Abstract
Background
Du Zhong (DZ), or Eucommiae Cortex, a traditional Chinese herbal medicine, has been used to treat osteoporosis. Although it has been reported that DZ can improve bone mass in ovariectomized rats, its pharmacological mechanisms in treating osteoporotic fractures (OPF) remain unclear.
Methods
In this study, we used a network pharmacological manner to explore its potential complicated mechanism in treating OPF. We obtained DZ compounds from TCMSP and BATMAN-TCM databases and collected potential targets of these compounds through target fishing based on TCMSP and BATMAN-TCM databases. Next, we collected the OPF targets by using CTD, GeneCards, OMIM, HPO, and GenCLiP 3 databases. And then the overlapping genes between DZ potential targets and OPF targets were used to build up the protein-protein interaction (PPI) network and to analyze their interactions and find out the big hub genes in this network. Subsequently, clusterProfiler package in R language was utilized to conduct the enrichment of Gene Ontology biological process and KEGG pathways.
Results
There were totally 93 active compounds and 916 related targets in DZ. After the enrichment analysis, we collected top 25 cellular biological processes and top 25 pathways based on the adjusted P value and found that the DZ anti-OPF effect was mainly associated with the regulation of ROS and inflammatory response. Furthermore, 64 hub genes in PPI network, such as MAPK1 (degree = 41), SRC (degree = 39), PIK3R1 (degree = 36), VEGFA (degree = 31), TP53 (degree = 29), EGFR (degree = 29), JUN (degree = 29), AGT (degree = 29), MAPK1, SRC, PIK3R1, VEGFA, and TP53, were considered as potential therapeutic targets, implying the underlying mechanisms of DZ acting on OPF.
Conclusion
We investigated the possible therapeutic mechanisms of DZ from a systemic perspective. These key targets and pathways provided promising directions for the future research to reveal the exact regulating mechanisms of DZ in treating OPF.
1. Introduction
Osteoporosis is a systemic bone disorder characterized by low bone density, poor bone quality, reduced bone strength, and an accompanying increased incidence of fractures [1]. The latest study noted that over 10 million people suffer from osteoporosis in the United States. Meanwhile, the treatment burden of osteoporosis, $22 billion in 2008, is expected to rise due to the consistently increasing aging population. Related osteoporosis treatments should be used to alleviate or reduce symptoms such as fractures, which could result in considerable cost savings [2]. Osteoporotic fractures (OPF) have serious consequences, such as declining functions and independence as well as increased morbidity and mortality [3, 4]. Hip fractures are considered to be the most expensive and debilitating of fractures because approximately 10 to 20% of patients will end up with a disability in the year following a hip fracture and half of patients will lose their independence. Osteoporotic fractures are also associated with motor function decline when occurring at other skeletal sites [5]. To curb bone loss, maintain bone mass, and decrease the risk of OPF, antiresorptive agents, anabolic agents, and bone mineral drugs have been widely used in clinical treatment [6]. However, it is noteworthy that almost all these antiosteoporosis treatments have limitations and side effects, such as increasing the risk of breast cancer, jaw necrosis, or atypical femur fracture [7, 8]. Therefore, alternative and safe intervention strategies may be promising.
Recently, traditional Chinese medicine (TCM) has attracted worldwide attention and has served as a main alternative treatment in East Asia, North America, and Europe due to its satisfactory curative effect, relatively low toxicity, and low cost [9–11]. TCM has been used in China for thousands of years, and it exerts a beneficial therapeutic effect and reduces side effects through multiple herb combinations to prevent and treat various diseases [12]. Meanwhile, TCM has increasingly been found to be effective in the treatment of osteoporosis [13, 14].
Du Zhong (DZ), or Eucommiae Cortex, is one of the most commonly used TCM herbs in the treatment of osteoporosis. DZ has been found to facilitate osteogenesis through activating osteoblasts and to inhibit osteolysis by suppressing osteoclast activity [15]. An animal experiment confirmed that Du Zhong Wan, consisting of Eucommiae Cortex and Radix Dipsaci, is capable of improving trabecular bone mineral density and bone biomechanical properties and thus plays a key role in the treatment of osteoporosis [16]. Although DZ exerted therapeutic effects against osteoporosis, the therapeutic mechanism of its exact is still unclear. Similar to TCM, DZ exerts its therapeutic efficacy by regulating multiple molecules in the human body. Therefore, it is still a major challenge to prove the mechanism of DZ through scientific trials that are used in Western medicine. Fortunately, the development of systems pharmacology offers researchers an alternative opportunity and an option to investigate the pharmacological mechanisms of TCM [17]. Recently, Wang et al. utilized a network pharmacology method to clarify the synergistic mechanism of Er-Xian decoction in treating osteoporosis [18]. Similarly, Gan et al. employed this systems pharmacology method to dissect the mechanisms of the therapeutic effect of Rhizoma Drynariae on osteoporosis [19].
In this network pharmacology work, we aim to comprehensively dissect the mechanisms of DZ in treating OPF. We collected related compounds of DZ from multiple databases and obtained the compounds' potential targets via target fishing. Then, we matched these targets with OPF-related targets that were collected from a multisource database. Next, using overlapping targets obtained from the previous process, we built a protein-protein network to analyze their internal interactions and then screened out the hub genes. Furthermore, we used the clusterProfiler package in R to conduct biological process and KEGG pathway enrichment analyses. The protocol of our experimental procedures is shown in Figure 1.
Figure 1.
The schematic map of the present study to investigate potential mechanisms of DZ in the OPF treatment.
2. Materials and Methods
2.1. Identification of Chemical Ingredients
To collect the active compounds of DZ, we utilized the Traditional Chinese Medicine System Pharmacology Database (TCMSP™, http://lsp.nwu.edu.cn/tcmsp.php) [20], which is a frequently used platform for systems pharmacology. The BATMAN-TCM platform (http://bionet.ncpsb.org/batman-tcm/) [21], one of the largest comprehensive TCM platforms, uses the similarity of known TCM drug–target interactions to predict potential compound–target interactions. In total, ninety-three chemical compounds were collected in this part.
2.2. Compound Screening and Target Prediction
2.2.1. OB Evaluation
Oral bioavailability (OB) refers to the percentage of an orally administered drug that reaches systemic circulation, and it is one of the most important pharmacokinetic profiles for drug screening. The TCMSP platform has adopted the OBioavail1.1 system, which integrates P450, 3A4, and P-glycoprotein information to obtain the OB value [22]. To determine the active ingredients used for further steps, we set the OB threshold at 30%.
2.2.2. DL Prediction
Drug-likeness (DL) is a qualitative index that represents the degree to which the target compound is “drug-like” and is used to remove chemically unsuitable compounds. TCMSP uses the Tanimoto similarity method to calculate the DL index by comparing the target compound to all 6511 molecules in the DrugBank database [23]. In this process, the compounds that did not meet the condition that DL ≥ 0.18 were removed.
2.2.3. BATMAN-TCM
To collect candidate compounds, we set the potential TAR score cutoff ≥20 and P-value cutoff <0.05.
2.2.4. Target Prediction of the Active Ingredients
The compounds have effects on the targets that induce them to exert their biological functions. Thus, we used the TCMSP and BATMAN-TCM platforms to predict the targets of active compounds.
2.3. Disease Target Identification by Multiple Databases
We used multiple databases to collect OPF-related targets, and the terms “Osteoporotic” and “Fracture” were used as the key words for the search. The databases in this step included the Comparative Toxicogenomics Database (CTD, http://ctdbase.org/) [24], GeneCards (https://www.genecards.org/) [25], OMIM (https://www.omim.org/) [26], HPO (https://hpo.jax.org/app/) [27], and GenCLiP3 (http://ci.smu.edu.cn/genclip3/analysis.php) [28]. In total, three thousand four hundred-thirteen OPF targets were found through these databases. Then, we constructed a Venn diagram to determine the overlap between OPF and the active compound targets because these overlapping targets may play a critical role in the treatment of OPF. These targets were analyzed by String (https://string-db.org/) [29], in which the minimum required interaction score was set at the highest confidence level (0.900), and then, the protein-protein interaction (PPI) data were exported.
2.4. Network Construction
2.4.1. Construction Method
Two main networks were built in this process, including the compound-hub gene network and the hub gene-pathway network. The target information was obtained from the KEGG pathway enrichment results. Cytoscape 3.6.2 (http://www.cytoscape.org/) was used for all the network visualization work. Cytoscape 3.6.2 is one of most powerful open-source software programs for constructing different networks visually [30]. However, there are several limitations faced by network pharmacology research. For example, questions including whether compounds activate or inhibit targets, how they exact their effects on targets, whether they use binding or catalysis, and so on remain unclear.
2.4.2. Topological Feature Definition
We used three parameters to describe and quantify the importance of nodes in these networks because nodes that bridge many edges with their neighbors are more likely to exert crucial mediating functions. (1) “Degree” means the number of edges shared with other nodes. Examining the node's degree is the most straightforward method of quantifying its centrality [31]. (2) “Betweenness centrality” is used to measure node centrality based on the shortest paths. A high-value node would play a more important role in the network because more regulation information will pass through it [32]. (3) “Closeness centrality” represents the mean distance between the node and all the other nodes in the network and is the reciprocal of the sum of the length of the shortest paths between a node and all the other nodes in the network. Therefore, the central node would be more likely to be close with all other nodes [33].
2.5. Biological Process and Pathway Enrichment Analysis
We used the clusterProfiler package in R (R 3.6.2 for Windows) to perform Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to identify biological processes and systemic involvement of pathways.
3. Results
3.1. Active Compounds of DZ
A total of 147 related compounds were found in DZ collected from the TCMSP database, and of these, 27 candidate compounds remained after the screening of ADME thresholds (OB ≥ 30%, DL ≥ 0.18). Using the BATMAN-TCM database, we obtained 73 active compounds that matched the filter criteria. In total, we collected 93 unique compounds.
3.2. Target Prediction and Analysis
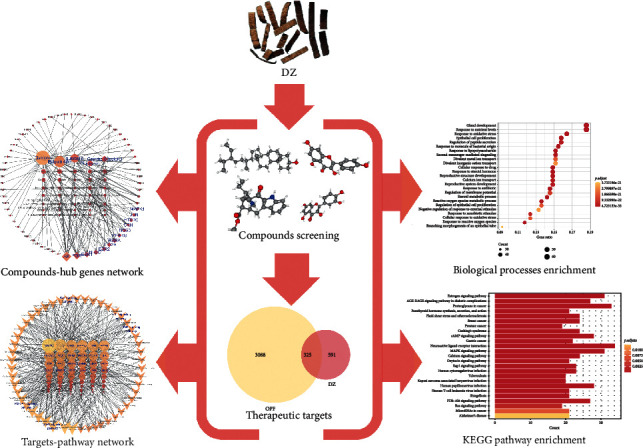
For these unique compounds, we obtained 961 unique related targets, 104 from TCMSP and 857 from BATMAN-TCM. We integrated the OPF genes that were obtained from multisource databases, including the CTD, GeneCards, OMIM, HPO, GenCLiP 3 databases, and a total of 3,834 related genes were collected. After construction of the Venn diagram, three hundred twenty-five overlapping targets between the related targets of DZ and OPF were selected as the key targets on which DZ exerts its anti-OPF effects (Figure 2).
Figure 2.
The Venn diagram for the targets of DZ and OPF. The overlap targets are the potential therapeutic genes for DZ when treating OPF.
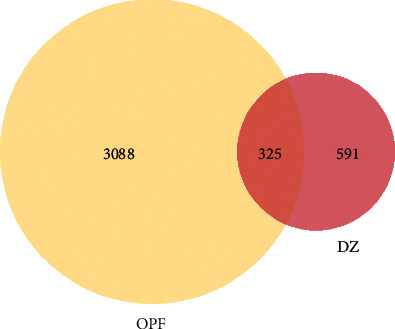
The data obtained from the String database were used to establish the PPI network for the 325 overlapping targets. In this network, there were 249 nodes and 1006 edges in total. Then, three main parameters, namely, “degree”, “betweenness”, and “closeness”, were used as filters to select key genes and to build the large hub nodes to determine the anti-OPF effect of DZ. The first screening threshold was degree ≥ 6, closeness ≥ 0.312, and betweenness ≥ 0.000, which resulted in 122 nodes and 745 edges. Then, these 72 key nodes were further screened with a second threshold consisting of degree ≥ 11, closeness ≥ 0.340, and betweenness ≥ 0.016, and 64 nodes and 414 edges remained after this screening (Figure 3). These nodes included MAPK1, SRC, PIK3R1, VEGFA, TP53, EGFR, JUN, AGT, IL6, EGF, MAPK8, FOS, F2, FN1, EDN1, TNF, PPBP, ESR1, NCOA1, CTNNB1, NR3C1, ALB, RXRA, CBL, INS, PRKACA, AHSG, TGFB1, HSPA8, AR, FGF23, GNAS, NFKB1, GAS6, PRKCD, AVPR2, APOA1, PDGFB, HSP90AB1, IL4, HSPA1A, IGF2, SIRT1, PPARG, PPARA, CDC42, PGR, IFNG, CST3, NCOA3, TAC1, PLCB1, NOS3, CCL5, FGF2, ESR2, STAT5A, WNT5A, HTR2C, HTR2A, CCL2, CRH, APOE, and CFD (Table 1). After sorting these 64 big hub nodes in descending order, we found that MAPK1 (degree = 41), SRC (degree = 39), PIK3R1 (degree = 36), VEGFA (degree = 31), TP53 (degree = 29), EGFR (degree = 29), JUN (degree = 29), AGT (degree = 29), IL6 (degree = 28), EGF (degree = 27), MAPK8 (degree = 26), FOS (degree = 25), and F2 (degree = 25) were the most important targets in this PPI network.
Figure 3.
The whole screening process for the PPI network through a topological method. In these images, the bigger size and brighter color represent higher DC value.
Table 1.
Information of 64 hub targets.
| UniProt ID | Gene symbol | Description | Degree |
|---|---|---|---|
| Q499G7 | MAPK1 | Mitogen-activated protein kinase 1 | 41 |
| P12931 | SRC | SRC proto-oncogene, non–receptor tyrosine kinase | 39 |
| P27986 | PIK3R1 | Phosphoinositide-3-kinase regulatory subunit 1 | 36 |
| P15692 | VEGFA | Vascular endothelial growth factor A | 31 |
| Q53GA5 | TP53 | Tumor protein p53 | 29 |
| Q504U8 | EGFR | Epidermal growth factor receptor | 29 |
| P05412 | JUN | Jun proto-oncogene | 29 |
| P01019 | AGT | Angiotensinogen | 29 |
| Q75MH2 | IL6 | Interleukin 6 | 28 |
| P01133 | EGF | Epidermal growth factor | 27 |
| P45983 | MAPK8 | Mitogen-activated protein kinase 8 | 26 |
| Q6FG41 | FOS | Fos proto-oncogene | 25 |
| P00734 | F2 | Coagulation factor II, thrombin | 25 |
| Q9UQS6 | FN1 | Fibronectin 1 | 24 |
| Q6FH53 | EDN1 | Endothelin 1 | 24 |
| Q5STB3 | TNF | Tumor necrosis factor | 24 |
| P02775 | PPBP | Pro-platelet basic protein | 24 |
| Q9UBT1 | ESR1 | Estrogen receptor 1 | 23 |
| Q15788 | NCOA1 | Nuclear receptor coactivator 1 | 23 |
| P35222 | CTNNB1 | Catenin beta 1 | 23 |
| P04150 | NR3C1 | Nuclear receptor subfamily 3 group C member 1 | 23 |
| P02768 | ALB | Albumin | 22 |
| Q6P3U7 | RXRA | Retinoid X receptor alpha | 21 |
| P22681 | CBL | Cbl proto-oncogene | 21 |
| P01308 | INS | Insulin | 20 |
| P17612 | PRKACA | Protein kinase cAMP-activated catalytic subunit alpha | 18 |
| P02765 | AHSG | Alpha 2-HS glycoprotein | 18 |
| P01137 | TGFB1 | Transforming growth factor beta 1 | 18 |
| V9HW22 | HSPA8 | Heat shock protein family A (Hsp70) member 8 | 17 |
| Q9NUA2 | AR | Androgen receptor | 17 |
| Q9GZV9 | FGF23 | Fibroblast growth factor 23 | 17 |
| Q5JWF2 | GNAS | GNAS complex locus | 16 |
| P19838 | NFKB1 | Nuclear factor kappa B subunit 1 | 16 |
| Q14393 | GAS6 | Growth arrest specific 6 | 15 |
| Q05655 | PRKCD | Protein kinase C delta | 15 |
| P30518 | AVPR2 | Arginine vasopressin receptor 2 | 15 |
| P02647 | APOA1 | Apolipoprotein A1 | 15 |
| P01127 | PDGFB | Platelet-derived growth factor subunit B | 15 |
| Q6PK50 | HSP90AB1 | Heat shock protein 90 alpha family class B member 1 | 14 |
| Q5FC01 | IL4 | Interleukin 4 | 14 |
| P0DMV9 | HSPA1A | Heat shock protein family A (Hsp70) member 1A | 14 |
| P01344 | IGF2 | Insulin-like growth factor 2 | 14 |
| Q96EB6 | SIRT1 | Sirtuin 1 | 13 |
| Q4W448 | PPARG | Peroxisome proliferator activated receptor gamma | 13 |
| Q07869 | PPARA | Peroxisome proliferator activated receptor alpha | 13 |
| P60953 | CDC42 | Cell division cycle 42 | 13 |
| P06401 | PGR | Progesterone receptor | 13 |
| P01579 | IFNG | Interferon gamma | 13 |
| P01034 | CST3 | Cystatin C | 13 |
| Q9Y6Q9 | NCOA3 | Nuclear receptor coactivator 3 | 12 |
| Q9Y494 | TAC1 | Tachykinin precursor 1 | 12 |
| Q9NQ66 | PLCB1 | Phospholipase C beta 1 | 12 |
| P29474 | NOS3 | Nitric oxide synthase 3 | 12 |
| P13501 | CCL5 | C-C motif chemokine ligand 5 | 12 |
| P09038 | FGF2 | Fibroblast growth factor 2 | 12 |
| Q92731 | ESR2 | Estrogen receptor 2 | 11 |
| Q59GY7 | STAT5A | Signal transducer and activator of transcription 5A | 11 |
| P41221 | WNT5A | Wnt family member 5A | 11 |
| P28335 | HTR2C | 5-Hydroxytryptamine receptor 2C | 11 |
| P28223 | HTR2A | 5-Hydroxytryptamine receptor 2A | 11 |
| P13500 | CCL2 | C-C motif chemokine ligand 2 | 11 |
| P06850 | CRH | Corticotropin releasing hormone | 11 |
| P02649 | APOE | Apolipoprotein E | 11 |
| P00746 | CFD | Complement factor D | 11 |
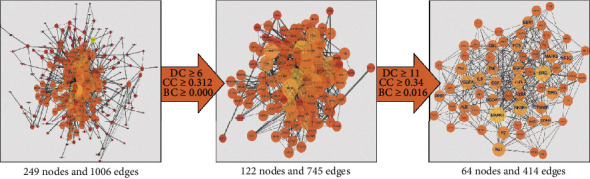
Then, we further built the big hub nodes-compound network (Figure 4) based on these 64 key targets. This network included 40 compound nodes and 64 large hub target nodes. Subsequently, we reordered these compound nodes in descending order of degree and found that Civetone was relevant to 25 large hub genes, Epiquinidine to 19 genes, quercetin to 13 genes, Gamma-Sitosterol to 7 genes, and Pinosylvin to 7 genes.
Figure 4.
The network for big hub genes-compounds connection. The diamond shape nodes are the hub genes and round ones represent compounds. And all nodes' color changes according to their degree value.
3.3. GO Biological Process Enrichment Analysis
After sorting the 343 biological process (BP) terms in ascending order of adjusted P value, we found that the top 25 biological processes (Figure 5) mainly concentrated on aspects of the reactive oxygen species metabolic process and its response to nutrient levels and epithelial cell proliferation. The following processes showed an enriched gene ratio: gland development (GO:0048732, 18.6%), response to nutrient levels (GO:0031667, 18.6%), response to a molecule of bacterial origin (GO:0002237, 15.5%), response to lipopolysaccharide (GO:0032496, 15.2%), response to an antibiotic (GO:0046677, 14.6%), reactive oxygen species metabolic process (GO:0072593, 13.6%), response to oxidative stress (GO:0006979, 16.4%), steroid metabolic process (GO:0008202, 14.2%), cellular response to drug (GO:0035690, 14.9%), epithelial cell proliferation (GO:0050673, 15.8%), response to a steroid hormone (GO:0048545, 14.9%), response to reactive oxygen species (GO:0000302, 11.8%), second-messenger-mediated signaling (GO:0019932, 15.2%), reproductive structure development (GO:0048608, 14.9%), calcium ion transport (GO:0006816, 14.9%), reproductive system development (GO:0061458, 14.9%), regulation of peptide secretion (GO:0002791, 15.8%), response to xenobiotic stimulus (GO:0009410, 12.4%), regulation of membrane potential (GO:0042391, 14.6%), cellular response to oxidative stress (GO:0034599, 12.4%), regulation of epithelial cell proliferation (GO:0050678, 13.6%), divalent metal ion transport (GO:0070838, 15.2%), divalent inorganic cation transport (GO:0072511, 15.2%), negative regulation of response to external stimulus (GO:0032102, 13.3%), and branching morphogenesis of an epithelial tube (GO:0048754, 9.3%). Based on these BP enrichment analyses, we found that the anti-OPF effect of DZ may result from its regulatory effect on the reactive oxygen species metabolic process, response to nutrient levels, and epithelial cell proliferation.
Figure 5.
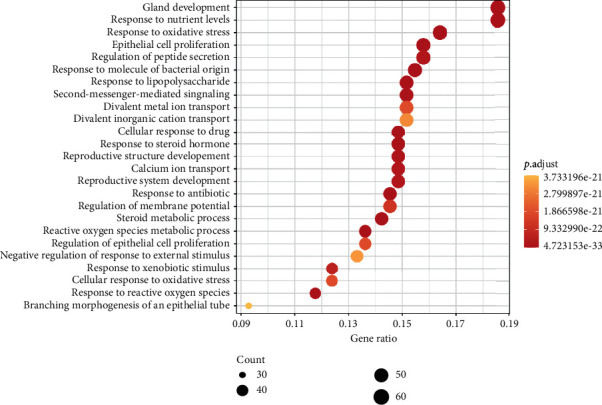
Top 25 processes of the biological process enrichment.
3.4. KEGG Pathway Enrichment Analysis
We conducted further KEGG pathway enrichment analyses of the 325 overlapping genes to determine the potential therapeutic mechanism of DZ for OPF. Then, we sorted 25 pathways based on the adjusted P value (Figure 6). These pathways were the AGE-RAGE signaling pathway in diabetic complications (hsa04933, 9.6%), the estrogen signaling pathway (hsa04915, 11.1%), proteoglycans in cancer (hsa05205, 11.8%), fluid shear stress and atherosclerosis (hsa05418, 8.6%), parathyroid hormone synthesis, secretion, and action (hsa04928, 7.5%), breast cancer (hsa05224, 8.6%), Cushing's syndrome (hsa04934, 8.6%), prostate cancer (hsa05215, 6.8%), the cAMP signaling pathway (hsa04024, 10%), morphine addiction (hsa05032, 6.4%), African trypanosomiasis (hsa05143, 4.3%), the GnRH signaling pathway (hsa04912, 6.4%), endocrine resistance (hsa01522, 6.4%), gastric cancer (hsa05226, 7.9%), neuroactive ligand-receptor interaction (hsa04080, 12.1%), the MAPK signaling pathway (hsa04010, 11.1%), the calcium signaling pathway (hsa04020, 8.6%), the oxytocin signaling pathway (hsa04921, 7.5%), malaria (hsa05144, 4.3%), Th17 cell differentiation (hsa04659, 6.1%), the HIF-1 signaling pathway (hsa04066, 6.1%), colorectal cancer (hsa05210, 5.4%), GnRH secretion (hsa04929, 4.6%), cortisol synthesis and secretion (hsa04927, 4.6%), and the TNF signaling pathway (hsa04668, 6.1%) (Table 2). We also simultaneously constructed the target-pathway network based on the DZ targets enriched in each pathway (Figure 7).
Figure 6.
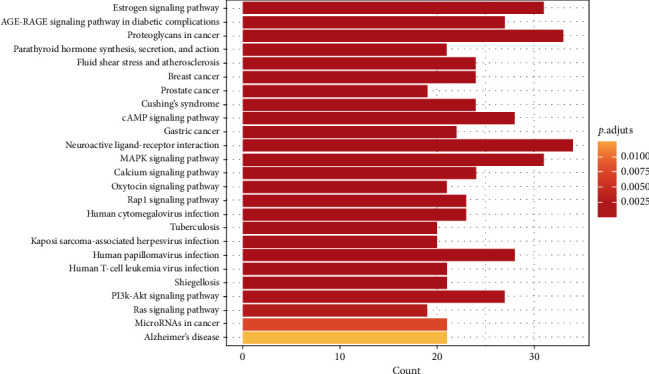
Top 25 pathways of the KEGG enrichment.
Table 2.
Information of 25 pathways.
| ID | Description | P. adjust | Count |
|---|---|---|---|
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 4.29750564649423e-15 | 27 |
| hsa04915 | Estrogen signaling pathway | 4.29750564649423e-15 | 31 |
| hsa05205 | Proteoglycans in cancer | 7.02978144058047e-12 | 33 |
| hsa05418 | Fluid shear stress and atherosclerosis | 3.50540676797616e-09 | 24 |
| hsa04928 | Parathyroid hormone synthesis, secretion, and action | 3.50540676797616e-09 | 21 |
| hsa05224 | Breast cancer | 8.65129662473866e-09 | 24 |
| hsa04934 | Cushing syndrome | 2.25061036469472e-08 | 24 |
| hsa05215 | Prostate cancer | 2.25061036469472e-08 | 19 |
| hsa04024 | cAMP signaling pathway | 4.25537215261066e-08 | 28 |
| hsa05032 | Morphine addiction | 4.35290458036493e-08 | 18 |
| hsa05143 | African trypanosomiasis | 5.25038311520481e-08 | 12 |
| hsa04912 | GnRH signaling pathway | 5.25038311520481e-08 | 18 |
| hsa01522 | Endocrine resistance | 1.16942061208876e-07 | 18 |
| hsa05226 | Gastric cancer | 1.53049773606446e-07 | 22 |
| hsa04080 | Neuroactive ligand-receptor interaction | 3.92997114777747e-07 | 34 |
| hsa04010 | MAPK signaling pathway | 4.75554213381079e-07 | 31 |
| hsa04020 | Calcium signaling pathway | 8.10557748077706e-07 | 24 |
| hsa04921 | Oxytocin signaling pathway | 1.09530352609987e-06 | 21 |
| hsa05144 | Malaria | 1.3497422679709e-06 | 12 |
| hsa04659 | Th17 cell differentiation | 1.90497387620456e-06 | 17 |
| hsa04066 | HIF-1 signaling pathway | 2.39525818923615e-06 | 17 |
| hsa05210 | Colorectal cancer | 2.67545458381123e-06 | 15 |
| hsa04929 | GnRH secretion | 2.67545458381123e-06 | 13 |
| hsa04927 | Cortisol synthesis and secretion | 3.01425725456316e-06 | 13 |
| hsa04668 | TNF signaling pathway | 3.01425725456316e-06 | 17 |
Figure 7.
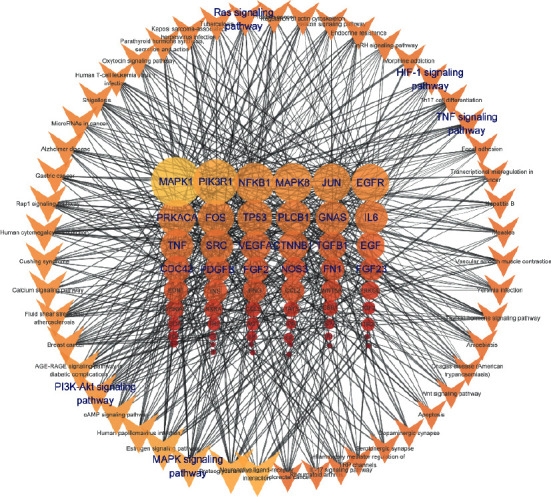
The targets-pathway network of DZ for treating OPF. The circle nodes represent big hub genes and the V-shape nodes represent the top 50 pathways. The nodes' size and colors are dependent on DC value.
4. Discussion
Osteoporosis is becoming a serious health burden due to the aging population. Therefore, prevention of osteoporosis and of osteoporotic fracture has considerable social and economic significance [34]. Drug therapy, such as bisphosphonates and denosumab, is a practical way to treat osteoporosis by inhibiting osteoclast development, formation, and survival [35, 36]. Although these antiresorptive drugs are effective in treating osteoporosis, the long-term use or high dosage of these drugs can cause serious side effects, such as atypical long bone fractures and jaw osteonecrosis [37, 38]. In terms of fracture healing, the use of bisphosphonates remains controversial because callus remodeling is impaired in testing on ovariectomized rats [39]. Furthermore, these antiresorptive drugs cannot rebuild bone that is lost in the progression of osteoporosis, so there is a need to find a method capable of promoting bone building [40].
DZ has been found to alleviate improved osteoblast activity and reverse bone loss in ovariectomized (OVX) mice, which may result from the interaction between its active compounds and related targets. Thus, we investigated the anti-OPF mechanism of DZ through a network pharmacology method. Based on the big hub genes-compounds network (Figure 4), we found that five key compounds, including Civetone, Epiquinidine, quercetin, Gamma-Sitosterol, and Pinosylvin, play dominant roles in this network. Therefore, these active compounds may lay the foundation for the promising anti-OPF effect of DZ.
In the present study, 325 common gene targets of DZ and OPF were identified and 64 hub genes were identified that may play critical roles in this treatment. Some anti-OPF effects of these genes have been confirmed by clinical trials or animal experiments. Chow et al. found that low-magnitude high-frequency vibration stimulation is able to promote fracture healing by positively regulating the impaired innate immune response (lower expression of TNF-α and IL-6) and promoting macrophage polarization in delayed osteoporotic fracture healing [41]. Wang et al. suggested that organic gallium can improve osteoporotic fracture healing by positively modulating the OPG/RANKL ratio and inhibiting the expression of TNF-α and IL-6 [42]. Zhang et al. found that 17β-estradiol, which is used in the clinic to prevent and treat postmenopausal women with osteoporotic fracture, can significantly downregulate ROS production and the expression of TNF-α and IL-1β, as well as the phosphorylation of ERK2 (MAPK1) [43]. Wang et al. suggested that ESR1 is a susceptibility gene for osteoporotic fracture in postmenopausal Chinese women [44]. De-Ugarte et al. found that miR-320a, which is known to target CTNNB1 of osteoblasts, is overexpressed in osteoporotic fractures [45]. Swanberg et al. found that elderly women who were carriers of the IFNG variant alleles had lower BMD and a lower risk of incident fracture [46]. Based on a large population-based cohort study, Rivadeneira et al. found that ESR2 variants were related to an increased risk of fragility fracture in postmenopausal women [47]. These gene targets play key roles in osteoblast differentiation and osteoclastogenesis during fracture repair. Moreover, the results presented in Figure 4 show that one active compound can interact with multiple genes, one gene with dicompound can interact with multiple genes, and one gene can interact with different compounds, which is in accordance with the modern drug theory of “multi-ingredient, multitarget” [48].
We conducted GO biological process and KEGG enrichment analyses of the overlapping genes to identify their functions. We found 44 (13.6%) genes involved in reactive oxygen species metabolic processes. Reactive oxygen species play key roles in regulating cell proliferation, apoptosis, migration, and differentiation [49], as well as in the regulation of osteoclasts and osteoblasts [50, 51]. EGFR is the overlapping gene between reactive oxygen species (ROS) and the PI3K-Akt signaling pathway (27 genes enriched, 9.6%) based on the results of the GOBP and KEGG pathway enrichment analyses. The PI3K-Akt signaling pathway was first found in tumor cells and proved to regulate numerous cell functions, and it has been proven to be related to the promotion of osteogenesis by upregulating the proliferation and differentiation of bone marrow mesenchymal stem cells [52–54]. Furthermore, activation of the EGFR-Akt-mTOR pathway can effectively protect osteoblasts against dexamethasone, which increases the level of ROS production in osteoblastic cells [55]. MAPK1 is a common gene in reactive oxygen species, the PI3K-Akt signaling pathway, the MAPK signaling pathway (31 genes enriched, 11.1%), and the TNF signaling pathway. MAPKs play important roles in cell proliferation, differentiation, and apoptosis, as well as in regulating inflammation [56]. ROS can lead to serious periodontal tissue destruction by promoting osteoclastic bone resorption, which is closely related to the protein expression of MAPKs and NF-κB [57]. Srinivasa et al. found that the facilitation of ROS on osteoclast differentiation and resorption can be reversed by inhibiting the NF-κB and calcineurin-NFAT pathways [58]. The activation of MAPKs also plays a key role in osteoclastogenesis induced by RANKL. However, these ROS-directed upregulations of MAPK and NF-κB signaling can be attenuated by strengthening the nuclear factor-erythroid 2-related factor 2 [59]. Therefore, the anti-OPF effect of DZ may depend on the regulatory effect on ROS through these key genes and pathways.
Forty-five (13.9%) genes were involved in the regulation of the inflammatory response. In vitro and in vivo studies have shown that the proinflammatory cytokines IL-6 and TNF-α are involved in the pathogenesis of osteoporosis and influence bone metabolism [60, 61]. Several studies have also found that high expression of inflammatory markers is closely related to increased bone loss [62–64] and that these markers are also risk factors for incidents of nontraumatic fractures. Women with increased expression of inflammatory markers have a 3-fold risk of hip fractures [65]. IL-6 and TNF-α are the overlapping genes between the regulation of inflammatory response (17 genes enriched, 6.1%) and the TNF signaling pathway (45 genes enriched, 13.9%) based on the results of the BP and KEGG pathway enrichment analyses. Ali Aydin et al. found higher expression levels of TNF- α and IL-6 in OVX model rats, and these cytokines reached their apex when a fracture occurred [66]. Moreover, these cytokines also increase the production of oxygen radicals, which can enhance osteoclastic activity while decreasing osteoblastic activity [67, 68]. Therefore, the anti-OPF effect of DZ may depend on the regulation of the inflammatory response through these key genes and pathways.
5. Conclusion
By utilizing network pharmacology, we investigated the potential targets of DZ and the underlying mechanism of its anti-OPF effects, which may be based on the regulation of ROS and the inflammatory response. According to the KEGG pathway enrichment results, we found that the PI3K-Akt signaling pathway, MAPK signaling pathway, and TNF signaling pathway may be the main pathways in treating OPF. Thus, we believe that the anti-OPF effect of DZ is mainly based on its direct or indirect regulation of the abovementioned potential targets and pathways and that DZ provides promising directions for future research, which is essential to reveal its exact regulatory mechanisms.
Abbreviations
- DZ:
Du Zhong
- OPF:
Osteoporotic fractures
- TCM:
Traditional Chinese medicine
- TCMSP:
Traditional Chinese Medicine System Pharmacology Database
- OB:
Oral bioavailability
- DL:
Drug-likeness
- CTD:
Comparative Toxicogenomics Database
- OMIM:
Online Mendelian Inheritance in Man
- HPO:
Human Phenotype Ontology
- PPI:
Protein-protein interaction
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- GO:
Gene Ontology
- BP:
Biological process
- OVX:
Ovariectomized
- ROS:
Reactive oxygen species.
Data Availability
All our main data used to support the findings of this study have been deposited in the Figshare repository (https://doi.org/10.6084/m9.figshare.12152607.v1).
Conflicts of Interest
The authors declare that they have no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Yongming Shuai and Fanhui Zeng conceived the idea of this article and supervised the research. Yongming Shuai performed the research, analyzed the data, and wrote the manuscript. Zhili Jiang and Qiuwen Yuan performed Target prediction and analysis as well as related enrichment processes. Shuqiang Tu and Fanhui Zeng participated in revising the data and improving manuscript writing. All authors reviewed the manuscript. And all authors read and approved the final version of the manuscript.
References
- 1.Camacho P. M., Petak S. M., Binkley N., et al. American association of clinical endocrinologists and American college of endocrinology clinical practice guidelines for the diagnosis and treatment of postmenopausal osteoporosis–2016. Endocrine Practice. 2016;22(4):1–42. doi: 10.4158/ep161435.10.4158/ep161435.glgl. [DOI] [PubMed] [Google Scholar]
- 2.Lewiecki E. M., Ortendahl J. D., Vanderpuye-Orgle J., et al. Healthcare policy changes in osteoporosis can improve outcomes and reduce costs in the United States. JBMR Plus. 2019;3 doi: 10.1002/jbm4.10192.e10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cauley J. A. Public health impact of osteoporosis. Journals of Gerontology Series A: Biomedical Sciences and Medical Sciences. 2013;68(10):1243–1251. doi: 10.1093/gerona/glt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clynes M. A., Harvey N. C., Curtis E. M., Fuggle N. R., Dennison E. M., Cooper C. The epidemiology of osteoporosis. British Medical Bulletin. 2020;133:105–117. doi: 10.1093/bmb/ldaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewiecki E. M., Chastek B., Sundquist K., et al. Osteoporotic fracture trends in a population of US managed care enrollees from 2007 to 2017. Osteoporosis International. 2020;31(7):1299–1304. doi: 10.1007/s00198-020-05334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letarouilly J.-G., Broux O., Clabaut A. New insights into the epigenetics of osteoporosis. Genomics. 2019;111(4):793–798. doi: 10.1016/j.ygeno.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Kharazmi M., Schilcher J., Hallberg P., Michaëlsson K. Bisphosphonate-associated atypical fractures of the femur: an update of the current evidence. Lakartidningen. 2019;116 [PubMed] [Google Scholar]
- 8.Tabatabaei-Malazy O., Salari P., Khashayar P., Larijani B. New horizons in treatment of osteoporosis. Daru Journal of Pharmaceutical Sciences. 2017;25(1) doi: 10.1186/s40199-017-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao L.-J., Tao R. Traditional Chinese medicine (TCM) therapy. Advances in Experimental Medicine and Biology. 2017;1010:261–280. doi: 10.1007/978-981-10-5562-1_13. [DOI] [PubMed] [Google Scholar]
- 10.Xu W., Towers A. D., Li P., Collet J.-P. Traditional Chinese medicine in cancer care: Perspectives and experiences of patients and professionals in China. European Journal of Cancer Care. 2006;15(4):397–403. doi: 10.1111/j.1365-2354.2006.00685.x. [DOI] [PubMed] [Google Scholar]
- 11.Sham T.-T., Chan C.-O., Wang Y.-H., Yang J.-M., Kam-Wah Mok D., Chan S.-W. A review on the traditional Chinese medicinal herbs and formulae with hypolipidemic effect. BioMed Research International. 2014;2014:1–21. doi: 10.1155/2014/925302.925302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P., Yang L.-p., Gong Y.-w. Application of systems biology technology in research of traditional Chinese medicine. Journal of Traditional Chinese Medicine. 2009;29(2):153–157. doi: 10.1016/s0254-6272(09)60054-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L. P., Wong K. Y., Yeung H. T., et al. Bone protective effects of danggui buxue tang alone and in combination with tamoxifen or raloxifene in vivo and in vitro. Frontiers in Pharmacology. 2018;9:p. 779. doi: 10.3389/fphar.2018.00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu F., Jiang J., Wang S., et al. An experimental research into the potential therapeutic effects of anti-osteoporosis decoction and Yougui pill on ovariectomy-induced osteoporosis. American Journal of Translational Research. 2019;11(9):6032–6039. [PMC free article] [PubMed] [Google Scholar]
- 15.Ha H., Ho J., Shin S., et al. Effects of Eucommiae Cortex on osteoblast-like cell proliferation and osteoclast inhibition. Archives of Pharmacal Research. 2003;26(11):929–936. doi: 10.1007/bf02980202. [DOI] [PubMed] [Google Scholar]
- 16.Li F., Yang X., Bi J., Yang Z., Zhang C. Antiosteoporotic activity of Du-Zhong-Wan water extract in ovariectomized rats. Pharmaceutical Biology. 2016;54(9):1857–1864. doi: 10.3109/13880209.2015.1133657. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W., Huai Y., Miao Z., Qian A., Wang Y. Systems pharmacology for investigation of the mechanisms of action of traditional chinese medicine in drug discovery. Frontiers in Pharmacology. 2019;10:p. 743. doi: 10.3389/fphar.2019.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang N., Xu P., Wang X., et al. Integrated pathological cell fishing and network pharmacology approach to investigate main active components of Er-Xian decotion for treating osteoporosis. Journal of Ethnopharmacology. 2019;241 doi: 10.1016/j.jep.2019.111977.111977 [DOI] [PubMed] [Google Scholar]
- 19.Gan D., Xu X., Chen D., Feng P., Xu Z. Network pharmacology-based pharmacological mechanism of the Chinese medicine rhizoma drynariae against osteoporosis. Medical Science Monitor. 2019;25:5700–5716. doi: 10.12659/msm.915170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ru J., Li P., Wang J., et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z., Guo F., Wang Y., et al. BATMAN-TCM: A bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Scientific Reports. 2016;6(1) doi: 10.1038/srep21146.21146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X., Zhang W., Huang C., et al. A novel chemometric method for the prediction of human oral bioavailability. International Journal of Molecular Sciences. 2012;13(6):6964–6982. doi: 10.3390/ijms13066964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao W., Xu X., Wang X., et al. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. Journal of Ethnopharmacology. 2013;145(1):1–10. doi: 10.1016/j.jep.2012.09.051. [DOI] [PubMed] [Google Scholar]
- 24.Grondin C. J., Davis A. P., Wiegers T. C., Wiegers J. A., Mattingly C. J. Accessing an expanded exposure science module at the comparative Toxicogenomics database. Environmental Health Perspectives. 2018;126(1) doi: 10.1289/ehp2873.014501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rebhan M., Chalifa-Caspi V., Prilusky J., Lancet D. GeneCards: A novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998;14(8):656–664. doi: 10.1093/bioinformatics/14.8.656. [DOI] [PubMed] [Google Scholar]
- 26.Amberger J. S., Hamosh A. Searching online mendelian inheritance in man (OMIM): A knowledgebase of human genes and genetic phenotypes. Current Protocols in Bioinformatics. 2017;58(1) doi: 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson P. N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The Human phenotype ontology: A tool for annotating and analyzing human hereditary disease. The American Journal of Human Genetics. 2008;83(5):610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J. H., Zhao L. F., Wang H. F., et al. GenCLiP 3: Mining human genes’ functions and regulatory networks from pubMed based on co-occurrences and natural language processing. Bioinformatics. 2019;36(6):1973–1975. doi: 10.1093/bioinformatics/btz807. [DOI] [PubMed] [Google Scholar]
- 29.Szklarczyk D., Morris J. H., Cook H., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P., Markiel A., Ozier O. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Missiuro P. V., Liu K., Zou L., et al. Information flow analysis of interactome networks. PLOS Computational Biology. 2009;5(4) doi: 10.1371/journal.pcbi.1000350.e1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raman K., Damaraju N., Joshi G. K. The organisational structure of protein networks: Revisiting the centrality-lethality hypothesis. Systems and Synthetic Biology. 2014;8(1):73–81. doi: 10.1007/s11693-013-9123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Y., Li M., Wang J., Pan Y., Wu F.-X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell P., Åkesson K., Chandran M., Cooper C., Ganda K., Schneider M. Implementation of models of care for secondary osteoporotic fracture prevention and orthogeriatric models of care for osteoporotic hip fracture. Best Practice & Research Clinical Rheumatology. 2016;30(3):536–558. doi: 10.1016/j.berh.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Russell R. G. G., Watts N. B., Ebetino F. H., Rogers M. J. Mechanisms of action of bisphosphonates: Similarities and differences and their potential influence on clinical efficacy. Osteoporosis International. 2008;19(6):733–759. doi: 10.1007/s00198-007-0540-8. [DOI] [PubMed] [Google Scholar]
- 36.Fukumoto S., Matsumoto T. Recent advances in the management of osteoporosis. F1000Research. 2017;6:p. 625. doi: 10.12688/f1000research.10682.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossini M., Adami G., Adami S., Viapiana O., Gatti D. Safety issues and adverse reactions with osteoporosis management. Expert Opinion on Drug Safety. 2016;15(3):321–332. doi: 10.1517/14740338.2016.1136287. [DOI] [PubMed] [Google Scholar]
- 38.Kharazmi M., Hallberg P., Warfvinge G., Michaëlsson K. Risk of a typical femoral fractures and osteonecrosis of the jaw associated with alendronate use compared with other oral bisphosphonates. Rheumatology. 2014;53(10):1911–1913. doi: 10.1093/rheumatology/keu286. [DOI] [PubMed] [Google Scholar]
- 39.Savaridas T., Wallace R. J., Salter D. M., Simpson A. H. R. W. Do bisphosphonates inhibit direct fracture healing? The Bone & Joint Journal. 2013;95-B(9):1263–1268. doi: 10.1302/0301-620x.95b9.31562. [DOI] [PubMed] [Google Scholar]
- 40.Wang J., Su K., Sang W., Li L., Ma S. Thiazide diuretics and the incidence of osteoporotic fracture: A systematic review and meta-analysis of cohort studies. Frontiers in Pharmacology. 2019;10:p. 1364. doi: 10.3389/fphar.2019.01364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chow S.-H., Chim Y.-N., Wang J., et al. Vibration treatment modulates macrophage polarisation and enhances early inflammatory response in oestrogen-deficient osteoporotic-fracture healing. European Cells and Materials. 2019;38:228–245. doi: 10.22203/ecm.v038a16. [DOI] [PubMed] [Google Scholar]
- 42.Wang J., He M., Wang G., Fu Q. Organic gallium treatment improves osteoporotic fracture healing through affecting the OPG/RANKL ratio and expression of serum inflammatory cytokines in ovariectomized rats. Biological Trace Element Research. 2018;183(2):270–279. doi: 10.1007/s12011-017-1123-y. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., He Y., Zong Y., et al. 17 -estradiol attenuates homocysteine-induced oxidative stress and inflammatory response as well as MAPKs cascade via activating PI3-K/Akt signal transduction pathway in Raw 264.7 cells. Acta Biochimica et Biophysica Sinica. 2015;47(2):65–72. doi: 10.1093/abbs/gmu124. [DOI] [PubMed] [Google Scholar]
- 44.Wang C., Zhang Z., Zhang H., et al. Susceptibility genes for osteoporotic fracture in postmenopausal Chinese women. Journal of Bone and Mineral Research. 2012;27(12):2582–2591. doi: 10.1002/jbmr.1711. [DOI] [PubMed] [Google Scholar]
- 45.De-Ugarte L., Yoskovitz G., Balcells S., et al. MiRNA profiling of whole trabecular bone: Identification of osteoporosis-related changes in MiRNAs in human hip bones. BMC Medical Genomics. 2015;8(1):p. 75. doi: 10.1186/s12920-015-0149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanberg M., McGuigan F. E., Ivaska K. K., Gerdhem P., Åkesson K. Polymorphisms in the inflammatory genes CIITA, CLEC16A and IFNG influence BMD, bone loss and fracture in elderly women. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0047964.e47964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivadeneira F., van Meurs J. B. J., Kant J., et al. Estrogen receptor β (ESR2) polymorphisms in interaction with estrogen receptor α (ESR1) and insulin-like growth factor I (IGF1) variants influence the risk of fracture in postmenopausal women. Journal of Bone and Mineral Research. 2006;21(9):1443–1456. doi: 10.1359/jbmr.060605. [DOI] [PubMed] [Google Scholar]
- 48.Sheng S., Wang J., Wang L., et al. Network pharmacology analyses of the antithrombotic pharmacological mechanism of Fufang Xueshuantong capsule with experimental support using disseminated intravascular coagulation rats. Journal of Ethnopharmacology. 2014;154(3):735–744. doi: 10.1016/j.jep.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 49.Domazetovic V., Marcucci G., Iantomasi T., Brandi M. L., Vincenzini M. T. Oxidative stress in bone remodeling: Role of antioxidants. Clinical Cases in Mineral and Bone Metabolism. 2017;14(2):209–216. doi: 10.11138/ccmbm/2017.14.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schröder K. NADPH oxidases in bone homeostasis and osteoporosis. Free Radical Biology and Medicine. 2019;132:67–72. doi: 10.1016/j.freeradbiomed.2018.08.036. [DOI] [PubMed] [Google Scholar]
- 51.Gan X., Huang S., Yu Q., Yu H., Yan S. S. Blockade of Drp1 rescues oxidative stress-induced osteoblast dysfunction. Biochemical and Biophysical Research Communications. 2015;468(4):719–725. doi: 10.1016/j.bbrc.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo G., Xu B., Huang Y. Icariside II promotes the osteogenic differentiation of canine bone marrow mesenchymal stem cells via the PI3K/AKT/mTOR/S6K1 signaling pathways. American Journal of Translational Research. 2017;9:2077–2087. [PMC free article] [PubMed] [Google Scholar]
- 53.Li G., Hu J., Chen H., et al. Enamel matrix derivative enhances the proliferation and osteogenic differentiation of human periodontal ligament stem cells on the titanium implant surface. Organogenesis. 2017;13(3):103–113. doi: 10.1080/15476278.2017.1331196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tong Y., Feng W., Wu Y., et al. Mechano-growth factor accelerates the proliferation and osteogenic differentiation of rabbit mesenchymal stem cells through the PI3K/AKT pathway. BMC Biochemistry. 2015;16(1):p. 1. doi: 10.1186/s12858-015-0031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan J.-b., Liu W., Zhu X.-h., et al. EGFR-AKT-mTOR activation mediates epiregulin-induced pleiotropic functions in cultured osteoblasts. Molecular and Cellular Biochemistry. 2015;398(1-2):105–113. doi: 10.1007/s11010-014-2210-4. [DOI] [PubMed] [Google Scholar]
- 56.Berghe W. V., Plaisance S., Boone E., et al. p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor-kappaB p65 transactivation mediated by tumor necrosis factor. Journal of Biological Chemistry. 1998;273(6):3285–3290. doi: 10.1074/jbc.273.6.3285. [DOI] [PubMed] [Google Scholar]
- 57.An Y., Zhang H., Wang C., et al. Activation of ROS/MAPK s/NF-κB/NLRP3 and inhibition of efferocytosis in osteoclast-mediated diabetic osteoporosis. The FASEB Journal. 2019;33(11):12515–12527. doi: 10.1096/fj.201802805rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiating L., Buyun J., Yinchang Z. Role of metformin on osteoblast differentiation in type 2 diabetes. Biomed Res Int. 2019;2019:6. doi: 10.1155/2019/9203934.9203934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thummuri D., Naidu V. G. M., Chaudhari P. Carnosic acid attenuates RANKL-induced oxidative stress and osteoclastogenesis via induction of Nrf2 and suppression of NF-κB and MAPK signalling. Journal of Molecular Medicine. 2017;95(10):1065–1076. doi: 10.1007/s00109-017-1553-1. [DOI] [PubMed] [Google Scholar]
- 60.Kimble R. B., Matayoshi A. B., Vannice J. L., Kung V. T., Williams C., Pacifici R. Simultaneous block of interleukin-1 and tumor necrosis factor is required to completely prevent bone loss in the early postovariectomy period. Endocrinology. 1995;136(7):3054–3061. doi: 10.1210/endo.136.7.7789332. [DOI] [PubMed] [Google Scholar]
- 61.Weitzmann M. N., Cenci S., Rifas L., Haug J., Dipersio J., Pacifici R. T cell activation induces human osteoclast formation via receptor activator of nuclear factor κB ligand-dependent and -independent mechanisms. Journal of Bone and Mineral Research. 2001;16(2):328–337. doi: 10.1359/jbmr.2001.16.2.328. [DOI] [PubMed] [Google Scholar]
- 62.Mosekilde L., Beck-Nielsen H., Sørensen O. H., et al. Hormonal replacement therapy reduces forearm fracture incidence in recent postmenopausal women - results of the Danish Osteoporosis Prevention Study. Maturitas. 2000;36(3):181–193. doi: 10.1016/s0378-5122(00)00158-4. [DOI] [PubMed] [Google Scholar]
- 63.Ding C., Parameswaran V., Udayan R., Burgess J., Jones G. Circulating levels of inflammatory markers predict change in bone mineral density and resorption in older adults: A longitudinal study. The Journal of Clinical Endocrinology & Metabolism. 2008;93(5):1952–1958. doi: 10.1210/jc.2007-2325. [DOI] [PubMed] [Google Scholar]
- 64.Gertz E. R., Silverman N. E., Wise K. S., et al. Contribution of serum inflammatory markers to changes in bone mineral content and density in postmenopausal women: A 1-year investigation. Journal of Clinical Densitometry. 2010;13(3):277–282. doi: 10.1016/j.jocd.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barbour K. E., Lui L.-Y., Ensrud K. E., et al. Inflammatory markers and risk of hip fracture in older white women: The study of osteoporotic fractures. Journal of Bone and Mineral Research. 2014;29(9):2057–2064. doi: 10.1002/jbmr.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aydin A., Halici Z., Albayrak A., et al. Treatment with carnitine enhances bone fracture healing under osteoporotic and/or inflammatory conditions. Basic & Clinical Pharmacology & Toxicology. Feb. 2015;117(3):173–179. doi: 10.1111/bcpt.12384. [DOI] [PubMed] [Google Scholar]
- 67.Mody N., Parhami F., Sarafian T. A., Demer L. L. Oxidative stress modulates osteoblastic differentiation of vascular and bone cells. Free Radical Biology and Medicine. 2001;31(4):509–519. doi: 10.1016/s0891-5849(01)00610-4. [DOI] [PubMed] [Google Scholar]
- 68.Watkins B., Lippman H. E., Le B. L., Li Y., Seifert M. F. Bioactive fatty acids: Role in bone biology and bone cell function. Progress in Lipid Research. 2001;40(1-2):125–148. doi: 10.1016/s0163-7827(00)00016-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All our main data used to support the findings of this study have been deposited in the Figshare repository (https://doi.org/10.6084/m9.figshare.12152607.v1).


