Abstract
In many lower animals, germ cell formation, migration, and maintenance depend on maternally provided determinants in germ plasm. In zebrafish, these processes have been extensively studied in terms of RNA-binding proteins and other coding genes. The role of small non-coding RNAs in the regulation of primordial germ cell (PGC) development remains largely unknown and poorly investigated, even though growing interests for the importance of miRNAs involved in a wide variety of biological processes. Here, we reported the role and mechanism of the germ plasm-specific miRNA miR-202-5p in PGC migration: (i) both maternal loss and knockdown of miR-202-5p impaired PGC migration indicated by the mislocalization and reduced number of PGCs; (ii) cdc42se1 was a direct target gene of miR-202-5p, and overexpression of Cdc42se1 in PGCs caused PGC migration defects similar to those observed in loss of miR-202-5p mutants; (iii) Cdc42se1 not only interacted with Cdc42 but also inhibited cdc42 transcription, and overexpression of Cdc42 could rescue PGC migration defects in Cdc42se1 overexpressed embryos. Thus, miR-202-5p regulates PGC migration by directly targeting and repressing Cdc42se1 to protect the expression of Cdc42, which interacts with actin to direct PGC migration.
Keywords: primordial germ cell, miR-202-5p, Cdc42se1, Cdc42, migration
Introduction
In most vertebrates, primordial germ cells (PGCs), the precursors of germ cells, segregate from somatic cells and migrate across the embryo to reach the site where they carry out their function (Raz, 2003; Tang et al., 2016). The event of PGC migration is precisely regulated as the long-distance migratory path across various developing tissues before reaching to the genital ridge (Richardson and Lehmann, 2010). There is a close correspondence between proper migration and PGC survival. Usually, the mislocalized PGC cells cannot maintain their PGC characteristics and are eventually eliminated to prevent germ cell tumors in most animals (Paksa and Raz, 2015).
In lower animals such as fruit fly, frog, and zebrafish, maternally supplied materials in germ plasm are indispensable for germ cell development (Extavour and Akam, 2003). It has been demonstrated that loss of germ plasm components led to PGC developmental defects (Raz, 2003; Richardson and Lehmann, 2010; Strome and Updike, 2015). For example, the RNA-binding protein Dnd1 binds with messenger RNAs (mRNAs), such as nos1, zeb1, and tdrd7, and prohibits miR-430 from associating with these mRNAs. Knockdown of Dnd1 would impair the motility of PGCs, which resulted in abnormal migration of PGCs and eventually caused the reduction or complete loss of PGC at early developing stage (Weidinger et al., 2003; Goudarzi et al., 2012; Gross-Thebing et al., 2017). In zygotic embryos with loss of vasa and tdrd12, PGCs could normally specify and migrate; however, germ cells failed to maintain and the mutant fish developed into infertile males (Hartung et al., 2014; Dai et al., 2017). In addition, the germline component nanos family played essential roles in germ stem cells and ovary development in zebrafish (Draper et al., 2007; Beer and Draper, 2013; Cao et al., 2019).
MicroRNAs (miRNAs) are a group of endogenous small non-coding RNAs, which usually bind to mRNAs of coding genes to direct their post-transcriptional repression in animals and plants (Bartel, 2009; Shenoy and Blelloch, 2014). Increasing evidences indicate that miRNAs are involved in the regulation of germ cell formation, migration, and maintenance (Hayashi et al., 2008; Shenoy and Blelloch, 2014; van den Driesche et al., 2014). In zebrafish, miR-430 played a robust role in PGC migration through the clearance of maternal stromal cell-derived factor 1 (SDF1) and its receptors (Staton et al., 2011). SDF1 is one of the master chemokine molecules, which guides PGC migration through germ-cell guidance receptors chemokine (C-X-C motif) receptor 4B (CXCR4B) and 7B (CXCR7B) (Doitsidou et al., 2002; Knaut et al., 2003; Boldajipour et al., 2008; Mizoguchi et al., 2008). PGC migration is regulated by transmembrane receptors that receive external chemoattractant signals, which are then translated to cytoskeletal change by effector molecules such as phospholipids and small GTPases (Richardson and Lehmann, 2010; Barton et al., 2016). A recent study indicated that expression of a dominant-negative form of small GTPase Cdc42 in the PGC led to round cell morphology, decreased formation of the membrane invaginations, and reduced blebbing (Goudarzi et al., 2017). However, the molecular mechanism underlying Cdc42 expression and regulation in PGCs was remained unclear in zebrafish.
Previously, we have identified a germ plasm-specific miRNA miR-202-5p, which was exclusively localized in PGCs during embryogenesis in zebrafish (Zhang et al., 2017). Here, we investigated the potential role and underlying mechanism of miR-202-5p in PGC migration. Our results demonstrated that miR-202-5p played a critical role in zebrafish PGC migration by maintaining the expression of Cdc42.
Results
miR-202 mutant zebrafish were fertile
To investigate the role of miR-202-5p in zebrafish germ cell development, we generated a miR-202-5p mutant line using Crispr/Cas9 gene-editing strategy. We observed five miR-202 mutants in F0 embryos injected with Cas9 mRNA and gRNA. The F0 embryos containing miR-202 mutant were raised to adulthood and crossed with wild-type (WT) zebrafish, and an 8-bp deletion (miR-202Δ8) mutant was chosen to establish the miR-202 mutant line for further analysis (Figure 1A). To confirm the deletion in miR-202 site, quantitative real-time PCR (qRT-PCR) was performed to check the expression of mature miR-202-5p. Indeed, no mature miR-202-5p was detected and the miR-202-3p level was also significantly decreased in the miR-202Δ8 mutant testis (Figure 1B). We observed that all miR-202 mutants developed normally, and the genotype of miR-202 mutants were inherited at the expected Mendelian ratio. There was no significant morphological difference among testes and ovaries of miR-202+/+, miR-202+/−, and miR-202−/− adults, in which normal spermatozoa and oocytes were observed (Figure 1C and D). Mating between miR-202−/− males and females produced offspring. Therefore, loss of miR-202 did not affect reproductive capacity in zebrafish.
Figure 1.
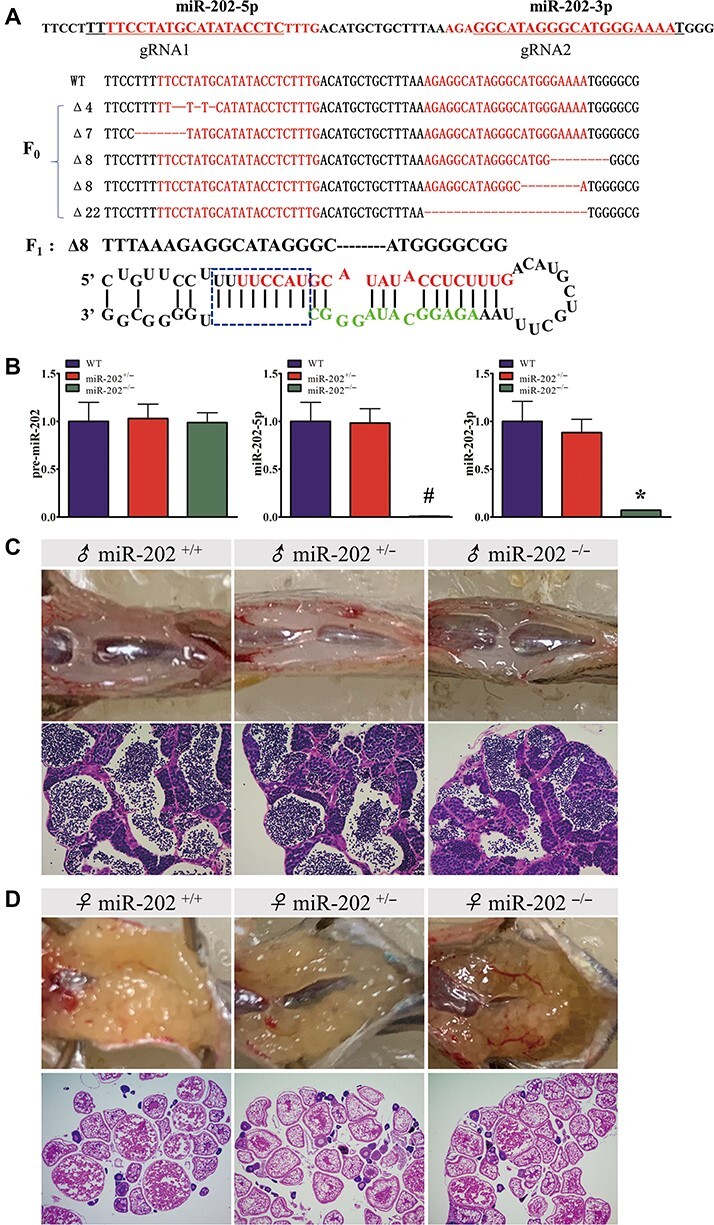
Analysis of gene expression and gonad structure in miR-202−/− adults. (A) Generation of miR-202 mutant by CRISPR/CAS9 genome editing. (B) qRT-PCR analysis of pre-miR-202, miR-202-5p, and miR-202-3p in miR-202+/+, miR-202+/−, and miR-202−/− testes. (C and D) Gonadal morphology and structure in miR-202+/+, miR-202+/−, and miR-202−/− adults. *P < 0.05; #, not detected.
Maternal loss of miR-202-5p impaired PGC migration
Subsequently, we investigated PGC development in miR-202 mutant embryos. The miR-202+/− zebrafish were crossed to produce F2 embryos that theoretically contained 25% miR-202−/− genotypic embryos. The F2 embryos developed normally, whose PGCs were visualized by whole-mount in situ hybridization (WISH) of vasa mRNA. WISH results indicated that there was no significant change in the number and localization of PGCs between F2 mutant and WT embryos, suggesting that zygotic loss of miR-202-5p did not affect PGC development (Figure 2A–C). Since miR-202-5p was a maternal factor, we further explored whether PGC development was influenced in maternal miR-202-5p mutant embryos. No miR-202-5p and little miR-202-3p were detected in 1-cell stage embryos, indicating that miR-202 was maternally deleted (Figure 2D). We analyzed PGC development in maternal loss of miR-202 (MmiR-202) mutant embryos. At dome stage when PGCs specified and initiated to migrate, there was no significant difference in the number and localization of PGC between WT and MmiR-202 embryos (Figure 2E and F). Whereas, at stage of 75% epiboly, PGCs were scatted widely and some mislocalized PGCs were found in MmiR-202 embryos (Figure 2G and H), and the number of mislocalized PGCs increased at 3-somite and prim 5 stages (Figure 2I–L). In comparison with WT embryos, the expression levels of PGC markers such as dnd, vasa, and nanos3 were significantly reduced during PGC migration in MmiR-202 embryos (Figure 2M). The number of PGCs in MmiR-202 embryos was significantly lower than that in WT embryos at corresponding stages during PGC migration (Figure 2N). Of the MmiR-202 embryos at prim 5 stage, 11.2% had normally developed PGCs, 56.3% had reduction of PGC only, 12.9% had PGC mislocalization only, and 19.6% displayed both PGC reduction and mislocalization (Figure 2O and P). To verify whether the PGC developmental defects in MmiR-202 embryos were caused by loss of miR-202-5p, miR-202-3p, or both, rescue experiments were performed in MmiR-202 embryos by injection of miR-202-5p and miR-202-3p mimics, respectively. As shown in Figure 2O, Q, and R, injection of miR-202-5p mimics rather than miR-202-3p mimics significantly attenuated PGC developmental defects in MmiR-202 embryos, suggesting that maternal loss of miR-202-5p was responsible for the PGC developmental defects in MmiR-202 embryos. To further demonstrate this, miR-202-5p knockdown was carried out by injecting miR-202-5p inhibitor into WT embryos, and the number and localization of PGC were assessed. The expression of PGC markers was decreased in miR-202-5p inhibitor-injected embryos (Supplementary Figure S1A–D), and similar defects including PGC mislocalization and reduction were found in miR-202-5p inhibitor-injected PGC-labelled transgenic embryos at prim 5 stage (Supplementary Figure S1E–L). There was a close correspondence between the PGC number reduction and the injection dosage of miR-202-5p inhibitor (Supplementary Figure S1M–P). Moreover, PGC mislocalization and number reduction were found in miR-202-5p inhibitor-injected embryos during PGC migration (Supplementary Figure S2). Thus, maternal loss or knockdown of miR-202-5p impaired PGC migration in zebrafish.
Figure 2.
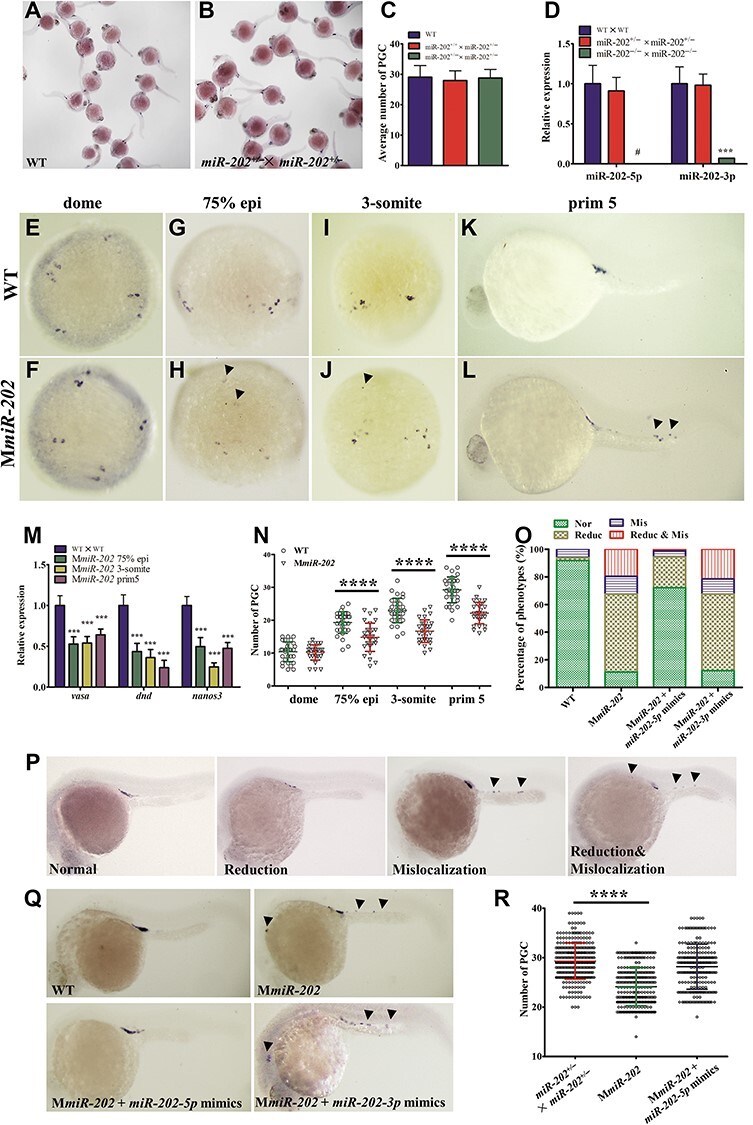
Maternal loss of miR-202-5p results in PGC reduction and mislocalization. (A and B) Whole-mount in situ hybridization of vasa in WT and F2 embryos. (C) Average number of PGCs in embryos produced from WT, miR-202+/+ × miR-202+/−, and miR-202+/− × miR-202+/− parents. (D) qRT-PCR analysis of miR-202-5p and miR-202-3p in 1-cell stage embryos produced from WT, miR-202+/− × miR-202+/−, and miR-202−/− × miR-202−/− parents. (E–L) Whole-mount in situ hybridization of vasa in WT and MmiR-202 embryos at dome, 75% epiboly (epi), 3-somite, and prim 5 stages. (M) qRT-PCR analysis of PGC markers in WT and MmiR-202 embryos from 75% epi, 3-somite, and prim5 stages. (N) PGC number in WT and MmiR-202 embryos. (O–Q) Statistical analysis of PGC phenotypes in WT, MmiR-202 embryos, and MmiR-202 embryos injected with miR-202-5p or miR-202-3p mimics at prim 5 stage (O) and their representative images (P and Q). Nor, normal; Mis, mislocalization; Reduc, reduction; Mis & Reduc, mislocalization and reduction. (R) PGC number in WT, MmiR-202 embryos, and MmiR-202 embryos injected with miR-202-5p mimics. Arrowheads indicate the mislocalized PGCs. **P < 0.01; ***P < 0.001; ****P < 0.0001; #, not detected.
miR-202-5p negatively regulated Cdc42se1 in PGC migration
miRNAs rarely produce functional protein products; they usually bind to mRNAs of target genes and initiate hierarchical biological events. To reveal the signaling pathway of miR-202-5p in PGC migration, potential target genes of miR-202-5p were bioinformatically predicted with PITA and miRanda (Supplementary Table S1). Among these candidates, cdc42se1 (cdc42 small effector 1) was one of the most promising candidates, whose 3’UTR contained a miR-202-5p binding site (Figure 3A). qRT-PCR results indicated that the embryonic endogenous cdc42se1 mRNA was decreased in response to overexpression of miR-202-5p mimics (Figure 3B). Since the expression of endogenous Cdc42se1 protein in embryos was almost undetectable (data not shown), pcDNA3.1(+)-Cdc42se1 plasmid containing a cdc42se1 open reading frame (ORF) that fused its own 3′UTR was co-transfected with control or miR-202-5p mimics in HEK293T cells. Western blotting showed that Cdc42se1 protein was significantly reduced in HEK293T cells in the presence of miR-202-5p mimics (Figure 3C). Furthermore, we analyzed the endogenous cdc42se1 mRNA and protein in MmiR-202 mutant individuals. In comparison with that in PGCs of WT embryos, the expression level of cdc42se1 mRNA in PGCs of MmiR-202 embryos was significantly upregulated (Figure 3D); the expression level of Cdc42se1 protein in MmiR-202 testis was upregulated (Figure 3E). Next, the direct interaction between miR-202-5p and cdc42se1 was examined by luciferase reporter assay by linking the 3′UTR of cdc42se1 to the C-terminus of luciferase in psiCheck2 vector. As shown in Figure 3F, the relative activity of luciferase fusing WT 3′UTR of Cdc42se1 was strongly repressed by miR-202-5p mimics, whereas mutation of the miR-202-5p binding site abolished this repression. Furthermore, we employed RNA reporter by fusing the red fluorescent protein (RFP) ORF with WT or miR-202-5p binding site-mutated 3′UTR of cdc42se1. Quantitative pixel-intensity analysis revealed a dramatically elevated level of RFP in PGCs when the miR-202-5p binding site was mutant (Figure 3G and H). These results demonstrated that cdc42se1 was a direct target gene of miR-202-5p.
Figure 3.
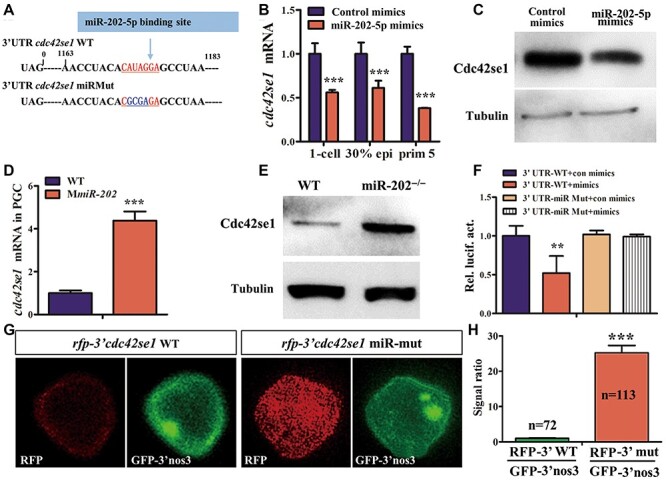
Cdc42se1 is a target gene of miR-202-5p. (A) The miR-202-5p binding site (red) in 3′UTR of cdc42se1 mRNA, and the mutant site is shown in blue. (B) qRT-PCR analysis of cdc42se1 mRNA in control or miR-202-5p mimic-injected embryos. (C) Western blotting analysis of Cdc42se1 protein in HEK293T cells co-transfected with pcDNA3.1(+)-Cdc42se1 plasmid containing the ORF of cdc42se1 fused with its own 3′UTR and control or miR-202-5p mimics. (D) qRT-PCR analysis of cdc42se1 mRNA in PGCs of WT and MmiR-202 embryos. (E) Western blotting analysis of Cdc42se1 protein level in WT and MmiR-202 testes. (F) Relative expression of luciferase activity in HEK293T cells transfected with psiCheck2-WT 3′UTR-Cdc42se1 (3′UTR-WT) or 3′UTR-miRMut and control mimics (con mimics) or miR-202-5p mimics (mimics). (G) Mutation of miR-202-5p binding site in cdc42se1 3′UTR caused increased expression of RFP in PGCs as compared to the control WT 3′UTR. The co-injected RNA (GFP-3′UTR of nos3) served as a control. (H) Quantitative representation of the normalized signal intensity in the experiment presented in G. **P < 0.01; ***P < 0.001.
Subsequently, we characterized the spatial and temporal expression patterns of cdc42se1. cdc42se1 was a maternal factor universally distributing at gastrulation stage; then, its expression was gradually decreased and mainly concentrated at the central neural system and the yolk syncytial layer (Supplementary Figure S3). To investigate the possible role of Cdc42se1 in PGC migration, loss and gain of function were performed by injection of cdc42se1 morpholino (MO) and mRNA, respectively. There was no observable defect of PGC development in cdc42se1 MO-injected embryos (Figure 4A and B). Whereas, significant number reduction and mislocalization of PGC were observed in cdc42se1 mRNA-injected embryos (Figure 4C and D), similar to the phenotype observed in maternal loss or knockdown of miR-202-5p embryos. Of the cdc42se1 mRNA-injected embryos, 20.6% had normally developed PGCs, 17.8% displayed reduction only, and 16.0% showed mislocalization only, but 45.6% displayed both reduction and mislocalization (Figure 4J). In cdc42se1 mRNA-injected embryos, co-injection of either cdc42se1 MO or miR-202-5p mimics almost completely rescued the PGC abnormalities (Figure 4E, F, and J). In addition, there was a strong relevance between cdc42se1 mRNA injection dosage and PGC number reduction (Figure 4G–I and K); there was no significant number reduction in embryos injected with 100 pg/embryo cdc42se1 mRNA, whereas significant number reduction was found when the injection dosage of cdc42se1 mRNA over 200 pg/embryo. Since overexpression of Cdc42se1 and knockdown of miR-202-5p caused similar phenotypes, we inferred that PGC developmental defects caused by knockdown of miR-202-5p might be due to an increase of Cdc42se1. To verify this, we co-injected cdc42se1 MO to repress the expression of Cdc42se1 in miR-202-5p inhibitor-injected embryos. However, the results showed that co-injection of cdc42se1 MO failed to rescue miR-202-5p inhibitor caused PGC reduction (Figure 4L). This might be because that cdc42se1 was one of the key target genes in miR-202-5p inhibitor-injected embryos, and other regulating factors might function in PGC migration.
Figure 4.
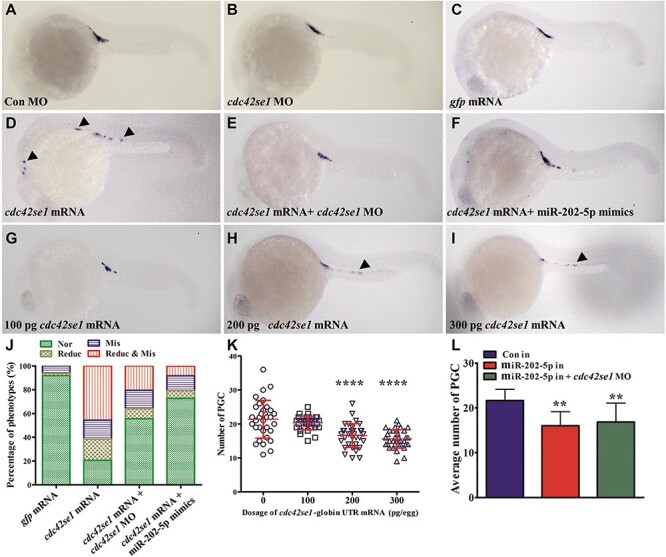
Overexpression of Cdc42se1 leads to PGC number reduction and mislocalization. (A–I) Representative images of prim 5 stage embryos injected with cdc42se1 mismatch control morpholino (Con MO; A), cdc42se1 MO (B), gfp mRNA (C), cdc42se1 mRNA (D), cdc42se1 mRNA and cdc42se1 MO (E), cdc42se1 mRNA and miR-202-5p mimics (F), 100 pg cdc42se1 mRNA (G), 200 pg cdc42se1 mRNA (H), and 300 pg cdc42se1 mRNA (I). Arrowheads indicate mislocalized PGCs. (J) The phenotypes of PGCs in embryos injected with gfp mRNA, cdc42se1 mRNA, cdc42se1 mRNA + cdc42se1 MO, and cdc42se1 mRNA + miR-202-5p mimics. (K) The number of PGCs in control embryos and embryos injected with different dosages of cdc42se1 mRNA at prim 5 stage. (L) The average number of PGCs in embryos injected with control inhibitor (in), miR-202-5p in, or miR-202-5p in + cdc42se1 MO. **P < 0.01; ****P < 0.0001.
Furthermore, we performed specific overexpression of Cdc42se1 in a small portion of PGC population by injecting Cdc42se1-RFP-UTR-nos3 mRNA into PGC-labelled transgenic embryos at 4-cell stage (Figure 5A). The results showed that overexpression of Cdc42se1 in PGCs resulted PGC mislocalization at prim 5 stage (Figure 5B–E), although some of Cdc42se1 overexpressed PGCs are still located at the genital ridge (Figure 5F). Of these mislocalized PGCs, some still possessed entire germ granule and maintained PGC identity (arrow in Figure 5E), whereas the remaining PGCs were degraded (asterisk in Figure 5D) or lost the PGC identity, germ granule (asterisk in Figure 5E). These data demonstrated that ectopic overexpression of Cdc42se1 in PGCs impaired PGC migration.
Figure 5.
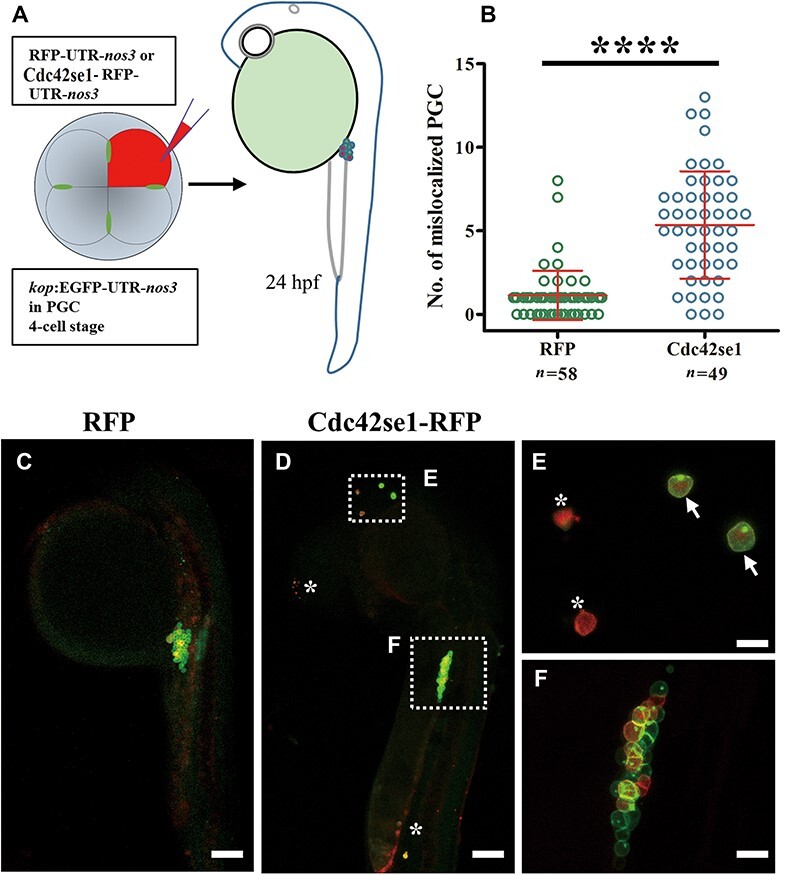
Overexpression of Cdc42se1 in PGCs leads to mislocalization. (A) Cdc42se1-RFP-UTR-nos3 mRNA was injected into 4-cell embryos of Tg1 kop:EGFP-UTR-nos3 line. (B) The number of mislocalized PGCs in embryos injected with RFP or Cdc42se1-RFP-UTR-nos3 mRNA. (C and D) Representative images of PGCs in embryos injected with RFP or Cdc42se1-RFP-UTR-nos3 at prim 5 stage. (E and F) The magnified images as indicated in D. Arrows indicate the mislocalized PGCs, and asterisks indicate the dead or transforming PGCs. ****P < 0.0001. Scale bar, 100 μm (E and F) and 20 μm (D).
Cdc42se1 negatively regulated Cdc42 in zebrafish PGC
In mammal, Cdc42se1 was a negative regulator of Cdc42 and involved in actin polymerization, cytoskeleton change, and cell motility (Pirone et al., 2000; Ching et al., 2005; Block et al., 2012). However, the physic relationship among Cdc42se1, Cdc42, and actin in zebrafish was remained unclear. To illustrate it, HEK293T cells were co-transfected with Myc-tagged zebrafish actin (Myc-actin) and Flag-tagged zebrafish Cdc42 (Flag-Cdc42), followed by co-immunoprecipitation (co-IP) assay with antibodies specific to the Flag tag. As shown in Figure 6A, the protein complex immunoprecipitated by anti-Flag antibody was also recognized by anti-Myc antibody, indicating that actin was physically associated with Cdc42. Similarly, co-IP assay demonstrated that Cdc42se1 interacted with both WT Cdc42 (wtCdc42) and activated mutant of Cdc42 (Cdc42Q61L) (Figure 6B and C). However, in HEK293T cells co-transfected with Myc-actin and Flag-Cdc42se1, the protein complex immunoprecipitated by anti-Flag was not recognized by anti-Myc antibody, suggesting that actin did not interact with Cdc42se1 (Supplementary Figure S4). These data indicated that Cdc42 separately interacted with actin and Cdc42se1 to form two complexes, Cdc42/actin and Cdc42/Cdc42se1.
Figure 6.
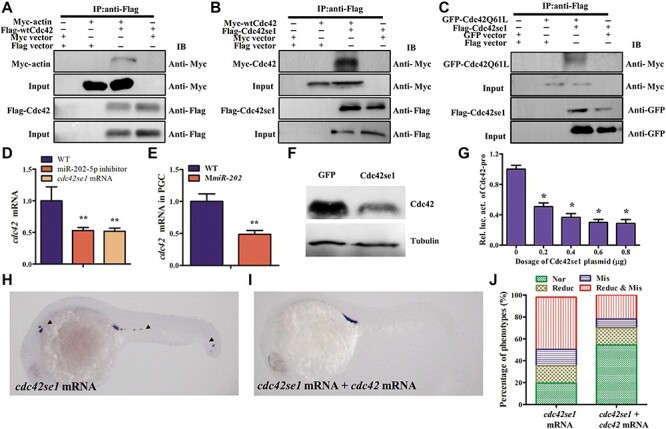
Cdc42se1 negatively regulates Cdc42 expression in PGCs. (A–C) The interactions among Cdc42se1, Cdc42, and actin were analyzed by co-IP. HEK293T cells were co-transfected with Myc-actin and Flag-Cdc42 (A), Myc-Cdc42 and Flag-Cdc42se1 (B), EGFP-Cdc42Q61L and Flag-Cdc42se1 (C) plasmids, and cells transfected pCMV-Myc, pCMV-Flag, or pEGFPN3 vector were used as controls. Twenty-four hours later, cells were immnoprecipitated with anti-Flag antibody, followed by analysis of the immunoprecipitates by western blotting. (D and E ) qRT-PCR analysis of the endogenous cdc42 mRNA in embryos injected with miR-202-5p inhibitor or cdc42se1 mRNA (D) or in PGCs of WT and MmiR-202 embryos (E). (F) The endogenous Cdc42 protein in cdc42se1 mRNA-injected embryos by western blotting. (G) Relative expression of luciferase activity driven by cdc42 promoter in HEK293T cells, which were co-transfected with different dosages of pcDNA3.1(+)-Cdc42se1 plasmid. (H and I) Representative images of embryos respectively injected with cdc42se1 mRNA (H) and cdc42se1 mRNA + cdc42 mRNA (I) by WISH analysis of vasa. (J) Statistical analysis of PGC phenotypes in embryos injected with gfp mRNA, cdc42se1 mRNA, and cdc42se1 + cdc42 mRNA. *P < 0.05; **P < 0.01.
We analyzed the endogenous Cdc42 expression in embryos injected with cdc42se1 mRNA or miR-202-5p inhibitor. The expression of both cdc42 mRNA and protein were significantly decreased in cdc42se1 mRNA or miR-202-5p inhibitor-injected embryos (Figure 6D and F). Furthermore, the endogenous cdc42 mRNA level in PGCs of MmiR-202 embryos was significantly decreased comparing with that in PGCs of WT embryos (Figure 6E). Since the expression level of cdc42 mRNA decreased, we analyzed whether Cdc42se1 downregulated cdc42 transcription by repressing cdc42 promoter activity. Indeed, the relative activity of luciferase driven by cdc42 promoter was strongly decreased by Cdc42se1 overexpression (Figure 6G). In zebrafish, overexpression of domain negative Cdc42 affected bleb formation and resulted in PGC mislocalization (Goudarzi et al., 2017), similar to that observed in Cdc42se1 overexpressed embryos. We proposed that the PGC developmental defects in Cdc42se1 overexpressed embryos might be due to a decrease of Cdc42. To test it, we co-injected cdc42 mRNA to rescue PGC migration in embryos injected with cdc42se1 mRNA. The results indicated that overexpression of Cdc42 significantly attenuated the PGC migration defects in Cdc42se1 overexpressed embryos (Figure 6H–J). In comparison with Cdc42se1 overexpressed embryos, upon co-injection of Cdc42, the percentage of embryos with normal PGCs increased to 54.4%, the percentage of embryos with only mislocalized PGCs decreased to 8.2%, and the percentage of embryos with both reduced number of PGCs and mislocalized PGCs decreased to 22.0% (Figure 6J). Therefore, Cdc42se1 negatively regulated Cdc42 in zebrafish PGCs.
Discussion
miRNAs were important regulators in PGC development in animals. In mouse, Dicer-deleted PGCs exhibited poor proliferation, indicating that miRNA biogenesis was required for mouse PGC development (Hayashi et al., 2008). LIN28-mediated repression of miRNA-let7 was required for BLIMP1 expression, a critical transcription factor for mouse PGC induction from ESC in vitro (Matzuk, 2009; West et al., 2009). The miR-290-295 cluster and let7 played opposing roles in mouse germ cell formation (Melton et al., 2010). In zebrafish, maternal and zygotic loss of dicer impaired PGC migration (Mishima et al., 2006). In zebrafish, the most widely and comprehensively studied miRNA associated with PGC development was the miR-430, a zygotic miRNA promoting deadenylation and clearance of maternal mRNAs (Giraldez et al., 2006). MiR-430 played robust roles in regulation of PGC migration by clearance of SDF1 ligands (sdf1a and sdf1b) and their receptors (cxcr4a, cxcr4b, cxcr7a, and cxcr7b). Repression or loss of miR-430-mediated clearance would expose otherwise buffered genetic lesions and impair PGC migration (Staton et al., 2011). Moreover, identification of the molecule module for bleb-based PGC motility revealed that PGC motility was controlled by a balance between miR-430 function and the action of germline-specific RNA-binding protein DND (Goudarzi et al., 2012).
Maternally supplied germ plasm components played indispensable roles in germ cell formation, migration, and maintenance in zebrafish (Richardson and Lehmann, 2010). To date, studies of germ plasm components in PGC development were largely focused in the functional role and molecular mechanism of coding genes in PGCs. For example, Dnd, Ca15b, Rgs14a, and Nanos3 were required for PGC survival and migration; Vasa and Piwi were essential for germ cell differentiation and maintenance (Koprunner et al., 2001; Houwing et al., 2007; Hartung et al., 2014; Paksa and Raz, 2015). Germ plasm-specific long non-coding RNAs (lncRNA) such as pgc and xlsirt were involved in germ cell migration and transcriptional silence in fruity fly and frog (Kloc and Etkin, 1994; Nakamura et al., 1996; Martinho et al., 2004). However, little was known about the role of maternal germ plasm associated lncRNAs and miRNAs and their functions in zebrafish PGC development. It has been found that miR-202-5p was gonad-specific miRNA in a broad of species. Mouse miR-202-5p was localized in Sertoli cells of primordial XY gonads and acted downstream of the testis-determining factor Sox9a, suggesting that miR-202-5p played an early role in testis development (Wainwright et al., 2013). Human miR-202-5p was expressed in Sertoli cells and possibly associated with spermatogenesis (Dabaja et al., 2015). Frog miR-202-5p was highly expressed in stage I–II oocytes (Armisen et al., 2009). MiR-202-5p was a germ plasm-specific miRNA (Zhang et al., 2017), suggesting a potential role of miR-202-5p in PGC development.
Herein, we used loss of function strategy to explore the function of miR-202-5p in PGC development. The miR-202 mutant line were fertile and produced functional eggs and sperms, suggesting that miR-202 was not primary for reproductive capacity in zebrafish (Figure 1). However, maternal loss or knockdown of miR-202-5p significantly impaired PGC migration (Figure 2; Supplementary Figures S1 and S2). The PGC developmental defects in MmiR-202 embryos, to a certain degree, was similar to that observed in MZdicer embryos (Mishima et al., 2006), demonstrating that miR-202-5p was a key miRNA in PGC development in zebrafish. Unlike other germ plasm-coding genes, such as vasa, tdrd12, and dnd, mutation of these did not affect PGC development during embryogenesis (Hartung et al., 2014; Dai et al., 2017; Li et al., 2017); however, their mutant lines ultimately developed into infertile ‘males’. These suggested that different types of germ plasm components played diverse roles in zebrafish reproduction. Since reproduction was critical for species continuation, various species had adopted diverse regulatory mechanisms, such as genetic compensation response (GCR), to ensure reproduction works if some abnormalities happened. Recently, it was reported that there are two different GCR strategies in zebrafish, indicating that the occurrence of some loss-of-function mutations would activate expression of other genes in the same family, which resulted in recessive phenotypes (El-Brolosy et al., 2019; Ma et al., 2019). It was uncertain whether there was GCR that activated the expression of functionally similar miRNA to protect the gonad development in miR-202 mutant line. In medaka, Gay et al. (2018) reported that miR-202-5p was mainly localized in ovarian granulosa cells. Deletion of miR-202 impaired early oogenesis/folliculogenesis and decreased the number of large follicles, ultimately leading to dramatically reduced female fecundity in medaka (Gay et al., 2018). The different cellular localization and function of miR-202-5p between zebrafish and medaka suggested that it might evolve diverse strategies for regulating reproduction in different fish species.
To clarify the mechanism by which miR-202-5p regulated PGC migration, target genes of miR-202-5p were bioinformatically predicted, and a critical miR-202-5p target gene cdc42se1 was identified (Figure 3). Previous studies showed that there was a weak interaction between cdc42se1 and Dnd in zebrafish (Chen et al., 2010). In this study, we demonstrated that overexpression of Cdc42se1 impaired PGC migration, similar to that observed in miR-202-5p knockdown or knockout embryos (Figures 4 and 5). However, knockdown of Cdc42se1 could not significantly rescue miR-202-5p inhibitor-mediated PGC development defects, indicating that cdc42se1 was just one of the critical dysregulated target genes of miR-202-5p. It was known that the PGC development was protected by transcriptional silence, which was regulated at the level of RNA polymerase II (pol II) activity and chromatin state (Strome and Updike, 2015). In Drosophila, the germ plasm-specific lncRNA pgc was important for PGC migration and maintenance by repression of RNA pol II-dependent transcription (Nakamura et al., 1996; Martinho et al., 2004). Prediction of target genes showed that numerous potential targets of miR-202-5p were associated with RNA pol II, translation, and chromatin structure (Supplementary Table S1), suggesting that other miR-202-5p targets might be involved in maintaining transcriptional silence in PGCs.
In zebrafish, small Rho-GTPase family, including Rac1, RhoA, and Cdc42, played essential roles in PGC migration. In migratory PGCs, Rac1 was responsible for formation of actin-rich structures and RhoA promoted retrograde actin flow, which were required for bleb formation (Ridley, 2015). Chemokine-guided germ cells within zebrafish embryos required the function of the small Rho GTPases Rac1 and RhoA (Kardash et al., 2010; Xu et al., 2012; Miranda-Rodriguez et al., 2017). Cdc42 was involved in cell cytoskeleton organization and cell migration (Heasman and Ridley, 2008) and controlled membrane invagination via the clathrin-independent pinocytic pathway (Sabharanjak et al., 2002). Goudarzi et al. (2017) reported that overexpression of an inactivated form of Cdc42 in PGCs impaired PGC membrane invagination and bleb formation and ultimately resulted in PGC mislocalization. In mouse, Cdc42se1 interacted with the activated form of Cdc42 through its N-terminal CRIB domain. Cdc42se1 blocked Cdc42-induced c-JNK activity and altered Cdc42-induced morphology change in COS1 cells and NIH-3T3 fibroblasts in a manner depending on Cdc42 binding (Pirone et al., 2000). In this study, we found that zebrafish Cdc42se1 interacted with both WT and activated mutant of Cdc42 (Figure 6B and C). Zebrafish and mouse Cdc42 homologues shared 99.5% identities, and the N-terminus of zebrafish and mouse Cdc42se1 homologues that contain the CRIB domain had 87.6% identities; therefore, it was possible that Cdc42se1 also negatively regulated Cdc42 activity in zebrafish. Moreover, Cdc42se1 reduced the endogenous Cdc42 expression by downregulating the activity of the Cdc42 promoter, and overexpression of the WT form of Cdc42 could rescue the PGC developmental defects in Cdc42se1 overexpressed embryos (Figure 6). Therefore, Cdc42se1 overexpression negatively regulated the expression of Cdc42 and thus impaired PGC migration. However, it remains unclear how Cdc42se1 affects Cdc42 transcription and whether Cdc42se1 affects Cdc42 activity in zebrafish.
In conclusion, we revealed a novel miRNA involved in the Cdc42 signaling pathway in PGC migration; germ plasm-specific miR-202-5p repressed the expression of the Cdc42-negative effector Cdc42se1 to ensure the sufficient expression level of Cdc42 in zebrafish PGC development.
Materials and methods
Ethics statement
All procedures with zebrafish were approved by the Ethics Committee of Sun Yat-sen University, and the methods were carried out in accordance with the approved guidelines.
Fish strains and cell lines
Zebrafish lines including AB line WT and PGC transgenic line Tg1 (Tg kop:EGFP-UTR-nos3) were purchased from the China Zebrafish Resource Center. Fish were raised at 28°C with 10 h darkness and 14 h light. All embryos were collected after natural spawning and staged as previously reported (Kimmel et al., 1995). HEK293T cells were maintained in DMEM supplemented with 10% FBS at 37°C in a 5% CO2 incubator.
Targeted gene disruption by CRISPR/Cas9
MiR-202-5p knockout by CRISPR/Cas9 was performed as previously described (Guo et al., 2017). Two guide RNAs (gRNAs) were designed with an online tool (http://crispr.mit.edu/), gRNA1 (GGAAAAAGGATACGTATATGG) and gRNA2 (GGCATAGGGCATGGGAAAATGGG). The gRNAs were transcribed with T7 RNA Polymerase Kit (Ambion). Cas9 mRNA (300 ng/μl) and gRNA (50 ng/μl) were injected into one-cell embryos, and the mutations were analyzed by sequencing.
Prediction of miR-202-5p target genes
Target genes of miR-202-5p were analyzed for sites complementary to the miR-202-5p seed sequence (UCCUAUG) using both PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html) and miRanda (http://www.microrna.org/microrna/home). Target genes predicted by both PITA and miRanda were considered as potential target genes of miR-202-5p (Supplementary Table S1).
Plasmid construction
The cDNA of cdc42se1 (ENSDARG00000023724, ZV11), including its ORF and 3′UTR, and ORF of cdc42 (ENSDARG00000044573, ZV11) were respectively inserted into the pCS2+ vector for mRNA synthesis by BamHI and XbaI. The 2000-bp fragment of the cdc42 promoter was inserted to the pGL3B vector to generate the pGL3-Cdc42-pro plasmid. The ORF of cdc42se1 and RFP and the 3′UTR of nos3 were inserted into the pCS2+ vector to generate the pCS2-Cdc42se1-RFP-UTR-nos3 plasmid. The ORFs of cdc42, actin (ENSDARG00000037746, ZV11), and cdc42se1 were respectively inserted into both pCMV-N-Flag and pCMV-N-Myc vectors by EcoRI and XhoI to generate Myc-actin, Myc-Cdc42, Flag-Cdc42se1, and Flag-Cdc42 plasmids. The ORF of cdc42se1 fused with its own 3′UTR was inserted into the pcDNA3.1(+) vector by EcoRI and NotI. The activated form of Cdc42 (Cdc42Q61L) was inserted into the pEGFPN3 vector by EcoRI and BamHI to generate the pEGFPN3-Cdc42Q61L (EGFP-Cdc42Q61L) plasmid. The 3′UTR of cdc42se1 was inserted into the psiCHECK2 vector. Then, the miR-202-5p binding site (CATAGGA) in the constructed WT plasmids were replaced to (CGCGAGA) by Hieff Mut™ Multi Site-Directed Mutagenesis Kit (Yeasen). Primers were list in Supplementary Table S2.
mRNA synthesis
For mRNA synthesis, ORFs were inserted into the pCS2+ vector. The plasmid was linearized with NotI and transcribed using SP6 mMESSAGE mMACHINE Kit (Ambion) according to the manufacturer’s instructions.
Isolation of PGC by FACS
GFP-3′UTR-nos3 mRNA (400 pg/embryo) was injected into 1-cell stage WT and MmiR-202 embryos. At 10 hpf, PGCs from 400 WT or MmiR-202 embryos were isolated by FACS. A total of 1200 PGCs were sorted and reverse-transcribed by Single Cell Sequence Specific Amplification Kit (Vazyme, P621-01). The cdc42se1 and cdc42 mRNA levels in PGCs of WT and MmiR-202 embryos were analyzed by qRT-PCR.
RNA isolation and qRT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The first-strand cDNA was synthesized using PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa). qRT-PCR analyses of cdc42se1, cdc42, and vasa were performed as previously described (Xiang et al., 2017). The relative gene expression was normalized with β-actin using 2−ΔΔCt methods. Expression analysis of miR-202-5p/miR-202-3p was performed as previously described (Zhang et al., 2017). Data were shown as mean ± SD from three independent experiments in triplicate. The primer sequences were listed in Supplementary Table S2.
WISH
The digoxin-labelled antisense probes of vasa and cdc42se1 were synthesized using the DIG RNA Labeling Mix with T7 RNA polymerase (Roche). WISH was performed as previously described (Thisse and Thisse, 2008; Zhang et al., 2017).
Dual luciferase reporter assay
HEK293T cells in 24 well plate were transiently co-transfected with 10 ng plasmid (WT or mutant psiCHECK2-3′UTR-Cdc42se1) and 25 nM miR-202-5p mimics or negative control with Lipofectamine 3000 (Invitrogen). The cells were lysed at 24 h post-transfection, and the luciferase activities were measured by a dual-luciferase reporter assay system (Promega). HEK293T cells in the 24-well plate were transfected with 250 ng pGL3B-Cdc42-pro plasmid and 25 ng pRL-TK, and cells transfected with the pGL3B vector and 25 ng pRL-TK were used as control. Furthermore, the cells were co-transfected with a total of 1 μg plasmids containing different amounts of pcDNA3.1(+)-Cdc42se1 plasmid/pcDNA3.1(+) vector (0.2/0.8, 0.4/0.6, 0.6/0.4, and 0.8/0.2 μg). The cells were lysed for luciferase assay after 24 h post-transfection. At least three independent experiments were performed.
Micro-injection
The miR-202-3p mimics, miR-202-5p inhibitor (a special modified antisense single RNA that could pair with miR-202-5p), mimics and corresponding negative controls were designed and purchased from RiboBio. cdc42se1 MO (CCGGAGAUGAGUGCGUUCUGGCAUA) targeting the start codon and its mismatch MO were designed from Gene Tools.
Three dosages of 10, 40, and 160 pg/embryo miR-202-5p inhibitor were respectively injected into 1-cell stage embryos, and embryos injected with equal amount of control inhibitors were used as control. To analyze the effect of miR-202-5p knockdown on PGC migration, embryos were respectively injected with 40 pg/egg miR-202-5p or control inhibitor and collected from dome to prim 5 stages, followed by whole-mount in situ hybridization of vasa.
The Cdc42se1-RFP-UTR-nos3 or rfp mRNA (300 pg/embryo) were injected in 4-cell embryos of the Tg1 line. The PGC migration was analyzed by a Zeiss LSM 880 confocal microscope.
Rescue experiments
To rescue miR-202-5p inhibitor (40 pg/embryo) knockdown embryos, embryos were co-injected with miR-202-5p mimics (40 pg/embryo) or cdc42se1 MO (4000 pg/embryo).
To rescue the PGC defects in MmiR-202 embryos, miR-202-5p, or miR-202-3p mimics (40 pg/embryo) were injected, and the PGC number and localization were analyzed. The experiment was independently performed in three repeats, and each group had >100 embryos.
Three dosages of cdc42se1 mRNA with its own 3′UTR (100, 200, 300 pg/embryo) were respectively injected into 1-cell stage embryos, which were collected for whole-mount in situ hybridization of vasa at prim 5 stage. To rescue Cdc42se1 overexpressed embryos, miR-202-5p mimics (40 pg/embryo), cdc42se1 MO (4000 pg/embryo), or cdc42 mRNA (150 pg/embryo) was injected into 1-cell stage embryos, respectively. Each experiment was independently performed in three repeats, and each group had >40 embryos.
PGC phenotype observation
To score PGC phenotypes, embryos were fixed at 24 hpf in 4% paraformaldehyde, and PGCs were labelled by in situ hybridization using the vasa anti-sense probe. Phenotypes of PGCs in each embryo were recorded as normal, number reduction only, mislocalization only or both reduction and mislocalization. To quantify the different phenotypes, embryos with over 25 PGCs at the genital ridge and with <3 ectopic localized PGCs were counted as normal; embryos with at least three mislocalized PGCs were counted as mislocalization; embryos with <10 PGCs were counted as number reduced. To determine the phenotype, sets of 30–50 embryos were scored. Standard deviations were calculated for multiple repetitions of each experiment.
Western blotting and co-IP assay
For analyzing miR-202-5p repressed Cdc42se1 expression in vitro, HEK293T cells were co-transfected with 5 μg pcDNA3.1(+)-Cdc42se1 plasmid and 100 nmol control mimics/miR-202-5p mimics. Forty-eight hours later, cells were lysed and analyzed by western blotting using anti-Cdc42se1 antibody.
For analyzing Cdc42 expression in vivo, 200 pg cdc42se1 mRNA was injected into 1-cell stage embryos, which were collected at 10 hpf for western blotting with anti-Cdc42 antibody. The testes from WT and MmiR-202 adults were collected and lysed for western blotting with anti-Cdc42 antibody.
For co-IP assay, HEK293T cells were co-transfected with 10 μg of different plasmid combinations as follows: Myc-Cdc42 and Flag-Cdc42se1, Myc-actin and Flag-Cdc42, Flag-Cdc42se1 and Myc-actin, Myc vector and Flag-Cdc42se1, Myc-Cdc42 and Flag vector, or Flag vector and Myc vector. Cells were lysed with lysis buffer (10 mM Tris–HCl [pH 7.5], 0.4 M NaCl, 1% NP-40, 0.4% Triton X-100, 0.2% sodium deoxycholate, 1 mM EDTA, protease inhibitors [Calbiochem]) at 48 h post-transfection. Western blotting and co-IP were performed as previously described (Jia et al., 2013). Anti-Flag antibody (M20008) and anti-Myc antibody (M20002) were purchased from Abmart; anti-α-tubulin antibody (ab15246) and anti-Cdc42 antibody (ab187643) were purchased from Abcam; anti-mouse IgG (7076S) was purchased from Cell Signaling Technology; anti-rabbit IgG (A0208) was purchased from Beyotime Technology.
Statistics analysis
All statistics were calculated using SPSS version 20. Differences between control and treatment groups were assessed by one-way ANOVA. P-values <0.05, < 0.01, < 0.001, and <0.0001 are identified with 1, 2, 3, and 4 asterisks, respectively. ns, P ≥ 0.05.
Supplementary Material
Funding
This work was supported by the National Natural Science Foundation of China (31771587 and 31970535), Fundamental Research Funds for the Central Universities (17lgpy66), the Pearl River S&T Nova Program of Guangzhou (201806010047), the Science and Technology Planning Project of Guangdong Province (2017A030303010), and the Guangdong Natural Science Foundation (2015A030308012).
Conflict of interest: none declared.
References
- Armisen, J., Gilchrist, M.J., Wilczynska, A., et al. (2009). Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res. 19, 1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, L.J., LeBlanc, M.G., and Lehmann, R. (2016). Finding their way: themes in germ cell migration. Curr. Opin. Cell Biol. 42, 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer, R.L., and Draper, B.W. (2013). Nanos3 maintains germline stem cells and expression of the conserved germline stem cell gene nanos2 in the zebrafish ovary. Dev. Biol. 374, 308–318. [DOI] [PubMed] [Google Scholar]
- Block, J., Breitsprecher, D., Kuhn, S., et al. (2012). FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 22, 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldajipour, B., Mahabaleshwar, H., Kardash, E., et al. (2008). Control of chemokine-guided cell migration by ligand sequestration. Cell 132, 463–473. [DOI] [PubMed] [Google Scholar]
- Cao, Z., Mao, X., and Luo, L. (2019). Germline stem cells drive ovary regeneration in zebrafish. Cell Rep. 26, 1709, 1709–1717.e3. [DOI] [PubMed] [Google Scholar]
- Chen, S., Zeng, M., Sun, H., et al. (2010). Zebrafish Dnd protein binds to 3′ UTR of geminin mRNA and regulates its expression. BMB Rep. 43, 438–444. [DOI] [PubMed] [Google Scholar]
- Ching, K.H., Kisailus, A.E., and Burbelo, P.D. (2005). The role of SPECs, small Cdc42-binding proteins, in F-actin accumulation at the immunological synapse. J. Biol. Chem. 280, 23660–23667. [DOI] [PubMed] [Google Scholar]
- Dabaja, A.A., Mielnik, A., Robinson, B.D., et al. (2015). Possible germ cell-Sertoli cell interactions are critical for establishing appropriate expression levels for the Sertoli cell-specific MicroRNA, miR-202-5p, in human testis. Basic Clin. Androl. 25, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X., Shu, Y., Lou, Q., et al. (2017). Tdrd 12 is essential for germ cell development and maintenance in zebrafish. Int. J. Mol. Sci. 18, 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou, M., Reichman-Fried, M., Stebler, J., et al. (2002). Guidance of primordial germ cell migration by the chemokine SDF-1. Cell 111, 647–659. [DOI] [PubMed] [Google Scholar]
- Draper, B.W., McCallum, C.M., and Moens, C.B. (2007). Nanos1 is required to maintain oocyte production in adult zebrafish. Dev. Biol. 305, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Driesche, S., Sharpe, R.M., Saunders, P.T., et al. (2014). Regulation of the germ stem cell niche as the foundation for adult spermatogenesis: a role for miRNAs? Semin. Cell Dev. Biol. 29, 76–83. [DOI] [PubMed] [Google Scholar]
- El-Brolosy, M.A., Kontarakis, Z., Rossi, A., et al. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour, C.G., and Akam, M. (2003). Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869–5884. [DOI] [PubMed] [Google Scholar]
- Gay, S., Bugeon, J., Bouchareb, A., et al. (2018). MiR-202 controls female fecundity by regulating medaka oogenesis. PLoS Genet. 14, e1007593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez, A.J., Mishima, Y., Rihel, J., et al. (2006). Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. [DOI] [PubMed] [Google Scholar]
- Goudarzi, M., Banisch, T.U., Mobin, M.B., et al. (2012). Identification and regulation of a molecular module for bleb-based cell motility. Dev. Cell 23, 210–218. [DOI] [PubMed] [Google Scholar]
- Goudarzi, M., Tarbashevich, K., Mildner, K., et al. (2017). Bleb expansion in migrating cells depends on supply of membrane from cell surface invaginations. Dev. Cell 43, 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Thebing, T., Yigit, S., Pfeiffer, J., et al. (2017). The vertebrate protein dead end maintains primordial germ cell fate by inhibiting somatic differentiation. Dev. Cell 43, 704–715. [DOI] [PubMed] [Google Scholar]
- Guo, W., Xie, B., Xiong, S., et al. (2017). miR-34a regulates sperm motility in zebrafish. Int. J. Mol. Sci. 18, 2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung, O., Forbes, M.M., and Marlow, F.L. (2014). Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol. Reprod. Dev. 81, 946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K., de Sousa Lopes, S.M.C., Kaneda, M., et al. (2008). MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One 3, e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman, S.J., and Ridley, A.J. (2008). Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690. [DOI] [PubMed] [Google Scholar]
- Houwing, S., Kamminga, L.M., Berezikov, E., et al. (2007). A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129, 69–82. [DOI] [PubMed] [Google Scholar]
- Jia, K.T., Wu, Y.Y., Liu, Z.Y., et al. (2013). Mandarin fish caveolin 1 interaction with major capsid protein of infectious spleen and kidney necrosis virus and its role in early stages of infection. J. Virol. 87, 3027–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardash, E., Reichman-Fried, M., Maitre, J.L., et al. (2010). A role for Rho GTPases and cell–cell adhesion in single-cell motility in vivo. Nat. Cell Biol. 12, 47–53. [DOI] [PubMed] [Google Scholar]
- Kimmel, C.B., Ballard, W.W., Kimmel, S.R., et al. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Kloc, M., and Etkin, L.D. (1994). Delocalization of Vg1 mRNA from the vegetal cortex in Xenopus oocytes after destruction of Xlsirt RNA. Science 265, 1101–1103. [DOI] [PubMed] [Google Scholar]
- Knaut, H., Werz, C., Geisler, R., et al. (2003). A zebrafish homologue of the chemokine receptor cxcr4 is a germ-cell guidance receptor. Nature 421, 279–282. [DOI] [PubMed] [Google Scholar]
- Koprunner, M., Thisse, C., Thisse, B., et al. (2001). A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 15, 2877–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Fujii, W., Naito, K., et al. (2017). Application of dead end-knockout zebrafish as recipients of germ cell transplantation. Mol. Reprod. Dev. 84, 1100–1111. [DOI] [PubMed] [Google Scholar]
- Ma, Z., Zhu, P., Shi, H., et al. (2019). PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263. [DOI] [PubMed] [Google Scholar]
- Martinho, R.G., Kunwar, P.S., Casanova, J., et al. (2004). A noncoding RNA is required for the repression of RNA polII-dependent transcription in primordial germ cells. Curr. Biol. 14, 159–165. [DOI] [PubMed] [Google Scholar]
- Matzuk, M.M. (2009). LIN28 lets BLIMP1 take the right course. Dev. Cell 17, 160–161. [DOI] [PubMed] [Google Scholar]
- Melton, C., Judson, R.L., and Blelloch, R. (2010). Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463, 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Rodriguez, J.R., Salas-Vidal, E., Lomeli, H., et al. (2017). RhoA/ROCK pathway activity is essential for the correct localization of the germ plasm mRNAs in zebrafish embryos. Dev. Biol. 421, 27–42. [DOI] [PubMed] [Google Scholar]
- Mishima, Y., Giraldez, A.J., Takeda, Y., et al. (2006). Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 16, 2135–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Verkade, H., Heath, J.K., et al. (2008). Sdf1/Cxcr4 signaling controls the dorsal migration of endodermal cells during zebrafish gastrulation. Development 135, 2521–2529. [DOI] [PubMed] [Google Scholar]
- Nakamura, A., Amikura, R., Mukai, M., et al. (1996). Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science 274, 2075–2079. [DOI] [PubMed] [Google Scholar]
- Paksa, A., and Raz, E. (2015). Zebrafish germ cells: motility and guided migration. Curr. Opin. Cell Biol. 36, 80–85. [DOI] [PubMed] [Google Scholar]
- Pirone, D.M., Fukuhara, S., Gutkind, J.S., et al. (2000). SPECs, small binding proteins for Cdc42. J. Biol. Chem. 275, 22650–22656. [DOI] [PubMed] [Google Scholar]
- Raz, E. (2003). Primordial germ-cell development: the zebrafish perspective. Nat. Rev. Genet. 4, 690–700. [DOI] [PubMed] [Google Scholar]
- Richardson, B.E., and Lehmann, R. (2010). Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat. Rev. Mol. Cell Biol. 11, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A.J. (2015). Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 36, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabharanjak, S., Sharma, P., Parton, R.G., et al. (2002). GPI-anchored proteins are delivered to recycling endosomes via a distinct Cdc42-regulated, clathrin-independent pinocytic pathway. Dev. Cell 2, 411–423. [DOI] [PubMed] [Google Scholar]
- Shenoy, A., and Blelloch, R.H. (2014). Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat. Rev. Mol. Cell Biol. 15, 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staton, A.A., Knaut, H., and Giraldez, A.J. (2011). miRNA regulation of sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat. Genet. 43, 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome, S., and Updike, D. (2015). Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol. 16, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W.W.C., Kobayashi, T., Irie, N., et al. (2016). Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 17, 585–600. [DOI] [PubMed] [Google Scholar]
- Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. [DOI] [PubMed] [Google Scholar]
- Wainwright, E.N., Jorgensen, J.S., Kim, Y., et al. (2013). SOX9 regulates microRNA miR-202-5p/3p expression during mouse testis differentiation. Biol. Reprod. 89, 34. [DOI] [PubMed] [Google Scholar]
- Weidinger, G., Stebler, J., Slanchev, K., et al. (2003). dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 13, 1429–1434. [DOI] [PubMed] [Google Scholar]
- West, J.A., Viswanathan, S.R., Yabuuchi, A., et al. (2009). A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460, 909–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Y., Liu, W., Jia, P., et al. (2017). Molecular characterization and expression analysis of interferon-gamma in black seabream Acanthopagrus schlegelii. Fish Shellfish Immunol. 70, 140–148. [DOI] [PubMed] [Google Scholar]
- Xu, H., Kardash, E., Chen, S., et al. (2012). Gβγ signaling controls the polarization of zebrafish primordial germ cells by regulating Rac activity. Development 139, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Liu, W., Jin, Y., et al. (2017). MiR-202-5p is a novel germ plasm-specific microRNA in zebrafish. Sci. Rep. 7, 7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


