Abstract.
Purpose: We present an original method for simulating realistic fetal neurosonography images specifically generating third-trimester pregnancy ultrasound images from second-trimester images. Our method was developed using unpaired data, as pairwise data were not available. We also report original insights on the general appearance differences between second- and third-trimester fetal head transventricular (TV) plane images.
Approach: We design a cycle-consistent adversarial network (Cycle-GAN) to simulate visually realistic third-trimester images from unpaired second- and third-trimester ultrasound images. Simulation realism is evaluated qualitatively by experienced sonographers who blindly graded real and simulated images. A quantitative evaluation is also performed whereby a validated deep-learning-based image recognition algorithm (ScanNav®) acts as the expert reference to allow hundreds of real and simulated images to be automatically analyzed and compared efficiently.
Results: Qualitative evaluation shows that the human expert cannot tell the difference between real and simulated third-trimester scan images. 84.2% of the simulated third-trimester images could not be distinguished from the real third-trimester images. As a quantitative baseline, on 3000 images, the visibility drop of the choroid, CSP, and mid-line falx between real second- and real third-trimester scans was computed by ScanNav® and found to be 72.5%, 61.5%, and 67%, respectively. The visibility drop of the same structures between real second-trimester and simulated third-trimester was found to be 77.5%, 57.7%, and 56.2%, respectively. Therefore, the real and simulated third-trimester images were consider to be visually similar to each other. Our evaluation also shows that the third-trimester simulation of a conventional GAN is much easier to distinguish, and the visibility drop of the structures is smaller than our proposed method.
Conclusions: The results confirm that it is possible to simulate realistic third-trimester images from second-trimester images using a modified Cycle-GAN, which may be useful for deep learning researchers with a restricted availability of third-trimester scans but with access to ample second trimester images. We also show convincing simulation improvements, both qualitatively and quantitatively, using the Cycle-GAN method compared with a conventional GAN. Finally, the use of a machine learning-based reference (in the case ScanNav®) for large-scale quantitative image analysis evaluation is also a first to our knowledge.
Keywords: second-trimester scan, third-trimester scan, transventricular plane, realistic simulation, cycle-consistent adversarial network, quantitative evaluation
1. Introduction
The second-trimester fetal anomaly scan (hereafter referred as the second-trimester scan) is an important part of pregnancy care and is routinely offered to pregnant women in high income countries between 18 and 22 weeks of gestation. A third-trimester growth ultrasound scan (hereafter referred to as the third-trimester scan) is increasingly recommended to improve the detection of breech presentation and small- and large-for-gestational age babies. This scan typically takes place between 32 and 36 weeks of gestation and is thought to reduce perinatal morbidity when linked to appropriate interventions, for example, induction of labor.1,2
In this paper, we specifically consider fetal neurosonography images. The second- and third-trimester fetal neurosonography images have quite different appearances due to brain maturation as well as changes in fetal size and skull ossification. Unlike second trimester scans, which have a standard guideline for image acquisition in most countries, there is no guideline for third-trimester scans. We are particularly interested in exploring whether third-trimester images can be simulated from the routine second-trimester images.
Simulating third-trimester scan images conditioned on second-trimester scan images has an advantage over simulating third-trimester scan images from scratch. The former approach focuses on simulating the appearance change between the two trimesters, in other words, it means that general fetal structure appearance is inherited from the second-trimester scan. Unlike some other medical imaging applications, pairwise data are not available for training due to changes in fetal alignment and presentation between second-trimester and third-trimester scans, as well as likely differences in ultrasound equipment and acquisition settings.3 In addition, taking the United Kingdom as one example, third-trimester scans are not currently universally offered to all pregnant women, so third-trimester scan data availability is limited.4 Indeed generating realistic third-trimester images from the abundance of second-trimester images to support third-trimester deep learning research was one of the original motivations of this work. Therefore, the approach taken in this paper is that image simulation is an image class translation problem in which paired image data do not exist but examples from each class are available.
This paper makes three primary contributions. First, we present a deep learning method to simulate third-trimester scan images of the fetal brain conditioned upon the appearance of second-trimester scan images. Second, to improve results, we modify design features of the cycle-consistent adversarial network (Cycle-GAN) architecture including the choice of loss function. The proposed architecture increases the training stability and achieves better performance relative to a conventional GAN. Finally, the third contribution is to our knowledge the first use of a machine learning-based reference for large-scale quantitative ultrasound image analysis evaluation.
2. Related Work
Medical image simulation is a developing research topic in medical imaging and has a wide range of potential applications. For example, it contributes to software-based tools for clinical training and procedure simulation, as well as generating synthetic data for deep learning algorithm research. Ultrasound simulation is difficult due to the significant variations in clinical ultrasound image appearance resulting from variations in acquisition, including image acquisition parameters and subject presentation. Traditional methods of ultrasound image simulation include physics-based ultrasound simulation and manikin-based simulation.5 These approaches have the disadvantages of either not being suitable for interactive use or being heavily dependent on the quality of material used in a physical phantom. Machine learning methods are in theory an attractive software-based alternative because they overcome these disadvantages by inferring the simulated images from a large amount of real image data.
Generative adversarial networks (GANs)6 are currently popular for simulated image generation in machine learning. However, the training stability of GANs is recognized as an issue because the generator and discriminator must be simultaneously trained in a mini-max game. A deep convolutional GAN architecture proposed in Ref. 7 was shown to provide high training stability in generating natural images. A convincing evidence that the network learns a hierarchical representation from object parts to scenes by testing on the LSUN, IMAGENET, and CIFAR-10 public image datasets was also presented in Ref. 7. A coupled GAN that learns joint distributions of multidomain images was proposed in Ref. 8. The architecture consists of a pair of GANs. The weights of the first few layers of both generative models, and the last few layers of both discriminator models were coupled together. This weight-sharing constraint enabled the learning of the joint distribution of images without tuples of corresponding images. To improve training stability of GANs, the least square GAN with a modified loss function that caused the generator to minimize the Pearson divergence between generated images and real images was proposed in Ref. 9.
There has been previous research on the generation of medical images conditioned upon some input factor. A conditional GAN structure that simulates ultrasound phantom images at given three-dimensional spatial locations was proposed in Ref. 10. A conditional GAN can also perform image-to-image translation with the goal to learn the mapping between an input image and an output image. It often requires paired training data that have one-to-one correspondence. In Ref. 11, a fully convolutional networks was designed to learn the parametric translation function for pairwise inputs to perform semantic segmentation. However, the creation of pairwise data is costly and not always available in practice. In our case, as noted earlier, we can hardly obtain pairwise matched examples of second-trimester scan images and third-trimester scan images due to the change in fetal presentation and image acquisition parameters across time.
However, deep learning approaches can be designed to learn a mapping between domains without pairwise labeled data. Cycle-GANs can be used to learn the mapping between two image domains without corresponding imagewise pairings.12 Cycle-GANs introduce an inverse mapping on top of the usual forward mapping to create a highly constrained environment. Cycle-GANs have been previously used for medical imaging simulation. The MR-to-CT image simulation is considered in Ref. 13. In this paper, the forward cycle consists of three separate CNNs. The first is trained to translate an input MR image into a CT image. The second one is trained to translate a synthesized CT image back to an MR image. The third CNN is trained to discriminate between synthesized and real CT images. The backward cycle is also trained to improve training stability. The results are evaluated using mean absolute error (MAE) and peak-signal-to-noise-ratio, provided labels are available for the used dataset.
An anatomy-aware synthesis framework proposed by Ref. 14 is used to achieve unpaired ultrasound to MRI image translation. The paper introduces the idea of “cross-modal attention” and uses a loss function for appearance and structural differences, respectively, to enhance the translation performance. Our proposed method uses a Cycle-GAN architecture to learn the mapping between a second-trimester and a third-trimester fetal neurosonography images.
3. Problem Definition
3.1. Clinical Fetal Ultrasound Second-Trimester and Third-Trimester Scan Appearance
The differences in visual appearance between the second- and third-trimester scans of the fetal head can be significant. To set the scene for the main contributions of the paper, in this section, we first discuss the key differences in appearance. We focus on the transventricular (TV) plane, which is used to estimate the head circumference (HC) and is one of three fetal biometry planes common to the second- and third-trimester imagings (the others being the abdominal circumference and femur length).
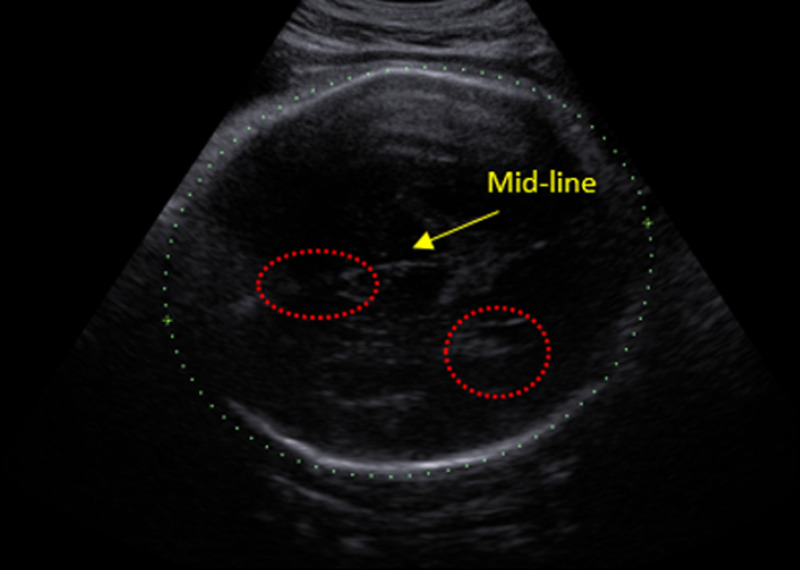
In the third trimester, the skull calcifies as the fetus matures, increasing the shadowing of the skull. As a result, it becomes more difficult to see internal structures of the brain. In international imaging guidelines for second-trimester imaging, such as the guidelines of the International Society of Ultrasound in Obstetrics & Gynecology (ISUOG)15 or the UK NHS Fetal Anomaly Screening Programme (FASP),16 sonographers are required to image the TV plane, which is defined as a symmetrical axial plane showing the cavum septi pellucidi (CSP), mid-line falx, and posterior horn of the lateral ventricle containing the choroid plexus. Figure 1 shows a typical second-trimester image of a standard TV imaging plane. The three key structures can be seen relatively clearly. However, the same rules are difficult to apply in the third-trimester since the visibility of intracranial structures is often poor, as shown in Fig. 2.
Fig. 1.
Representative example of a second-trimester ultrasound scan image at the level of the TV plane. The mid-line falx, CSP, and choroid/ventricle are the key structures that define this biometry plane.
Fig. 2.
Representative example of a third-trimester ultrasound scan image of the TV plane. The CSP and choroid/ventricle are not visible. The mid-line falx is barely visible to the eye (compare with Fig. 1).
In this example, only about 30% of the mid-line falx is visible by eye, and almost none of the CSP and choroid/ventricle can be seen. After manually inspecting hundreds of second- and third-trimester scan images, it is evident that the major appearance difference between second- and third-trimester images is the potential lack of visibility of certain key structures in the third-trimester TV plane image. This observation is further explored in a quantitative way in the next subsection.
3.2. Quantitative Metrics for Image Simulation Quality
In general, it is very difficult to quantitatively evaluate image simulation tasks. For instance, taking a classic computer vision application of painting style translation, a simulation of painting style translation from Leonardo da Vinci to Vincent Willem van Gogh cannot be assessed quantitatively as there is no ground truth. Similarly, in our case, the second and third trimesters are not acquired under identical imaging conditions. This is due to variations in anatomical presentation, fetus position, and ultrasound image settings across time. Hence, for a given second-trimester image, we do not have the ground truth equivalent third-trimester image (and vice versa). Our simulation is expected to show the general characteristics of a third-trimester TV plane, but not being similar to any specific third-trimester images. Therefore, using a numerical evaluation, such as MAE or mean squared error is also not applicable to our problem. For a general GAN image generation task, the Frechet Inception Distance has been proposed to measure the resolution and diversity of the generated images.17 However, diversity is not applicable in our case as each simulated image is conditioned on a specific input and all simulations are within a single class. We need to design a quantitative metric that quantifies the visibility of diagnostically important structures in a third-trimester ultrasound image.
ScanNav® (Intelligent Ultrasound Ltd., Milton Park, Abingdon, United Kingdom)18 is a precommercial deep learning-based automatic image analysis software application that assesses the quality of second-trimester scans according to the UK FASP guidelines. It is able to accurately detect different structures of a standard plane in large scale. It had been designed and trained by analzsing thousands of clinical second-trimester ultrasound images to label and grade images. Details of the underpinning algorithms and data used to build the quality assessment model are proprietary. However, we assume that the quality of software-derived annotation labels is at the same level as a typical experienced sonographer. This assumption is confirmed to be valid for our data later in this article. Under this assumption, ScanNav® is treated as equivalent to an experienced sonographer in ultrasound image classifications. It can assess and compare hundreds of real and simulated second-trimester scans efficiently and quickly (which is something that is tedious and costly to achieve if the task is performed by human experts). As ScanNav® has been designed for the task of assessing second-trimester scans, we need to validate its ability to assess third-trimester images. We drew insight from the experience of human sonographers in assessing third-trimester images since there are currently no official standards for third-trimester scan imaging. As a result, in practice human sonographers adapt their knowledge of how second-trimester scans are defined to third-trimester scans and relax some of the assumptions. For instance, the mid-line falx in third-trimester is typically less visible than a second-trimester scan. In addition, the CSP and choroid/ventricle are not always visible either. With this observational insight, we applied ScanNav® to provide numerical values to these qualitative comments as described next.
First, ScanNav® was tested on the second-trimester scan images from a large hospital image dataset.19 The full image resolution was . Then, 1500 separate second-trimester images that were not used in training were selected from the dataset and tested against the FASP TV plane criteria, with the result reported in Table 1, column 2. We define this as the “reference performance.” 1500 third-trimester images were then tested against the same FASP TV plane criteria using ScanNav®. The detection rate of key structures was computed for both the test second- and third-trimester images that are reported in Table 1. The detection rate indicates the ability to find a particular structure in the TV plane. The difference between second- and third-trimester structure detection rates is reported in Table 1, column 3.
Table 1.
ScanNav®TV plane criteria acceptance rate (as a percentage) for anomaly and growth scans.
| Second trimester (%) | Third trimester (%) | Difference (%) | |
|---|---|---|---|
| Magnification | 100.0 | 98.6 | 1.4 |
| Choroid/ventricle | 94.8 | 7.6 | 87.2 |
| CSP | 76.2 | 22.3 | 53.9 |
| Mid-line falx | 91.7 | 52.2 | 39.5 |
Note: The numbers in bold indicate the difference is big and significant, especially compared to the magnification difference.
Table 1 shows a significant drop in the detection rates of the choroid/ventricle, CSP, and mid-line falx between the second-trimester and third-trimester scan images. Visual inspection of a random subset of the third-trimester images shows that these structures are indeed often not visible in the third-trimester images. In the case of the mid-line falx, it is often present but sometimes is broken in appearance, whereas second-trimester scan criteria require it to be continuous. Finally, note that the magnification criterion is satisfied in both cases, indicating that the images are sufficiently zoomed for measurement in both cases.
This analysis has provided insight into the appearance characteristics of real third-trimester scan images and established a quantitative baseline to show how ScanNav® interprets real third-trimester scans. In Sec. 5, we use ScanNav® to assess the realism of simulated third-trimester scan images in a quantitative manner.
4. Third-Trimester Transventricular Scan Simulation Method
4.1. Data, Preprocessing, and Data Augmentation
We used TV images from a clinical dataset collected at the Oxford University Hospitals NHS Trust during the period of 2011 to 2013. We randomly selected a subset of images from this dataset. This subset contains 800 second-trimester (gestational age 18 to 22 weeks) and 700 third-trimester (gestational age 33 to 36 weeks) TV images (1500 images in total). 1400 of these images (700 each of second-trimester and third-trimester images) were used to train a deep learning model. The remaining 100 second-trimester images were set aside for testing. The second-trimester and third-trimester scan images were acquired using the same ultrasound machine model. There were 378 unique anonymous IDs included in this study. We did not know how many second- and third-trimester images were from the same fetus since all participants in this study were anonymous for each of their visits. To remove redundant information (e.g., text and user interface), each image was manually cropped around the HC. Two cropping methods were considered. For the first approach, a fixed size bounding box was directly applied to crop the original image. The coordinates of the bounding box were fixed and decided by manual inspection. In the second approach, each image was cropped with a bounding box placed around the skull. In this case, the fetal head is always in the center of the cropped image. In both cases, cropped images were downscaled and resized to to accommodate the GPU memory constraints of a GTX 1080 Ti. For data augmentation, a 10-deg rotation in a random direction was applied once to every training image. The effect of this data augmentation is evaluated in Sec. 5.
Figure 3 shows typical example images from the two modes of cropping. Images on the top row of Fig. 3 (fixed sized bounding box) can be seen to have a greater variability in appearance.
Fig. 3.
(a) Fixed size bounding box cropping for second-trimester scan images. (b) Bounding box cropping around the skull on second-trimester scan images. The top and bottom are cropped from the same image in each case. Cropped images have been resized to the same size of .
4.2. Cycle-GAN Architecture Design
Figure 4 shows a schematic of the proposed fetal ultrasound simulation Cycle-GAN-based architecture. and represent the training inputs: unpaired second-trimester and third-trimester scan images, respectively. In the forward cycle, the generator takes the input and generates “fake” third-trimester image . The discriminator compares the simulated third-trimester image and the “real” image and tries to distinguish them to its best ability (via a chosen cost function). There is a backpropagation from to to improve the quality of the generated image. The backward cycle uses the simulated third-trimester image as input. The generator generates the reconstructed second-trimester image conditioned on with the discriminator . The objective function includes a generator and discriminator loss for both the forward and the backward cycle and a cycle consistency loss. The cycle consistency loss is calculated to ensure the forward and backward generators learn the inverse mapping such that depends on .
Fig. 4.
Schematic structure of the fetal ultrasound simulation Cycle-GAN. Black represents the forward cycle and blue represents the backward cycle. The simulated third-trimester image in the red box is the result.
Details of the network structure are given in Fig. 5. The generator network consists of a U-net structure with downsampling and upsampling layers.20 The U-net consists of expansive pathways that combine feature information through upsampling and concatenation with high-resolution features. Each convolution is followed by a rectified linear unit (ReLU) and instance normalisation; therefore, it is an asymmetrical 12 layer U-net. The discriminator network acts like a 12-layers CNN. It takes the third-trimester scan images as input. It contains four downsampling layers with an initial filter number of 64. Each downsampling layer has a stride with a leaky ReLU activation function and instance normalisation. There are two fully connected layers followed by a dropout layer between them to minimize overfitting. The kernel size is for both the generator and the discriminator.
Fig. 5.
The network structure of our Cycle-GAN. (a) The generator network and (b) the discriminator network. The dotted line represents repeated layer combination of down/upsampling layer, activation function, and normalization.
For all experiments, an Adam optimizer is used with a learning rate of 0.0002, , and . As the experimental result shows that the batch size does not have significant impact on the training, we choose a batch size of 1 for both the generator and discriminator. We set the network training at 1500 epochs. The network converges, and we found no appreciable increase in visual quality for the generated image after 1000 epochs.
4.3. Least Square Loss
Reference 21 has previously demonstrated the effectiveness of using a least squares loss function for the discriminator in GANs to increase training stability. When the discriminator is too strong in a traditional GAN formulation, the generator receives diminishing gradients, which impedes training. Using a least square loss, gradient backpropagation through the generator is maintained even if the discriminator acts as a perfect classifier. The minimax objective function for least-squares GANs is defined as
| (1) |
Here and represent the discriminator and generator, respectively, represents the third-trimester scan, and represents the second-trimester scan. The labels for the fake and real data are and , respectively. The objective value for the generator is . Mao found that the Pearson divergence between real and generated datasets is minimized if and . We therefore set , , and in our network. Alternatively, we could have set , in line with the traditional GAN loss scheme. However, in preliminary experiments (not reported), we found that minimizing the Pearson divergence between the real data set and generated images increases the image clarity and generator convergence rate; hence it has been used in this paper.
5. Experiment and Results
5.1. Performance Using Different Cropping Methods
First we evaluate the effect of the two cropping methods introduced in Sec. 4.1. When sonographers try to determine the difference between second- and third-trimester images, they focus on the internal structure change inside the skull. Thus, we might hypothesize that skull-based cropping will yield a better performance in the Cycle-GAN than fixed bounding box cropping.
Figure 6 shows typical results for the proposed method using fixed bounding box cropped inputs. In each row, “original ” is the second-trimester image input to the generator . “Translated ” is the simulated third-trimester scan image using an loss. “Reconstructed ” is the backward prediction (simulation) of the second-trimester image from the backward cycle , which is subsequently used in a cycle-consistency loss for the Cycle-GAN to ensure consistency between the original input image and the reconstruction. Comparing the original and translated , the latter has a significant decrease in visibility of internal structures, typically the CSP and ventricles.
Fig. 6.
Examples of temporal translation from second-trimester scan images (original ) to third-trimester scan images (translated ) using fixed boundary cropped inputs.
Figure 7 shows typical results using skull-cropped images. Comparing with Fig. 6, observe that the simulated images are almost identical to the original inputs with little difference in appearance of the ventricles, CSP, and mid-line appearance. We attribute the decrease in the quality of simulation to the limited variability in the skull-cropped images. In testing, we use the fixed bounding box cropped inputs for quality simulations.
Fig. 7.
Examples of temporal translation from anomaly scan images (original ) to growth scan images (translated ) using flexible boundary inputs.
5.2. Qualitative Analysis
Figure 8 shows some examples of real third-trimester scans (top row) and simulated third-trimester scans (bottom row) using the fixed bounding-box cropping. We found the visibility of structures in the real third-trimester scan images and the simulated third-trimester scan images to be similar in columns 1, 2, and 4. However, column 3 shows a failure case of the generator in which the simulation model fails to fully grasp the concept of the fan-shaped-beam, thus leading to boundary image artifacts.
Fig. 8.
(a) The real third-trimester scan images from ground-truth. (b) Simulated third-trimester scan images. Simulated images are picked randomly from the test results.
Real second- and third-trimester scan images were mixed with the simulated third-trimester scan images in different ways for qualitative evaluation by experts. Although the images were presented to sonographers at a lower spatial resolution than typical clinical practices, they claim that it did not affect their ability to distinguish the real and simulated images as the appearance of brain structure is clear enough. Three experiments were conducted as described next.
Experiment 1: Since the proposed method uses second-trimester scan images to simulate third-trimester scan images, it is important to verify that human testers can distinguish between second-trimester and third-trimester images at the resolution of . We expected to see a high accuracy due to the visible differences of key brain structures. To test this hypothesis, we randomly selected 20 images consisting of a combination of real second-trimester and real third-trimester scans. These images were presented to eight human experts, and they were asked to say whether they were a second- or third-trimester image. The average accuracy and standard deviation of correct prediction was computed. The human experts who participated in the experiment had fetal ultrasound image analysis experience ranging from 1 to 5 years. The average correct prediction accuracy for all testers was 93.5% with a standard deviation of 1.46%.
Experiment 2: It was designed to verify the visual quality of the simulated images. Twenty images consisting of a combination of real and simulated third-trimester scan images were presented to human testers in a random order. The testers had no knowledge of the number of real or simulated images within the 20 images. Again, there were eight human experts participating in the experiment. When each image was presented, the human expert had to make a binary decision of whether an image was real or fake.
In this case, intuitively, the overall prediction accuracy should tend toward 50% if the image simulation is perfect. This is because a perfect simulation could be classified as either real or fake as the human experts cannot identify the differences between the real and the simulated ones; it could be equally selected as either class. Statistically, if a mix of hundreds of simulated and real images were considered, the best average accuracy of a human expert should be around 50%. Results from this experiment are summarized in Table 2, in which the true positive rate (TPR), true negative rate (TNR), and Accuracy (Acc) are reported. Five out of eight testers scored an accuracy higher than 50%. On average, the prediction accuracy was . It was observed that the strongest visual cues that human experts used to differentiate between simulated and real images were geometric distortions to the fan-shaped borders of ultrasound images and not the anatomy of the fetal skull itself. Tester 8, the most experienced clinical sonographer, achieved 100% sensitivity and an overall prediction accuracy of 80%.
Table 2.
Assessment of real and simulated third-trimester images of the proposed method.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Average | |
|---|---|---|---|---|---|---|---|---|---|
| TPR (%) | 77.8 | 44.4 | 44.4 | 77.8 | 66.7 | 66.7 | 33.3 | 100 | 63.9 |
| TNR (%) | 54.5 | 72.7 | 36.4 | 63.6 | 36.4 | 63.6 | 54.5 | 63.6 | 55.7 |
| Acc. (%) | 65 | 60 | 40 | 70 | 50 | 65 | 45 | 80 | 59.4 |
Experiment 3: We further evaluated whether the simulated third-trimester images were visually different from real second-trimester ones. This experiment considered pairwise combinations of one real second-trimester scan and one corresponding simulated third-trimester scan image. The two images within the image pair were presented in random order. There were a total number of 20 images (10 image pairs). There were six testers for this experiment. Each tester was asked to classify which image was the third-trimester scan and which was the second-trimester scan. In this binary classification task, positive indicates the real second-trimester scan and negative indicates the simulated third-trimester scan. The average prediction accuracy was 91.2% and the standard deviation was 3.9%. Ideally, both the TPR and TNR should be as high as possible. Table 3 shows that a high accuracy was achieved in distinguishing the original second-trimester scan from the simulated third-trimester scan image. The average prediction accuracy was 84.2% with standard deviation of 12.4%.
Table 3.
Test result of real second-trimester scan (positive) and simulated third-trimester scan images (negative).
| 1 | 2 | 3 | 4 | 5 | 6 | Average | |
|---|---|---|---|---|---|---|---|
| TPR (%) | 100 | 100 | 90 | 90 | 100 | 100 | 96.7 |
| TNR (%) | 100 | 100 | 60 | 60 | 60 | 50 | 71.7 |
| Acc. (%) | 100 | 100 | 75 | 75 | 80 | 75 | 84.2 |
In summary:
-
•
Experiment 1 shows that the image readers in the subsequent experiments were qualified to perform fetal ultrasound image recognition.
-
•
Experiment 2 shows that the simulated third-trimester images were very close in visual appearance to real third-trimester images as the overall evaluation accuracy was close to 50%, which means that the human experts cannot tell which one is real.
-
•
Finally, experiment 3 further proves that the simulated third-trimester images are close in appearance to the real ones and distinguishable from their second-trimester input.
5.3. Quantitative Analysis
Quantitative evaluation was performed using the ScanNav® software introduced in Sec. 3.2. In this evaluation, third-trimester image simulations were considered successful if the detection rate of the choroid/ventricle, CSP, and mid-line falx were similar to those found for a real third-trimester scan. However, the clinical dataset used to build the simulation model was from a different data source than the one used to generate Table 1. Therefore, the first step was to apply ScanNav® using the training dataset of our proposed Cycle-GAN for preliminary testing. Having done this, the detection rate for key structures was calculated for the simulated third-trimester images generated from three different inputs: fixed bounding box crop, skull bounding box crop, and skull bounding box crop with data augmentation.
Table 4 shows the structure detection rate of ScanNav® applied on the training dataset of our proposed Cycle-GAN. Rows 1 and 2 show the detection rate of the real second-trimester and real third-trimester scans, respectively. The detection rate drop between the two is regarded as the base line for the quantitative comparison. It shows that the detection rate difference was 72.5%, 61.5%, and 67.0% for the ventricle, CSP, and mid-line falx, respectively. The trend of decreasing detection rate between second-trimester and third-trimester scan is consistent with the result of Table 1. It shows consistent performance by directly applying ScanNav® on our dataset. The minor structure detection rate differences between Tables 1 and 4 can be attributed to two factors. First, different ultrasound machines and sonographers were used for each analysis. Second, in the former case full resolution images were used, and in our case cropped and resized images were used.
Table 4.
Key structure detection rate of ScanNav for the real second-trimester inputs and simulated third-trimester scans.
| Choroid/ventricle (%) | CSP (%) | Mid-line falx (%) | |
|---|---|---|---|
| Real second-trimester scana | 85.0 | 69.0 | 92.5 |
| Real third-trimester scan | 12.5 | 7.5 | 25.5 |
| Simulated result using skull bounding box with data augmentation | 78.8 | 18.8 | 85.0 |
| Simulated result using skull bounding box crop | 42.5 | 18.8 | 77.5 |
| Simulated result using fixed bounding box crop | 7.5 | 11.3 | 36.3 |
Real second-trimester scans selected had a clear visualization of the choroid, the ventricle, the CSP, and the mid-line falx according to the FASP guidelines.
Table 4 rows 3 to 5 show the difference in detection rate compared with row 1. The simulated result would be concluded as valid if the detection rate drop is significant and similar compared with the rate drop between rows 1 and 2. All results are reported with a test data size of 100 images. The worst simulation result is in row 3 (simulation using skull bounding box with data augmentation). The best simulation with the most significant detection rate drop selected by ScanNav® is in row 5 (simulation with fixed bounding box crop). It had the highest detection rate drop from 85.0% to 7.5% for the ventricle and from 69.0% to 11.3% for the CSP. The mid-line detection rate drop in row 5 is lower than the real third-trimester scan in row 2. It has the highest detection rate drop among the three simulations from 92.5% to 36.3%. All three key structures have similar detection rate to real third-trimester images. Moreover, this result agrees with the human assessment in the qualitative evaluation in Sec. 5.2. The table also shows that using the skull bounding box crop has a worse performance than using the fixed bounding box crop. This is because placing the skull in the center of the image and resizing the cropped image reduces appearance variation of the input. It is much more difficult for Cycle-GAN to learn the characteristics of the images in this case. The detection rate decrease for both skull cropping variants is much lower than the base line (real third-trimester scan in row 2).
In summary, simulated third-trimester images generated with a fixed bounding box crop input were found to give the most similar structure visibility detection rate to real third-trimester scan images. By quantitatively comparing the visibility of key structures inside the head, we have shown that the method that employs a fixed crop bounding box without data argumentation to simulate third-trimester images has the most similar structure detection rate to that found with real third-trimester images. The proposed quantitative evaluation method has allowed us to successfully quantify appearance differences between real second-trimester and simulated third-trimester images. The quantitative evaluation also complements and supports the qualitative analysis findings.
5.4. Comparison with a Conventional GAN
In this section, we justify our chosen architecture in comparison with a conventional GAN. A conventional GAN6 consists of a single generator and discriminator pair. We implemented a conventional GAN and empirically determined the parameters that gave the best visual result. Specifically, the convolutional layers in the discriminator were set to and . There were two types of convolutional layer settings in the generator. The first one was , , which was used for upsampling. The second one was , , which was used for smoothing purpose between the first type of layers. “LeakyReLU” and batch normalization were applied to all convolution layers. In a conventional GAN, Gaussian noise is used as input in the generator. Thus, the simulated images were not conditioned on the second-trimester scan. Figure 9 shows a comparison of simulated images generated using a conventional GAN and our proposed method with a fixed size bounding box crop.
Fig. 9.
(a) Simulated third-trimester scan from conventional GAN. (b) Simulated third-trimester scan images from the proposed Cycle-GAN.
Simulated images from a conventional GAN were found to have lower visual quality compared with the Cycle-GAN network by eye. First, we observed checkerboard artifacts in the conventional GAN simulated images, which is shown clearly in all three images of the top row in Fig. 9. Second, images 1 and 3 in the top row of Fig. 9 show that the shape of the simulated skull is irregular, which is easily recognisable as “fake.” These types of artifacts were not observed with the Cycle-GAN model.
Table 5 shows the experiment 2 analysis on the simulated images from a conventional GAN. We compared the simulated third-trimester images with real third-trimester images. The results show that the average TPR, TNR, and accuracy are all lower than our proposed Cycle-GAN method. Recall that 50% is the optimal accuracy if a simulation is perfect. A higher number in accuracy indicates that the simulated images were easier to distinguish by visual cues. Therefore, we can conclude that the simulated images generated by the conventional GAN are visually less “real” than those generated using the Cycle-GAN method. Table 6 shows the conventional GAN results using experiment 3 in which testers were asked to distinguish between real second-trimester and simulated third-trimester images. The high accuracy in this evaluation indicates an easy separation of the real second-trimester and simulated third-trimester images. However, it is because of the low simulation quality of the GAN rather than the natural differences between second- and third-trimester TV plane.
Table 5.
Assessment of real and simulated images produced by a conventional GAN.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Average | |
|---|---|---|---|---|---|---|---|---|---|
| TPR (%) | 77.8 | 88.9 | 66.7 | 100.0 | 88.9 | 100.0 | 55.6 | 100.0 | 84.7 |
| TNR (%) | 72.7 | 72.7 | 81.8 | 90.9 | 72.7 | 100.0 | 81.8 | 81.8 | 81.8 |
| Acc. (%) | 75.0 | 80.0 | 75.0 | 95.0 | 80.0 | 100.0 | 70.0 | 90.0 | 83.1 |
Table 6.
Assessment of real second-trimester scan and simulated third-trimester scan images produced by a conventional GAN.
| 1 | 2 | 3 | 4 | 5 | 6 | Average | |
|---|---|---|---|---|---|---|---|
| TPR (%) | 77.8 | 77.8 | 66.7 | 100.0 | 88.9 | 88.0 | 83.2 |
| TNR (%) | 90.9 | 81.8 | 90.1 | 72.7 | 72.7 | 72.7 | 80.2 |
| Acc. (%) | 85.0 | 80.0 | 80.0 | 85.0 | 80.0 | 80.0 | 81.7 |
6. Conclusions
We have proposed a Cycle-GAN architecture with least square loss to simulate fetal ultrasound third-trimester TV images conditioned on second-trimester TV ultrasound images. We generated visually acceptable TV images as assessed by human experts. A qualitative evaluation shows that simulated third-trimester scan images are similar in appearance to the real ones so as to be indistinguishable when judged by observers trained to recognize images. Quantitative evaluation is often difficult for image generation tasks. A quantitative evaluation method is proposed that employs a deep learning-based algorithm to check that the quality of simulated images is similar to real images defined in terms of detectability of key structures. To our knowledge, this is the first time that a deep-learning tool has been used for quantitative evaluation in this way in medical imaging. The quantitative evaluation shows that the detectability of structures in simulated images is similar to the detectability observed in real third-trimester images. Finally, we have shown that our method gives superior results than that generated using a conventional GAN architecture.
Acknowledgments
The authors acknowledge the ERC (ERC-ADG-2015 694581, project PULSE), EPSRC (EP/MO13774/1, EP/L505316/1), and the NIHR Biomedical Research Centre funding scheme.
Biographies
Yangdi Xu graduated from the University of Bristol and specialized in machine learning and computer vision. His research focused on modeling human daily routine/activity patterns from images and videos. As a research associate at the University of Oxford, his research focuses on ultrasound image analysis using deep-learning models. His project collaborates with the John Redcliff Hospital and involves but is not limited to obstetric ultrasound image simulations and gaze pattern modeling of sonographers.
Lok Hin Lee is a DPhil candidate at the Institute of Biomedical Imaging, University of Oxford. He is the recipient of the Croucher Scholarship for Doctoral Study. His doctoral research focuses on increasing the availability and ease of use of ultrasound imaging in resource-constrained areas by using deep-learning techniques to aid diagnosis, and he is in active collaboration with clinicians from the John Radcliffe Hospital.
Biographies of the other authors are not available.
Disclosures
ScanNav® is a precommercial software application of Intelligent Ultrasound Ltd. The version used in this study will not necessarily be the version used in any future product or research study. JAN and AP are cofounders of and consultants to Intelligent Ultrasound Ltd. JAN suggested the use of ScanNav® to support the evaluation of the simulations in this study. The other authors declare that there are no conflicts of interest related to this article.
Contributor Information
Yangdi Xu, Email: yangdidixu@gmail.com.
Lok Hin Lee, Email: lokhin.lee@hertford.ox.ac.uk.
Lior Drukker, Email: lior.drukker@wrh.ox.ac.uk.
Mohammad Yaqub, Email: mohammad.yaqub@intelligentultrasound.com.
Aris T. Papageorghiou, Email: aris.papageorghiou@wrh.ox.ac.uk.
Alison J. Noble, Email: alison.noble@eng.ox.ac.uk.
References
- 1.Henrichs J., et al. , “Effectiveness and cost-effectiveness of routine third trimester ultrasound screening for intrauterine growth restriction: study protocol of a nationwide stepped wedge cluster-randomized trial in The Netherlands (the Iris Study),” BMC Pregnancy Childbirth 16(1), 310 (2016). 10.1186/s12884-016-1104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sovio U., et al. , “Screening for fetal growth restriction with universal third trimester ultrasonography in nulliparous women in the pregnancy outcome prediction (POP) study: a prospective cohort study,” The Lancet 386(10008), 2089–2097 (2015). 10.1016/S0140-6736(15)00131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drukker L., et al. , “Safety indices of ultrasound: adherence to recommendations and awareness during routine obstetric ultrasound scanning,” Ultraschall Med. 41(2), 138–145 (2020). 10.1055/a-1074-0722 [DOI] [PubMed] [Google Scholar]
- 4.Drukker L., et al. , “How often do we incidentally find a fetal abnormality at the routine third-trimester growth scan? A population-based study,” Am. J. Obstet. Gynecol. (2020). 10.1016/j.ajog.2020.05.052 [DOI] [PubMed] [Google Scholar]
- 5.Cant R. P., Cooper S. J., “Simulation-based learning in nurse education: systematic review,” J. Adv. Nursing 66(1), 3–15 (2010). 10.1111/j.1365-2648.2009.05240.x [DOI] [PubMed] [Google Scholar]
- 6.Goodfellow I., et al. , “Generative adversarial nets,” in Adv. Neural Inf. Process. Syst., pp. 2672–2680 (2014). [Google Scholar]
- 7.Radford A., Metz L., Chintala S., “Unsupervised representation learning with deep convolutional generative adversarial networks,” CoRR (2016). [Google Scholar]
- 8.Liu M.-Y., Tuzel O., “Coupled generative adversarial networks,” in Adv. Neural Inf. Process. Syst., pp. 469–477 (2016). [Google Scholar]
- 9.Mao X., et al. , “Least squares generative adversarial networks,” in Proc. IEEE Int. Conf. Comput. Vision, pp. 2794–2802 (2017). 10.1109/ICCV.2017.304 [DOI] [Google Scholar]
- 10.Hu Y., et al. , “Freehand ultrasound image simulation with spatially-conditioned generative adversarial networks,” Lect. Notes Comput. Sci. 10555, 105–115 (2017). 10.1007/978-3-319-67564-0_11 [DOI] [Google Scholar]
- 11.Long J., Shelhamer E., Darrell T., “Fully convolutional networks for semantic segmentation,” in Proc. IEEE Conf. Comput. Vision and Pattern Recognit., pp. 3431–3440 (2015). 10.1109/CVPR.2015.7298965 [DOI] [PubMed] [Google Scholar]
- 12.Zhu J.-Y., et al. , “Unpaired image-to-image translation using cycle-consistent adversarial networks,” in Proc. IEEE Int. Conf. Comput. Vision, pp. 2223–2232 (2017). 10.1109/ICCV.2017.244 [DOI] [Google Scholar]
- 13.Wolterink J. M., et al. , “MR-to-CT synthesis using cycle-consistent generative adversarial networks,” in 31st Conf. Neural Inf. Process. Syst. (2017). [Google Scholar]
- 14.Jiao J., et al. , “Anatomy-aware self-supervised fetal MRI synthesis from unpaired ultrasound images,” Lect. Notes Comput. Sci. 11861, 178–186 (2019). 10.1007/978-3-030-32692-0_21 [DOI] [Google Scholar]
- 15.Salomon L., et al. , “Practice guidelines for performance of the routine mid-trimester fetal ultrasound scan,” Ultrasound Obstet. Gynecol. 37(1), 116–126 (2011). 10.1002/uog.8831 [DOI] [PubMed] [Google Scholar]
- 16.England P. H., “Fetal anomaly screening programme: programme handbook” (2015).
- 17.Salimans T., et al. , “Improved techniques for training GANs,” in Adv. Neural Inf. Process. Syst., pp. 2234–2242 (2016). [Google Scholar]
- 18.Yaqub M., et al. , “Op01. 10: auditing the quality of ultrasound images using an AI solution: scannav® for fetal second trimester ultrasound scans,” Ultrasound Obstet. Gynecol. 54, 87 (2019). 10.1002/uog.2065630620419 [DOI] [Google Scholar]
- 19.Yaqub M., et al. , “Quality-improvement program for ultrasound-based fetal anatomy screening using large-scale clinical audit,” Ultrasound Obstet. Gynecol. 54(2), 239–245 (2019). 10.1002/uog.20144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronneberger O., Fischer P., Brox T., “U-net: convolutional networks for biomedical image segmentation,” Lect. Notes Comput. Sci. 9351, 234–241 (2015). 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 21.Mao X., et al. , “On the effectiveness of least squares generative adversarial networks,” IEEE Trans. Pattern Anal. Mach. Intell. 41, 2947–2960 (2019). 10.1109/TPAMI.2018.2872043 [DOI] [PubMed] [Google Scholar]