| HQ quality | |||||
| Cohort CRC-FCHQ-FC |
Cohort CRC-FHQ FOLFIRI |
||||
| Training No. (%) | Validation No. (%) | Training No. (%) | Validation No. (%) | ||
| Patients, no. | 78 | 38 | 78 | 51 | 288 |
| Gender | |||||
| Male | 50 (64.1) | 29 (76.3) | 51 (64.1) | 31 (60.8) | 184 (63.9) |
| Female | 28 (35.9) | 9 (23.7) | 27 (34.6) | 20 (39.2) | 104 (36.1) |
| Primary site | |||||
| Right colon | 20 (25.6) | 14 (36.8) | 20 (25.6) | 17 (33.3) | 70 (24.3) |
| Left colon | 58 (74.4) | 24 (63.2) | 58 (74.4) | 34 (66.7) | 217 (75.3) |
| Prior adjuvant chemotherapy | |||||
| Yes | 9 (11.5) | 6 (15.8) | 10 (12.8) | 6 (11.8) | 40 (13.9) |
| No | 69 (88.5) | 32 (84.2) | 68 (87.2) | 45 (88.2) | 248 (86.1) |
| Leucocytes | |||||
| Low | 65 (83.3) | 35 (92.1) | 65 (83.3) | 34 (66.7) | 234 (81.3) |
| High | 12 (15.4) | 3 (7.9) | 10 (12.8) | 15 (29.4) | 45 (15.6) |
| LDH baseline | |||||
| Low | 41 (52.6) | 18 (47.4) | 28 (35.9) | 21 (41.2) | 147 (51.0) |
| High | 29 (37.2) | 15 (39.5) | 36 (46.2) | 21 (41.2) | 111 (38.5) |
| Unknown | 8 (10.3) | 5 (13.2) | 14 (17.9) | 9 (17.6) | 30 (10.4) |
| Maximum staining intensity | |||||
| Low | 13 (16.7) | 1 (2.6) | 12 (15.4) | 7 (13.7) | 48 (16.7) |
| High | 65 (83.3) | 37 (97.4) | 65 (83.3) | 44 (86.3) | 238 (82.6) |
| Histology of primary tumor | |||||
| Adenocarcinoma | 72 (92.3) | 35 (92.1) | 76 (97.4) | 50 (98.0) | 283 (98.3) |
| Mucinous adenocarcinoma | 6 (7.7) | 3 (7.9) | 2 (2.6) | 1 (2.0) | 5 (1.7) |
| Histological differentiation | |||||
| Well differentiated | 10 (12.8) | 4 (10.5) | 15 (19.2) | 7 (13.7) | 33 (11.5) |
| Moderately differentiated | 51 (65.4) | 24 (63.2) | 50 (64.1) | 33 (64.7) | 191 (66.3) |
| Poorly differentiated | 15 (19.2) | 6 (15.8) | 12 (15.4) | 8 (15.7) | 44 (15.3) |
| KRAS BRAF mutation status | |||||
| KRAS mutant | 25 (32.1) | 19 (50.0) | 26 (33.3) | 17 (33.3) | 75 (26.0) |
| KRAS and BRAF WT | 50 (64.1) | 15 (39.5) | 46 (59.0) | 31 (60.8) | 129 (44.8) |
| BRAF mutant | 3 (3.8) | 4 (10.5) | 6 (7.7) | 3 (5.9) | 8 (2.8) |
| Unknown | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 76 (26.4) |
| Target lesions at baseline, no. | |||||
| 0 | 0 (0.0) | 1 (2.6) | 0 (0.0) | 1 (2.0) | 2 (0.7) |
| 1 | 5 (6.4) | 5 (13.2) | 12 (15.4) | 3 (5.9) | 31 (10.8) |
| 2 | 9 (11.5) | 10 (26.3) | 13 (16.7) | 11 (21.6) | 45 (15.6) |
| 3 | 14 (17.9) | 7 (18.5) | 18 (23.1) | 11 (21.6) | 44 (15.3) |
| 4 | 13 (16.7) | 4 (10.5) | 5 (6.4) | 6 (11.8) | 43 (14.9) |
| 5 | 24 (30.8) | 7 (18.5) | 16 (20.5) | 10 (19.6) | 70 (24.3) |
| >5 | 14 (17.9) | 4 (10.5) | 14 (17.9) | 10 (19.6) | 53 (18.4) |
| Nontarget lesions at baseline, no. | |||||
| 0 | 17 (21.8) | 16 (42.1) | 20 (25.6) | 16 (31.4) | 77 (26.7) |
| 1 | 35 (44.9) | 8 (21.1) | 25 (32.1) | 16 (31.4) | 90 (31.3) |
| 2 | 10 (12.8) | 4 (10.5) | 22 (28.2) | 11 (21.6) | 50 (17.4) |
| 3 | 6 (7.7) | 6 (15.8) | 6 (7.7) | 4 (7.8) | 19 (6.6) |
| 4 | 2 (2.6) | 1 (2.6) | 2 (2.6) | 1 (2.0) | 13 (4.5) |
| 5 | 4 (5.1) | 2 (5.3) | 2 (2.6) | 1 (2.0) | 18 (6.3) |
| >5 | 4 (5.1) | 1 (2.6) | 1 (1.3) | 2 (3.9) | 21 (7.3) |
| Only liver metastasis | 57 (73.1) | 27 (71.1) | 49 (62.8) | 37 (72.5) | 211 (73.3) |
| Only liver and lung metastasis | 63 (80.8) | 33 (86.8) | 73 (93.6) | 45 (88.2) | 253 (87.8) |
Liver metastatic patients included in the HQ cohorts have similar characteristics. The only difference is the quality of the CT scan acquisition protocol and reconstruction settings. The distribution of clinical characteristics for each subgroup of patients was compared using analysis of variance and χ2 (P values) and did not demonstrate any statistically significant difference between the cohorts. CT = computed tomography; CRC = colorectal cancer; F = FOLFIRI; FC = FOLFIRI+cetuximab; HQ = High computed tomography quality; SD = Standard computed tomography quality.
We have evaluated the quality of CT-scan acquisition. More information about CT-scan characteristics, CT-scan quality (14–16), and radiomics quality score (17) can be found in Supplementary Methods (available online). Our goal was to create the signature in a dataset with high signal-to-noise ratio because of high imaging quality (HQ dataset) and evaluate if it can be more universally applicable to a dataset with standard quality (SD).
Using the nomenclature (tumor type) – (treatment) – (quality of the dataset), the two cohorts (CRC-FC, CRC-F) were divided into four datasets and patients (patients) were randomly assigned to training and validation sets using a 2:1 ratio (CRC-FCHQ: 78:38 patients, CRC-FCSD: 124:62 patients, CRC-FHQ: 78:51 patients, CRC-FSD: 158:78 patients) (Table 1; Figure 1).
Table 1.
Patient characteristics and cohorts in the CRYSTAL trial (NCT00154102)
| Treatment arm | Cohort FOLFIRI plus cetuximab | Cohort FOLFIRI alone |
|---|---|---|
| No. | Dataset CRC-FCHQ: 116 patients | Dataset CRC-FHQ: 129 patients |
| Dataset CRC-FCSD: 186 patients | Dataset CRC-FSD: 236 patients | |
| Tumor characteristics | ||
| Tumor type | CRC | CRC |
| Stage | Advanced | Advanced |
| Lesion(s) segmented | Liver metastases | Liver metastases |
| Clinical trial characteristics | ||
| Biomarker | KRAS* WT mutational status | KRAS* WT mutational status |
| Treatment regimen | ||
| Chemotherapy | FOLFIRI | FOLFIRI |
| Anti-EGFR | Cetuximab (anti-EGFR mAb) | None |
| Primary endpoint | ||
| Outcome | OS, PFS | OS, PFS |
| Sensitivity to anti-EGFR mAb | OS benefit compared with FOLFIRI alone | N/A |
| Time of completion of CT scans | Baseline and 8 weeks | Baseline or 8 weeks |
| Clinical trial no. | NCT00154102 | NCT00154102 |
KRAS status and not RAS status was evaluated post hoc in the CRYSTAL clinical trial. CRC = colorectal cancer; F = FOLFIRI; FC = FOLFIRI+cetuximab; HQ = High computed tomography quality; SD = Standard computed tomography quality; mAb = monoclonal antibody.
Reference Standard: Sensitivity vs Resistance to Systemic Cancer Therapy
The signature was trained and validated to predict tumor sensitivity to systemic anticancer treatment. All cancer patients were divided into two groups: sensitive to treatment and not sensitive (resistant) to treatment (Supplementary Methods, available online).
In mCRC patients (datasets CRC-FCHQ, CRC-FCSD, CRC-FHQ, CRC-FSD), the reference standard to determine tumor sensitivity to treatment was OS. Patients were considered resistant if they died before 17.7 months (median OS in the training set) and were considered sensitive otherwise. The failure event for OS was defined as death due to any cause. Survival time was measured from the date of random assignment to the date of death or last follow-up (censored observation). No included patients were censored before 17.7 months: all patients were followed beyond 17.7 months unless they died due to any cause.
Predictors Used in the Signature: Radiomics and Deep-Learning Features
In total, 3499 quantitative image features including 1757 radiomics features and 1742 pretrained deep-learning features were used to characterize the early CT changes in tumor phenotype. The full details of lesion segmentation and feature extractions can be found in the Supplementary Methods (available online).
To reduce overfitting, a radiomics signature integrated up to four of these features. These four were those ranked highest by the machine learning algorithm in terms of prediction importance (18) using the best predictive model developed in the training set of the discovery cohort CRC-FCHQ. The full details of model building can be found in the Supplementary Methods (available online).
Signature Building in Cohort CRC-FCHQ
In the training set of the discovery cohort (CRC-FCHQ), we developed a multivariable prediction model (ie, the signature, to predict tumor sensitivity to treatment) (see Figure 2). In the implementation, a “coarse” to “fine” strategy was developed to select optimal features from the large number of extracted quantitative image features to build the signature. The coarse selection approach consists of three processes (reproducibility analysis, redundancy analysis, and feature ranking) aiming to screen many nonreproducible, redundant, and noninformative candidate features. The fine selection approach consists of a “forward” search and feature combination, aiming to select optimal features and build the final optimal model.
Signature Calibration in Cohorts CRC-FCSD, CRC-FHQ, and CRC-FSD
The four imaging biomarkers identified in the signature developed in the training set of CRC-FCHQ were computed in the training sets of the cohorts CRC-FCSD, CRC-FHQ, and CRC-FSD. The signature developed in the training set of CRC-FCHQ was transferred and calibrated to predict tumor sensitivity to treatment in the training sets of the cohorts CRC-FCSD, CRC-FHQ, and CRC-FSD as described in Supplementary Methods (available online).
Validation of the Signature
The primary endpoint was to evaluate the performance of the signature to predict tumor sensitivity to treatment. The performance was measured using area under the receiver operating characteristic curve (AUC) in the validation sets of the four cohorts consisting of patients that were not used for training.
The methodology used for secondary endpoints is described in Supplementary Methods I.6.1 (available online).
Statistical Analysis
Statistical analysis was conducted using Matlab (version 2016 b; Mathworks, Natick, MA) and SPSS (version 23.0; IBM; Armonk, NY). AUC and AUC’s 95% confidence interval (CI) were used to indicate classification performance. Analysis of variance was performed to compare continuous variables, and χ2 test was performed to compare categorical variables. Cox regression was used to investigate the effect of survival variables, and log-rank test was used to compare survival times of two groups. A P less than .05 was used to determine statistical significance. All tests were two-sided.
Results
Participants
Table 2 shows that no statistically significant differences were detected by the post hoc exploratory tests performed between cohorts CRC-FCHQ, CRC-FCSD, CRC-FHQ, and CRC-FSD and between training and validation sets for each treatment arm. Supplementary Figures 2 and 3 (available online) show tumor imaging-phenotyping using unsupervised analysis. Supplementary Table 3 (available online) shows that features were different between HQ and SD datasets; hence there was a need for a calibration in each cohort.
Performance of the Signature in Cohort CRC-FCHQ
Of 116 patients, 95 patients died during the follow-up. The median follow-up duration was 21.8 months overall and 46.7 months (range = 22.4–55.0) in censored patients. All patients with censored survival data had follow-up longer than 17.7 months and belonged to the low-risk group.
In the training set, the best predictive model was built using a Random Forest algorithm (18) and 4 Radiomics features (Figures 3). The performance of the signature to diagnose treatment-sensitive tumors was AUC = 0.83 (95% CI = 0.75 to 0.92); sensitivity = 0.77, and specificity = 0.85. 4 Radiomics features were included in the signature (Table 3;Figures 3): Shape SI4, LoG Z Entropy, LoG X Entropy, and GTDM Contrast. These four features were transferred to all cohorts. Shape SI4 characterizes the local curvatures in surface shape of a three-dimensional object. Entropy is a metric of the disorder or heterogeneity of the signal that is computed after the application of a Gaussian smoothing filter to reduce noise and a Laplacian operator to highlight regions of rapid gray-level change in images. Gray Tone Difference Matrix features describe visual properties of texture based on gray-tone difference between a pixel and its neighborhood. Definitions are presented in Supplementary Methods (available online).
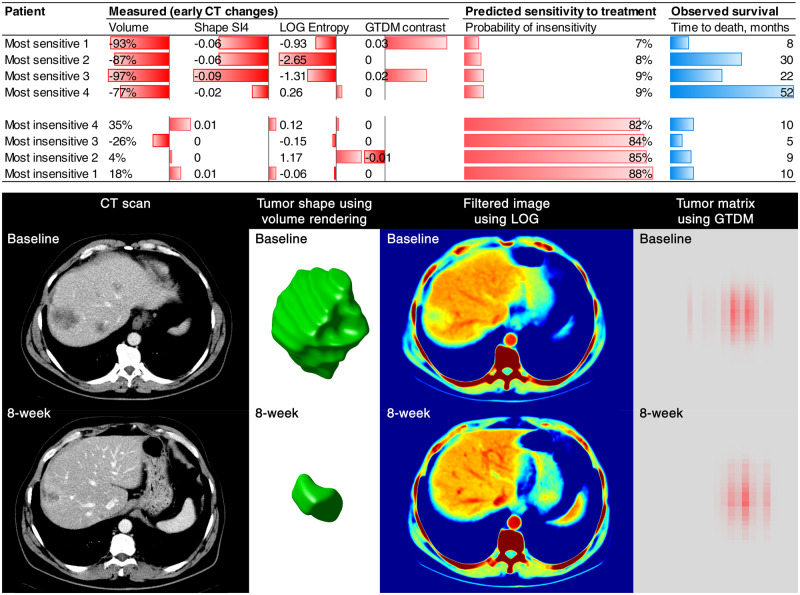
Figure 3.
Visual representation of the four imaging features included in the signature. The changes in the radiomics features in the patients with the lowest and the highest probability of insensitivity to treatment according to the radiomics signature are presented on this graph (most sensitive 1–4 vs most insensitive 1–4). The changes in tumor imaging phenotype of the patient “most sensitive 1” is displayed below. As demonstrated, CT-scan images are transformed to other mathematical spaces for feature extraction, for example, CT image is transformed to LOG space for computing the entropy value (spatial heterogeneity), and tumor pixels within segmentation contour are transformed to GTDM matrix for computing the contrast value.
Table 3.
Performance of the AI signature in the validation sets*
| Analysis | Anti-EGFR treatment |
No anti-EGFR treatment |
||
|---|---|---|---|---|
| FOLFIRI + cetuximab |
FOLFIRI only |
|||
| mCRC |
mCRC |
|||
| CRC-FCHQ | CRC-FCSD | CRC-FHQ | CRC-FSD | |
| Reference standard | OS | OS | OS | OS |
| No. patients | ||||
| Total | 116 | 186 | 129 | 236 |
| Training | 78 | 124 | 78 | 159 |
| Sensitive | 48 | 71 | 42 | 85 |
| Resistant | 30 | 53 | 36 | 74 |
| Validation | 38 | 62 | 51 | 78 |
| Sensitive | 23 | 34 | 32 | 42 |
| Resistant | 15 | 28 | 19 | 36 |
| Signature | ||||
| Algorithm | Random Forest | Random Forest | Random Forest | Random Forest |
| Features | Shape SI4 | Shape SI4 | Shape SI4 | Shape SI4 |
| LoG Z Entropy | LoG Z Entropy | LoG Z Entropy | LoG Z Entropy | |
| GTDM Contrast | GTDM Contrast | GTDM Contrast | GTDM Contrast | |
| LoG X Entropy | LoG X Entropy | LoG X Entropy | LoG X Entropy | |
| Performance | ||||
| Tumor sensitivity | OS >17.7 mo | OS >17.7 mo | OS >17.7 mo | OS >17.7 mo |
| AUC, training | 0.83 (95% CI = 0.75 to 0.95) | 0.84 (95% CI = 0.76 to 0.89) | 0.75 (95% CI = 0.63 to 0.85) | 0.75 (95% CI = 0.67 to 0.82) |
| AUC, validation | 0.80 (95% CI = 0.69 to 0.94) | 0.72 (95% CI = 0.59 to 0.83) | 0.59 (95% CI = 0.44 to 0.72) | 0.55 (95% CI = 0.43 to 0.66) |
| Association with OS | ||||
| Cox regression | ||||
| Hazard ratio | 44.3 (95% CI = 6.4 to 307.7) | 6.5 (95% CI = 1.8 to 23.6) | 1.9 (95% CI = 0.4 to 8.4) | 0.96 (95% CI = 0.2 to 4.3) |
| Death | 31/38 | 53/62 | 42/51 | 69/78 |
| P | .0001 | .005 | .43 | .96 |
The performance of the AI signature to detect treatment sensitivity was evaluated in all cohorts. AI = Artificial Intelligence; AUC = Area Under the Curve; CT = computed tomography; CRC = colorectal cancer; F = FOLFIRI; FC = FOLFIRI+cetuximab; HQ = High computed tomography quality; SD = Standard computed tomography quality.
In the validation set, the performance of the signature (AUC = 0.80, 95% CI = 0.69 to 0.94, sensitivity = 0.80, and specificity = 0.78) statistically significantly (two-sided P < .001) outperformed both KRAS mutational status at baseline (AUC = 0.67, P < .001) and 8-week tumor shrinkage (AUC = 0.75 using either RECIST 1.1-like unidimensional tumor shrinkage or volumetric tumor shrinkage) (P < .001 using bootstrap; Supplementary Figure 6, available online). There was no biological or clinical phenotype of failure (Supplementary Table 2, available online).
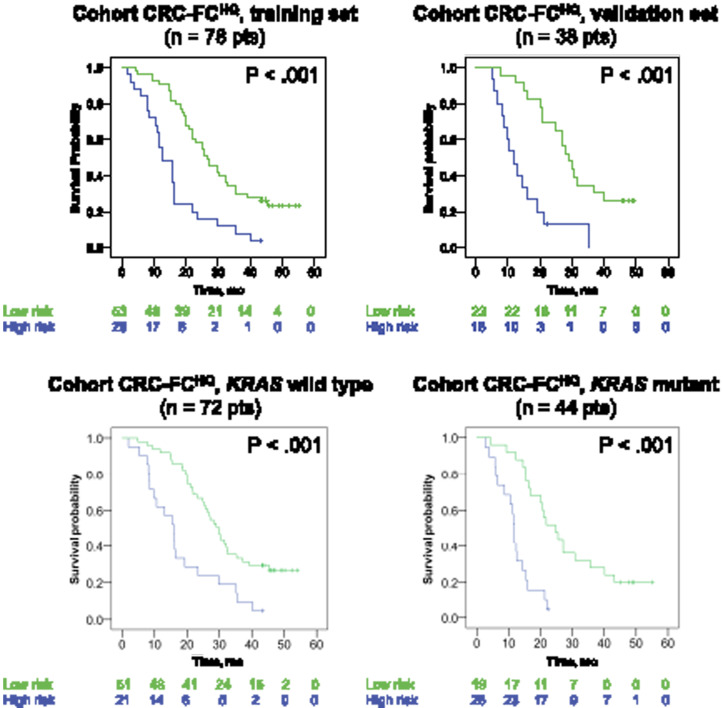
In the cohort CRC-FCHQ, the signature showed good prognostic discrimination for OS (P < .001) by univariate Cox regression analysis. High-risk signature (signature > 0.5) was associated with shorter OS than patients with low-risk signature in the training set, the validation set, KRAS wild-type patients, and KRAS mutant tumors (Figure 4) (P < .001). The signature measured at 8 weeks in KRAS wild-type patients can be used to predict survival at any time (nomogram, Supplementary Figure 7, available online). The signature provided incremental value over existing benchmarks for the prediction of OS because univariate Cox regression analyses adjusted for KRAS mutational status demonstrated that the signature had a statistically significantly better fit for the prediction of patients’ OS than tumor shrinkage (P < .001) (Supplementary Figure 6; Supplementary Tables 1, 3, and 4, available online).
Figure 4.
Risk stratification using the signature in the discovery cohort. The discovery cohort included mCRC patients treated with FOLFIRI+ cetuximab. Kaplan-Meier graphs depicting overall survival in patients stratified at high-risk (signature >0.5) or low-risk (signature ≤0.5) at 8 weeks by the signature. P values are based on dataset-stratified two-sided log-rank tests. A, B) Survival probability in the training and in the validation sets. C, D) Survival probability in KRAS wild-type and KRAS mutant groups. CRC = colorectal cancer; F = FOLFIRI; FC = FOLFIRI+cetuximab; HQ= High computed tomography quality; SD = Standard computed tomography quality.
Generalizability of the Signature to CRC-FCSD, CRC-FHQ, and CRC-FSD
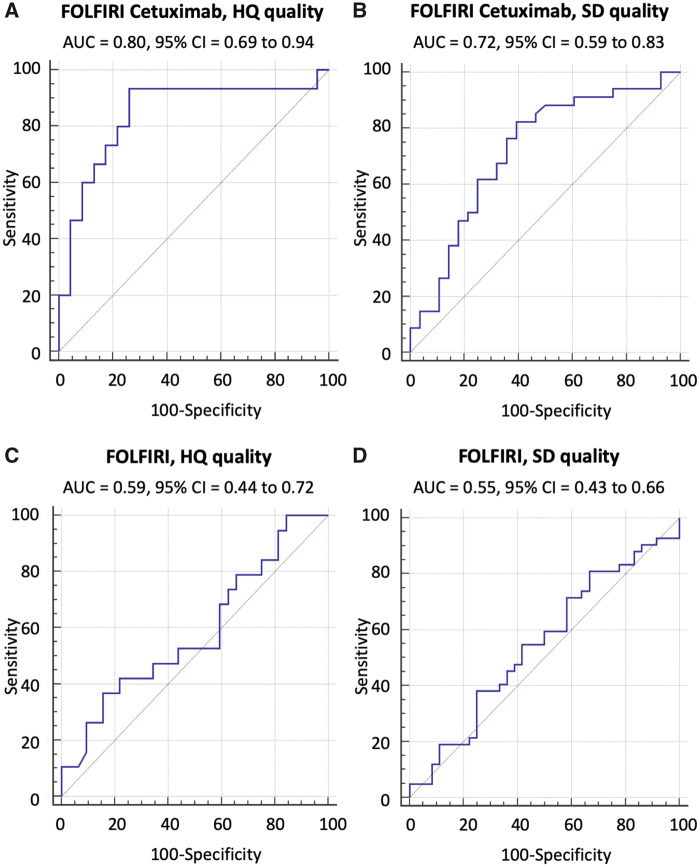
The Random Forest algorithm (18) using the 4 Radiomics features signature developed and used in the CRC-FCHQ analysis (Supplementary Figure 4, available online) was applied to the other training datasets for calibration. The performance of the signature (Table 3; Figure 5; Supplementary Figure 8; Supplementary Tables 5 and 6, available online) in the validation sets consisting of unseen data was calculated. AUC was 0.72 (95% CI = 0.59 to 0.83), sensitivity = 0.82, and specificity = 0.61 in CRC-FCSD. AUC was 0.59 (95% CI = 0.44 to 0.72), sensitivity = 0.37, and specificity = 0.84 in CRC-FHQ. AUC was 0.55 (95% CI = 0.43 to 0.66), sensitivity = 0.81, and specificity = 0.33 in CRC-FSD.
Figure 5.
Performance of the signature in the validation sets of the three independent testing cohorts. CT quality was classified as high (HQ) or standard (SD). The Random Forest algorithm using the 4 Radiomics features signature developed and used in the FCHQ analysis was applied to the other training data sets for calibration. Then, its performance was validated in the four validation sets. Area under the receiver operating characteristic curve (AUC) and AUC’s 95% confidence interval (CI) were used to indicate classification performance. The signature (AUC [95CI]) used temporal decrease in tumor spatial heterogeneity plus boundary infiltration to successfully predict sensitivity to anti-EGFR therapy (FCHQ [A], FCSD [B]) but failed with chemotherapy (FHQ [C], FSD [D]). F = FOLFIRI; FC = FOLFIRI+cetuximab; HQ= High computed tomography quality; SD = Standard computed tomography quality.
Discussion
Using standard-of-care CT scans acquired in multicenter clinical trials, AI successfully identified on-treatment imaging biomarkers associated with tumor sensitivity to anti-EGFR therapy in CRC patients. These four imaging biomarkers enabled development of a signature, which offered high sensitivity and specificity, to guide the clinical decision to continue EGFR targeted therapies. The innovation of our work compared with the existing literature was that the signature was dynamic, generalized to four datasets acquired in multicenter studies, used a limited subset of features to reduce overfitting, and considered image quality according to new quality control tools (17,19,20). Because it is based on routinely acquired CT scans, such a tool could be widely incorporated into clinical practice at minimal cost to detect EGFR-resistant tumors due to clonal acquisition of resistance mechanisms through alternate pathways and downstream events.
The signature features can be understood as noninvasive in vivo surrogates of treatment-induced biological changes and generally fall into two categories: indicators of heterogeneity of the metastatic burden and evaluation of tumor-parenchyma interactions. In patients with both KRAS wild-type or mutated tumors, the imaging features identified by our signature provided incremental value over 8-week tumor shrinkage (6) in a multifactorial prediction model. Although KRAS wild-type status at baseline was associated with better outcome to treatment, adding this mutational status to the signature did not contribute to the incremental value. The capacity of the complete RAS mutation status to improve predictability was not assessed in our research. Although this finding is consistent with the literature suggesting that on-treatment markers of efficacy are better predictors of outcome than baseline variables, it also indicates that imaging biomarkers can overcome two weaknesses associated with the use of a single genomic biomarker to predict outcome to targeted therapies: tumor heterogeneity and pathways complexity.
Because CT scans can provide information regarding the entire tumor burden and be repeated noninvasively, they are better suited to address the spatial and temporal heterogeneity of tumors than genomic analysis of tissue from a single biopsy site. First, although KRAS mutations are early driving events in colorectal tumorigenesis and progression, substantial spatial heterogeneity exists in KRAS status. Despite good concordance between the primary tumors and matched distant metastases, KRAS status shows 8% intratumoral heterogeneity in the primary tumor, up to 10% variability between the primary tumor and metastases, and up to 12% heterogeneity among multiple metastatic lesions (21). Second, the limitations of genomic biomarkers are further demonstrated by the fact that failure of anti-EGFR therapy is observed in one-half of the patients with wild-type RAS CRC. Although downstream mutations in genes (eg, BRAF and PI3KCA mutations) are not predictive biomarkers for anti-EGFR therapy, they seem to predict a poor prognosis independent of treatment. Third, temporal heterogeneity may also emerge from the inherent dynamic evolution and adaptation of clones acquiring distinct resistance mechanisms, especially in the presence of selection pressure from anti-EGFR targeted therapies (22).
Decrease in heterogeneity of the metastatic burden (eg, entropy) on CT scan following cetuximab treatment was the best predictor of tumor sensitivity to anti-EGFR therapies. The signature can be understood as an AI analysis of CT image heterogeneity (23) capturing a key hallmark of cancer (24): the heterogeneity of an expanding aberrant and distorted neo-vasculature supplying tumor growth. These macroscopic patterns of neovascularization are marked by heterogeneous and excessive blood flow, microhemorrhage, and leakiness, which produce a heterogeneous accumulation of iodine on contrast-enhanced CT scans. Radiomics signature can therefore capture neovascularization patterns (14,25–27) associated with reduced treatment efficacy (25,28), poor outcome because of reduced drug delivery (29–32), hypoxia, and promotion of immune evasion, tumor progression, and metastasis (24). From a broader perspective, there is a rationale that even basic CT features such as size changes are associated with pathologic response (ie, the percentages of residual tumor) and OS in mCRC patients treated with chemotherapy regimens containing molecular targeted therapies targeting such as anti-EGFR or anti-VEGF (33).
The second weakness of the single genomic biomarker is the complexity of cancer pathways. Following blockade of oncogenic pathways by targeted therapies, the Darwinian selection process will tend to select cancer cells that exploit alternative pathways. AI analysis of CT scans can identify signatures associated with downstream events linked to favorable outcomes with anti-EGFR therapy. Several tumor biopsy–driven clinical trials (13) demonstrated that inhibition of the EGF receptor activation alone did not confirm clinical benefit in mCRC and other solid tumors. This is explained by the selection of clones with downstream mutations allowing the tumor to continue to proliferate (eg, EGFR, then RAS, then RAF or PI3K). A radiomics signature can dynamically assess features associated with efficient blockade of the full range of oncogenetic pathways, and their interconnections promise to accelerate the development of molecularly targeted therapies.
Tumor shape (eg, shape SI4) and boundary sharpness (eg, Sigmoid Slope Energy) features were strongly associated with OS and capture complex patterns of macroscopic changes at tumor-liver interfaces in mCRC that reflect both infiltrative tumor growth and perfusion change. A pushing interface in which tumor cells directly compress the liver has a poorer prognosis than interfaces in which a desmoplastic lymphocytic stroma separates the tumor cells from the liver or in which tumor cells replace liver parenchyma without pushing or desmoplastic reaction (34). Conversely, the presence of a peritumoral contrast-enhancement rim region surrounding liver tumors is biologically relevant as a reflection of recruitment of hepatic artery branches by vascular endothelial growth factor and/or a reduced peritumoral portal flow (mechanical tumor compression and/or vessel lumen-narrowing secondary to fixed leukocyte adherence) (16,35–40).
There are several innovative aspects of our work. Although AI techniques are increasingly proposed to aid drug discovery and clinical imaging, none have yet been identified and robustly validated in heterogeneous datasets such as large multicenter clinical trials. Our innovative methodology (41–43) lies in combining the deep-learning feature set (ImageNet pretrained VGG-16) and radiomics feature set (engineered features) and applying them using a dynamic approach (two time points) to response assessment in cancer therapies. Our results suggest that the performance of imaging-based response assessment could progressively increase by moving from unidimensional size, to volume (44), and finally by integrating radiomics features. Another innovation—compared with other radiomic signatures in CRC patients (45–47)—is that we tightly controlled the quality (17) of image acquisition through robust reconstruction settings (15), reproducible segmentation (16), and selection of optimal contrast enhancement at the portal venous phase (14). This was by design to increase the signal-to-noise ratio because of the heterogeneity in acquisition protocols and image quality in a landmark historical multicenter clinical trial with “real-life” heterogeneous image quality. Finally, we reduced the risks of type I error and overfitting by using only four imaging biomarkers in the signature and by evaluating the generalizability of these imaging biomarkers to four validation sets.
This work has limitations but opens numerous perspectives and opportunities for improvement. First, future research could assess the performance of CT scan evaluation earlier than 8 weeks or pool data from multiple timepoints and modeling the growth and decay of radiomics features for all available patient scans beyond week 8. Second, before prospectively implementing this decision tool in RAS wild-type patients, we expect to calibrate the signature to all generations of CT scanners and fine-tune convolutional neural network models in external and larger datasets (48). Third, we will evaluate the possible temporal within-patient heterogeneity of responses to identify therapy-resistant clones. Fourth, there is a need to determine if these imaging biomarkers uncover a prognostic effect rather than predictive (to anti-EGFR therapy), specifically in certain subpopulations. As an ancillary study, AI could not identify a phenotype associated with the sensitivity of tumors in the FOLFIRI-only group in a clinically significant manner. Because CRC is known to evolve by a reiterative process of genetic diversification and clonal evolution, a targeted therapy efficient in only some patients might indeed produce greater interpatient variability in terms of depth of response than chemotherapy (49). This is explained by the fact that sensitive tumors will be more homogeneous than insensitive tumors because resistant mutation events will develop heterogeneous resistant tumor niches.
Temporal changes in the spatial heterogeneity of metastatic tumor burden were linked to outcome. Heterogeneity was defined by the conversion of a per-lesion phenotype to a per-patient phenotype. First, we calculated the spatial heterogeneity between neighboring voxels in each metastatic lesion. Then, we aggregated all metastatic lesions through a weighted average. Of note, each liver metastatic lesions may theoretically behave differently from metastatic disease in peritoneum and lungs. Nonetheless, several facts support our methodology. First, liver metastasis affects greater than one-half of CRC patients (1). Second, a modeling framework (22)—considering resistance development and clonal selection—demonstrated similar dynamics among individual lesions within the same tumor site location and different dynamics in only 35% of patients with lesions in different anatomic locations (22).
Although there was no apparent selection bias, the signature was designed for CRC patients with liver metastases with known KRAS status in a landmark trial (48–51). The architecture of the liver parenchyma differs from other tissues such as the lung parenchyma. Therefore, some imaging features (ie, shape and boundary sharpness features) might intuitively need to be calibrated for each malignant site, whereas surrogates of heterogeneity may generalize. We anticipate that features identified in this historical landmark clinical trial can be easily generalized and fine-tuned to analyze newer datasets. First, features were generalizable from the HQ cohort to patients with SD quality imaging settings. Second, response evaluation on CT scan has not fundamentally changed over the last decades since new technologies focused on increasing the speed and reducing radiation dose (50). The main advantage of using an “old” trial is the extended follow-up of patients that allowed only 18% of data to be censored for the OS. Most newer clinical trial datasets are not yet mature or available from sponsors; nevertheless, groups are actively attempting to make these publicly available (51). In conclusion, AI-derived support tools could give clinicians an early prediction of the success of treatment with FC using conventional standard-of-care CT scans.
Funding
The authors acknowledge financial support from the National Cancer Institute (U01 CA140207, U01 CA225431) and imaging data received from Merck KGaA, Darmstadt, Germany. LD’s work was funded by grants from Fondation Philanthropia and Fondation Nuovo-Soldati.
Notes
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
PE is an employee of Merck KGaA, which has the marketing rights to cetuximab in many countries. LHS and BZ receive royalties from Varian Medical Systems. The other authors have no disclosures.
Supplementary Material
References
- 1. Van Cutsem E, Cervantes A, Nordlinger B,. et al. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(suppl 3):iii1–9. [DOI] [PubMed] [Google Scholar]
- 2. Schwartz LH, Mazumdar M, Brown W, et al. Variability in response assessment in solid tumors: effect of number of lesions chosen for measurement. Clin Cancer Res. 2003;9(12):4318–4323. [PubMed] [Google Scholar]
- 3. Oxnard GR, Zhao B, Sima CS, et al. Variability of lung tumor measurements on repeat computed tomography scans taken within 15 minutes. J Clin Oncol. 2011;29(23):3114–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao B, James LP, Moskowitz CS, et al. Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Radiology. 2009;252(1):263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao B, Tan Y, Bell DJ, et al. Exploring intra- and inter-reader variability in uni-dimensional, bi-dimensional, and volumetric measurements of solid tumors on CT scans reconstructed at different slice intervals. Eur J Radiol. 2013;82(6):959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Piessevaux H, Buyse M, Schlichting M, et al. Use of early tumor shrinkage to predict long-term outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol. 2013;31(30):3764–3775. [DOI] [PubMed] [Google Scholar]
- 7. Fillon M. Making sense of the mountains of new cancer data. J Natl Cancer Inst. 2017;109(2):djx020. doi: 10.1093/jnci/djx020. [DOI] [PubMed] [Google Scholar]
- 8. Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–1417. [DOI] [PubMed] [Google Scholar]
- 9. Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–1765. [DOI] [PubMed] [Google Scholar]
- 10. Van Cutsem E, Lenz HJ, Kohne CH, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol. 2015;33(7):692–700. [DOI] [PubMed] [Google Scholar]
- 11. Van Cutsem E, Kohne CH, Lang I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29(15):2011–2019. [DOI] [PubMed] [Google Scholar]
- 12. Qin S, Li J, Wang L, et al. Efficacy and tolerability of First-Line Cetuximab Plus Leucovorin, Fluorouracil, and Oxaliplatin (FOLFOX-4) versus FOLFOX-4 in patients with RAS wild-type metastatic colorectal cancer: the open-label, randomized, phase III TAILOR trial. J Clinc Oncol. 2018;36(30):3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23(11):2513–2520. [DOI] [PubMed] [Google Scholar]
- 14. Dercle L, Lu L, Lichtenstein P, et al. Impact of variability in portal venous phase acquisition timing in tumor density measurement and treatment response assessment: metastatic colorectal cancer as a paradigm. J Clin Oncol Clinical Cancer Informatics. 2017;1:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao B, Tan Y, Tsai W-Y, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6(1):23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang Q, Lu L, Dercle L, et al. Interobserver variability in tumor contouring affects the use of radiomics to predict mutational status. J Med Imaging. 2017;5(01):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lambin P, Leijenaar RT, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. [DOI] [PubMed] [Google Scholar]
- 18. Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 19. Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180–1191. [DOI] [PubMed] [Google Scholar]
- 20. Limkin E, Sun R, Dercle L, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28(6):1191–1206. [DOI] [PubMed] [Google Scholar]
- 21. Han CB, Li F, Ma JT, et al. Concordant KRAS mutations in primary and metastatic colorectal cancer tissue specimens: a meta-analysis and systematic review. Cancer Invest. 2012;30(10):741–747. [DOI] [PubMed] [Google Scholar]
- 22. Terranova N, Girard P, Ioannou K, et al. Assessing similarity among individual tumor size lesion dynamics: the CICIL methodology. CPT Pharmacometrics Syst Pharmacol. 2018;7(4):228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dercle L, Ammari S, Bateson M, et al. Limits of radiomic-based entropy as a surrogate of tumor heterogeneity: ROI-area, acquisition protocol and tissue site exert substantial influence. Sci Rep. 2017;7(1):7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanahan D, Weinberg RA.. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 25. Choi H, Charnsangavej C, Faria SC, et al. Correlation of computed tomography and positron emission tomography in patients with metastatic gastrointestinal stromal tumor treated at a single institution with imatinib mesylate: proposal of new computed tomography response criteria. J Clin Oncol. 2007;25(13):1753–1759. [DOI] [PubMed] [Google Scholar]
- 26. Choi H, Charnsangavej C, de Castro Faria S, et al. CT evaluation of the response of gastrointestinal stromal tumors after imatinib mesylate treatment: a quantitative analysis correlated with FDG PET findings. AJR Am J Roentgenol. 2004;183(6):1619–1628. [DOI] [PubMed] [Google Scholar]
- 27. Benjamin RS, Choi H, Macapinlac HA, et al. We should desist using RECIST, at least in GIST. J Clin Oncol. 2007;25(13):1760–1764. [DOI] [PubMed] [Google Scholar]
- 28. van der Veldt AA, Meijerink MR, van den Eertwegh AJ, et al. Choi response criteria for early prediction of clinical outcome in patients with metastatic renal cell cancer treated with sunitinib. Br J Cancer. 2010;102(5):803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith AD, Shah SN, Rini BI, et al. Morphology, Attenuation, Size, and Structure (MASS) criteria: assessing response and predicting clinical outcome in metastatic renal cell carcinoma on antiangiogenic targeted therapy. AJR Am J Roentgenol. 2010;194(6):1470–1478. [DOI] [PubMed] [Google Scholar]
- 30. Smith AD, Lieber ML, Shah SN.. Assessing tumor response and detecting recurrence in metastatic renal cell carcinoma on targeted therapy: importance of size and attenuation on contrast-enhanced CT. AJR Am J Roentgenol. 2010;194(1):157–165. [DOI] [PubMed] [Google Scholar]
- 31. Krajewski KM, Guo M, Van den Abbeele AD, et al. Comparison of four early posttherapy imaging changes (EPTIC; RECIST 1.0, tumor shrinkage, computed tomography tumor density, Choi criteria) in assessing outcome to vascular endothelial growth factor-targeted therapy in patients with advanced renal cell carcinoma. Eur Urol. 2011;59(5):856–862. [DOI] [PubMed] [Google Scholar]
- 32. Ammari S, Thiam R, Cuenod CA, et al. Radiological evaluation of response to treatment: application to metastatic renal cancers receiving anti-angiogenic treatment. Diagn Interv Imaging. 2014;95(6):527–539. [DOI] [PubMed] [Google Scholar]
- 33. Chun YS, Vauthey JN, Boonsirikamchai P, et al. Association of computed tomography morphologic criteria with pathologic response and survival in patients treated with bevacizumab for colorectal liver metastases. JAMA. 2009;302(21):2338–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van den Eynden GG, Bird NC, Majeed AW, et al. The histological growth pattern of colorectal cancer liver metastases has prognostic value. Clin Exp Metastasis. 2012;29(6):541–549. [DOI] [PubMed] [Google Scholar]
- 35. Ridge JA, Bading J, Gelbard A, et al. Perfusion of colorectal hepatic metastases. Cancer. 1987;59(9):1547–1553. [DOI] [PubMed] [Google Scholar]
- 36. Meijerink MR, van Waesberghe JHT, van Schaik C, et al. Perfusion CT and US of colorectal cancer liver metastases: a correlative study of two dynamic imaging modalities. Ultrasound Med Biol. 2010;36(10):1626–1636. [DOI] [PubMed] [Google Scholar]
- 37. Meijerink MR, van Waesberghe JHT, van der Weide L, et al. Total-liver-volume perfusion CT using 3-D image fusion to improve detection and characterization of liver metastases. Eur Radiol. 2008;18(10):2345–2354. [DOI] [PubMed] [Google Scholar]
- 38. Tsushima Y, Funabasama S, Sanada S, et al. Development of perfusion CT software for personal computers. Acad Radiol. 2002;9(8):922–926. [DOI] [PubMed] [Google Scholar]
- 39. Tsushima Y, Funabasama S, Aoki J, et al. Quantitative perfusion map of malignant liver tumors, created from dynamic computed tomography data. Acad Radiol. 2004;11(2):215–223. [DOI] [PubMed] [Google Scholar]
- 40. Kruskal JB, Thomas P, Kane RA, et al. Hepatic perfusion changes in mice livers with developing colorectal cancer metastases. Radiology. 2004;231(2):482–490. [DOI] [PubMed] [Google Scholar]
- 41. Yasaka K, Akai H, Abe O, et al. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology. 2018;286(3):887–896. [DOI] [PubMed] [Google Scholar]
- 42. Hamm CA, Wang CJ, Savic LJ, et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29(7):3338–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao W, An X, Cong L, Lyu C, Zhou Q, Guo R. Application of deep learning in quantitative analysis of 2-dimensional ultrasound imaging of nonalcoholic fatty liver disease. J Ultrasound Med. 2020;39(1):51–59. [DOI] [PubMed] [Google Scholar]
- 44. Zhao B, Oxnard GR, Moskowitz CS, et al. A pilot study of volume measurement as a method of tumor response evaluation to aid biomarker development. Clin Cancer Res. 2010;16(18):4647–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ganeshan B, Miles KA, Young RC, et al. Hepatic enhancement in colorectal cancer: texture analysis correlates with hepatic hemodynamics and patient survival. Acad Radiol. 2007;14(12):1520–1530. [DOI] [PubMed] [Google Scholar]
- 46. Liang C, Huan Y, He L, et al. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget. 2016;7(21):31401–31412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. [DOI] [PubMed] [Google Scholar]
- 48. LeCun Y, Bengio Y, Hinton G.. Deep learning. Nature. 2015;521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- 49. Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jensen CT, Wagner-Bartak NA, Vu LN, et al. Detection of colorectal hepatic metastases is superior at standard radiation dose CT versus reduced dose CT. Radiology. 2019;290(2):400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dercle L, Connors DE, Tang Y, et al. Vol-PACT: a foundation for the NIH public-private partnership that supports sharing of clinical trial data for the development of improved imaging biomarkers in oncology. JCO Clin Cancer Inform. 2018;2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.