Abstract
Background:
EGFR/c-Met activation/amplification and co-expression, mTOR upregulation/activation, and Akt/Wnt signaling upregulation have been individually associated with more aggressive disease and characterized as potential prognostic markers for lung cancer patients.
Methods:
Tumors obtained from 109 participants with stage I–IV non-small cell lung cancer (NSCLC) were studied for EGFR/c-Met co-localization as well as for total and active forms of EGFR, c-Met, mTOR, S6K, beta-catenin, and Axin2. Slides were graded by two independent blinded pathologists using a validated scoring system. Protein expression profile correlations were assessed using Pearson correlation and Spearman’s rho. Prognosis was assessed using Kaplan–Meier analysis.
Results:
Protein expression profile analysis revealed significant correlations between EGFR/p-EGFR (p = 0.0412) and p-mTOR/S6K (p = 0.0044). Co-localization of p-EGFR/p-c-Met was associated with increased p-mTOR (p = 0.0006), S6K (p = 0.0018), and p-S6K (p < 0.0001) expression. In contrast, active beta-catenin was not positively correlated with EGFR/c-Met nor any activated proteins. Axin2, a negative regulator of the Wnt pathway, was correlated with EGFR, p-EGFR, p-mTOR, p-S6K, EGFR/c-Met co-localization, and p-EGFR/p-c-Met co-localization (all p-values <0.03). Kaplan–Meier analysis revealed shorter median survival in participants with high expression of Axin2, total beta-catenin, total/p-S6K, total/p-mTOR, EGFR, and EGFR/c-Met co-localization compared with low expression. After controlling for stage of disease at diagnosis, subjects with late-stage disease demonstrated shorter median survival when exhibiting high co-expression of EGFR/c-Met (8.1 month versus 22.3 month, p = 0.050), mTOR (6.7 month versus 22.3 month, p = 0.002), and p-mTOR (8.1 month versus 25.4 month, p = 0.004) compared with low levels.
Conclusions:
These findings suggest that increased EGFR/c-Met signaling is correlated with upregulated mTOR/S6K signaling, which may in turn be associated with shorter median survival in late-stage NSCLC.
Keywords: biomarker, EGFR/c-Met, mTOR, non-small cell lung cancer, prognosis
Introduction
Lung cancer is the leading cause of cancer mortality worldwide. In 2019, a projected 142,670 people in the United States died from lung cancer, more deaths than those attributed to colon, breast, prostate, and ovarian cancers combined.1 Among diagnosed lung cancer cases, 80–90% are classified as non-small cell lung cancer (NSCLC), the form of lung cancer investigated in this study.2 Approximately 75% of NSCLC cases are locally advanced or metastatic (stage III–IV) at the time of diagnosis, obviating the role of surgery.3 Over the past decade, the development of targeted tyrosine kinase inhibitor (TKI) therapies and guideline-directed screening for prognostic and predictive molecular markers has led to a significant improvement in progression-free survival. Specifically, routine testing for EGFR, ALK, ROS1, BRAF, MET, ERBB2 (HER2), KRAS, and RET is recommended by the International Association for the Study of Lung Cancer (IASLC) and the Association for Molecular Pathology.4–16 Nevertheless, NSCLC-attributed mortality rates remain high.1,17 In addition, studies have elucidated the role of upregulation of molecular signaling pathways, including EGFR, c-Met, mTOR, and Akt/Wnt, in the pathogenesis of NSCLC.18,19
c-Met, also known as the hepatocyte growth factor receptor (HGFR), is a membrane-bound Receptor Tyrosine Kinase (RTK) that is coded for by the MET gene. When amplified/activated, c-Met plays a role in tumor growth, angiogenesis, and metastasis.20,21 In addition, amplified c-Met expression has been correlated with poor prognosis in other tumor types, including liver, gastric, breast, and brain malignancies.21 Activation of c-Met signaling has also been associated with upregulation of a variety of downstream signaling pathways, including the RAS/MAPK, PI3K/Akt, and Wnt pathways.22–24 In NSCLC, co-localization of c-Met and epidermal growth factor receptor (EGFR) has been shown to have a synergistic effect on cell proliferation, downstream activation of signal transduction, and may indicate a more aggressive tumor phenotype that results in worse prognosis.25
The mammalian target of rapamycin/S6 protein kinase (mTOR/S6K) pathway is a key intracellular regulator of cell growth, survival, migration, and invasion in NSCLC.26,27 Increased mTOR/S6K activity has been associated with poor clinical prognosis in early-stage NSCLC.18,19,26,28 In both pre-clinical studies and clinical trials, the inhibition of mTOR has demonstrated potential benefit when employed to treat NSCLC.29,30 Clinical trials have also been conducted to test the efficacy of mTOR inhibitors in combination therapy. Therefore, the study of multiple target proteins found in tumors for use in combination could help improve patient prognosis.31
The Wnt pathway proteins consist of highly conserved glycoproteins that induce downstream signaling upon binding to the activated Frizzled transmembrane receptor, a mechanism of action which stabilizes and inhibits the degradation of beta-catenin.32 Active beta-catenin is the nuclear, signaling form of beta-catenin that is transcriptionally active and unphosphorylated at serine 37 and threonine 41.33,34 Stabilization of beta-catenin leads to transcriptional activation of genes involved in cellular proliferation.32,35 Axin2, a negative regulator of beta-catenin, functions by facilitating the phosphorylation of beta-catenin and thereby tagging it for ubiquitin-mediated destruction.32 Various studies have implicated increased nuclear beta-catenin in both the oncogenesis and pathogenesis of NSCLC, particularly in lung tumors with EGFR mutations.35,36 In addition, in-vitro studies indicate that upregulation of Akt/Wnt signaling as well as mTOR activation could be associated with more aggressive disease phenotypes in patients with NSCLC.18,19
Correlations in protein expression and activation patterns between EGFR, c-Met, EGFR/c-Met co-localization, mTOR, S6K, beta-catenin, and Axin2 in lung tumors are not well established. Currently, few studies have investigated the roles of the Wnt pathway; Axin2, a negative regulator of the Wnt pathway; and EGFR/c-Met co-expression on activation of the downstream mTOR pathway. Furthermore, the fundamental role of mTOR in late-stage NSCLC prognosis is not clearly defined. In response to these deficiencies, this novel study aimed to evaluate correlations in protein expression and activation patterns between EGFR, c-Met, EGFR/c-Met co-localization, mTOR, S6K, beta-catenin, and Axin2 to determine if these signaling pathways were concomitantly upregulated in vivo in a cohort of NSCLC tumors collected at the time of diagnosis and if they have prognostic value with regard to median survival time.
Materials and methods
Case selection, tissue acquisition, and exclusion criteria
All patients at participating institutions with a diagnosis of stage I–IV NSCLC were considered for participation in the study. Samples were obtained in accordance with approval from the University of Illinois College of Medicine at Rockford Institutional Review Board (IRB) under approval number 351597-11. Written informed consent was obtained from all patients prior to their inclusion in the study. This consent form emphasized how participation in the study was voluntary and all tissue samples were de-identified to ensure patient confidentiality. Patients were excluded from participating if they declined to consent to the use of their tissue. A total of 109 patients was identified. All samples used in this study were collected from Winnebago County in Illinois, which has a 14% higher age-adjusted mortality rate (2016) due to lung cancer compared with the national rate and a 20% higher average annual age-adjusted incidence rate (2011–2015) of lung cancer compared with the national rate.37,38
Tissue de-identification and study blinding
A research coordinator not involved in staining, grading, or data analysis was responsible for de-identifying the tissue blocks and labeling them with randomly generated numerical study codes. All investigators involved in processing, staining, and grading of the tissues were blinded to all patient data and outcomes until all cases were scored and re-associated with the de-identified outcome data and clinical treatment for statistical analysis.
Tissue preparation
Tissues were fixed in 10% neutral buffered formalin for a minimum of 6 h and dehydrated in increasing Ethanol (EtOH) concentrations, followed by infiltration with xylene and paraffin. Paraffin blocks were prepared by being embedded in hot wax and then cooled on a cold plate. Fresh slides were sectioned from the coded, paraffin-embedded tissue blocks using a microtome to cut 4-micron thick sections, which were mounted on glass slides.
Immunohistochemistry staining protocol
In brief, protein expression of interest was examined retrospectively in NSCLC patients through immunohistochemistry (IHC) using formalin-fixed paraffin embedded tumor samples and individual probes, as described earlier.39 Samples were double-stained during assessment of co-localization. Positive and negative staining controls used for each protein marker are listed in Supplemental Data Table S1. Positive control tissues used were known to express the targeted protein while negative controls were obtained through omission of primary antibody on a positive control tissue.
Reagents for IHC staining
Avidin/biotin blocking kit (# SP-2001), normal horse serum (# S-2000), normal goat serum (# S-1000), VECTASTAIN Elite ABC kit (# PK-6100), VECTASTAIN ABC-AP kit (# AK-5000), ImmPACT DAB peroxidase substrate (# SK-4105), and VECTOR red alkaline phosphatase substrate kit (# SK-5100) were obtained from Vector Laboratories (Burlingame, CA, USA). Hematoxylin was obtained from Sigma-Aldrich (St Louis, MO, USA). Permount was obtained from Fisher Scientific (Pittsburgh, PA, USA). Antibodies used were Horseradish Peroxidase (HRP)-conjugated anti-EGFR (1:2000 dilution) Thermo Fisher Scientific (Rockford, IL, USA) cat.# PA1-1110S-HRP, Anti-p-c-Met (1:1000 dilution) Life Technologies (Grand Island, NY, USA) cat.# 3077S, biotinylated anti-mouse IgG (1:250 dilution) Vector Laboratories (Burlingame, CA, USA) cat.# BA-2000, and biotinylated anti-rabbit IgG (1:250 dilution) Vector Laboratories (Burlingame, CA, USA) cat.# BA-1000. Remaining antibodies obtained from Cell Signaling Technology (Danvers, MA, USA) were the following: Anti-p-EGFR (1:3200 dilution) cat.# 2236S, Anti-c-Met (1:300 dilution) cat.# 8198P, Anti-mTOR (1:50 dilution) cat.# 2983, Anti-p-mTOR (1:100 dilution) cat.# 2976S, Anti-p70 S6K (1:400 dilution) CST cat.# 2708S, and Anti-pp70 S6K (1:150 dilution) cat.# 9234S. All products were stored and used according to manufacturers’ instructions.
IHC staining of control tissues
Positive control tissues were known to express the target protein, while negative controls omitted primary antibody on positive control tissue. In addition, cells in the normal areas of the lung which do not stain were also used as an internal negative control. Positive controls were placenta for EGFR, EGF-treated H2170 cells for p-EGFR, liver/kidney for c-Met, HGF-treated H2170 cells for p-c-Met, validated NSCLC tissue for mTOR, EGF-treated H2170 cells for p-mTOR, liver/spleen/validated NSCLC tissue for S6K, and HGF-treated H2170 cells for p-S6K. H2170 cell line protein expression patterns were verified by western blotting, as described earlier.18
Grading of stained slides
Staining intensity was graded by two blinded pathologists on a 0–3 scale, with 0 = no staining, 1 = weak staining, 2 = moderate staining, and 3 = strong staining. The grading pathologist also recorded the percentage area of tumor cells staining positively for each corresponding intensity score. The overall graded score was obtained by multiplying the intensity scores (from 0 to 3) by the corresponding percentage of positively stained cells (from 0% to 100%) with a scale ranging from 0 to 300. The equation for obtaining the grading score is as follows: %Tumor Grade 3*3 + %Tumor Grade 2*2 + %Tumor Grade 1*1 + %Tumor Grade 0*0, as described earlier.40
Statistical analysis
Two-tailed t-tests and chi-square tests were utilized to assess demographic differences between high and low protein expression groups for each protein target. Distributions were assessed for normality with Kolmogorov–Smirnov and Shapiro–Wilks tests. To analyze the correlations between the various protein targets and their expression profiles, Pearson correlation coefficients (rp) and Spearman’s rho coefficients (rs) were computed. Data for the protein expression profile correlations were based on the grading score from the blinded pathologists (0 = no expression, 1–100 = low expression, 101–200 = moderate expression, 201–300 = high expression). To investigate the relationships between protein expression, activation, and patient prognosis, patient survival was tracked from the time of diagnosis to 5 years or until death, whichever came first. Kaplan–Meier (KM) and generalized Wilcoxon methods were used to assess prognosis and survival comparing high and low expression groups for each protein. Again, data were transformed into binary variables for each protein target based on the median grading score for that protein, with a score of the median or greater representing high expression and a score less than the median representing low expression. A further sub-analysis was performed to control for the effect of stage on prognosis. To do so, the KM analysis for each target protein was stratified by stage at the time of diagnosis (defined as early-stage = stage I–IIIa and late-stage = stage IIIb–IV). Values with a p ⩽ 0.05 were considered statistically significant.
Results
Demographics
Table 1 presents the demographics information for tissues obtained from the 109 participants in this study. The table includes information on patient sex, race, smoking status, smoking history, histology, cancer stage at diagnosis, and treatment received (Table 1). When comparing demographic characteristics between high and low expression groups for each protein of interest, we found several significant differences. Late-stage (stage IIIb–IV) patients were more likely than early-stage (stage I–IIIa) patients to show high expression of EGFR (p = 0.012), mTOR (p = 0.047), S6K (p < 0.001), total beta-catenin (p = 0.002), Axin2 (p = 0.020), p-mTOR (p = 0.010), and p-S6K (p < 0.001) (Supplemental Data Table S2). In addition, surgical resection was more likely with low p-EGFR/p-c-Met co-localization (p = 0.024) and low expression levels of EGFR (p = 0.006), S6K (p < 0.001), total beta-catenin (p = 0.003), Axin2 (p < 0.001), p-mTOR (p = 0.018), and p-S6K (p < 0.001) (Supplemental Data Table S2). Age was significantly lower for the high expression group than the low expression group for p-c-Met (63.6 versus 69.1 years old, respectively; p = 0.049). In summary, we found that the group with high levels of protein expression and activation tended to have a more advanced stage at the time of NSCLC diagnosis and was less likely to have undergone a surgical resection.
Table 1.
Demographics table for NSCLC patients.
| Sex | Total | Percentage |
|---|---|---|
| Male | 50 | 45.9% |
| Female | 58 | 53.2% |
| Unknown | 1 | 0.9% |
| Race | ||
| White | 102 | 93.6% |
| Non-white | 4 | 3.7% |
| Unknown | 3 | 2.7% |
| Smoking status | ||
| Current | 45 | 41.3% |
| Former | 48 | 44.0% |
| Never | 9 | 8.3% |
| Unknown | 7 | 6.4% |
| Smoking history | ||
| >30 pack-years | 58 | 53.2% |
| <30 pack-years | 28 | 25.7% |
| Never | 9 | 8.3% |
| Unknown | 14 | 12.8% |
| Histology | ||
| Adenocarcinoma | 94 | 86.2% |
| Mixed adenosquamous | 4 | 3.7% |
| Squamous | 6 | 5.5% |
| Bronchioalveolar/other | 5 | 4.6% |
| Cancer stage | ||
| Stage I | 22 | 20.2% |
| Stage II | 8 | 7.3% |
| Stage III | 21 | 19.3% |
| Stage IV | 54 | 49.5% |
| Unknown | 4 | 3.7% |
| Treatment | ||
| Received | 91 | 83.5% |
| Not received | 18 | 16.5% |
NSCLC, non-small cell lung cancer.
Sample distribution
Protein expression/activity levels were not normally distributed, as indicated by both the Kolmogirov–Smirnov and the Shapiro–Wilk tests.
Protein expression patterns and profile correlations
In general, most of the proteins were expressed over a broad range, from a minimum of 0 to a maximum of 300. Total beta-catenin was more heavily expressed in all samples, with a minimum expression of 100 and maximum of 300. p-c-Met and p-mTOR had a minimum expression of 0 and a maximum expression of 280. Mean expression was higher for S6K and p-S6K than mTOR and p-mTOR, a possible result of signal amplification and the capacity of one molecule of p-mTOR to activate multiple p-S6Ks. Standard deviation was lower on average for the total proteins, with larger standard deviations seen in activated/phosphorylated proteins. S6K and p-S6K demonstrated relatively higher levels of expression with the lowest variation. As S6K was the furthest downstream marker assayed in this study, this result might represent a confluence of various cell signaling pathways converging on this signaling protein (Figure 1).
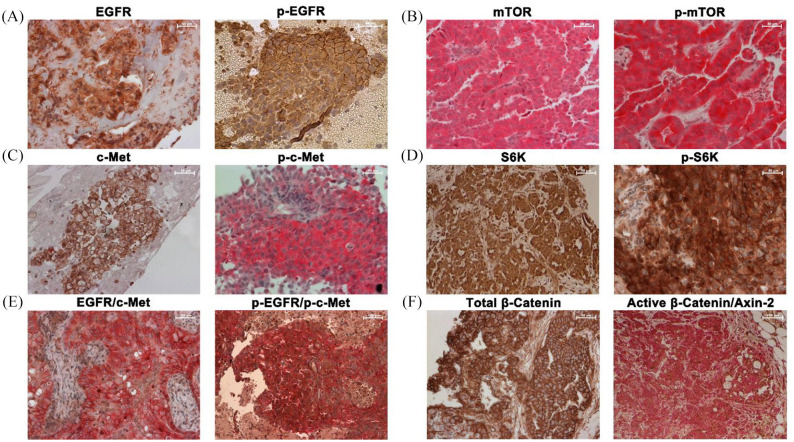
Figure 1.
Immunohistochemistry of different biomarkers in tissue biopsies of patients with NSCLC. (a) EGFR and p-EGFR. (b) mTOR and p-mTOR. (c) c-Met and p-c-Met. (d) S6K and p-S6K. (e) Double staining for EGFR/c-Met and p-EGFR/p-c-Met. EGFR/p-EGFR is brown, c-Met/p-c-Met is red. (f) Total beta-catenin and double staining of active beta-catenin/Axin2. Beta-catenin is brown and Axin2 is red. NSCLC, non-small cell lung cancer.
We observed that EGFR expression was correlated with downstream mTOR expression and p-S6K activity (rp = 0.304, p = 0.008; rp = 0.479, p < 0.0001, respectively), while p-EGFR expression was correlated with downstream p-mTOR and p-S6K activity (rp = 0.311, p = 0.0067; rp = 0.505, p < 0.0001, respectively). EGFR and p-EGFR expression were also correlated with Axin2 (rp = 0.473, p < 0.0001; rp = 0.391, p = 0.0005, respectively). In addition, p-EGFR/p-c-Met co-localization was correlated with increased S6K expression and p-mTOR/p-S6K activation (rp = 0.407, p = 0.0018; rp = 0.436, p = 0.0006; rp = 0.530, p < 0.0001, respectively).
Total beta-catenin was correlated with total and active EGFR and S6K as well as total Axin2 (all rp ⩾ 0.290 and all p-values ⩽0.0117). Active beta-catenin expression was correlated with both c-Met and Axin2 (rp = −0.283, p = 0.014; rp = −0.375, p = 0.0008, respectively), which may suggest c-Met could play a role in activating beta-catenin and subsequently stimulating its negative regulator, Axin2, as a negative feedback loop.41 Finally, the strong correlations of Axin2 with the majority of total and active EGFR-mTOR-S6K signaling proteins (all rp ⩾ 0.308 and all p-values <0.05) may imply that activation of the EGFR-mTOR signaling axis may lead to downregulation of Wnt signaling through the negative regulatory effects of Axin2 (Table 2).
Table 2.
Spearman and Pearson correlations between selected biomarkers.
| EGFR | c-Met | S6K | mTOR | Total β-Catenin | Axin-2 | p-EGFR | p-c-Met | p-S6K | p-mTOR | Active β-Catenin |
EGFR/ c-Met co-localization |
p-EGFR/ p-c-Met co-localization |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EGFR |
rs
rp |
0.277 p = 0.0186 |
0.302 p = 0.0085 |
0.326 p = 0.0043 |
0.419 p = 0.0002 |
0.460 p < 0.0001 |
|||||||
| c-Met |
rs
rp |
0.235 p = 0.0427 |
0.860 p < 0.0001 |
||||||||||
| S6K |
rs
rp |
0.297 p = 0.0096 |
0.431 p = 0.0002 |
0.255 p = 0.0295 |
0.236 p = 0.0442 |
0.467 p < 0.0001 |
0.335 p = 0.0033 |
0.302 p = 0.0193 |
0.451 p = 0.0005 |
||||
| mTOR | 0.304 p = 0.0080 |
rs
rp |
0.289 p = 0.0120 |
||||||||||
| Total β-catenin | 0.331 p = 0.0037 |
0.290 p = 0.0117 |
rs
rp |
0.432 p = 0.0001 |
0.424 p = 0.0002 |
0.537 p < 0.0001 |
|||||||
| Axin-2 | 0.473 p < 0.0001 |
0.365 p = 0.0016 |
0.444 p < 0.0001 |
rs
rp |
0.370 p = 0.0011 |
0.549 p < 0.0001 |
0.314 p = 0.0061 |
-0.376 p = 0.0008 |
0.331 p = 0.0098 |
0.360 p = 0.0055 |
|||
| p-EGFR | 0.238 p = 0.0412 |
0.393 p = 0.0005 |
0.391 p = 0.0005 |
rs
rp |
0.477 p < 0.0001 |
0.273 p = 0.0179 |
0.553 p < 0.0001 |
||||||
| p-c-Met |
rs
rp |
0.720 p < 0.0001 |
|||||||||||
| p-S6K | 0.479 p < 0.0001 |
0.405 p = 0.0003 |
0.522 p < 0.0001 |
0.570 p < 0.0001 |
0.505 p < 0.0001 |
rs
rp |
0.373 p = 0.0008 |
0.540 p < 0.0001 |
|||||
| p-mTOR | 0.326 p = 0.0044 |
0.308 p = 0.0072 |
0.311 p = 0.0067 |
0.369 p = 0.008 |
rs
rp |
0.417 p = 0.0008 |
0.465 p = 0.0002 |
||||||
| Active β-Catenin |
-0.283 p = 0.0140 |
0.289 p = 0.0119 |
-0.375 p = 0.0008 |
rs
rp |
-0.325 p = 0.0114 |
||||||||
| EGFR/ c-Met co-localization |
0.789 p < 0.0001 |
0.286 p = 0.0268 |
0.289 p = 0.0239 |
-0.393 p = 0.0019 |
rs
rp |
||||||||
| p-EGFR/ p-c-Met co-localization |
0.407 p = 0.0018 |
0.320 p = 0.0143 |
0.495 p < 0.0001 |
0.691 p < 0.0001 |
0.530 p < 0.0001 |
0.436 p = 0.0006 |
rs
rp |
Statistically significant Spearman correlation coefficients, rs, (upper diagonal) and statistically significant Pearson correlation coefficients, rp, (lower diagonal) for the relationships among the EGFR, c-Met, mTOR-S6K, and Wnt pathway proteins. Pearson correlations were used to assess the linear associations of the proteins. As some of the relationships were non-linear, Spearman’s Rho, which can detect both linear and logarithmic relationships, was used to assess correlations between different biomarkers. p-values (two-tailed) for each statistically significant coefficient are provided in red.
Clinical outcomes and protein expression in patients not stratified for cancer stage
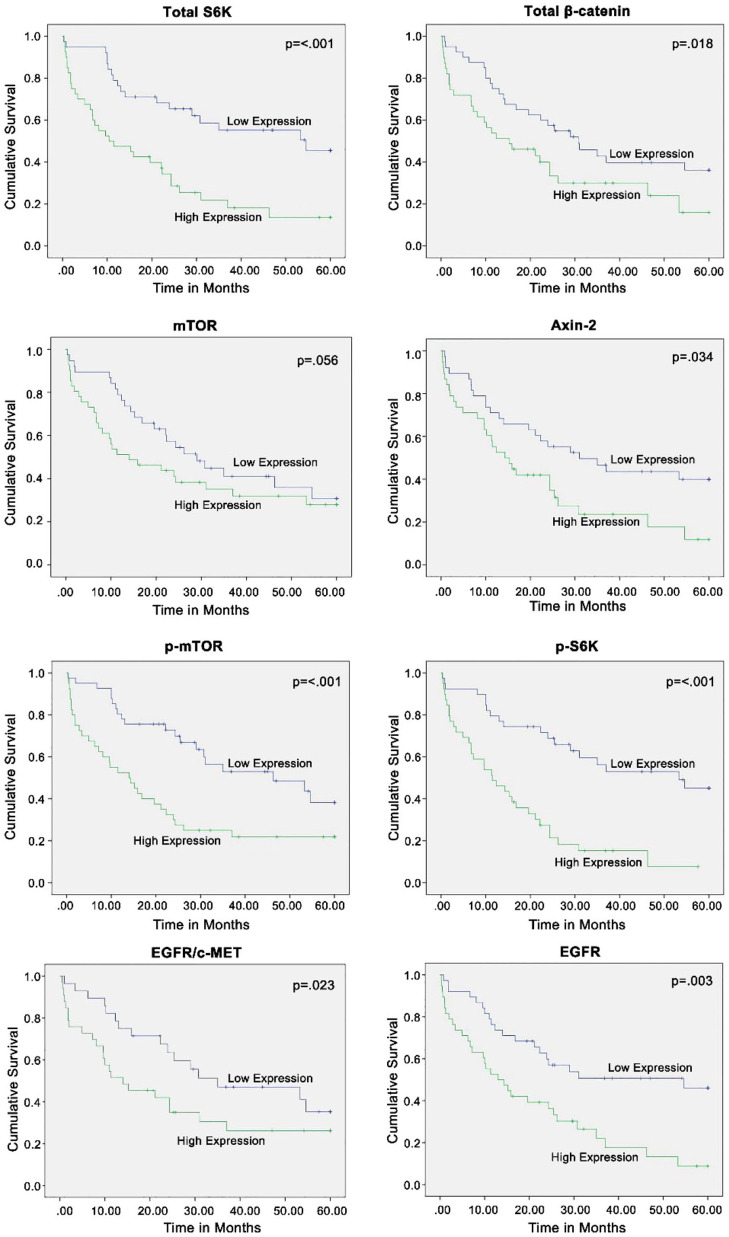
KM analysis comparing high and low expression groups revealed that the elevation in expression and activation of several proteins at the time of diagnosis was associated with significantly worse patient prognosis. In particular, for the following proteins, high expression was associated with shorter median survival compared with low expression: EGFR (13.0 months versus 54.6 months, respectively; p = 0.003); mTOR (14.0 months versus 29.0 months, respectively; p = 0.056); p-mTOR (14.0 months versus 46.3 months, respectively; p < 0.001); S6K (10.4 months versus 54.6 months, respectively; p < 0.001); p-S6K (11.5 months versus 53.3 months, respectively; p < 0.001); beta-catenin (15.2 months versus 30.8 months, respectively; p = 0.018); Axin2 (14.3 months versus 31.0 months, respectively; p = 0.034); and EGFR/c-Met co-localization (14.0 months versus 35.0 months, respectively; p = 0.023) (Figure 2). Contrarily, elevated expression and activation of c-Met, p-EGFR, p-c-Met, active beta-catenin, and p-EGFR/p-c-Met co-localization was not associated with a significant difference in median survival time compared with those with low expression (Supplemental Data Figure S1).
Figure 2.
Kaplan–Meier survival analyses not stratified by cancer staging demonstrated that increased expression levels of total S6K, total beta-catenin, Axin2, p-mTOR, p-S6K, EGFR/c-Met co-expression, and EGFR were significantly associated with shorter median survival. Median expression level of each biomarker was chosen as the cut-off between the high and low expression groups. Equality of survival distributions for the two expression groups were studied using the log rank test. p ⩽ 0.05 is significant.
In summary, amplified expression of EGFR and elevated co-localization of EGFR/c-Met, along with amplified expression and activation of the mTOR-S6K pathway at the time of diagnosis, were associated with worse prognosis. Increased total beta-catenin expression was also associated with worse prognosis. Axin2, which may serve as an indicator of downregulated active beta-catenin, was similarly associated with a worse prognosis.
Early versus late-stage disease analysis
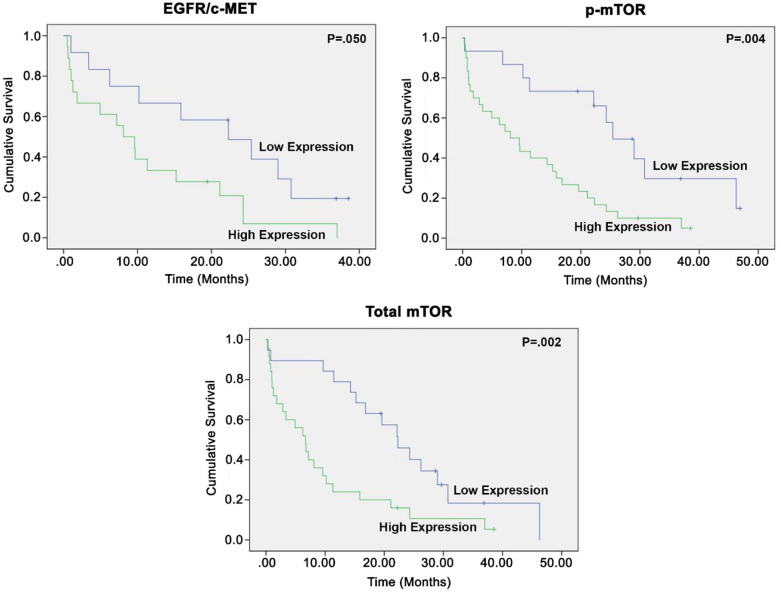
In response to the concurrent observations that patients with high expression levels were more likely to be diagnosed at late stages and less likely to undergo surgical resection, we performed a KM analysis of the survival data. To do this analysis, we stratified survival into early (I–IIIa) and late (IIIb–IV) disease categories, depending on stage at the time of diagnosis. Patients with late-stage disease who had high levels of mTOR and p-mTOR protein expression had shorter median survival compared with those with late-stage disease and lower levels of protein expression (mTOR: 6.7 versus 22.3 months, p = 0.002; p-mTOR: 8.1 versus 25.4 months, p = 0.004, respectively) (Figure 3). Patients with late-stage disease who had increased EGFR/c-Met co-localization also had shorter median survival compared with those with late-stage disease and low levels of co-localization (8.1 versus 22.3 months, p = 0.050) (Figure 3). In early-stage disease, non-significant differences in patient survival were observed between the same high and low expression groups as described previously (Supplemental Data Figure S2). This may be due to a lack of significant activation of these signaling pathways in early-stage disease.
Figure 3.
Kaplan–Meier survival analyses stratified by stage IIIB–IV NSCLC demonstrated that increased co-expression of EGFR/c-Met and increased expression of p-mTOR and total mTOR were significantly associated with shorter median survival. Median expression level of each biomarker was chosen as the cut-off between the high and low expression groups. Equality of survival distributions for the two expression groups were studied using the log rank test. p ⩽ 0.05 is significant. NSCLS, non-small cell lung cancer.
In summary, after controlling for stage of disease at diagnosis, high levels of mTOR expression, p-mTOR activation, and EGFR/c-Met co-localization were associated with a significantly shorter median survival compared with those with lower expression/activation/co-localization.
Discussion
In general, the diagnosis of NSCLC correlates with poor prognosis.42 The current availabilities of targeted TKI therapy and guideline-directed screening for molecular markers have led to a significant improvement in progression-free survival.9,10,13,14 Despite an advanced understanding of NSCLC tumor biology and treatment, NSCLC-attributed mortality rates remain high.1,17 This observation has prompted further investigation into the tumor biology of NSCLC, and multiple mechanisms which appear to play a role in NSCLC pathogenesis and resistance to TKIs have been elucidated here.4,6–8,12,15,16
We observed that EGFR and p-EGFR expression were correlated with downstream expression and activity of mTOR-S6K and Axin2. Interestingly, p-EGFR/p-c-Met co-localization was strongly correlated with increased mTOR-S6K activation. This implies that signaling through co-localized activated EGFR/c-Met may be a potent activator of the mTOR-S6K pathway.18,19 Total beta-catenin was correlated with various proteins in the EGFR and mTOR-S6K pathways in addition to Axin2, a negative regulator of the Wnt signaling pathway.41 Finally, strong correlations of Axin2 with the majority of total and active EGFR-mTOR-S6K signaling proteins may imply that activation of this signaling axis may lead to downregulation of Wnt signaling through the negative regulatory effects of Axin2 and by promotion of phosphorylation and degradation of beta-catenin. These associations suggest that in vivo p-EGFR/p-c-Met co-localization may be associated with increased downstream signaling via activation of the mTOR-S6K signaling pathway, particularly in patients with advanced disease at time of diagnosis and in those who lack EGFR tyrosine kinase domain mutations/ALK rearrangements.
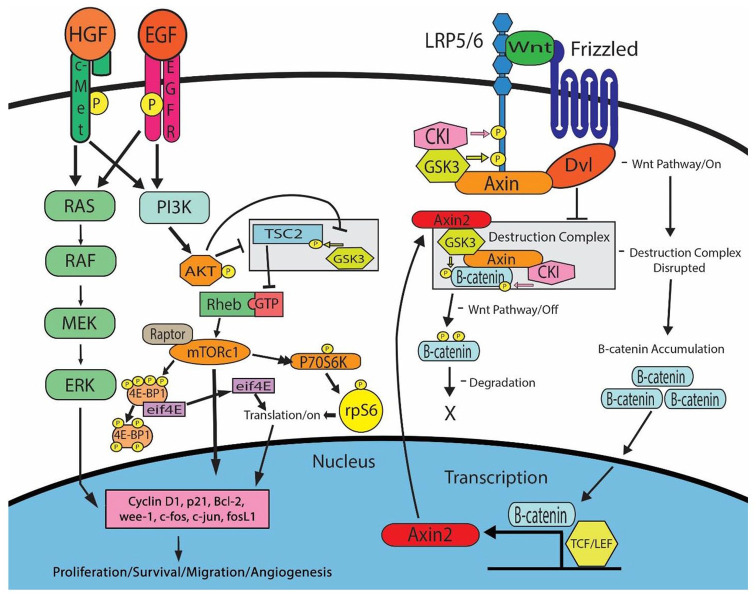
Furthermore, studies suggest that the mTOR pathway may play a key role in NSCLC pathogenesis.12,15 Increased downstream mTOR activity has been associated with EGFR, PI3K/Akt, and c-Met as well as various other signaling proteins (Figure 4).6,15,43 This proposed mechanism is consistent with data published from the The Cancer Genome Atlas (TCGA) database evaluating tumors across multiple cancer types, including more than 500 lung adenocarcinomas, which suggests that EGFR, PI3K, and Wnt pathways play a significant role in oncogenic signaling utilizing crosstalk through positive and negative regulators, including Axin2 and GSK-3.44 Our previous mechanistic studies in H2170 and H358 cell lines made resistant to EGFR and c-MET tyrosine kinase inhibitors found 2–8-fold upregulation of mTOR (p-S6K, and p-mTOR) and Wnt (active beta-catenin) signaling proteins.18 Additional studies supporting our proposed mechanism were completed in H2170 and H1975 TKI-resistant cell lines and found key Wnt and mTOR proteins to be differentially modulated, including upregulation of active beta-catenin and p-GSK-3β in H2170 cells.19 In H1975 cells combination treatment with an mTOR inhibitor (everolimus) and erlotinib resulted in synergistic cell growth inhibition.19 In clinical studies, patients with early-stage disease, high mTOR activity, and histological evidence of angioinvasion were reported to have poor prognosis.26 Here, we observed that patients across all stages of NSCLC who expressed high levels of EGFR, mTOR, p-mTOR, S6K, p-S6K, total beta-catenin, Axin2, and EGFR/c-Met co-localization at the time of diagnosis had a significantly worse prognosis compared with those with low expression levels. Alternatively, those with higher expression and activation of the previously mentioned proteins had an increased likelihood of advanced disease diagnosis and were less likely to undergo surgical resection, two potential contributions to worse prognosis. Even after controlling for stage at diagnosis, patients with elevated levels of mTOR and p-mTOR and co-localized EGFR/c-Met were still found to have a worse prognosis.
Figure 4.
A proposed mechanism for the EGFR/c-Met-mTOR/S6K signaling axis in NSCLC. Both EGFR and c-Met expression can lead to activation of the PI3K, mTOR, and MAPK pathways, resulting in increased cell proliferation, survival, migration, and angiogenesis through downstream targets such as Cyclin D1, p21, Bcl-2, wee-1, c-fos, c-jun and fosL1.45–49 In addition, Wnt pathway activation leads to the recruitment of Axin2, CKI, and GSK-3, allowing for nuclear accumulation of active beta-catenin and enhanced transcription. Finally, it is shown that upregulation of Axin2, a negative regulator of the Wnt signaling pathway, can bind to GSK3 and lead to phosphorylation and degradation of beta-catenin. NSCLC, non-small cell lung cancer.
The association of mTOR activity with poor prognosis may be due to a variety of mechanisms. The mTOR protein itself is a well-characterized mitogenic signaling protein that regulates downstream molecules such as cyclin D, an integral player in cell cycle progression at the G1-S checkpoint.50 Therefore, no matter the stage, a NSCLC tumor expressing increased levels of total and active mTOR may be inherently more capable of proliferation and thus be more clinically aggressive. Conversely, it may be possible that amplified protein expression or activation is a mere reflection of disease that has already advanced. The mTOR pathway also lies downstream from several proteins involved in proliferation and survival, as well as other mediator proteins, which utilize mTOR signaling to achieve their effects.51,52 This suggests that mTOR activity, specifically at the time of diagnosis, may reflect the downstream summation of multiple upregulated signaling pathways. If true, this could explain why some studies have found no correlation between increased mTOR and prognosis, while others have.53,54 It is also a possibility that upstream, activating mediators of the mTOR pathway drive disease phenotypes by activating alternative or complementary pathways. By way of this reasoning, perhaps elevated EGFR/c-Met co-localization leading to upregulated mTOR signaling is associated with worse prognosis at the time of diagnosis, whereas alternative upstream pathways leading to mTOR may not necessarily be so. Currently, few studies have evaluated whether, in the presence of increased mTOR-S6K, variations in signaling proteins upstream of mTOR-S6K are better predictors of prognosis. Our study has investigated this question.
As seen in the results of this study, EGFR expression and activation, when co-expressed with c-Met, demonstrated a statistically significant positive correlation with mTOR activation. This suggests that the mTOR pathway acts as a downstream mediator through which EGFR signaling plays a role in NSCLC pathogenesis.6,7,12,15 This model is further supported by the observation that expression of p-S6K, a downstream mTOR pathway protein, is positively correlated with EGFR expression and activation. It is also notable that studies have observed EGFR-c-MET heterodimerization,55 co-precipitation56 and also that both c-Met (61%) and EGFR (80%) are highly expressed in NSCLC tumors.39,57 Earlier studies have also shown a synergistic effect on inhibition of proliferation in the presence of EGFR and c-MET inhibitors leading to synergistic inhibition of downstream signaling.25 Therefore, double IHC staining showing the co-localization of p-EGFR and p-c-MET in patient tumors is suggestive of crosstalk and increased activation of both receptor tyrosine kinases. In addition, the correlation of increased p-EGFR/p-c-MET with upregulation of downstream p-mTOR and p-S6K in human tissues offers clinical data to support our prior mechanistic pre-clinical studies reporting that EGFR and c-MET may signal through mTOR/S6K to mediate their oncogenic potential.18
Additional studies should be done to characterize mTOR expression profiles completely in a larger NSCLC cohort and determine their clinical relevance. More studies are also needed to validate prospectively whether mTOR and p-mTOR testing in advanced NSCLC may be a viable prognostic biomarker for clinicians at diagnosis. Furthermore, as targeted mTOR inhibitors become available for the standard of care treatment of NSCLC, future studies may elucidate whether testing for mTOR and p-mTOR may provide a level of personalized medicine through predicting which patients are likely to benefit from mTOR inhibitor therapy. Finally, among the biomarkers investigated, molecular alterations have been found in EGFR, c-MET, Axin-2, and beta-catenin.58–61 In the future, it would be interesting to study if molecular alterations contribute to modulation in expression of these biomarkers.
Conclusion
We conclude that EGFR/c-Met co-expression and the correlated activation of the mTOR pathway represents a possible mechanism for a more aggressive NSCLC tumor phenotype and may be clinically associated with shorter median survival in late-stage NSCLC patients (Figure 3). In addition, Axin2 may also be a predictive biomarker for lung cancer prognosis.
Supplemental Material
Supplemental material, Supplementary_Data_Final for EGFR/c-Met and mTOR signaling are predictors of survival in non-small cell lung cancer by Zachary D Crees, Caleb Shearrow, Leo Lin, Jennifer Girard, Kavin Arasi, Aayush Bhoraskar, Joseph Berei, Adam Eckburg, Austin D. Anderson, Christian Garcia, Ariana Munger, Sunil Palani, Thomas J Smith, Shylendra B Sreenivassappa, Connie Vitali, Odile David and Neelu Puri in Therapeutic Advances in Medical Oncology
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number R21CA158965-01A1(http://www.nih.gov) and Community Grant from the Community Foundation of Northern Illinois (Grant ID # I3736) to Neelu Puri. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ORCID iDs: Joseph Berei  https://orcid.org/0000-0002-8278-128X
https://orcid.org/0000-0002-8278-128X
Adam Eckburg  https://orcid.org/0000-0002-1180-6240
https://orcid.org/0000-0002-1180-6240
Christian Garcia  https://orcid.org/0000-0001-6365-1079
https://orcid.org/0000-0001-6365-1079
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Zachary D Crees, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Caleb Shearrow, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Leo Lin, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Jennifer Girard, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Kavin Arasi, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Aayush Bhoraskar, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Joseph Berei, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Adam Eckburg, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Austin D. Anderson, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA
Christian Garcia, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Ariana Munger, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Sunil Palani, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, IL, USA.
Thomas J Smith, College of Education, Northern Illinois University, Dekalb, IL, USA.
Shylendra B Sreenivassappa, Department of Hematology/Oncology, OSF Saint Anthony Medical Center, Rockford, IL, USA.
Connie Vitali, Department of Pathology, University of Illinois College of Medicine at Rockford IL, USA.
Odile David, Department of Pathology, University of Illinois College of Medicine at Chicago, IL, USA.
Neelu Puri, Department of Biomedical Sciences, University of Illinois College of Medicine at Rockford, 1601 Parkview Avenue, Room Number E-632, Rockford, IL 61107, USA.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. [DOI] [PubMed] [Google Scholar]
- 2. Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018; 29: iv192–iv237. [DOI] [PubMed] [Google Scholar]
- 3. Blandin Knight S, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol 2017; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res 2006; 12: 6494–6501. [DOI] [PubMed] [Google Scholar]
- 5. Desai A, Menon SP, Dy GK. Alterations in genes other than EGFR/ALK/ROS1 in non-small cell lung cancer: trials and treatment options. Cancer Biol Med 2016; 13: 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dobashi Y, Suzuki S, Kimura M, et al. Paradigm of kinase-driven pathway downstream of epidermal growth factor receptor/Akt in human lung carcinomas. Hum Pathol 2011; 42: 214–226. [DOI] [PubMed] [Google Scholar]
- 7. Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002; 110: 163–175. [DOI] [PubMed] [Google Scholar]
- 8. Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005; 352: 786–792. [DOI] [PubMed] [Google Scholar]
- 9. Leung L, Mok TS, Loong H. Combining chemotherapy with epidermal growth factor receptor inhibition in advanced non-small cell lung cancer. Ther Adv Med Oncol 2012; 4: 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018; 142: 321–346. [DOI] [PubMed] [Google Scholar]
- 11. Nan X, Xie C, Yu X, et al. EGFR TKI as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer. Oncotarget 2017; 8: 75712–75726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Price DJ, Grove JR, Calvo V, et al. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science 1992; 257: 973–977. [DOI] [PubMed] [Google Scholar]
- 13. Riely GJ, Politi KA, Miller VA, et al. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res 2006; 12: 7232–7241. [DOI] [PubMed] [Google Scholar]
- 14. Sattler M, Abidoye O, Salgia R. EGFR-targeted therapeutics: focus on SCCHN and NSCLC. Sci World J 2008; 8: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang L, Yue W, Zhang L, et al. mTOR and PTEN expression in non-small cell lung cancer: analysis by real-time fluorescence quantitative polymerase chain reaction and immunohistochemistry. Surg Today 2012; 42: 419–425. [DOI] [PubMed] [Google Scholar]
- 16. Xu Y, Liu H, Chen J, et al. Acquired resistance of lung adenocarcinoma to EGFR-tyrosine kinase inhibitors gefitinib and erlotinib. Cancer Biol Ther 2010; 9: 572–582. [DOI] [PubMed] [Google Scholar]
- 17. Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 2008; 14: 2895–2899. [DOI] [PubMed] [Google Scholar]
- 18. Fong JT, Jacobs RJ, Moravec DN, et al. Alternative signaling pathways as potential therapeutic targets for overcoming EGFR and c-Met inhibitor resistance in non-small cell lung cancer. PLoS One 2013; 8: e78398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Botting GM, Rastogi I, Chhabra G, et al. Mechanism of resistance and novel targets mediating resistance to EGFR and c-Met tyrosine kinase inhibitors in non-small cell lung cancer. PLoS One 2015; 10: e0136155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson M, Koukoulis G, Kochhar K, et al. Selective tumorigenesis in non-parenchymal liver epithelial cell lines by hepatocyte growth factor transfection. Cancer Lett 1995; 96: 37–48. [DOI] [PubMed] [Google Scholar]
- 21. Birchmeier C, Birchmeier W, Gherardi E, et al. Met, metastasis, motility and more. Nat Rev Mol 2003; 4: 915–925. [DOI] [PubMed] [Google Scholar]
- 22. Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 1995; 80: 179–185. [DOI] [PubMed] [Google Scholar]
- 23. Graziani A, Gramaglia D, Cantley LC, et al. The tyrosine-phosphorylated hepatocyte growth factor/scatter factor receptor associates with phosphatidylinositol 3-kinase. J Biol Chem 1991; 266: 22087–22090. [PubMed] [Google Scholar]
- 24. Monga SP, Mars WM, Pediaditakis P, et al. Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes. Cancer Res 2002; 62: 2064–2071. [PubMed] [Google Scholar]
- 25. Puri N, Salgia R. Synergism of EGFR and c-Met pathways, cross-talk and inhibition, in non-small cell lung cancer. J Carcinog 2008; 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gately K, Al-Alao B, Dhillon T, et al. Overexpression of the mammalian target of rapamycin (mTOR) and angioinvasion are poor prognostic factors in early stage NSCLC: a verification study. Lung Cancer 2012; 75: 217–222. [DOI] [PubMed] [Google Scholar]
- 27. Poomakkoth N, Issa A, Abdulrahman N, et al. p90 Ribosomal S6 kinase: a potential therapeutic target in lung cancer. J Transl Med 2016; 14: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu ZX, Sun RF, Mo XM, et al. The p70S6K specific inhibitor PF-4708671 impedes non-small cell lung cancer growth. PLoS One 2016; 11: e0147185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Don AS, Zheng XF. Recent clinical trials of mTOR-targeted cancer therapies. Rev Recent Clin Trials 2011; 6: 24–35. [DOI] [PubMed] [Google Scholar]
- 30. Xie J, Wang X, Proud CG. mTOR inhibitors in cancer therapy. F1000 Res 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waqar SN, Baggstrom MQ, Morgensztern D, et al. A phase I trial of temsirolimus and pemetrexed in patients with advanced non-small cell lung cancer. Chemotherapy 2016; 61: 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell 2012; 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- 33. Maher MT, Flozak AS, Hartsell AM, et al. Issues associated with assessing nuclear localization of N-terminally unphosphorylated beta-catenin with monoclonal antibody 8E7. Biol Direct 2009; 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Persad A, Venkateswaran G, Hao L, et al. Active beta-catenin is regulated by the PTEN/PI3 kinase pathway: a role for protein phosphatase PP2A. Genes Cancer 2016; 7: 368–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakayama S, Sng N, Carretero J, et al. beta-catenin contributes to lung tumor development induced by EGFR mutations. Cancer Res 2014; 74: 5891–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suzuki M, Shigematsu H, Nakajima T, et al. Synchronous alterations of Wnt and epidermal growth factor receptor signaling pathways through aberrant methylation and mutation in non small cell lung cancer. Clin Cancer Res 2007; 13: 6087–6092. [DOI] [PubMed] [Google Scholar]
- 37. Illinois Department of Public Health. County Cancer Incidence Report Illinois, 2020. http://www.idph.state.il.us/cancer/statistics.htm (accessed 28 May 2020).
- 38. Centers for Disease Control and Prevention. Leading cancer cases and deaths, male and female, 2020. http://wonder.cdc.gov. (accessed 28 May 2020).
- 39. Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res 2005; 65: 1479–1488. [DOI] [PubMed] [Google Scholar]
- 40. Nitadori J, Ishii G, Tsuta K, et al. Immunohistochemical differential diagnosis between large cell neuroendocrine carcinoma and small cell carcinoma by tissue microarray analysis with a large antibody panel. Am J Clin Pathol 2006; 125: 682–692. [DOI] [PubMed] [Google Scholar]
- 41. Jho EH, Zhang T, Domon C, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 2002; 22: 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cetin K, Ettinger DS, Hei YJ, et al. Survival by histologic subtype in stage IV nonsmall cell lung cancer based on data from the surveillance, epidemiology and end results program. Clin Epidemiol 2011; 3: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vicary GW, Roman J. Targeting the mammalian target of rapamycin in lung cancer. Am J Med Sci 2016; 352: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 2018; 173: 321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Law M, Forrester E, Chytil A, et al. Rapamycin disrupts cyclin/cyclin-dependent kinase/p21/proliferating cell nuclear antigen complexes and cyclin D1 reverses rapamycin action by stabilizing these complexes. Cancer Res 2006; 66: 1070–1080. [DOI] [PubMed] [Google Scholar]
- 46. Grimaldi A, Balestrieri ML, D’Onofrio N, et al. The synergistic effect of everolimus and chloroquine on endothelial cell number reduction is paralleled by increased apoptosis and reduced autophagy occurrence. PLoS One 2013; 8: e79658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 2017; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aleem E, Arceci RJ. Targeting cell cycle regulators in hematologic malignancies. Front Cell Dev Biol 2015; 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lau EY, Lo J, Cheng BY, et al. Cancer-associated fibroblasts regulate tumor-initiating cell plasticity in hepatocellular carcinoma through c-Met/FRA1/HEY1 signaling. Cell Rep 2016; 15: 1175–1189. [DOI] [PubMed] [Google Scholar]
- 50. Foster DA, Yellen P, Xu L, et al. Regulation of G1 cell cycle progression: distinguishing the restriction point from a nutrient-sensing cell growth checkpoint(s). Genes Cancer 2010; 1: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cargnello M, Tcherkezian J, Roux PP. The expanding role of mTOR in cancer cell growth and proliferation. Mutagenesis 2015; 30: 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li L, Liu D, Qiu ZX, et al. The prognostic role of mTOR and p-mTOR for survival in non-small cell lung cancer: a systematic review and meta-analysis. PLoS One 2015; 10: e0116771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dhillon T, Mauri FA, Bellezza G, et al. Overexpression of the mammalian target of rapamycin: a novel biomarker for poor survival in resected early stage non-small cell lung cancer. J Thorac Oncol 2010; 5: 314–319. [DOI] [PubMed] [Google Scholar]
- 55. Ortiz-Zapater E, Lee RW, Owen W, et al. MET-EGFR dimerization in lung adenocarcinoma is dependent on EGFR mutations and altered by MET kinase inhibition. PLoS One 2017; 12: e0170798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jo M, Stolz DB, Esplen JE, et al. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem 2000; 275: 8806–8011. [DOI] [PubMed] [Google Scholar]
- 57. Kanematsu T, Yano S, Uehara H, et al. Phosphorylation, but not overexpression, of epidermal growth factor receptor is associated with poor prognosis of non-small cell lung cancer patients. Oncol Res 2003; 13: 289–298. [DOI] [PubMed] [Google Scholar]
- 58. Saigi M, McLeer-Florin A, Pros E, et al. Genetic screening and molecular characterization of MET alterations in non-small cell lung cancer. Clin Transl Oncol 2018; 20: 881–888. [DOI] [PubMed] [Google Scholar]
- 59. Doglioni C, Piccinin S, Demontis S, et al. Alterations of beta-catenin pathway in non-melanoma skin tumors: loss of alpha-ABC nuclear reactivity correlates with the presence of beta-catenin gene mutation. Am J Clin Pathol 2003; 163: 2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cooper WA, Lam DC, O’Toole SA, et al. Molecular biology of lung cancer. J Thorac Dis 2013; 5 (Suppl. 5): S479–S490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanzaki H, Ouchida M, Hanafusa H, et al. Single nucleotide polymorphism of the AXIN2 gene is preferentially associated with human lung cancer risk in a Japanese population. Int J Mol Med 2006; 18: 279–284. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_Data_Final for EGFR/c-Met and mTOR signaling are predictors of survival in non-small cell lung cancer by Zachary D Crees, Caleb Shearrow, Leo Lin, Jennifer Girard, Kavin Arasi, Aayush Bhoraskar, Joseph Berei, Adam Eckburg, Austin D. Anderson, Christian Garcia, Ariana Munger, Sunil Palani, Thomas J Smith, Shylendra B Sreenivassappa, Connie Vitali, Odile David and Neelu Puri in Therapeutic Advances in Medical Oncology