Abstract
Nasopharyngeal flocked swabs placed in viral transport media (VTM) are the preferred collection methodology for respiratory virus testing. Due to the rapid depletion of available reagents and swabs, we have validated an alternative swab placed in phosphate-buffered saline (PBS) for use in respiratory virus testing in a SARS-CoV-2 real-time polymerase chain reaction assay and a multiplexed respiratory virus panel. We collected nasopharyngeal (NP) swabs and oropharyngeal (OP) swabs from 10 healthy volunteers. Flocked swabs were placed in VTM and alternative swabs in PBS. In this feasibility study, we show that NP collection is better for detection of human material than OP collection, as measured by significantly lower RNase P gene cycle threshold values, and that a Dacron polyester swab in PBS shows equivalent detection of SARS-CoV-2 and RSV to a flocked swab in VTM in contrived specimens. Diluted SARS-CoV-2–positive patient specimens are detectable for up to 72 h at 4 °C.
Keywords: SARS-CoV-2, 2019-nCOV, COVID-19, Alternative reagents
1. Introduction
Following its emergence in late 2019, severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) causing the associated coronavirus disease 2019 (COVID-19) has reached pandemic proportions. In order to monitor viral spread through the population, it is critical that testing facilities are able to detect this novel coronavirus. Viral detection has overwhelmingly been taken over by molecular methods, through the use of polymerase chain reaction (PCR) assays and multiplexed syndromic platforms. For this novel coronavirus, clinical microbiology and public health laboratories in the United States have relied on commercially developed emergency use authorizations (EUAs) as a response to the SARS-CoV-2 pandemic (FDA, 2020). For these EUA assays and patients with respiratory symptoms, it is commonplace to utilize nasopharyngeal (NP) collection with a nylon-tipped flocked swab to promote the collection of cell-associated virus. However, in response to the dramatic increase in global and nationwide testing, there has been a shortage of reagents available to testing laboratories. The current reagents approved for viral testing include flocked swabs placed into viral transport media (VTM), and a shortage could have the undesired effect to halt testing for COVID-19 and other respiratory illnesses (CDC, 2020). This feasibility study was conducted in response to the nationwide shortage in collection reagents during the pandemic caused by SARS-CoV-2 (ASM, 2020). In this study, we show that a Dacron polyester swab in phosphate-buffered saline (PBS) can be substituted for a flocked swab in VTM for both NP and oropharyngeal (OP) collection and that PBS is a suitable medium to allow for the detection of both SARS-COV-2 and RSV in contrived specimens and diluted known SARS-CoV-2–positive patient specimens over time.
2. Materials and methods
2.1. Detection of human RNase P gene internal control using the SARS-CoV-2 RT-PCR assay
In response to the SARS-CoV-2 pandemic, the Centers for Disease Control and Prevention (CDC) 2019 novel Coronavirus (2019-nCoV) Real-Time PCR Diagnostic Panel has received US Food and Drug Administration (FDA) approval and has been adopted by the National Institutes of Health (NIH) Clinical Center, herein referred to as the SARS-CoV-2 RT-PCR assay (CDC, 2020; FDA, 2020). The SARS-CoV-2 RT-PCR assay is used to screen NP and OP swab specimens in VTM and BAL for SARS-CoV-2 from patients with respiratory symptoms. The SARS-CoV-2 RT-PCR assay implemented at NIH Clinical Center utilizes the easyMAG automated nucleic acid extractor (bioMérieux, Marcy l'Etoile, France). A Taqman assay using 2 primer/probe sets is used to detect 2 distinct regions of the N gene (nucleocapsid protein, referred to as N1 and N2) and an internal control to detect the human RNase P (RP1) gene present in all specimen collections. This assay was performed on the ABI 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA).
In order to determine the suitability of the Dacron polyester swab (Thermo Fisher Scientific, 149601J, Waltham, MA) in PBS at pH 7.2 (KD Medical, RGF-3190, Columbia, MD) compared to the flocked nylon swab (Becton Dickinson, BD B220252, Franklin Lakes, NJ) in VTM (Thermo Fisher Scientific, R12500 MicroTest M4 Tube, Waitham, MA) for NP and OP collection, each swab combination was used for an NP and an OP collection on 10 healthy volunteers to give a total of 40 samples. Healthy volunteers consented to have replicate swabs tested for RP1, N1, and N2 gene with the SARS-CoV-2 RT-PCR assay and respiratory virus testing with the BioFire FilmArray Respiratory Pathogen Panel. Collection was performed by trained personnel wearing N95 masks, eye protection, gloves, and disposable gowns. Specimens were transported in a secondary labeled container. Each swab was placed into 3 mL of media (PBS or VTM) and stored on ice for transport to the clinical laboratory for testing. Upon receipt, a portion of each specimen was aliquoted in a biosafety cabinet and stored at 4 °C. To test for the adequate collection of human material during collection, 1 aliquot of each specimen was processed at 24 h and 72 h for nucleic acid extraction. NP and OP specimens from both comparison conditions (flocked swab in VTM; Dacron swab in PBS) were extracted and subjected to RT-PCR looking for amplification of the RNase P gene.
2.2. Detection of RSV in contrived specimens using the BioFire FilmArray Respiratory Pathogen Panel
To test for the ability of the transport medium to adequately preserve the virus prior to detection, 2 aliquots of each NP collection were spiked with a stock of cultured RSV to generate contrived specimens. The RSV stock was stored at −70 °C at 10,000 times the limit of detection (LOD) for the BioFire FilmArray Respiratory Panel (bioMérieux, Marcy l'Etoile, France), which was previously defined as 2 Tissue Culture Infectious Dose 50/mL (TCID50/mL). Handling of RSV and healthy volunteer specimens was done in a biosafety cabinet while wearing an N95 mask. The day of collection, the 10,000×-LOD stock was diluted 1:10 in PBS and 1:100 into the aliquot to give a final RSV concentration of 10×-LOD. The stored RSV was diluted once in PBS to 10×-LOD and tested immediately to confirm detectability of the virus. At 24 and 72 h after collection and storage at 4 °C, 300 μL of contrived specimen was tested using the BioFire FilmArray Respiratory Panel. One aliquot of each specimen was tested at 24 h to account for potential real-time storage and turnaround of specimens collected 1 day and tested the next, and the remaining was tested at 72 h to test the longevity of sample storage.
2.3. Detection of heat-inactivated SARS-CoV-2 viral particles in contrived specimens using the using the SARS-CoV-2 RT-PCR assay
With the use of a biosafety cabinet and standard precautions, 3 paired specimens from healthy volunteers were spiked with heat-inactivated SARS-CoV-2 viral particles. While SARS-CoV-2 isolation and propagation require a Biosafety Level 3 (BSL-3) laboratory and BSL-3 practices, heat treatment of SARS-CoV-2 renders the virus noninfectious, and the heat-inactivated form is considered a BSL-1 agent per the manufacturer's instructions. Heat-inactivated SARS-CoV-2 (ATCC, VR-1986HK, Manassas, VA) was diluted to a final concentration of 2 × 103 times the established LOD used for the SARS-CoV-2 EUA assay detection of N1 and N2 genes. The LOD was previously determined by our laboratory to be 0.5 genome copies/μL using digital droplet PCR. Medium was either the VTM from which a flocked nylon swab from a NP collection had been inserted or PBS from which a Dacron polyester from a NP collection had been inserted. Three aliquots of 250 μL were made of the contrived specimens containing SARS-CoV-2 and were stored at 4 °C. Nucleic acid extraction and PCR were performed as described in the SARS-CoV-2 RT-PCR assay at time 0, 24 h, and 72 h after spiking (FDA, 2020).
2.4. Detection of virus obtained from specimens from SARS-CoV-2–positive patient samples diluted in media obtained from healthy volunteers
VTM from NP swabs were obtained from patients with known SARS-CoV-2–positive PCRs that had cycle threshold (Ct) values of 15–32 for the N1 gene target and 17–36 for the N2 gene target. This allowed for testing the detection of strong (Ct 15–24) and weakly (Ct > 30) positive specimens prior to dilution (Mitchell et al., 2020). The known SARS-CoV-2–positive samples were identified from patients that had been sampled with the standard collection method: an NP collection from a flocked nylon swab placed into VTM. Each sample was stored at 4 °C until it was diluted 1:10 in pooled media from NP collections from healthy volunteers. Media used for dilution were either VTM into which a collected flocked nylon swab had been inserted or PBS into which a collected Dacron polyester swab had been inserted. All diluted samples were stored at 4 °C. SARS-CoV-2 PCR was then performed at time 0, 24 h, and 72 h to determine whether SARS-CoV-2 remained detectable in media obtained using the new collection method.
2.5. Equivalence and stability
Equivalence and stability of RP1 gene detection were measured using a 3-way analysis of variance (ANOVA) with Tukey's multiple-comparisons test to compare the effect of variation of 3 factors: time (24 versus 72 h), collection type (NP versus OP), and swab/media (flocked swab/VTM versus Dacron swab/PBS). Our goal of equivalence for detection of the N1, N2, and RP1 genes was concordant results within 1 Ct value. Similarly, our intent to measure stability when comparing the time points of each collection type was a range within 1 Ct value. Final determination of equivalence and stability was based on consistent trends between comparisons. For specimens diluted 1:10 (n = 1 per condition), equivalence and stability were defined as the comparison of paired specimens between VTM and PBS and between time points, respectively, within 1 Ct value. RSV detection through the BioFire FilmArray platform was qualitative in nature. Therefore, equivalence between VTM and PBS and stability between 24 and 72 h were defined as 90% qualitative concordance in detection of RSV in the same collection type (i.e., NP, OP).
3. Results
3.1. Polyester Dacron swab in PBS shows equivalent performance to a flocked swab in VTM and stability up to 72 h for the detection of RP1 using the SARS-CoV-2 RT-PCR assay
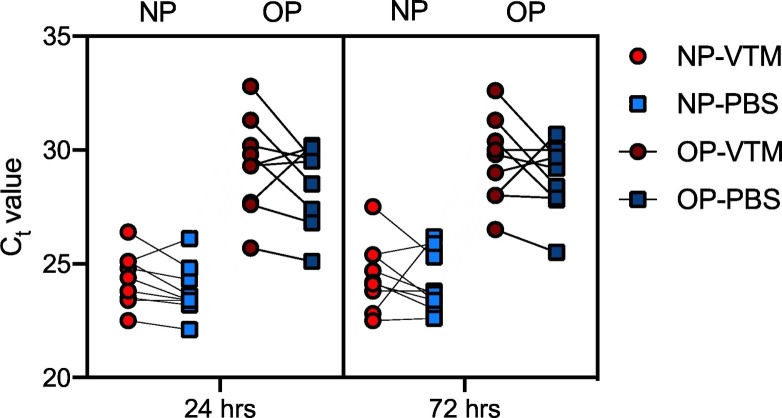
The human internal control gene RP1 was detectable in all collection conditions, with the exception of 1 failed extraction of an NP collection (VTM/flocked swab) at 72 h (Supplementary Table 1). The Dacron swab in PBS showed equivalent performance in the detection of the RP1 gene when compared to the currently used flocked swab in VTM for NP and OP collection methods at 24 and 72 h (Fig. 1). Moreover, the signal was comparable between 24- and 72- h time points, indicating that the viral target was stable in both VTM and PBS when stored at 4 °C (Fig. 1). The human RP1 gene was not measured at 0 h, and thus, stability cannot be determined between 0 and 24 h. These data show that Ct values are influenced by the collection method (NP versus OP). Data here show that a lower Ct value is attainable with the NP collection when compared to OP collection regardless of the swab/media used for specimen storage or the time point tested.
Fig. 1.
Dacron polyester swab in PBS shows equivalent performance and stability up to 72 h compared to a flocked nylon swab in VTM. Nasopharyngeal and oropharyngeal collections were obtained using a flocked nylon swab placed into VTM (red, circle) or a Dacron polyester swab and placed into PBS (blue, square). Ct values for detection of the human RNase P (RP1) gene were measured by RT-PCR from 10 healthy volunteers at 24 h (light red, light blue) or 72 h (dark red, dark blue). Each of 8 collection condition and time point combinations was tested on 10 healthy volunteers, with paired comparisons shown between flocked swab/VTM and Dacron swab/PBS collection methods. A 3-way ANOVA with Tukey's multiple-comparisons test was performed to compare Ct values between NP and OP collection types in each swab/media combination at 24 and 72 h. Multiple comparisons show that at both 24 and 72 h, the Ct values between NP-VTM and OP-VTM and NP-PBS and OP-PBS differ significantly, with all 4 comparisons giving P < 0.0001. Thus, the collection type (NP versus OP) led to a significant source of variation (P < 0.0001).
3.2. PBS shows equivalent performance to VTM and stability at 24 and 72 h for the detection of RSV using the BioFire FilmArray Respiratory Pathogen Panel
Here, the feasibility of using PBS for viral detection was determined by using contrived specimens spiked with live RSV virus at 10×-LOD. A 10× RSV-LOD dilution in PBS was tested immediately and was detected by the BioFire FilmArray Respiratory Pathogen Panel (data not shown). At 24 h, RSV was detected in 9 of 10 spiked VTM samples and 10 out of 10 spiked PBS samples (Table 1 ). Additionally, at 72 h, RSV was detected in 9 out of 10 spiked VTM samples and all 10 spiked PBS samples (Table 1). Thus, RSV was detected in all spiked PBS media but was not detected in 2 specimens of spiked VTM. These data suggest that PBS is both equivalent in performance to VTM and stable between 24 and 72 h, as it meets the definition of 90% qualitative concordance between media and time points.
Table 1.
PBS shows equivalent performance to VTM and stability at 24 and 72 h for the detection of RSV using the BioFire FilmArray Respiratory Pathogen Panel. NP collections from flocked swabs in VTM and Dacron swabs in PBS (n = 10 each) were spiked with 10×-LOD of RSV to give a final concentration of 20 TCID50/mL. The results for RSV on the Respiratory Pathogen Panel were detected (+) or not detected (−).
| Type | Vehicle/swab | Time point | BioFire FilmArray RP Panel results | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NP | VTM/flocked | 24 h | − | + | + | + | + | + | + | + | + | + |
| 72 h | + | + | + | + | + | + | − | + | + | + | ||
| NP | PBS/Dacron | 24 h | + | + | + | + | + | + | + | + | + | + |
| 72 h | + | + | + | + | + | + | + | + | + | + | ||
3.3. PBS shows equivalent detection of heat-inactivated SARS-CoV-2 viral particles compared to VTM using the SARS-CoV-2 RT-PCR assay
Heat-inactivated SARS-CoV-2 viral particles were detected in spiked healthy volunteer media following NP collection at time 0, 24 h, and 72 h after spiking (Supplementary Table 2; Fig. 2). One sample at 72 h in VTM had no amplification for any of the targets and thus no Ct value for N1, N2, or RP1, consistent with an extraction failure. In response to this failed extraction, 2 additional samples were spiked and extracted for PCR at 24 and 72 h to give a final n = 3 (time 0) and n = 5 (24 h, 72 h). Two paired N2 gene Ct values had a difference of 1.80 and 1.85, and 2 paired RP1 data points differed by 2.83 and 3.99 (Supplementary Table 2). However, other absolute differences were consistently closer to or less than 1 Ct value (N1, n = 12; N2, n = 10; RP1, n = 10), demonstrating that paired specimens were similarly detected in VTM and PBS (Supplementary Table 2; Fig. 2). Accordingly, equivalence standards were met for the all gene targets (N1, N2, RP1) in the SARS-CoV-2 real-time PCR assay. To compare stability, each target Ct value was compared between time 0, 24 h, and 72 h. Curiously, the N1 target appeared to have a decreasing Ct value over time, and this trend was consistent between VTM and PBS (Supplementary Table 2). However, the N2 and RP1 gene targets had stable Ct values over the time points, ranging within roughly 1 Ct value (Supplementary Table 2).
Fig. 2.
PBS and VTM show equivalent detection of heat-inactivated SARS-CoV-2 and human material. Nasopharyngeal collections were obtained using a flocked swab placed into VTM (solid symbols) or a Dacron polyester swab and placed into PBS (open symbols). Ct values for detection of the N1 (green) and N2 (blue) nucleocapsid gene segments and internal control RP1 (magenta) gene were measured at time 0 (n = 3), 24 h (n = 5), and 72 h (n = 5) from media spiked with heat-inactivated SARS-CoV-2. One sample in the VTM 72-h condition failed the nucleic acid extraction step. Each symbol represents the Ct value from 1 sample, with VTM and PBS paired symbols connected by lines, represented as time 0 (gray dashed line), 24 h (black dotted line), and 72 h (black solid line).
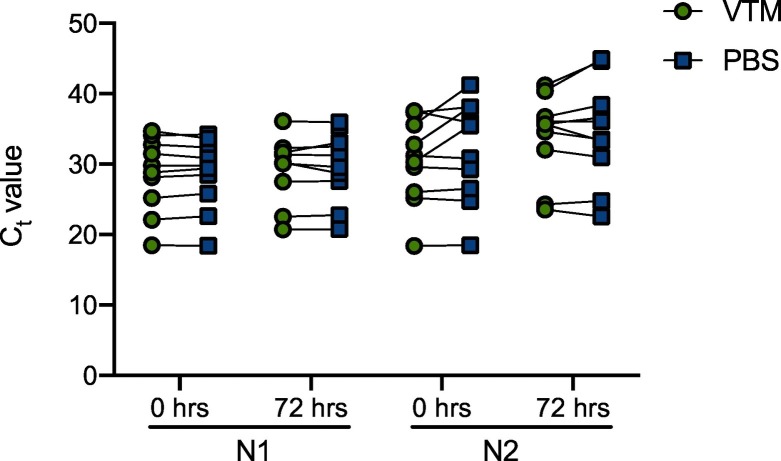
3.4. PBS shows similar detection of diluted positive SARS-CoV-2 patient specimens compared to VTM using the SARS-CoV-2 RT-PCR assay
Known positive samples that were diluted 1:10 in healthy volunteer media (VTM or PBS) following NP collection (flocked or Dacron swabs, respectively) had viral RNA detectable immediately after dilution and for up to 72 h after dilution. Ct values for viral targets N1 and N2 increased approximately 3 Ct values in the diluted samples (Supplementary Table 3), consistent with a 1:10 dilution (Mitchell et al., 2020). The RP1 gene Ct values were similar across all time points in diluted PBS and VTM (data not shown). All paired specimens remained positive by PCR after 72 h, with the exception of 1 specimen diluted in VTM which did not amplify the N1 target. Known SARS-CoV-2 samples diluted 1:10 in VTM or PBS were similar but did not show equivalence, as defined as +/− 1 Ct value between paired samples (Supplementary Table 3; Fig. 3). In particular, specimens with higher original Ct values (lower detectable viral RNA) had greater variability in the detection in VTM and PBS, although there were no clear trends. Moreover, between VTM- and PBS-diluted specimens, there was consistently greater variation in Ct values in the N2 gene (Supplementary Table 3; Fig. 3). Both VTM and PBS did not show stability in Ct values (defined as +/− 1 Ct value) between time points for the detection of N1 and N2 gene targets (Supplementary Table 3; Fig. 3). Although some did meet this requirement, many did not.
Fig. 3.
PBS shows similar detection of diluted positive SARS-CoV-2 patient specimens compared to VTM using the SARS-CoV-2 real-time PCR assay. Ten known positive SARS-CoV-2 patient specimens with varying Ct values were diluted 1:10 into VTM from a flocked swab NP collection (green circles) or PBS from a Dacron swab NP collection (blue squares). Paired Ct values from each diluted sample are shown for the N1 and N2 genes at 0 h and 72 h after dilution.
4. Discussion
In response to the reagent shortage faced by laboratories across the country in response to the SARS-CoV-2 pandemic, this feasibility study aimed to identify replacement components with which to continue respiratory virus testing. Our results show the suitability and equivalence of using Dacron polyester swabs in PBS for efficient collection by NP and OP collection methods, representing a change in both swab type and transport media type. Though a previous study had compared a change in both swab and solution simultaneously, the method also used a nasal sample instead of a nasopharyngeal sample, which may have contributed to a lower sensitivity (Péré et al., 2020). Recent studies have had success addressing the paucity of reagent availability by utilizing various collection methods, including nasal samples via mid-turbinate or anterior nasal collections (Moran et al., 2020; Péré et al., 2020; Rhoads et al., 2020; Vermeriren et al., 2020). By maintaining nasopharyngeal collection in this study, this alternative collection method upholds the current CDC recommendation for nasopharyngeal collection, especially in asymptomatic patients (CDC, 2020). Moreover, our results show that nasopharyngeal collection allows for more sensitive detection (lower Ct values) of human material by RT-PCR than oropharyngeal collection. This may be due to an ability to collect more cells from the nasopharynx than the oropharynx, possibly because of the narrower cavity of the nasopharynx, and/or the absence of saliva. Additionally, the volunteers reported discomfort during the NP collection from the Dacron swab, which is larger than the flocked swab, and could be considered a drawback for this specific Dacron swab.
Our findings are consistent with previous conclusions that a lack of available VTM can be replaced with the common laboratory reagent PBS for detection of SARS-CoV-2 using a commercially developed molecular assay (Cobas® SARS-CoV-2; Roche Diagnostics, Indianapolis, IN) (Rodino et al., 2020). Herein, we also show that PBS is able to support viral detection of RSV and SARS-CoV-2 from independently collected and contrived specimens with molecular detection methods including multiplexed BioFire Respiratory Pathogen Panel and the SARS-CoV-2 real-time PCR assay, respectively. Although RSV was detected in all PBS-based specimens, RSV was not detected in 1 VTM specimen at 24 h and 1 VTM specimen at 72 h. This may indicate variation in preparation of the contrived samples near the LOD or reflect the inherent sensitivity of the BioFire platform. Live RSV alone was chosen because it was readily available in the laboratory at an exact concentration. RSV LOD determinations on the BioFire platform had previously been done in our laboratory, and therefore, we could address sensitivity of this respiratory virus in PBS near the LOD. In addition to obscuring SARS-CoV-2 testing, reagent shortages may also interrupt collection of specimens for testing of other respiratory viruses. These findings suggest that PBS may be validated as a transport medium for molecular detection of other respiratory viruses.
Lastly, we established that both heat-inactivated SARS-CoV-2 and virus diluted from SARS-CoV-2–positive patient specimens are stable in PBS for up to 72 h at 4 °C. A single specimen of heat-inactivated SARS-CoV-2 spiked into VTM with a flocked swab (old methodology) had an undetectable result for all 3 PCR targets (N1, N2, and RP1) at the 72-h time point only. We believe this was due to a single RNA extraction failure, as the RP1 gene was detected from the same volunteer at a different time point. However, we were unable to repeat the assay on the same contrived specimen due to limited availability of remaining media from that healthy volunteer. In order to address this shortcoming, we retrospectively added 2 more data points to this 72-h time point and the earlier 24- h time point. We observed greater variability in Ct values from patient specimens diluted in healthy volunteer matrix for the N2 target (maximum delta Ct of 5.62) compared to the N1 target for which the highest difference in Ct values was 1.55. Diluted specimens with higher baseline N2 Ct values showed greater variation. It has been suggested that diluting specimens with Ct values equal or greater to 31 may approach the LOD of the test (Mitchell et al., 2020). Importantly, the SARS-CoV-2 virus remained detectable in PBS after storage for 72 h at 4 °C despite the differences in Ct values.
For unknown reasons, the N1 gene appears to trend toward a lower Ct count over time in heat-inactivated SARS-CoV-2 viral particles spiked into both VTM and PBS media obtained from healthy volunteers. This trend was not seen in the N2 or RP1 gene, nor with 1:10 diluted clinical samples positive for SARS-CoV-2. Importantly, the qualitative detection of heat-inactivated SARS-CoV-2 was present in all of the PBS media at all time points. While this study is limited by the absence of a positive volunteers directly sampled by the new and old methodologies, the final feasibility of a new methodology utilizing Dacron polyester swabs in PBS was presented from known SARS-CoV-2–positive patient specimens, even starting with low levels of virus and diluted 1:10 in media from NP collections. The PBS samples remained SARS-CoV-2 positive after 1:10 dilution for up to 72 h after refrigerated storage and showed equivalent performance (within 1 Ct value) via paired comparison to the existing VTM media being used. While we found that substituting PBS for VTM is useful for molecular detection using PCR, it may not be appropriate for viral culture if that were required.
These results come at a critical moment. SARS-CoV-2 testing must continue using reagents that allow for equivalent testing performance, and we have shown here that commonly stocked laboratory items, including Dacron polyester swabs and PBS, may be substituted for viral detection after appropriate equivalence studies are conducted.
The following are the supplementary data related to this article.
RNase P gene Ct values. Ct values obtained from the human RP1 gene internal control from NP and OP collections of flocked swabs placed into VTM (VTM/FL) or Dacron swabs placed into PBS (PBS/DA) after 24 h and 72 h of storage at 4 °C (n = 10 each condition/time point). Average ± standard deviation is shown for RP1 at each time point/collection type. Stability for each vehicle/swab collection type compares 24 h to 72 h, and equivalence compares VTM and PBS between time points. The specimen labeled (−) failed the nucleic acid extraction and did not have a Ct value. All samples were negative for SARS-CoV-2 gene targets N1 and N2.
PBS shows equivalent detection of heat-inactivated SARS-CoV-2 viral particles compared to VTM using the SARS-CoV-2 RT-PCR assay. Ct values obtained from the SARS-CoV-2 N1 and N2 nucleocapsid genes and human RP1 gene internal control from NP collections of flocked swabs placed into VTM or Dacron swabs placed into PBS, immediately after (n = 3) spiking with heat-inactivated SARS-CoV-2 viral particles at 2000×-LOD to give a final concentration of 1000 genome copies/μL, or after 24 h (n = 5) and 72 h (n = 5) of storage at 4 °C. Average ± standard deviation is shown for N1, N2, and RP1 detection for each collection type at each time point. The absolute difference in Ct values from paired specimens is shown in the far right.
PBS shows equivalent detection of diluted positive SARS-CoV-2 patient specimens compared to VTM using the SARS-CoV-2 RT-PCR assay. NP swabs were collected from healthy volunteers in respective media and used to dilute known SARS-CoV-2 specimens 1:10, where the leftmost column shows the original N1 and N2 Ct values for reference. Each condition was paired at time 0 (time of dilution) and 72 h after dilution and storage at 4 °C. The absolute difference in Ct values from paired specimens for N1 and N2 is shown in the far right.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH Clinical Center, Department of Laboratory Medicine. We are grateful to the laboratory of Michael Memoli for supplying us with live RSV virus for this experiment. We thank the healthy volunteers for allowing repeat NP and OP specimen collection. We also acknowledge the laboratory technologists, staff, nurses, and physicians working diligently in response to the SARS-CoV-2 pandemic.
References
- ASM ASM expresses concern about coronavirus test reagent shortages. 2020, March 10. https://www.asm.org/Articles/Policy/2020/March/ASM-Expresses-Concern-about-Test-Reagent-Shortages Available from:
- CDC Interim guidelines for collecting, handling, and testing clinical specimens from persons for coronavirus disease 2019 (COVID-19) 2020, April 14. https://www.cdc.gov/coronavirus/2019-ncov/lab/lab-biosafety-guidelines.html Available from:
- FDA Letter of authorization. 2020, March 15. https://www.fda.gov/media/134919/download Available from:
- Mitchell S., St. George K., Rhoads D., Butler-Wu S., Dharmarha V., McNult P. Verification procedure for commercial tests with Emergency Use Authorization for the detection of SARS-CoV-2 RNA American Society of Microbiology. 2020, April 3. https://asm.org/ASM/media/Protocol-Images/ASM_EUA_verification_040220_FINAL.pdf Available from: [DOI] [PMC free article] [PubMed]
- Moran A., Beavis K.G., Mautshek S.M., Ciaglia C., Francois N., Tesic V. The detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche 2 cobas SARS-CoV-2 assays. J Clin Microbiol. 2020 doi: 10.1128/JCM.00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péré H, Podglajen I, Wack M, Flamarion E, Mirault T, Goudot G, Hauw-Berlemont C, Le L, Caudron E, Carrabin S, Rodary J, Ribeyre T, Bélec L, Veyer D. 2020. Nasal swab sampling for SARS-CoV-2: a convenient alternative in time of nasopharyngeal swab shortage. J Clin Microbiol. Doi: 10.1128/JCM.00721-20:JCM.00721-20. [DOI] [PMC free article] [PubMed]
- Rhoads D.D., Cherian S.S., Roman K., Stempak L.M., Schmotzer C.L., Sadri N. Comparison of Abbott ID Now, Diasorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol. 2020 doi: 10.1128/JCM.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino K.G., Espy M.J., Buckwalter S.P., Walchak R.C., Germer J.J., Fernholz E. Evaluation of saline, phosphate buffered saline and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J Clin Microbiol. 2020 doi: 10.1128/jcm.00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeriren C., Marchand-Senécal X., Sheldrake E., Bulier D., Smieja M., Chong S. Comparison of Copan Eswab and FLOQswab for COVID-19 PCR diagnosis: working around a supply shortage. J Clin Microbiol. 2020 doi: 10.1128/JCM.00669-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNase P gene Ct values. Ct values obtained from the human RP1 gene internal control from NP and OP collections of flocked swabs placed into VTM (VTM/FL) or Dacron swabs placed into PBS (PBS/DA) after 24 h and 72 h of storage at 4 °C (n = 10 each condition/time point). Average ± standard deviation is shown for RP1 at each time point/collection type. Stability for each vehicle/swab collection type compares 24 h to 72 h, and equivalence compares VTM and PBS between time points. The specimen labeled (−) failed the nucleic acid extraction and did not have a Ct value. All samples were negative for SARS-CoV-2 gene targets N1 and N2.
PBS shows equivalent detection of heat-inactivated SARS-CoV-2 viral particles compared to VTM using the SARS-CoV-2 RT-PCR assay. Ct values obtained from the SARS-CoV-2 N1 and N2 nucleocapsid genes and human RP1 gene internal control from NP collections of flocked swabs placed into VTM or Dacron swabs placed into PBS, immediately after (n = 3) spiking with heat-inactivated SARS-CoV-2 viral particles at 2000×-LOD to give a final concentration of 1000 genome copies/μL, or after 24 h (n = 5) and 72 h (n = 5) of storage at 4 °C. Average ± standard deviation is shown for N1, N2, and RP1 detection for each collection type at each time point. The absolute difference in Ct values from paired specimens is shown in the far right.
PBS shows equivalent detection of diluted positive SARS-CoV-2 patient specimens compared to VTM using the SARS-CoV-2 RT-PCR assay. NP swabs were collected from healthy volunteers in respective media and used to dilute known SARS-CoV-2 specimens 1:10, where the leftmost column shows the original N1 and N2 Ct values for reference. Each condition was paired at time 0 (time of dilution) and 72 h after dilution and storage at 4 °C. The absolute difference in Ct values from paired specimens for N1 and N2 is shown in the far right.