Abstract
Breast cancer is the second leading cause of cancer-related deaths in women in the United States. The triple-negative breast cancer (TNBC) subtype associates with higher rates of relapse, shorter overall survival, and aggressive metastatic disease. Hormone therapy is ineffective against TNBC, leaving patients with limited therapeutic options. Mammalian orthoreovirus (reovirus) preferentially infects and kills transformed cells, and a genetically engineered reassortant reovirus infects and kills TNBC cells more efficiently than prototypical strains. Reovirus oncolytic efficacy is further augmented by combination with topoisomerase inhibitors, including the frontline chemotherapeutic doxorubicin. However, long-term doxorubicin use correlates with toxicity to healthy tissues. Here, we conjugated doxorubicin to reovirus (reo-dox) to control drug delivery and enhance reovirus-mediated oncolysis. Our data indicate that conjugation does not impair viral biology and enhances reovirus oncolytic capacity in TNBC cells. Reo-dox infection promotes innate immune activation, and crosslinked doxorubicin retains DNA-damaging properties within infected cells. Importantly, reovirus and reo-dox significantly reduce primary TNBC tumor burden in vivo, with greater reduction in metastatic burden after reo-dox inoculation. Together, these data demonstrate that crosslinking chemotherapeutic agents to oncolytic viruses facilitates functional drug delivery to cells targeted by the virus, making it a viable approach for combination therapy against TNBC.
Keywords: oncolytics, virus, triple-negative breast cancer, doxorubicin, reovirus, cell death, DNA damage, drug delivery, virus-host interactions
Graphical Abstract

The chemotherapeutic drug doxorubicin was conjugated to oncolytic reovirus (reo-dox) to control drug delivery and enhance viral-mediated oncolysis of cancer cells. Conjugation of the drug to the virus does not impair viral biology, enhances reovirus oncolytic capacity, and retains the damaging properties of doxorubicin.
Introduction
Breast cancer is most prevalent among new cancer diagnoses and the second leading cause of cancer-related deaths in women in the United States.1 Breast cancer subtypes are categorized according to the surface expression of three receptor proteins: estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). The triple-negative breast cancer (TNBC) subtype characteristically lacks expression of ER, PR, and HER2. TNBC accounts for approximately 12% of breast tumors,2,3 and among metastatic breast cancers, TNBC exhibits higher rates of relapse and shorter overall survival compared to other subtypes of breast cancer.4,5
Treatment of TNBCs is largely limited to chemotherapy, radiation therapy, and surgery. Hormone therapies, which have been efficacious in the treatment of other types of breast cancer, are inadequate in TNBC due to the lack of the hormone therapy targets, ER, PR, and HER2.6,7 TNBCs are initially more sensitive to chemotherapy compared to other breast cancer subtypes, but patients with residual disease after neoadjuvant chemotherapy and surgery suffer higher rates of mortality.4,5 Thus, there is great need for efficacious, targeted therapies to improve therapeutic outcomes in TNBC.
The concept that viral infection can correlate with decreased tumor burden has existed nearly as long as knowledge about viruses.8 Recent developments in genetic engineering of viruses has allowed the development of oncolytic viruses with increased recognition of receptors overexpressed in tumor tissue9,10 and viruses that encode or package suicide or pro-apoptotic genes or agents for delivery to cancer cells.11,12 Viruses can also be manipulated to upregulate antigen presentation and T cell antitumor response.13,14 Despite ongoing efforts, talimogene laherparepvec (T-Vec, OncoVEXGM-CSF, Imlygic), an attenuated and genetically engineered herpes simplex virus (HSV) that overexpresses granulocyte-macrophage colony-stimulating factor (GM-CSF), is the only oncolytic virus that has been approved for clinical use in the United States and Europe (ClinicalTrials.gov: NCT00769704).15, 16, 17, 18
Mammalian orthoreovirus (reovirus) is a nonenveloped, segmented double-stranded RNA (dsRNA) virus.19 Most humans are infected with reovirus during childhood, but infection rarely causes disease.20 Reovirus preferentially replicates in transformed cells, making it an attractive oncolytic agent.21 Tropism to cancer cells is linked to many factors, including overexpression of the proteinaceous virus receptor junctional adhesion molecule A (JAM-A),22,23 dysregulated Ras signaling,24 downregulated dsRNA-dependent protein kinase R (PKR),25 and increased expression of cathepsins.26, 27, 28 A laboratory-adapted reovirus is in clinical trials to test its efficacy against several cancers,29, 30, 31, 32 but its efficacy against TNBC has not been extensively investigated.
We previously generated a reassortant reovirus (henceforward referred to as reovirus) with enhanced infectivity and cytotoxic properties in TNBC cells.33 The oncolytic properties of reovirus in TNBC are enhanced when combined with topoisomerase inhibitors topotecan, etoposide, and doxorubicin (dox).33 Dox is a topoisomerase II inhibitor that is a frontline chemotherapeutic for many cancers. Dox intercalates with DNA, inhibiting topoisomerase II binding with DNA and strand religation.34 Dox is a potent anticancer agent, but its clinical use is limited by severe systemic toxicity and association with cardiomyopathy, especially during long-term treatment.6,35,36 Improving the specificity of cytotoxic agents to cancer cells can increase their efficacy and improve quality of care for patients.37
In this study, we show that dox conjugation to reovirus (reo-dox) enhances cytotoxicity to TNBC cells without impairing reovirus biology. We demonstrate that infection with reo-dox elicits an innate immune response to the virus and promotes DNA damage resulting in activation of the double-strand break response pathway, indicative of dox bioactivity. Additionally, in a murine TNBC model, reo-dox dramatically reduced primary tumor and metastatic burden compared to dox-treated animals. Together, our data indicate that crosslinking small molecules to oncolytic reovirus can serve as a platform for efficacious, targeted delivery of drugs and oncolytic virus to infected cells, limiting the progression of aggressive metastatic breast cancer.
Results
Doxorubicin Conjugation to Reovirus Enhances Cytotoxicity in TNBC Cells
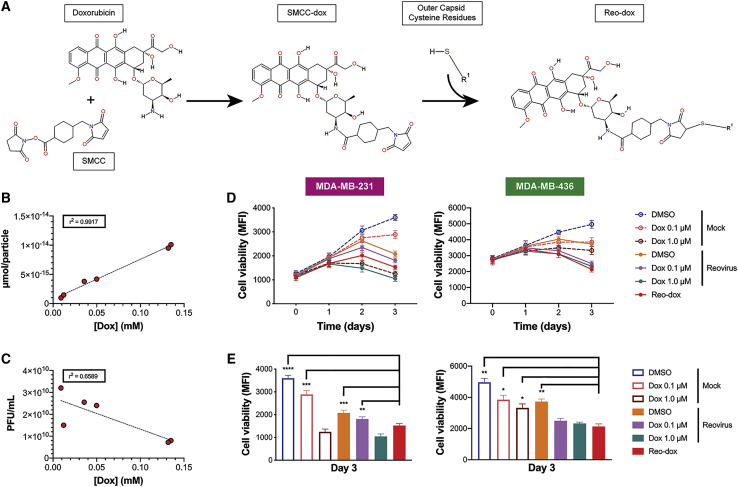
We previously identified that infecting TNBC cells with a genetically engineered reovirus in the presence of doxorubicin (dox) yields additive cytotoxicity.33 Dox is an effective chemotherapy for the treatment of TNBC,6 but toxicity to healthy cells and tissues limits its clinical utility.35,36 Conjugation of small molecule fluorescent dyes to reovirus to generate fluorescently-labeled viral particles has little impact on virus biology or cargo function.38, 39, 40 To minimize off-tumor effects of dox and selectively deliver the drug to virus-infected cells, we conjugated dox to reovirus (reo-dox) using the heterobifunctional covalent crosslinker succinimidyl 4-(n-maleimidomethyl)cyclohexane-1-carboxylate (SMCC; Figure 1A). To determine the concentration of dox crosslinked to reovirus, we measured dox absorbance at 480 nm by ultraviolet-visible spectroscopy (Figure 1B; Table S1). On average, 5.01 10−15 μmol of dox are present on one reovirus particle. Dox concentration positively correlates with μmol of dox per reovirus particle with an r2 value of 0.9917 (Figure 1B) and negatively correlates with viral titer with an r2 value of 0.6589 (Figure 1C), indicating that higher concentrations of crosslinked dox dampen reovirus infectivity. These data indicate that dox can be successfully conjugated to reovirus using SMCC with minimal impact on the infective properties of the virus.
Figure 1.
Doxorubicin Conjugation to Reovirus Enhances Viral Cytotoxicity in TNBC Cells
(A) Chemistry of doxorubicin conjugation to reovirus. The lone primary amine of doxorubicin reacts with the succinimide functional group of succinimidyl 4-(n-maleimidomethyl) cyclohexane-1-carboxylate (SMCC) to form SMCC-dox. Cysteine residues on viral capsid proteins (R1) react with the maleimide functional group of SMCC-dox, yielding a final crosslinked product or doxorubicin bound to reovirus (reo-dox). (B and C) UV-vis spectroscopy was performed on reo-dox preparations (Table S1). (B) Doxorubicin concentration was correlated with the amount of drug per reovirus particle and (C) viral titer. r2 values are presented for six independently labeled reo-dox preparations. (D and E) TNBC cells were pretreated with vehicle (DMSO) or doxorubicin. Cells were infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell. (D) Cell viability was measured over 3 days post infection. (E) Cell viability at 3 dpi from (D). Data represent the mean of four independent experiments. Error bars, SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001 by one-way ANOVA for reo-dox compared to all conditions.
To determine the cytotoxic properties of reo-dox in TNBC cells, we pretreated MDA-MB-231 and MDA-MB-436 cells (both of the mesenchymal stem-like [MSL] cellular subtype41) with vehicle (DMSO) or increasing concentrations of dox and infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell (Figure 1D). In MDA-MB-231 cells, reo-dox (red) significantly reduced viability by day 3 post infection compared to reovirus alone (orange) and reovirus infection after 0.1 μM dox pretreatment (violet; Figure 1E). Reo-dox also impaired cell viability with faster kinetics than virus alone or virus infection after 0.1 μM dox. In MDA-MB-436 cells, reovirus infection alone induced mild cytotoxicity, and pretreatment with 0.1 or 1.0 μM dox followed by reovirus infection enhanced viral cytotoxicity. Infection with reo-dox reduced MDA-MB-436 cell viability to similar levels as reovirus infection of dox-pretreated cells and significantly reduced viability compared to cells treated with dox alone or reovirus infection alone (Figures 1D and 1E). These data indicate that infection of TNBC cells with reo-dox yields greater cytotoxicity than virus alone.
Dox Conjugation Does Not Affect Reovirus Replication Kinetics
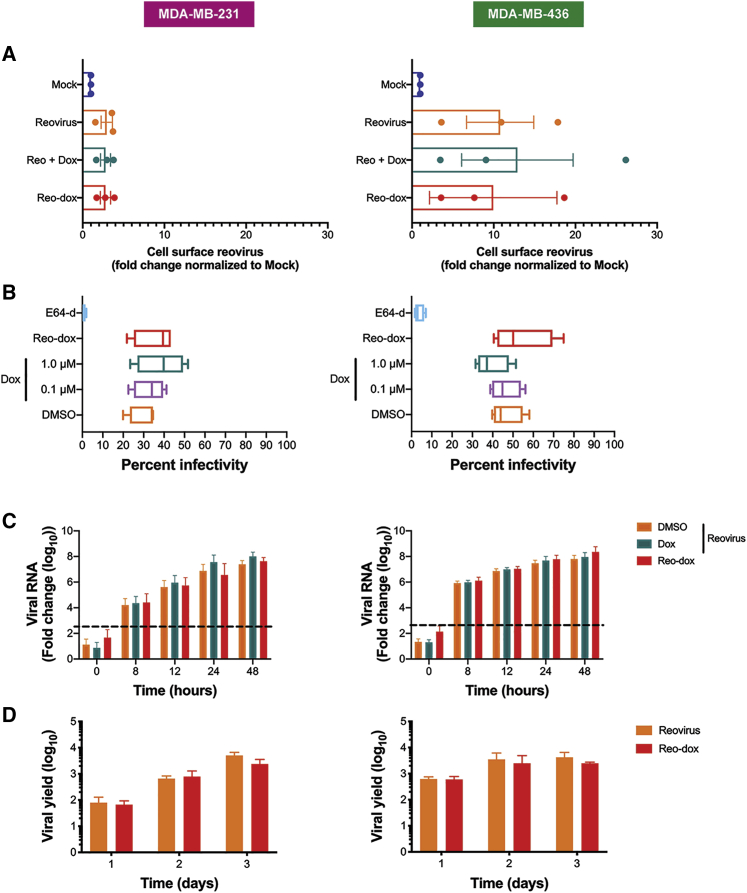
To evaluate the effect of dox conjugation on reovirus biology, we evaluated reo-dox attachment, infectivity, and replication in TNBC cells. Reovirus cell attachment is mediated by a strength-adhesion mechanism in which the viral attachment fiber σ1 binds cell-surface carbohydrate and proteinaceous receptor JAM-A or NgR1.22,42 To investigate whether dox conjugation altered the ability of reovirus to attach to TNBC cells, we pretreated MDA-MB-231 and MDA-MB-436 cells with vehicle (DMSO) or dox, adsorbed with mock, reovirus, or reo-dox at an MOI of 1 × 105 particles/cell at 4°C, and assessed for cell surface reovirus by flow cytometry using indirect immunofluorescence with reovirus-specific antiserum (Figures 2A and S1A). In both cell lines, cell-surface reovirus and the percent of cells with virus were similar in cells adsorbed with reovirus alone, reovirus pretreated with dox, or reo-dox. Interestingly, 3–4 times more reovirus bound to MDA-MB-436 cells than MDA-MB-231 cells. This is likely due to different levels of cell-surface JAM-A. These data indicate that dox conjugation to reovirus does not affect the ability of reovirus to attach to TNBC cells.
Figure 2.
Reo-dox Has Similar Attachment, Infectivity, and Replication Kinetics as Reovirus, But Enhanced Cytotoxicity in TNBC Cells
Results for MDA-MB-231 cells are displayed on left and MDA-MB-436 cells are displayed on right for all panels. (A) TNBC cells were pretreated with vehicle (DMSO) or doxorubicin and adsorbed with mock, reovirus, or reo-dox at an MOI of 1 × 105 particles/cell for 1 h at 4°C. Cells were assessed for cell surface reovirus by flow cytometry using indirect immunofluorescence. (B) TNBC cells were pretreated with vehicle (DMSO), E64-d, or doxorubicin and infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell for 18 h. Infectivity was assessed by indirect immunofluorescence. (C) TNBC cells were pretreated with vehicle (DMSO) or doxorubicin and infected with mock, reovirus, or reo-dox, and qPCR was performed to assess mRNA levels of reovirus S1 gene. Dashed line represents background baseline levels observed in mock. Data are shown as fold change normalized to a housekeeping gene. (D) TNBC cells were adsorbed with reovirus or reo-dox at an MOI of 10 PFU/cell over a 3 day time course. Viral titers were assessed by plaque assay on L929 mouse fibroblasts and viral yield was calculated as fold increase in titer compared to day 0. Data represent the means of three (A) or four (B–D) independent experiments. Error bars, SEM.
To test whether dox conjugation to reovirus impacted reovirus infectivity, we pretreated MDA-MB-231 cells and MDA-MB-436 cells with vehicle (DMSO), dox, or the cysteine protease inhibitor E64-d, which blocks reovirus cell entry by preventing proteolytic processing of virions during endocytosis.43 Cells were infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell, and assessed for infectivity after 18 h by indirect immunofluorescence using reovirus-specific antiserum (Figure 2B). Similar to previous observations,33 1.0 μM dox pretreatment of MDA-MB-231 cells slightly enhanced reovirus infectivity, whereas 1.0 μM dox pretreatment of MDA-MB-436 cells slightly decreased reovirus infectivity. Reo-dox infectivity was slightly higher or similar to virus alone or virus following dox pretreatment in both MDA-MB-231 and MDA-MB-436 cells (Figure 2B). These data indicate that reo-dox can efficiently infect TNBC cells and that crosslinking dox to virus has negligible effects on infectivity.
To determine whether crosslinking dox to reovirus alters viral replication in TNBC cells, we pretreated MDA-MB-231 and MDA-MB-436 cells with vehicle (DMSO) or 1.0 μM dox, infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell, and assessed for viral replication by qPCR over a 2 day time course of infection (Figure 2C). In parallel, cells were adsorbed with mock, reovirus, or reo-dox at an MOI of 10 PFU/cell and assessed for replication by plaque assay on L929 mouse fibroblasts over a 3 day time course of infection (Figures 2D and S1B). Reo-dox replicated to similar levels with similar kinetics as reovirus alone or in in the context of dox-pretreated cells. Viral RNA levels in MDA-MB-436 cells were 10-fold higher over the first day of infection than in MDA-MB-231 cells (Figure 2C). Viral yield was similar in cells infected with reo-dox compared to cells infected with reovirus alone (Figure 2D). Peak titers reached 108 PFU/mL by 2 days post infection (dpi) in MDA-MB-436 cells and 3 dpi in MDA-MB-231 cells (Figure S1B), corroborating the finding that MDA-MB-436 cells support faster replication kinetics. These data indicate that infection of MDA-MB-436 cells is more efficient than MDA-MB-231 cells correlating with differences in attachment. Further, dox conjugation to reovirus does not alter virus attachment and infection kinetics compared to reovirus alone.
Dox Conjugation to Reovirus Does Not Alter Innate Immune Response to Virus Infection
Reovirus infection of MDA-MB-231 cells induces interferon lambda 1 (IFNL1) transcription and cytokine production with no transcription of IFNB1.33 To investigate the IFN response to reo-dox infection in TNBC cells, we treated MDA-MB-231 (Figure 3A) and MDA-MB-436 (Figure 3B) cells with vehicle (DMSO) or 1.0 μM dox and infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell for 0–48 h and assessed IFNL1, IFNB1, and IFNG mRNA levels by qPCR. In MDA-MB-231 cells, there was no detectable IFNB1, but IFNL1 RNA levels were 100-fold higher by 8 h post infection (hpi) under all conditions with reovirus, peaking at 24 hpi, compared to uninfected cells. Reovirus infection following dox pretreatment (teal) resulted in slightly greater IFNL1 RNA levels. Dox treatment alone induced IFNL1 transcription by 48 hpi, consistent with previous findings.33 Dox treatment alone or followed by reovirus induced elevated IFNG RNA levels by 12 hpi, peaking by 48 hpi. Reovirus infection following dox pretreatment yielded 10-fold greater IFNG RNA levels than dox treatment alone by 48 hpi. Reo-dox infection yielded increased IFNG RNA levels by 24 hpi, peaking at 48 hpi, though to a lesser extent than free dox alone or with reovirus infection. In MDA-MB-436 cells, IFNL1 and IFNB1 RNA were detected under all conditions with reovirus as early as 8 hpi. Whereas IFNB1 RNA remained at similar levels throughout the times tested, IFNL1 RNA levels increased and peaked at 24 hpi. Dox treatment alone induced transcription of IFNL1 and IFNB1 RNA by 24 h post treatment, peaking by 48 hpi. Interestingly, IFNL1, but not IFNB1, RNA levels increased over time with vehicle treatment. IFNG RNA was detected following dox treatment in the presence or absence of reovirus and in reo-dox infected cells. IFNG RNA was 10-fold higher in cells infected with reovirus after dox treatment compared to vehicle-treated cells at 0 hpi, with levels increasing over the course of the infection. In MDA-MB-436 cells infected in the absence of dox, little IFNG RNA was detected. The lower levels of IFNG after reo-dox infection in both cell lines likely reflects a delay in dox delivery to infected cells compared to the addition of dox directly to cells and suggest that a lower concentration of dox is delivered on a per-cell basis (Table S1).
Figure 3.
Reo-Dox Induces Type-III IFN in TNBC Cells
(A) MDA-MB-231 and (B) MDA-MB-436 cells were pretreated with vehicle (DMSO) or doxorubicin and infected with mock, reovirus, or reo-dox. RNA was isolated from cells at times shown and qPCR was performed to assess mRNA levels of IFNL1, IFNB1, and IFNG. Data are shown as fold change to DMSO mock at 0 h for four independent experiments. Error bars, SEM. Levels of IFN-λ, IFN-β (MDA-MB-436 only), and IFN-γ in cell supernatants were detected by ELISA in (C) MDA-MB-231 and (D) MDA-MB-436 cells. Data are shown as pg/mL of IFN for four independent experiments. Error bars, SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001 by two-way ANOVA. Open ended brackets indicate multiple comparisons to one condition, doubled-sided brackets indicate comparison between two conditions only.
To determine whether transcriptional upregulation of IFNL1, IFNB1, and IFNG results in increased secreted IFN proteins, supernatants from MDA-MB-231 and MDA-MB-436 cells pretreated with vehicle (DMSO) or dox and infected with mock, reovirus, or reo-dox were assessed for IFN-λ, IFN-β, and IFN-γ by ELISA (Figures 3C and 3D). In MDA-MB-231 cells, IFN-λ was only detected after infection with reovirus, reovirus following dox pretreatment, or reo-dox (Figure 3C). Following reo-dox infection, IFN-λ was detected by12 hpi, peaking at 24 hpi. In cells infected with reovirus after dox pretreatment, IFN-λ was not detected until 24 hpi with levels increasing at 48 hpi. IFN-λ levels were greater in cells infected in the presence of dox (following pretreatment or conjugated to reovirus) than reovirus alone. IFNB1 RNA was not detected, so we did not test for secreted IFN-β levels in these cells. In MDA-MB-436 cells, reovirus infection induced robust IFN-λ and IFN-β secretion, with detectable levels at 12 hpi and increasing through the course of infection (Figure 3D). Levels of detected IFNλ were 5–10× those of IFN-β. IFN-λ and IFN-β levels were greater in infected cells in the presence of dox than virus alone. Although IFNL1 RNA was detected by 24 and 48 h after treatment with vehicle and dox in the absence of virus, secreted cytokine was not detected in DMSO-treated cells, and low levels of IFN-λ were detected in dox-treated cells.
In both MDA-MB-231 and MDA-MB-436 cells, all conditions promoted low levels of IFN-γ throughout the experiment despite changes in RNA levels. IFN-γ levels in MDA-MB-436 cells were about 10× those observed in MDA-MB-231 cells (Figures 3C and 3D). Together, these data suggest a context-dependent innate immune response to reovirus infection. In both MDA-MB-231 and MDA-MB-436 cells, infection resulted in robust type III IFN production with some additive enhancement conferred by the presence of dox, whether in soluble or crosslinked form. Interestingly, IFN-λ and -β have minimal to no direct effect on TNBC cell viability, despite substantial activation of signaling pathways responsive to the cytokines (Figure S2).33
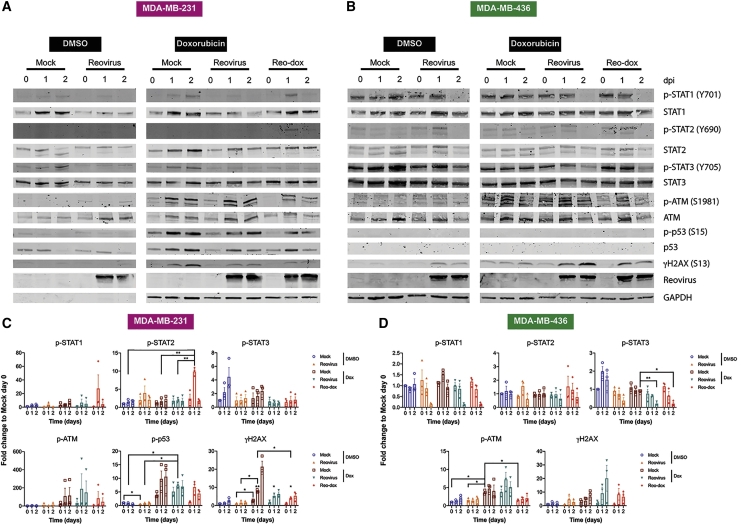
To determine whether MDA-MB-231 and MDA-MB-436 cells respond to IFN in the context of dox treatment and reovirus infection, we pretreated cells with vehicle (DMSO) or dox, infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell, and assessed the activation status of STAT1 and STAT2 by immunoblot over a 2-day time course of infection (Figures 4 and S3). In MDA-MB-231, phospho-STAT1 (p-STAT1) levels were slightly elevated in cells treated with dox in the absence of reovirus, and reovirus infection in the presence or absence of dox decreased p-STAT1 levels. Infection of MDA-MB-231 cells with reo-dox induced robust levels of p-STAT1 and p-STAT2 at 1 dpi. In MDA-MB-436 cells, p-STAT1 levels were consistently high with the exception of a small increase in p-STAT1 levels 1 day following dox treatment in the absence of reovirus and reovirus infection leading to decreased p-STAT1 levels at 2 dpi. Infection in the presence or absence of dox had little effect on p-STAT2 levels, with no p-STAT2 detected at 2 dpi. Infection of MDA-MB-436 cells with reo-dox did not significantly increase p-STAT1 or p-STAT2 levels as observed in MDA-MB-231 cells.
Figure 4.
Reo-Dox Activates DNA Damage Response Pathways and Modulates Innate Immune Activity in TNBC Cells
(A and C) MDA-MB-231 and (B and D) MDA-MB-436 cells were treated with vehicle (DMSO) or doxorubicin and infected with reovirus or reo-dox at an MOI of 100 PFU/cell. (A and B) Whole cell lysates for 0, 1, and 2 dpi were resolved by SDS-PAGE and immunoblotted with antibodies specific for phosphorylated and total STAT1, STAT2, STAT3, ATM, p53, H2AX, reovirus, and GAPDH. Residues recognized by phosphorylation-specific antibodies are shown in parentheses. Blots are representative of three independent experiments. (C and D) Quantitative densitometry was performed on all phosphorylated and total proteins. Data represent means of three independent experiments normalized to respective mock day 0 values. Error bars, SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01 by two-way ANOVA. ∗ above γH2AX day 1 bar graphs in (C) indicate comparisons between reovirus plus dox or reo-dox and DMSO mock on the same day.
Activated STAT3 is associated with epithelial-to-mesenchymal transition, survival, and proliferation, and is constitutively activated in 40% of breast cancers.44 While interleukin-6 (IL-6) is the primary agonist for STAT3 activation, type I and type III IFN can activate STAT3. In MDA-MB-231 and MDA-MB-436 cells, p-STAT3 levels decreased in the presence of dox alone, and reovirus alone or combined with soluble or conjugated dox decreased p-STAT3 to a greater degree than drug alone. These data indicate that while dox dampens the activation of STAT3, reovirus infection potently inhibits the activation of this protein.
Crosslinking Dox to Reovirus Does Not Affect the DNA Damaging Properties of Dox
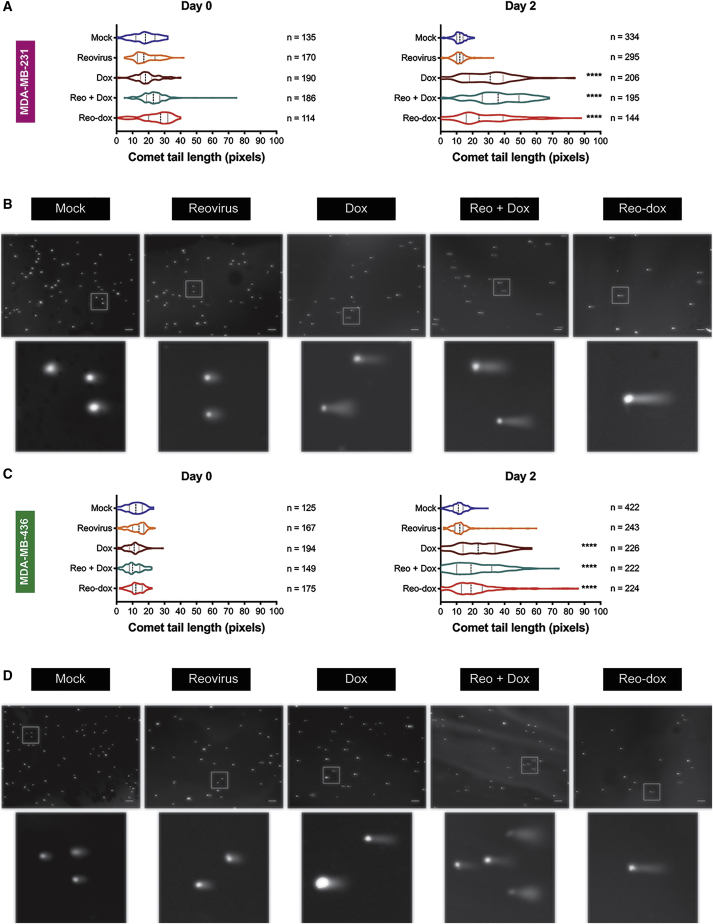
To determine whether the DNA-damaging properties of dox remain intact when conjugated to reovirus, we treated MDA-MB-231 and MDA-MB-436 cells with vehicle (DMSO) or dox, infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell, and analyzed for DNA damage by single-cell electrophoresis comet assay (Figure 5). In both cells, dox treatment, reovirus infection after dox pretreatment, and reo-dox infection resulted in greater DNA fragmentation than mock or reovirus infection alone by 2 dpi. Unconjugated dox-induced DNA damage was greater in MDA-MB-231 cells (Figure 5A) than in MDA-MB-436 cells (Figure 5C), corroborating cell viability data (Figure 1D).
Figure 5.
Reo-Dox Infection of TNBC Cells Induces DNA Double-Strand Breaks
(A and B) MDA-MB-231 and (C and D) MDA-MB-436 cells were pretreated with vehicle (DMSO) or doxorubicin and infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell. DNA double-strand break damage was assessed by single cell electrophoresis (comet assay) on 0 and 2 dpi. Chromatin was visualized by epifluorescence using DAPI staining. (A and C) Comet tail length was measured for imaged cells. Data represented as violin plots. Median and upper and lower quartiles are presented as dotted lines within violins. ∗∗∗∗p ≤ 0.0001 compared to mock and reovirus by one-way ANOVA for reo-dox compared to all conditions. (B and D) Representative images of comets on 2 dpi. Scale bar, 200 μm. Inset highlights boxed cells. Data are representative of two independent experiments.
H2A histone family member X (H2AX) is phosphorylated (γH2AX) by ataxia telangiectasia mutated (ATM) in response to DNA double-strand breaks.45,46 ATM and p53 are phosphorylated as part of the response to double-strand breaks.47,48 To assess whether dox-induced DNA damage promotes activation of the double-strand break response, we assessed the activation levels of ATM, p53, and H2AX in MDA-MB-231 and MDA-MB-436 cells in the presence or absence of dox and reovirus or reo-dox infection (Figure 4). MDA-MB-436 cells do not express full-length p53 due to a nonsense frameshift mutation at position E204 in p53.49 Reovirus infection alone did not induce activation of ATM, but γH2AX levels in MDA-MB-436 cells were elevated by 1 and 2 dpi. Treatment with dox alone, infection of dox-treated cells, and reo-dox infection led to robust increase of levels of p-ATM, p-p53, and γH2AX in MDA-MB-231 cells and p-ATM and γH2AX in MDA-MB-436 cells. Reovirus infection dampened dox-induced H2AX phosphorylation in MDA-MB-231 cells, but dox-induced γH2AX levels increased after reovirus infection of MDA-MB-436 cells, suggesting some cell-specific effects by reovirus on H2AX phosphorylation. DNA damage response was enhanced by reo-dox infection, though to a lesser extent as exogenous dox treatment, correlating with less DNA fragmentation observed by comet assay. The lower levels of DNA damage response activity detected biochemically in MDA-MB-436 cells correlate with higher basal ATM phosphorylation (Figure 4), lower levels of measured DNA fragmentation (Figure 5), and less cytotoxic effects of dox in these cells (Figures 1C and 1D). Together, these data indicate that reovirus does not interfere with dox-induced DNA damage response, and crosslinking dox to reovirus does not impair the pharmacological properties of dox.
Reovirus and Reo-Dox Reduce 4T1 Murine Tumors and Metastases In Vivo
The 4T1 murine mammary carcinoma cell line is a widely used TNBC model.50, 51, 52, 53 To determine whether the 4T1 cell line is a viable model for reovirus and reo-dox oncolysis, we infected cells pretreated in the absence or presence of dox with mock, reovirus, or reo-dox at increasing MOIs and assessed infectivity (Figure 6A). At all MOIs tested, reo-dox and reovirus infectivity were similar and infection after dox pretreatment enhanced reovirus infectivity to varying degrees depending on the MOI, mimicking the effect observed in MDA-MB-231 cells (Figure 2B).33 To assess the impact of reovirus infection on cell viability, we treated 4T1 cells with DMSO or dox, or infected with mock or increasing MOIs of reovirus or reo-dox, and cell viability was measured over a 6 day time course of infection (Figure 6B). At all MOIs tested, reovirus and reo-dox infection negatively affected cell viability in a dose-dependent manner, with reo-dox impairing cell viability with faster kinetics than reovirus alone. Infection with reo-dox at MOIs of 50 and 100 PFU/cell resulted in similar cell viability levels as 10 μM dox treatment alone at day 6. Together, these data indicate that reovirus and reo-dox can efficiently infect and impair cell viability of 4T1 cells in vitro and that 4T1 cells are sensitive to dox.
Figure 6.
Reo-Dox and Reovirus Infect and Kill 4T1 Cells In Vitro and In Vivo, and Reduce 4T1 Cell Metastatic Potential to the Lungs
(A) 4T1 cells were pretreated with vehicle (DMSO) or doxorubicin and infected with mock, reovirus, or reo-dox. Infectivity was assessed after 20 h by indirect immunofluorescence using reovirus-specific antiserum. (B) 4T1 cells were treated with vehicle (DMSO) or doxorubicin, or they were infected with reovirus or reo-dox at increasing MOIs. Cell viability was assessed at times shown (left) and day 6 alone for statistical analysis (right). (A and B) Data represent mean of four independent experiments. Error bars, SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001 by one-way ANOVA. ∗ above bar graphs in (B) indicate comparisons between reovirus and reo-dox at the same MOI. (C) Female, 8-week-old BALB/c mice were challenged with 5 × 104 4T1 cells via subcutaneous injection in the hind flank. At day 10 and 14 post challenge (arrows in D and E), mice were treated with PBS, 54.4 μg/mL doxorubicin, or 5 × 108 PFU of reovirus or reo-dox. The experimental endpoint was 21 days post tumor challenge. (D) Percent change in weight of mice was calculated as the weight on day of measurement normalized to weight at day 5. (E) Tumor area was measured at days indicated by data points. ∗∗p < 0.01, reovirus and reo-dox compared to PBS by two-way ANOVA. (C–E) n = 5 mice per treatment group. Error bars, SEM.
To assess the ability of reovirus and reo-dox to impair TNBC tumor growth and metastasis in vivo, we challenged female BALB/c mice with 5 × 104 4T1 cells subcutaneously in the hind flank and administered PBS, 54.4 μg/mL dox, or 5 × 108 PFU reovirus or reo-dox intratumorally on days 10 and 14 post tumor challenge (Figure 6C). Mouse weight (Figure 6D) and primary tumor area (Figure 6E) were assessed through 21 days post tumor challenge. Treatments had minimal effect on mouse weight. Animals treated with dox (pink) or inoculated with reovirus (teal) had a greater reduction in weight from day 10 to 14 compared to PBS (black) or reo-dox (violet), possibly indicating lower toxicity of the initial dose (Figure 6D). Intratumoral delivery of reovirus and reo-dox significantly impaired tumor growth, with tumors being ∼60% smaller compared to PBS treatment and ∼50% smaller than dox treatment by endpoint (Figure 6E). These data indicate that intratumoral reovirus and reo-dox can limit tumor growth at the primary site.
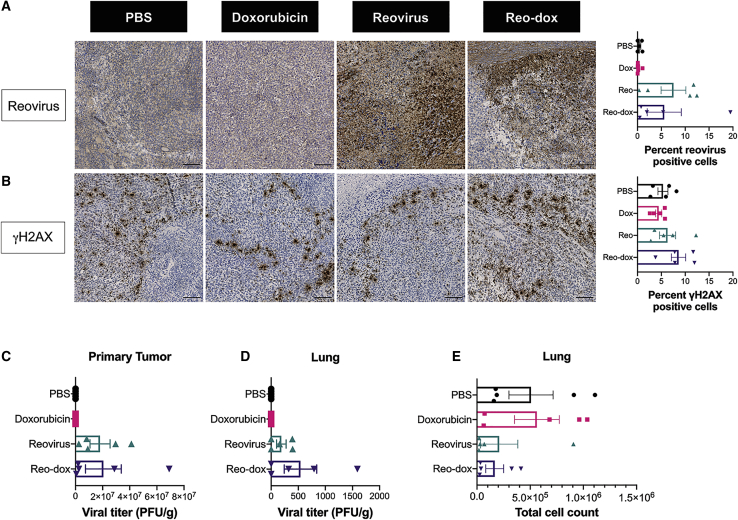
To determine the extent of viral antigen and DNA damage in the primary tumors, reovirus antigen and γH2AX levels were assessed by immunohistochemistry (IHC; Figures 7 and S4). Reovirus antigen staining was detected only in tumors infected with reovirus or reo-dox, with viral antigen detected in 7.5% (reovirus) and 5.6% (reo-dox) of cells in the tumor (Figure 7A). γH2AX staining was only slightly elevated in reo-dox-infected tumors, with 1.5–2 times more γH2AX-positive cells than any other condition (Figure 7B). To determine the levels of viral replication at primary and metastatic (lung) tumor sites, we assessed viral titers by plaque assay (Figures 7C and 7D). Viral titers were similar at the primary tumor site for animals inoculated with reovirus or reo-dox (Figure 7C), reflecting that observed by IHC staining (Figure 7A). To determine whether virus and 4T1 cells were present in the lungs, a site of TNBC metastasis, we assessed viral titers (Figure 7D) and 4T1 cell numbers (Figure 7E) at this site. The number of 4T1 cells in the lungs was determined by clonogenic assay using 6-thioguanine selection (Figure 7E).52,54 In the lungs, reovirus levels were approximately 3 times greater in mice that received reo-dox than those inoculated with reovirus. In PBS-treated mice, a median of 1.85 × 105 4T1 cells was present in the lungs, and dox treatment did not significantly impact the number of 4T1 cells. Although not statistically significant, infection with reovirus reduced the median number of 4T1 cells per lung by ∼80%–85%, and infection with reo-dox reduced the median number of 4T1 cells per lung by ∼95%. Together, these data indicate that reovirus and reo-dox replicate to a similar extent within the primary tumor and that reo-dox can promote DNA damage in the primary tumor site. Further, reovirus and reo-dox replicate at the metastatic site. These data also suggest that reovirus and reo-dox limit the metastatic potential of TNBC cells in vivo. The results of work presented here show that crosslinking dox to reovirus is a viable alternative for the codelivery of oncolytic virus and anti-neoplastic drugs to tumor sites, with potential benefits at primary and metastatic tumor sites.
Figure 7.
Reovirus Antigen and γH2AX Are Detected in 4T1 Tumors Infected with Reo-Dox
Primary tumor tissues were assessed for (A) reovirus antigen and (B) γH2AX levels by indirect immunohistochemistry. (A and B) Inset images enlarged from representative whole tissue scans in Figure S4. Scale bars, 100 μm. Percent of cells in whole tissue scans positive for reovirus antigen and γH2AX presented on right. Bar graphs represent mean of representative tissue from each mouse. n = 5. Error bars, SEM (C) Titers for reovirus or reo-dox present in primary tumor tissue and (D) lungs were assessed by plaque assay on L929 mouse fibroblasts. (E) Total number of metastatic 4T1 cells in lungs were counted. (C–E) n = 5. Error bars, SEM.
Discussion
Reovirus is a prime platform for the development of enhanced oncolytic virotherapies. Reovirus preferentially infects and replicates in tumor cells,21 can be delivered via intratumoral and intravenous administration,19 and can be combined with genotoxic and immunogenic agents that enhance oncolysis.32,33,55, 56, 57, 58, 59, 60 A lab-adapted serotype 3 reovirus is in phase I–III clinical trials (ClinicalTrials.gov identifier NCT01656538),18 though success has been limited and evaluation of reovirus as an oncolytic in breast cancers is minimal.29, 30, 31, 32 For this study, we used a genetically engineered reassortant reovirus (r2Reovirus), which more efficiently infects and promotes greater cytotoxic effects in TNBC cells compared to prototypical strains, including the reovirus that is currently in clinical trials.33 In TNBC cells, topoisomerase inhibitors augment reovirus infectivity, cytotoxicity, and innate immune activation.33 In this study, we show that conjugation of doxorubicin (dox) to oncolytic reovirus (reo-dox) using a heterobifunctional covalent crosslinker allows delivery of bioactive dox to cells, promoting enhanced cytotoxicity through the effects of the drug and the virus.
We estimate that reo-dox carries an average of 3,000 molecules of dox per viral particle (Table S1). Crosslinking fluorescent molecules to reovirus using succinimidyl esters preferentially labels reovirus outer capsid structural proteins λ2, μ1, σ1, and σ3.38 Crystal structures of reovirus structural proteins μ1 and σ3 and reovirus virions suggest the presence of multiple solvent-exposed cysteine residues,61 indicating that the estimation of dox molecules per virion is biologically feasible. Here we show that dox conjugation on reovirus has minimal effects on virus biology.
Reo-dox attaches to cells to similar levels compared to unlabeled reovirus, suggesting that dox conjugation does not hinder reovirus attachment fiber σ1 binding to cell surface carbohydrate or proteinaceous receptor JAM-A.22,23,62,63 Based on the σ1 crystal structure, a single cysteine residue is hidden within the monomeric tertiary structure,64 suggesting that dox is unlikely to be crosslinked to σ1. Although we did not observe a difference in overall levels of reo-dox attached to cells compared to unlabeled reovirus, we observed a small reduction in the percentage of cells with attached reo-dox compared to unlabeled virus (Figure S1). It is possible that this is the result of SMCC-dox causing some aggregation of viral particles, resulting in similar number of viral particles attached to cells, but fewer cells with virus. Reovirus attachment to MDA-MB-436 cells was 3–5× that observed in MDA-MB-231 cells, likely due to higher cell surface levels of JAM-A.
Reo-dox efficiently infects and replicates in TNBC cells. Paralleling that observed by attachment, reovirus infects MDA-MB-436 cells more efficiently than MDA-MB-231 cells. Pretreatment of MDA-MB-231 cells with 1 μM dox slightly increases infectivity, similar to previous observations.33 Intriguingly, pretreatment of MDA-MB-436 cells with 1 μM dox slightly reduced infectivity despite MDA-MB-436 cells being less sensitive to the cytotoxic effects of dox. It is not known how exogenous dox effects these alterations to reovirus infectivity. In contrast to pretreatment of cells with dox, reovirus infectivity after dox crosslinking is largely unaffected. Reo-dox likely delivers dox to cells after attachment and endocytic uptake, whereas exogenous dox can affect cells before virus attachment, endocytic uptake, and transport of the virus. Thus, the effects of dox on reovirus infectivity likely impact a post-attachment step in the virus replication cycle. Reo-dox also exhibits similar replication kinetics as unlabeled virus in MDA-MB-231 cells and MDA-MB-436 cells. Viral RNA levels and viral yield of unlabeled reovirus and reo-dox are greater in MDA-MB-436 cells than MDA-MB-231 cells at 1 dpi, indicating that MDA-MB-436 cells are more permissive to reovirus infection than MDA-MB-231 cells.
Conjugation of dox to reovirus does not significantly impact virus biology, but reo-dox elicits more robust cytotoxicity with faster kinetics than unlabeled reovirus in MDA-MB-231 and MDA-MB-436 cells. While exogenous 1 μM dox with and without reovirus impairs cell viability to slightly greater levels than reo-dox in MDA-MB-231 cells, reo-dox performs as well as exogenous dox with and without virus in MDA-MB-436 cells (Figure 1D). The differences observed between combination and conjugation treatment in MDA-MB-231 cells are likely due to the lower effective dose of dox delivered via conjugation and that only cells initially infected with reo-dox receive the drug, with progeny unlabeled virus being responsible for subsequent cytotoxicity. While reo-dox likely delivers a lower effective dose of the drug per cell, the delivery is also controlled and contained to only those cells taking up virus. Moreover, there is likely a proportion of cells that take up virions that do not result in productive infection but still receive the drug.
Virus infection can induce potent innate immune responses, and dox conjugation to reovirus does not hinder the TNBC response to virus infection. Reovirus and reo-dox infection of MDA-MB-231 cells upregulate transcription of IFNL1 but not IFNB1 (Figure 3A).33 In MDA-MB-436 cells, reovirus and reo-dox infection induce transcription of both IFNL1 and IFNB1 (Figure 3B), but only IFN-λ is secreted at high levels by both TNBC cell lines (Figures 3C and 3D). Little is known about the role of type III-IFN on TNBC cell biology. Treatment of MDA-MB-231 and MDA-MB-436 cells with recombinant IFN-β or IFN-λ results in robust STAT1 and STAT2 activation with little effect on cellular proliferation (Figure S2),33 suggesting that both cell lines express the cognate receptors for type-I and -III IFNs. Gut mucosal epithelial cells depend on type-III IFN to protect against viral pathogens.65,66 An antiviral role for IFN-λ has been described in other tissues and cell lineages including human placental trophoblast protection against Zika virus67 and in vitro human cervical epithelial cell resistance to dengue virus.68 IFN-λ in murine mammary epithelial cells recruits CD4+ T cells to the tumor microenvironment.69 TNBC cells secrete high levels of IFN-λ in response to reovirus alone and when combined with topoisomerase inhibitors.33 As such, it is possible that increased IFN-λ secretion by reo-dox-infected TNBC cells promotes a more favorable environment for recruiting immune modulatory cells to the site of infection.
Despite the upregulated levels of IFNL1 and secreted IFN-λ, only reo-dox infection of MDA-MB-231 cells robustly activated STAT1 and STAT2 (Figure 4). It is possible that reovirus infection of MDA-MB-231 cells in the presence of topoisomerase inhibitors can favor STAT1 and STAT2 activation. We previously observed that pretreatment of MDA-MB-231 cells with the topoisomerase I inhibitor topotecan followed by reovirus infection induces a similar STAT1 and STAT2 activation phenotype.33 Delivery of dox after reo-dox attachment may afford an optimal timing for cells to mount robust innate immune and DNA damage responses. MDA-MB-436 cells exhibit basal activation of STAT1, STAT2, and STAT3. This activity may be related to MDA-MB-436 cells having higher basal levels of secreted IFN-γ than MDA-MB-231 cells (Figure 3). IFN-γ primarily signals through STAT1 but can also activate other STAT proteins.70 Despite robust secretion of IFN-λ after reovirus infection, STAT activity decreases by 2 dpi.
One critical question was whether crosslinking dox to reovirus affects the pharmacological properties of the drug. We show that cells infected with reo-dox exhibit clear evidence of DNA damage and response, with the number of cells harboring DNA double-strand breaks increasing by 2 dpi to comparable levels as dox treatment (Figure 5). Reo-dox also induces robust levels of γH2AX and activated ATM in both cell lines, as well as phosphorylated p53 in MDA-MB-231 cells (Figure 4). While the mechanism by which dox is released from the viral particle remains unclear, we hypothesize that proteolytic processing during endocytosis of the virus results in dox being released from the viral particle. It is possible that extracellular proteases secreted by some TNBC that can interact with the virus71 may facilitate removal of SMCC-dox from viral particles before endocytic uptake. While SMCC forms an uncleavable thioether bond,72 SMCC and dox bind through succinimidyl ester chemistry yielding an amide bond. As such, dox may undergo cleavage by amidases or other amide-targeting enzymes delivered to late endosomes, restoring the primary amine of dox and releasing it from viral protein or peptides to interact with nuclear DNA.73, 74, 75
This study is the first to evaluate the effects of this particular reassortant reovirus and reo-dox in an in vivo model of TNBC. Reovirus and reo-dox significantly reduce tumor area compared to PBS control and greatly enhance tumor reduction compared to dox treatment (Figure 6). Viral antigen staining and assessment of viral titers of tumors indicate that viable virus is present in primary tumors over a week after inoculation (Figure 7). Replicating virus at the tumor site likely contributes to virus-mediated tumor regression. Shrinking primary tumors are accompanied by diminished levels of metastasized 4T1 cells in the lungs with both reovirus alone and reo-dox. Reo-dox inoculation results in greater reduction in metastatic 4T1 cells and up to 3× more actively replicating virus in lung tissue than reovirus alone (Figure 7D). Metastatic TNBC PDX models can exhibit elevated transcription of oxidative phosphorylation metabolism (OXPHOS) genes compared to primary tumors.76 Inhibition of OXPHOS significantly reduces micrometastatic seeding of lungs in MDA-MB-231 and 4T1 in vivo models, indicating a dependence on the shift from glycolytic to OXPHOS metabolism for TNBC cell metastasis.76 It is possible that reovirus infection disrupts mitochondrial membrane potential, interfering with mitochondrial enzymes involved in electron transport chain and limiting metastatic potential. Interestingly, cancer cells shifted toward OXPHOS metabolism exhibit increased sensitivity to antineoplastic agents like dox.77,78 As such, cells with greater metastatic potential due to a metabolic shift to OXPHOS may be more susceptible to cytotoxic effects of reo-dox. It remains to be seen whether the reduced metastatic burden in the lungs is primarily due to enhanced 4T1 cell death in primary tumors or oncolytic activity in the metastatic site. Alternatively, an antiviral immune response like that elicited in vitro in human cell lines may potentiate anti-tumor responses in the tumor microenvironment in mice inoculated with reo-dox.
In this study, we have shown that crosslinking dox to reovirus is a viable alternative to deliver chemotherapeutic agents in combination with oncolytic viruses. This approach mitigates off-target effects of small molecule therapeutics by selectively delivering the drug to cells and tissues targeted by the oncolytic virus. Drug-virus conjugation also enhances the anti-neoplastic effects of chemotherapeutic agents and oncolytic viruses by simultaneously delivering both agents to the same cell. This study presents evidence that reovirus conjugated with a genotoxic drug is a promising advancement in metastatic TNBC therapy.
Materials and Methods
Cells, Virus, and Antibodies
MDA-MB-231 cells (gift from Jennifer Pietenpol, Vanderbilt University) and MDA-MB-436 cells (ATCC HTB-130) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Life Technologies) and 100 U per mL penicillin and streptomycin (Life Technologies). Spinner-adapted L929 cells (Terry Dermody, University of Pittsburgh) were grown in Joklik’s modified minimal essential medium (MEM) with 5% FBS, 2 mM L-glutamine (Life Technologies), penicillin and streptomycin, and 0.25 mg per mL amphotericin B (Life Technologies). 4T1 cells (gift from Periasamy Selvaraj, Emory University) were grown in DMEM supplemented with 10% FBS, 100 U per mL penicillin and streptomycin, 5 mM L-glutamine, and 5 mM HEPES buffer (Life Technologies).
Working stocks of reassortant reovirus were prepared by plaque purification and passage in L929 cells.79 Purified virions were prepared using second-passage L929 cell lysate stocks. Virus was purified from infected cell lysates by Vertrel XF (TMC Industries) extraction and CsCl gradient centrifugation.80 The band corresponding to the density of reovirus particles (1.36 g/cm3) was collected and dialyzed exhaustively against virion storage buffer (150 mM NaCl, 15 mM MgCl 2, 10 mM Tris-HCl [pH 7.4]). The reovirus particle concentration was determined from the equivalence of 1 unit of optical density at 260 nm to 2.1 × 1012 particles.81 Viral titers were determined by plaque assay using L929 cells.82
Reovirus polyclonal rabbit antiserum raised against reovirus strains T1L and T3D was purified83 and cross-adsorbed for MDA-MB-231 or 4T1 cells. Secondary IRDye 680 and 800 antibodies (Li-Cor Biosciences) and goat anti-rabbit A488 (Life Technologies) were used.
Doxorubicin Conjugation to Reovirus
10 mM doxorubicin was diluted to 7.5 mM in PBS. Doxorubicin was mixed with 3.7 mg/mL succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC) in a 10:1 ratio and incubated at room temperature for 30 min. Doxorubicin and SMCC solution was dialyzed against cold PBS for 1 h at 4°C. Desalted SMCC-doxorubicin solution was measured for doxorubicin concentration by UV-vis spectroscopy on a Nanodrop Nd-8000 (Thermo Fisher Scientific). Absorbance at 480 nm (A480) was assessed and compared to a doxorubicin gradient standard curve. 3 × 1012 particles of reovirus were diluted in PBS up to 100 μL and combined with 400 μL of 500 μM SMCC-dox. Reovirus plus SMCC-dox solution was incubated for 30 min at room temperature with agitation on a tube revolver (Thermo Fisher Scientific) on reciprocating setting. The solution was dialyzed exhaustively against cold PBS overnight at 4°C.
Cell Viability Assay
To determine the effect of reo-dox on cell viability, we pretreated MDA-MB-231 and MDA-MB-436 cells with increasing concentrations of doxorubicin for 1 h at 37°C. Reovirus or reo-dox was added to cells at an MOI of 100 PFU/cell, and incubated in the presence of doxorubicin for 0–3 days. 4T1 cells were treated with vehicle (DMSO) or 10 μM doxorubicin or infected with reovirus or reo-dox at MOIs of 10, 50, and 100 PFU/cell for 0–3 days. Presto Blue (Invitrogen) was added at each time point for 30 min at 37°C and fluorescence (540 nm excitation/590 nm emission) was measured with a Synergy HT or Synergy H1 plate reader (Biotek).
Flow Cytometric Analysis of Cell-Surface Reovirus
MDA-MB-231 and MDA-MB-436 cells were treated with vehicle or 1 μM doxorubicin for 30 min at 37°C and then 4°C for 30 min. Media was removed and cells were adsorbed with reovirus or reo-dox at an MOI of 1 × 105 particles/cell in OMEM (GIBCO) for 1 h at 4°C. Cells were washed with PBS, detached with Cellstripper (Cellgro) for 10 min at 37°C, quenched and washed with PBS containing 2% FBS. Surface reovirus antigen was stained using reovirus-specific antiserum and Alexa Fluor 488 secondary antibody (A488, Thermo Fisher Scientific). Cells were fixed in 1% EM-grade paraformaldehyde (Electron Microscopy Sciences). Mean fluorescence intensity (MFI) and percent reovirus-positive cells were assessed using a CytoFLEX flow cytometer (Beckman Coulter) and quantified using FlowJo software.
Reovirus Infectivity Assay
Reo-dox infectivity was assessed by indirect immunofluorescence assay. MDA-MB-231, MDA-MB-436, and 4T1 cells were pretreated with vehicle (DMSO), 2.0 μM or 0.2 μM doxorubicin, or 8 μM E64-d for 1 h at 37°C. Reovirus or reo-dox was added to cells and incubated for 18 h at 37°C. Cells were fixed with ice-cold methanol for at least 30 min. Methanol was removed and cells were washed twice with PBS and blocked with PBS containing 1% BSA for 15 min at room temperature. Cells were stained with reovirus-specific polyclonal antiserum (1:2,000) for 1 h at room temperature, washed twice with PBS, stained with A488 (1:1,000) for 1 h at room temperature, stained with 0.5 ng/mL 4'6'-diamidino-2-phenylindole (DAPI) for 5 min at room temperature, and washed twice with PBS. Immunofluorescence was detected using a Lionheart FX Automated Microscope (Biotek) with a 4×-PLFL phase objective (NA 0.13), and percent infectivity was determined (reovirus-positive cells/DAPI-positive cells) using Gen5 software (Biotek).
Reovirus Replication Assay
MDA-MB-231 and MDA-MB-436 cells were adsorbed with reovirus or reo-dox at an MOI of 10 PFU/cell in OMEM (GIBCO) for 1 h at room temperature, washed with PBS, and incubated for 0–3 days with complete media at 37°C. Cells were freeze-thawed three times and viral titers were determined by plaque assay using L929 cells. Viral yields were calculated by dividing viral titers by the viral titer from day 0.
qPCR Assessment of Type-I and -III Interferon Transcript Levels
MDA-MB-231 and MDA-MB-436 cells were treated with DMSO or 2 μM doxorubicin for 1 h at 37°C, infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell, and incubated for 0, 8, 12, 24, and 48 h. RNA was isolated using a QIAGEN RNeasy kit with on-column DNase digestion. cDNAs were generated using 500 ng of RNA and random primers with the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) in a SimpliAmp Thermal Cycler (Thermo Fisher Scientific). cDNA was diluted 1:5 in nuclease-free water and qPCR reactions were performed in MicroAmp Fast Optical 96-Well Reaction Plates (Applied Biosystems) using Primetime qPCR assays (IDT) for IFNB1 (GenBank: NM_002176), IFNL1 (GenBank: NM_172140), HPRT1 (GenBank: NM_000194), and a custom assay for the reovirus S1 gene segment (Probe: 5′-/56-FAM/TCAATGCTG/ZEN/TCGAACCACGAGTTGA/3IABkFQ/-3′; Primer 1: 5′-CGAGTCAGGTCACGCAATTA-3′; Primer 2: 5′-GGATGTTCGTCCAGTGAGATTAG-3′) using a 7500 Fast Real-Time PCR System (Applied Biosystems) and accompanying software to analyze qPCR data.
IFN ELISA
MDA-MB-231and MDA-MB-436 cells were treated with DMSO or 2 μM doxorubicin for 1 h at 37°C, infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell, and incubated for 0, 8, 12, 24, and 48 h. Cell supernatants were collected and levels of IFN-λ or IFN-β were determined with the Human IFN-λ 1/3 and Human IFN-β DuoSet ELISA kits (R&D Systems). Plates were read on a Synergy HT or Synergy H1 plate reader (Biotek) using 450 nm for sample detection and 540 nm for wavelength correction.
Immunoblotting for DNA Damage Response and Innate Immune Molecules
MDA-MB-231 or MDA-MB-436 cells were treated with DMSO or 2 μM doxorubicin for 1 h at 37°C, infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell, and incubated for 0–2 days at 37°C. To assess the ability of IFNs to stimulate immune signaling, we treated MDA-MB-436 cells with 10 and 100 ng/mL of IFN-λ or 100 and 1,000 IU/mL IFN-β for 1 h at 37°C. Whole cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% sodium dodecyl sulfate, 0.1% sodium deoxycholate) and fresh Protease Inhibitor Cocktail (P8340, Sigma-Aldrich), Phosphatase Inhibitor Cocktail 2 (P5726, Sigma-Aldrich), 1 mM sodium vanadate, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and protein concentration was determined using the DC protein assay (Bio-Rad). Whole cell lysates were resolved by SDS-PAGE in 4%–20% gradient Mini-PROTEAN TGX gels (Bio-Rad) and transferred to 0.2 μm pore size nitrocellulose membranes (Bio-Rad). Membranes were incubated for 1 h in blocking buffer (Tris-buffered saline [TBS] with 5% powdered milk), incubated with primary antibodies specific for phospho-ATM (S1981, clone 10H11.E12, #4526), -p53(S15, #9284), -H2AX (S139, #2577), -STAT1 (Y701, clone D4A7 #7649), -STAT2 (Y690, clone D3P2P, #88410), -STAT3 (Y705, clone D3A7, #9145), total ATM (clone D2E2, #2873), p53 (clone 1C12, #2524), STAT1 (clone D3A7, #9145), STAT2 (clone D9J7L, #72604), STAT3 (clone 124H6, #9139), and GAPDH (clone GA1R, MA5-15738), and reovirus polyclonal antiserum overnight at 4°C. Antibodies are from Cell Signaling Technology except for GAPDH (Thermo Fisher Scientific). Membranes were washed with TBS-T (TBS with 0.1% Tween 20), incubated with secondary antibodies conjugated to IRDye 680 or IRDye 800, and imaged using a Li-Cor Odyssey CLx and processed in ImageStudio (LI-COR Biosciences).
Comet Assay
Protocol adapted from Olive and Banáth.84 MDA-MB-231 and MDA-MB-436 cells were pretreated with vehicle (DMSO) or 2 μM doxorubicin for 1 h at 37°C. Without changing media, cells were infected with mock, reovirus, or reo-dox at an MOI of 100 PFU/cell in an equal volume of media for 1 h at 37°C. At 0 and 2 days post infection, trypsin was added and cells were diluted to 2 × 104 cells/mL in cold PBS. 1% low melting point agarose (Lonza) in pure water was added to each cell solution in a 3:1 ratio. Agarose-cell solution was dispensed on a glass microscope slide, and a glass coverslip was placed on top of the agarose solution. Slides were allowed to gel at 4°C for 10 min, coverslips were removed, and slides were incubated for 10 min at 4°C. Slides were arranged in single layers in 150 mm culture dishes, lysis buffer (2% sarkosyl, 0.5 M Na2 EDTA, 0.5 mg/mL proteinase K) were added to each slide, and slides were incubated for 18 h at 37°C. Gels were washed three times for 20 min with 1× TBE (tris-HCl, borate, EDTA) at room temperature. Slides were electrophoresed in TBE for 25 min at 0.6 V/cm (12 V on a Bio-Rad Wide Mini-Sub Cell GT gel rig), then washed with water before submerging in 1:10,000 DAPI in water for 20 min with occasional rocking. Slides were washed with water and imaged on an epifluorescent microscope. Cells were scored with the Comet Score software.
In Vivo 4T1 Model
Female BALB/c mice, 6–8 weeks of age, were purchased from Jackson Laboratories and were maintained in accordance with IACUC approved institutional guidelines and protocols. Mice were housed in static racks with Micro-Isolator housing with individual water and food supplies. 4T1 breast cancer cells were inoculated subcutaneously on the hind flank with 5 × 104 cells in 100 μL of PBS. Mice were monitored for tumor growth and weight loss and were euthanized if tumors became ulcerated or reached >2 cm2 or lost 25% of their initial weight following IACUC protocols. Doxorubicin was delivered intratumorally at a concentration of 0.14 mg/kg in 50 μL of PBS, and reovirus or reo-dox was delivered intratumorally at a concentration of 5 × 108 PFU in 50 μL of PBS once tumors reached 7 mm in diameter. After inoculation, mice were housed in Animal Biocontainment Level 2 (ABSL2) rooms and handled following the Division of Animal Resources (DAR) guidelines based on the Biosafety in Microbiological and Biomedical Laboratories (BMBL) 5th edition guidelines.
Immunohistochemistry on 4T1 Tissue Samples
Tissue samples were fixed in 10% formalin for 24 h. Formalin was aspirated from samples and tissues were stored in 70% ethanol until processed for paraffin embedding and slide mounting (Emory Cancer Tissue and Pathology Shared Resource Core). Mounted tumor tissues were stained for γH2AX (Cell Signaling, S139, clone 20E3 #9781) at 1:480 and reovirus using polyclonal antibody crossadsorbed in L929 mouse fibroblasts and 4T1 murine mammary adenocarcinoma cells. Reovirus antibody was used at 1:8,000. Stained tissues were scanned using an Olympus Nanozoomer whole-slide scanner (Emory Cancer Tissue and Pathology Shared Resource Core). Images were analyzed using QuPath software85 with the following scripts:
Reovirus polyclonal antibody detection was run on manually selected whole tissues:
setImageType(‘BRIGHTFIELD_H_DAB’);
setColorDeconvolutionStains(‘{”Name”: “H-DAB default,” “Stain 1”: “Hematoxylin,” “Values 1”: “0.65111 0.70119 0.29049 “, “Stain 2”: “DAB,” “Values 2”: “0.26917 0.56824 0.77759 “, “Background”: ” 255 255 255 “}');
runPlugin(‘qupath.imagej.detect.nuclei.PositiveCellDetection’, ‘{”detectionImageBrightfield”: “Hematoxylin OD,” “requestedPixelSizeMicrons”: 0.5, “backgroundRadiusMicrons”: 8.0, “medianRadiusMicrons”: 0.0, “sigmaMicrons”: 1.5, “minAreaMicrons”: 2.0, “maxAreaMicrons”: 400.0, “threshold”: 0.1, “maxBackground”: 2.0, “watershedPostProcess”: true, “excludeDAB”: false, “cellExpansionMicrons”: 5.0, “includeNuclei”: true, “smoothBoundaries”: true, “makeMeasurements”: true, “thresholdCompartment”: “Cell: DAB OD mean,” “thresholdPositive1”: 0.2, “thresholdPositive2”: 0.4, “thresholdPositive3”: 0.6, “singleThreshold”: false}’);
γH2AX
setImageType(‘BRIGHTFIELD_H_DAB’);
setColorDeconvolutionStains(‘{”Name”: “H-DAB default,” “Stain 1”: “Hematoxylin,” “Values 1”: “0.65111 0.70119 0.29049 “, “Stain 2”: “DAB,” “Values 2”: “0.26917 0.56824 0.77759 “, “Background”: ” 255 255 255 “}');
runPlugin(‘qupath.imagej.detect.tissue.SimpleTissueDetection2′, ‘{”threshold”: 212, “requestedPixelSizeMicrons”: 5.0, “minAreaMicrons”: 10000.0, “maxHoleAreaMicrons”: 1000000.0, “darkBackground”: false, “smoothImage”: true, “medianCleanup”: true, “dilateBoundaries”: false, “smoothCoordinates”: true, “excludeOnBoundary”: false, “singleAnnotation”: true}’);
selectAnnotations();
runPlugin(‘qupath.imagej.detect.nuclei.PositiveCellDetection’, ‘{”detectionImageBrightfield”: “Hematoxylin OD,” “requestedPixelSizeMicrons”: 0.5, “backgroundRadiusMicrons”: 8.0, “medianRadiusMicrons”: 0.0, “sigmaMicrons”: 2.0, “minAreaMicrons”: 2.0, “maxAreaMicrons”: 400.0, “threshold”: 0.1, “maxBackground”: 2.0, “watershedPostProcess”: true, “excludeDAB”: false, “cellExpansionMicrons”: 5.0, “includeNuclei”: true, “smoothBoundaries”: true, “makeMeasurements”: true, “thresholdCompartment”: “Nucleus: DAB OD mean,” “thresholdPositive1”: 0.3, “thresholdPositive2”: 0.4, “thresholdPositive3”: 0.5, “singleThreshold”: false}’);
Clonogenic Assay for Metastatic 4T1 Cells in Lungs
Mice were euthanized on day 21 post tumor challenge. Lungs were digested and processed to a single cell suspension using collagenase type IV (Sigma-Aldrich) for 3 h at 37°C with constant motion. Homogenates were strained using a 70 μm cell strainer. Cells were washed and suspended in DMEM with 10% FBS (Hyclone) containing 6-thioguanine (Sigma-Aldrich). Serial dilutions were made and cultures were grown in a 6-well plate at 37°C with 5% CO2 until the first well reached confluence. Cell counts were obtained using a hemocytometer.
Statistical Analysis
Mean values for triplicate and quadruplicate experiments were compared using one or two-way analysis of variance (ANOVA) with Dunnett’s or Tukey’s multiple-comparisons test (Graph Pad Prism). p values of < 0.05 were considered statistically significant. For clarity, comparisons lacking statistical significance are not annotated.
Author Contributions
J.T.L.B., L.E.M., R.M.R.S., and B.A.M. designed in vitro and in vivo experiments. J.T.L.B., L.E.M., and R.M.R.S. contributed to carrying out experiments and data analysis. J.T.L.B. and B.A.M. wrote the manuscript and prepared figures. P.S. provided in vivo study support.
Conflicts of Interest
All authors concur with the submission and declare that they have no conflicts of interest other than the reassortant virus used in this study being part of International Patent Application No PCT/US2019/036151.
Acknowledgments
We thank Dr. Curtis Henry, Emily Greene, and Allison Wolf for critical revision and feedback during manuscript preparation. This work was supported by funding from the Children's Healthcare of Atlanta and the Pediatric Research Institute, Winship Comprehensive Cancer Institute #IRG-14-188-01 from the American Cancer Society, and the National Institutes of Health (R01 AI146260 to B.A.M.), NCI/NIH R01 grant (R01 CA202763 to P.S.), and Diversity Supplement grant (R01 CA202763-S to L.E.M.). Flow cytometry experiments were performed in the Emory Pediatrics Flow Cytometry Core (UL1TR002378). Imaging was performed at the Emory Integrated Cellular Imaging Core (2P30CA138292-04 and the Emory Pediatrics Institute). Mouse tissue histology and imaging was performed by the Cancer Tissue and Pathology shared resource of Winship Cancer Institute of Emory University and NIH/NCI (P30CA138292). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.08.008.
Supplemental Information
References
- 1.American Cancer Society . 2020. Cancer Facts & Figures 2020. (Atlanta: American Cancer Society) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Cancer Society . 2019. Breast Cancer Facts & Figures 2019-2020. (Atlanta: American Cancer Society) [Google Scholar]
- 3.NAACCR.org Available from: https://naaccr.org.
- 4.Haffty B.G., Yang Q., Reiss M., Kearney T., Higgins S.A., Weidhaas J., Harris L., Hait W., Toppmeyer D. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J. Clin. Oncol. 2006;24:5652–5657. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 5.Dent R., Trudeau M., Pritchard K.I., Hanna W.M., Kahn H.K., Sawka C.A., Lickley L.A., Rawlinson E., Sun P., Narod S.A. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 6.Wahba H.A., El-Hadaad H.A. Current approaches in treatment of triple-negative breast cancer. Cancer Biol. Med. 2015;12:106–116. doi: 10.7497/j.issn.2095-3941.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeichner S.B., Terawaki H., Gogineni K. A Review of Systemic Treatment in Metastatic Triple-Negative Breast Cancer. Breast Cancer (Auckl.) 2016;10:25–36. doi: 10.4137/BCBCR.S32783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dock G. The Influence of Complicating Diseases Upon Leukemia. Am. J. Med. Sci. 1904;127:563–592. [Google Scholar]
- 9.Dmitriev I., Krasnykh V., Miller C.R., Wang M., Kashentseva E., Mikheeva G., Belousova N., Curiel D.T. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 1998;72:9706–9713. doi: 10.1128/jvi.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morizono K., Xie Y., Ringpis G.E., Johnson M., Nassanian H., Lee B., Wu L., Chen I.S. Lentiviral vector retargeting to P-glycoprotein on metastatic melanoma through intravenous injection. Nat. Med. 2005;11:346–352. doi: 10.1038/nm1192. [DOI] [PubMed] [Google Scholar]
- 11.Doronin K., Toth K., Kuppuswamy M., Ward P., Tollefson A.E., Wold W.S. Tumor-specific, replication-competent adenovirus vectors overexpressing the adenovirus death protein. J. Virol. 2000;74:6147–6155. doi: 10.1128/jvi.74.13.6147-6155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirvinen M., Rajecki M., Kapanen M., Parviainen S., Rouvinen-Lagerström N., Diaconu I., Nokisalmi P., Tenhunen M., Hemminki A., Cerullo V. Immunological effects of a tumor necrosis factor alpha-armed oncolytic adenovirus. Hum. Gene Ther. 2015;26:134–144. doi: 10.1089/hum.2014.069. [DOI] [PubMed] [Google Scholar]
- 13.Li J.L., Liu H.L., Zhang X.R., Xu J.P., Hu W.K., Liang M., Chen S.Y., Hu F., Chu D.T. A phase I trial of intratumoral administration of recombinant oncolytic adenovirus overexpressing HSP70 in advanced solid tumor patients. Gene Ther. 2009;16:376–382. doi: 10.1038/gt.2008.179. [DOI] [PubMed] [Google Scholar]
- 14.Burke J.M., Lamm D.L., Meng M.V., Nemunaitis J.J., Stephenson J.J., Arseneau J.C., Aimi J., Lerner S., Yeung A.W., Kazarian T. A first in human phase 1 study of CG0070, a GM-CSF expressing oncolytic adenovirus, for the treatment of nonmuscle invasive bladder cancer. J. Urol. 2012;188:2391–2397. doi: 10.1016/j.juro.2012.07.097. [DOI] [PubMed] [Google Scholar]
- 15.Wolters Kluwer Health FDA Approves First Oncolytic Virus Therapy Imlygic for Melanoma. Oncology Times. 2015;37:36. [Google Scholar]
- 16.Franke V., Berger D.M.S., Klop W.M.C., van der Hiel B., van de Wiel B.A., Ter Meulen S., Wouters M.W.J.M., van Houdt W.J., van Akkooi A.C.J. High response rates for T-VEC in early metastatic melanoma (stage IIIB/C-IVM1a) Int. J. Cancer. 2019;145:974–978. doi: 10.1002/ijc.32172. [DOI] [PubMed] [Google Scholar]
- 17.Andtbacka R.H.I., Collichio F., Harrington K.J., Middleton M.R., Downey G., Ӧhrling K., Kaufman H.L. Final analyses of OPTiM: a randomized phase III trial of talimogene laherparepvec versus granulocyte-macrophage colony-stimulating factor in unresectable stage III-IV melanoma. J. Immunother. Cancer. 2019;7:145. doi: 10.1186/s40425-019-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ClinicalTrials.gov (2018). Available from: https://clinicaltrials.gov.
- 19.Dermody T.S., Parker J., Sherry B. Orthoreoviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Sixth Edition. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 1304–1346. [Google Scholar]
- 20.Selb B., Weber B. A study of human reovirus IgG and IgA antibodies by ELISA and western blot. J. Virol. Methods. 1994;47:15–25. doi: 10.1016/0166-0934(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 21.Duncan M.R., Stanish S.M., Cox D.C. Differential sensitivity of normal and transformed human cells to reovirus infection. J. Virol. 1978;28:444–449. doi: 10.1128/jvi.28.2.444-449.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton E.S., Forrest J.C., Connolly J.L., Chappell J.D., Liu Y., Schnell F.J., Nusrat A., Parkos C.A., Dermody T.S. Junction adhesion molecule is a receptor for reovirus. Cell. 2001;104:441–451. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- 23.Kirchner E., Guglielmi K.M., Strauss H.M., Dermody T.S., Stehle T. Structure of reovirus sigma1 in complex with its receptor junctional adhesion molecule-A. PLoS Pathog. 2008;4:e1000235. doi: 10.1371/journal.ppat.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norman K.L., Hirasawa K., Yang A.D., Shields M.A., Lee P.W. Reovirus oncolysis: the Ras/RalGEF/p38 pathway dictates host cell permissiveness to reovirus infection. Proc. Natl. Acad. Sci. USA. 2004;101:11099–11104. doi: 10.1073/pnas.0404310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strong J.E., Coffey M.C., Tang D., Sabinin P., Lee P.W. The molecular basis of viral oncolysis: usurpation of the Ras signaling pathway by reovirus. EMBO J. 1998;17:3351–3362. doi: 10.1093/emboj/17.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcato P., Shmulevitz M., Pan D., Stoltz D., Lee P.W. Ras transformation mediates reovirus oncolysis by enhancing virus uncoating, particle infectivity, and apoptosis-dependent release. Mol. Ther. 2007;15:1522–1530. doi: 10.1038/sj.mt.6300179. [DOI] [PubMed] [Google Scholar]
- 27.Nouh M.A., Mohamed M.M., El-Shinawi M., Shaalan M.A., Cavallo-Medved D., Khaled H.M., Sloane B.F. Cathepsin B: a potential prognostic marker for inflammatory breast cancer. J. Transl. Med. 2011;9:1. doi: 10.1186/1479-5876-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasiljeva O., Korovin M., Gajda M., Brodoefel H., Bojic L., Krüger A., Schurigt U., Sevenich L., Turk B., Peters C., Reinheckel T. Reduced tumour cell proliferation and delayed development of high-grade mammary carcinomas in cathepsin B-deficient mice. Oncogene. 2008;27:4191–4199. doi: 10.1038/onc.2008.59. [DOI] [PubMed] [Google Scholar]
- 29.Villalona-Calero M.A., Lam E., Otterson G.A., Zhao W., Timmons M., Subramaniam D., Hade E.M., Gill G.M., Coffey M., Selvaggi G. Oncolytic reovirus in combination with chemotherapy in metastatic or recurrent non-small cell lung cancer patients with KRAS-activated tumors. Cancer. 2016;122:875–883. doi: 10.1002/cncr.29856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lolkema M.P., Arkenau H.T., Harrington K., Roxburgh P., Morrison R., Roulstone V., Twigger K., Coffey M., Mettinger K., Gill G. A phase I study of the combination of intravenous reovirus type 3 Dearing and gemcitabine in patients with advanced cancer. Clin. Cancer Res. 2011;17:581–588. doi: 10.1158/1078-0432.CCR-10-2159. [DOI] [PubMed] [Google Scholar]
- 31.Harrington K.J., Karapanagiotou E.M., Roulstone V., Twigger K.R., White C.L., Vidal L., Beirne D., Prestwich R., Newbold K., Ahmed M. Two-stage phase I dose-escalation study of intratumoral reovirus type 3 dearing and palliative radiotherapy in patients with advanced cancers. Clin. Cancer Res. 2010;16:3067–3077. doi: 10.1158/1078-0432.CCR-10-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajani K., Parrish C., Kottke T., Thompson J., Zaidi S., Ilett L., Shim K.G., Diaz R.M., Pandha H., Harrington K. Combination Therapy With Reovirus and Anti-PD-1 Blockade Controls Tumor Growth Through Innate and Adaptive Immune Responses. Mol. Ther. 2016;24:166–174. doi: 10.1038/mt.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodríguez Stewart R.M., Berry J.T.L., Berger A.K., Yoon S.B., Hirsch A.L., Guberman J.A., Patel N.B., Tharp G.K., Bosinger S.E., Mainou B.A. Enhanced Killing of Triple-Negative Breast Cancer Cells by Reassortant Reovirus and Topoisomerase Inhibitors. J. Virol. 2019;93:e01411-19. doi: 10.1128/JVI.01411-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tewey K.M., Rowe T.C., Yang L., Halligan B.D., Liu L.F. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 35.Buzdar A.U., Marcus C., Smith T.L., Blumenschein G.R. Early and delayed clinical cardiotoxicity of doxorubicin. Cancer. 1985;55:2761–2765. doi: 10.1002/1097-0142(19850615)55:12<2761::aid-cncr2820551206>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 36.Johnson-Arbor K., Dubey R. StatPearls; Treasure Island, FL: 2020. Doxorubicin. [PubMed] [Google Scholar]
- 37.Birrer M.J., Moore K.N., Betella I., Bates R.C. Antibody-Drug Conjugate-Based Therapeutics: State of the Science. J. Natl. Cancer Inst. 2019;111:538–549. doi: 10.1093/jnci/djz035. [DOI] [PubMed] [Google Scholar]
- 38.Fecek R.J., Busch R., Lin H., Pal K., Cunningham C.A., Cuff C.F. Production of Alexa Fluor 488-labeled reovirus and characterization of target cell binding, competence, and immunogenicity of labeled virions. J. Immunol. Methods. 2006;314:30–37. doi: 10.1016/j.jim.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Huang S., Wang H., Carroll C.A., Hayes S.J., Weintraub S.T., Serwer P. Analysis of proteins stained by Alexa dyes. Electrophoresis. 2004;25:779–784. doi: 10.1002/elps.200305723. [DOI] [PubMed] [Google Scholar]
- 40.Mainou B.A., Zamora P.F., Ashbrook A.W., Dorset D.C., Kim K.S., Dermody T.S. Reovirus cell entry requires functional microtubules. MBio. 2013;4:e00405-13. doi: 10.1128/mBio.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann B.D., Bauer J.A., Chen X., Sanders M.E., Chakravarthy A.B., Shyr Y., Pietenpol J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konopka-Anstadt J.L., Mainou B.A., Sutherland D.M., Sekine Y., Strittmatter S.M., Dermody T.S. The Nogo receptor NgR1 mediates infection by mammalian reovirus. Cell Host Microbe. 2014;15:681–691. doi: 10.1016/j.chom.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baer G.S., Dermody T.S. Mutations in reovirus outer-capsid protein sigma3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 1997;71:4921–4928. doi: 10.1128/jvi.71.7.4921-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez J.V., Febbo P.G., Ramaswamy S., Loda M., Richardson A., Frank D.A. Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer Res. 2005;65:5054–5062. doi: 10.1158/0008-5472.CAN-04-4281. [DOI] [PubMed] [Google Scholar]
- 45.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 46.Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 47.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 48.Canman C.E., Lim D.S., Cimprich K.A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M.B., Siliciano J.D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 49.Cancer Cell Line Encyclopedia Available from: https://portals.broadinstitute.org/ccle.
- 50.Pulaski B.A., Ostrand-Rosenberg S. Reduction of established spontaneous mammary carcinoma metastases following immunotherapy with major histocompatibility complex class II and B7.1 cell-based tumor vaccines. Cancer Res. 1998;58:1486–1493. [PubMed] [Google Scholar]
- 51.Pulaski B.A., Terman D.S., Khan S., Muller E., Ostrand-Rosenberg S. Cooperativity of Staphylococcal aureus enterotoxin B superantigen, major histocompatibility complex class II, and CD80 for immunotherapy of advanced spontaneous metastases in a clinically relevant postoperative mouse breast cancer model. Cancer Res. 2000;60:2710–2715. [PubMed] [Google Scholar]
- 52.Pulaski B.A., Ostrand-Rosenberg S. Mouse 4T1 breast tumor model. Curr. Protoc. Immunol. 2001;Chapter 20:Unit 20.2. doi: 10.1002/0471142735.im2002s39. [DOI] [PubMed] [Google Scholar]
- 53.Kaur P., Nagaraja G.M., Zheng H., Gizachew D., Galukande M., Krishnan S., Asea A. A mouse model for triple-negative breast cancer tumor-initiating cells (TNBC-TICs) exhibits similar aggressive phenotype to the human disease. BMC Cancer. 2012;12:120. doi: 10.1186/1471-2407-12-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franken N.A., Rodermond H.M., Stap J., Haveman J., van Bree C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 55.Pan D., Marcato P., Ahn D.G., Gujar S., Pan L.Z., Shmulevitz M., Lee P.W. Activation of p53 by chemotherapeutic agents enhances reovirus oncolysis. PLoS ONE. 2013;8:e54006. doi: 10.1371/journal.pone.0054006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roulstone V., Twigger K., Zaidi S., Pencavel T., Kyula J.N., White C., McLaughlin M., Seth R., Karapanagiotou E.M., Mansfield D. Synergistic cytotoxicity of oncolytic reovirus in combination with cisplatin-paclitaxel doublet chemotherapy. Gene Ther. 2013;20:521–528. doi: 10.1038/gt.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinemann L., Simpson G.R., Boxall A., Kottke T., Relph K.L., Vile R., Melcher A., Prestwich R., Harrington K.J., Morgan R., Pandha H.S. Synergistic effects of oncolytic reovirus and docetaxel chemotherapy in prostate cancer. BMC Cancer. 2011;11:221. doi: 10.1186/1471-2407-11-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sei S., Mussio J.K., Yang Q.E., Nagashima K., Parchment R.E., Coffey M.C., Shoemaker R.H., Tomaszewski J.E. Synergistic antitumor activity of oncolytic reovirus and chemotherapeutic agents in non-small cell lung cancer cells. Mol. Cancer. 2009;8:47. doi: 10.1186/1476-4598-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelly K.R., Espitia C.M., Zhao W., Wu K., Visconte V., Anwer F., Calton C.M., Carew J.S., Nawrocki S.T. Oncolytic reovirus sensitizes multiple myeloma cells to anti-PD-L1 therapy. Leukemia. 2018;32:230–233. doi: 10.1038/leu.2017.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samson A., Scott K.J., Taggart D., West E.J., Wilson E., Nuovo G.J., Thomson S., Corns R., Mathew R.K., Fuller M.J. Intravenous delivery of oncolytic reovirus to brain tumor patients immunologically primes for subsequent checkpoint blockade. Sci. Transl. Med. 2018;10:eaam7577. doi: 10.1126/scitranslmed.aam7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liemann S., Chandran K., Baker T.S., Nibert M.L., Harrison S.C. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reiss K., Stencel J.E., Liu Y., Blaum B.S., Reiter D.M., Feizi T., Dermody T.S., Stehle T. The GM2 glycan serves as a functional coreceptor for serotype 1 reovirus. PLoS Pathog. 2012;8:e1003078. doi: 10.1371/journal.ppat.1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reiter D.M., Frierson J.M., Halvorson E.E., Kobayashi T., Dermody T.S., Stehle T. Crystal structure of reovirus attachment protein σ1 in complex with sialylated oligosaccharides. PLoS Pathog. 2011;7:e1002166. doi: 10.1371/journal.ppat.1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dietrich M.H., Ogden K.M., Katen S.P., Reiss K., Sutherland D.M., Carnahan R.H., Goff M., Cooper T., Dermody T.S., Stehle T. Structural Insights into Reovirus σ1 Interactions with Two Neutralizing Antibodies. J. Virol. 2017;91:e01621-16. doi: 10.1128/JVI.01621-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peterson S.T., Kennedy E.A., Brigleb P.H., Taylor G.M., Urbanek K., Bricker T.L., Lee S., Shin H., Dermody T.S., Boon A.C.M., Baldridge M.T. Disruption of type III interferon genes Ifnl2 and Ifnl3 recapitulates loss of the type III IFN receptor in the mucosal antiviral response. J. Virol. 2019;93:e01073-19. doi: 10.1128/JVI.01073-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mordstein M., Neugebauer E., Ditt V., Jessen B., Rieger T., Falcone V., Sorgeloos F., Ehl S., Mayer D., Kochs G. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bayer A., Lennemann N.J., Ouyang Y., Bramley J.C., Morosky S., Marques E.T., Jr., Cherry S., Sadovsky Y., Coyne C.B. Type III Interferons Produced by Human Placental Trophoblasts Confer Protection against Zika Virus Infection. Cell Host Microbe. 2016;19:705–712. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palma-Ocampo H.K., Flores-Alonso J.C., Vallejo-Ruiz V., Reyes-Leyva J., Flores-Mendoza L., Herrera-Camacho I., Rosas-Murrieta N.H., Santos-López G. Interferon lambda inhibits dengue virus replication in epithelial cells. Virol. J. 2015;12:150. doi: 10.1186/s12985-015-0383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burkart C., Arimoto K., Tang T., Cong X., Xiao N., Liu Y.C., Kotenko S.V., Ellies L.G., Zhang D.E. Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-λ and elevated secretion of Cxcl10. EMBO Mol. Med. 2013;5:1035–1050. doi: 10.1002/emmm.201201864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qing Y., Stark G.R. Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J. Biol. Chem. 2004;279:41679–41685. doi: 10.1074/jbc.M406413200. [DOI] [PubMed] [Google Scholar]
- 71.Fernandes J.P., Cristi F., Eaton H.E., Chen P., Haeflinger S., Bernard I., Hitt M.M., Shmulevitz M. Breast Tumor-Associated Metalloproteases Restrict Reovirus Oncolysis by Cleaving the σ1 Cell Attachment Protein and Can Be Overcome by Mutation of σ1. J. Virol. 2019;93:e01380-19. doi: 10.1128/JVI.01380-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang Y., Li S., Wang X., He B., He B., Dai W., Zhang H., Wang X., Wang Y., Zhou D., Zhang Q. A Nanosystem of Amphiphilic Oligopeptide-Drug Conjugate Actualizing Both αvβ3 Targeting and Reduction-Triggered Release for Maytansinoid. Theranostics. 2017;7:3306–3318. doi: 10.7150/thno.20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hermanson G.T. Third Edition. Elsevier; 2013. The Reactions of Bioconjugation. Bioconjugate Techniques. [Google Scholar]
- 74.Pillay C.S., Elliott E., Dennison C. Endolysosomal proteolysis and its regulation. Biochem. J. 2002;363:417–429. doi: 10.1042/0264-6021:3630417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Authier F., Posner B.I., Bergeron J.J. Endosomal proteolysis of internalized proteins. FEBS Lett. 1996;389:55–60. doi: 10.1016/0014-5793(96)00368-7. [DOI] [PubMed] [Google Scholar]
- 76.Davis R.T., Blake K., Ma D., Gabra M.B.I., Hernandez G.A., Phung A.T., Yang Y., Maurer D., Lefebvre A.E.Y.T., Alshetaiwi H. Transcriptional diversity and bioenergetic shift in human breast cancer metastasis revealed by single-cell RNA sequencing. Nat. Cell Biol. 2020;22:310–320. doi: 10.1038/s41556-020-0477-0. [DOI] [PubMed] [Google Scholar]
- 77.Yadav N., Kumar S., Marlowe T., Chaudhary A.K., Kumar R., Wang J., O’Malley J., Boland P.M., Jayanthi S., Kumar T.K. Oxidative phosphorylation-dependent regulation of cancer cell apoptosis in response to anticancer agents. Cell Death Dis. 2015;6:e1969. doi: 10.1038/cddis.2015.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alam S.R., Wallrabe H., Svindrych Z., Chaudhary A.K., Christopher K.G., Chandra D., Periasamy A. Investigation of Mitochondrial Metabolic Response to Doxorubicin in Prostate Cancer Cells: An NADH, FAD and Tryptophan FLIM Assay. Sci. Rep. 2017;7:10451. doi: 10.1038/s41598-017-10856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kobayashi T., Antar A.A., Boehme K.W., Danthi P., Eby E.A., Guglielmi K.M., Holm G.H., Johnson E.M., Maginnis M.S., Naik S. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe. 2007;1:147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Furlong D.B., Nibert M.L., Fields B.N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith R.E., Zweerink H.J., Joklik W.K. Polypeptide components of virions, top component and cores of reovirus type 3. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 82.Virgin H.W., Dermody T.S., Tyler K.L. Cellular and humoral immunity to reovirus infection. Curr. Top. Microbiol. Immunol. 1998;233:147–161. doi: 10.1007/978-3-642-72095-6_8. [DOI] [PubMed] [Google Scholar]
- 83.Virgin H.W., 4th, Bassel-Duby R., Fields B.N., Tyler K.L. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing) J. Virol. 1988;62:4594–4604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Olive P.L., Banáth J.P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 85.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.