Abstract
To develop non-opioid therapies for postoperative incisional pain, we must understand its underlying molecular mechanisms. In this study, we assessed global gene expression changes in dorsal root ganglia neurons in a model of incisional pain to identify pertinent molecular pathways. Male, Sprague–Dawley rats underwent infiltration of 1% capsaicin or vehicle into the plantar hind paw (n = 6–9/group) 30 min before plantar incision. Twenty-four hours after incision or sham (control) surgery, lumbar L4–L6 dorsal root ganglias were collected from rats pretreated with vehicle or capsaicin. RNA was isolated and sequenced by next generation sequencing. The genes were then annotated to functional networks using a knowledge-based database, Ingenuity Pathway Analysis. In rats pretreated with vehicle, plantar incision caused robust hyperalgesia, up-regulated 36 genes and downregulated 90 genes in dorsal root ganglias one day after plantar incision. Capsaicin pretreatment attenuated pain behaviors, caused localized denervation of the dermis and epidermis, and prevented the incision-induced changes in 99 of 126 genes. The pathway analyses showed altered gene networks related to increased pro-inflammatory and decreased anti-inflammatory responses in dorsal root ganglias. Insulin-like growth factor signaling was identified as one of the major gene networks involved in the development of incisional pain. Expression of insulin-like growth factor -2 and IGFBP6 in dorsal root ganglia were independently validated with quantitative real-time polymerase chain reaction. We discovered a distinct subset of dorsal root ganglia genes and three key signaling pathways that are altered 24 h after plantar incision but are unchanged when incision was made after capsaicin infiltration in the skin. Further exploration of molecular mechanisms of incisional pain may yield novel therapeutic targets.
Keywords: Dorsal root ganglia, insulin-like growth factor, incisional pain, postoperative pain, transcriptome
Introduction
Management of acute incisional pain is an essential component of perioperative care. Approximately 234 million major surgical procedures are performed every year worldwide,1 and approximately 20% of these are performed in the United States alone.2 Uncontrolled or postoperative pain is reported by approximately 50% of surgical patients.3,4 Opioids are the mainstay for perioperative pain management. However, they produce an array of harmful side effects including nausea,5 tolerance (requirement of an increased dose for analgesia),6 hyperalgesia,7 constipation,5 ileus,8 urinary retention,8 respiratory depression,5 and death in overdose cases.9 Importantly, patients receiving an opioid prescription after surgery were 44% more likely to become long-term opioid users compared with those who did not receive opioids.10,11 Thus, there is a need to develop non-opioid approaches for managing incisional pain.
The development of non-opioid therapies for incisional pain requires a comprehensive understanding of the underlying molecular mechanisms. For peripheral mechanisms of incisional pain, it is critical to identify molecular changes in primary afferent neurons that innervate incised tissues. In this regard, next-generation high-throughput sequencing (NGS) technology have been used to evaluate the gene expression profiles in several preclinical pain models12–16 but not in incisional pain. Because sensory neurons from different tissues respond uniquely to injury,17–19 it is necessary to assess incisional pain-related changes in gene expression.
A rat model of incisional pain has been extensively characterized, which includes behavioral phenotypes, nociceptor sensitization, and pharmacologic modulation.20–25 Additionally, our lab and others have reported that intra-plantar infiltration of capsaicin attenuates spontaneous pain behaviors and heat hyperalgesia.26,27 This phenomenon provides a unique opportunity to identify neural transcriptomes that are specifically induced during incisional pain. For example, incision-induced gene regulatory changes that are sensitive to capsaicin pretreatment are likely involved in the development of pain after incision, whereas those which are insensitive to capsaicin pretreatment are likely involved in processes unrelated to pain.
We, therefore, determined the effect of incisional pain on the transcriptional profile of the dorsal root ganglia (DRG) by performing whole-transcriptome next-generation RNA sequencing. We hypothesized that the acute incisional injury will dramatically alter gene transcription in the rat DRGs, and that we would identify novel pain pathways, which may yield novel therapeutic targets.
Methods
Animals
Adult male Sprague–Dawley rats (250–300 g) purchased from Charles River (USA) were used in this study. Two rats were housed together in one Plexiglass cage and all animals were maintained on a 12 h light/dark cycle with free access to food and water. All procedures were approved by the Institutional Animal Care and Use Committees at the University of Minnesota.
Surgical procedures
Rats were anesthetized with isoflurane for intra-plantar injection of capsaicin, vehicle, and plantar incision. Briefly, rats were placed in an induction chamber using 5% isoflurane in room air followed by 2% to 3% isoflurane via a nose cone after loss of the righting reflex. Once adequately anesthetized, the intended surgical site for incision (mid-plantar region of the right hind paw) was infiltrated subcutaneously with 200 µL of either 1% capsaicin (10 mg/mL) or vehicle (80% saline, 10% ethanol, 10% Tween-80 (Sigma-Aldrich, St. Louis, MO)) using a 1 mL syringe with a 25 G needle. Surgery consisted of a 20 mm longitudinal incision made through the skin and fascia of the plantar hind paw as previously described.23,25 The flexor digitorum brevis muscle was then elevated, stressed, and incised longitudinally with the muscle origin and insertion remaining intact. The skin was closed with two mattress 5–0 silk sutures. Animals were housed on soft bedding and allowed to recover. Sutures were removed on the third post-operative day. Sham control rats underwent anesthesia but not surgical incision.
Measurement of spontaneous foot lifting behaviors
Spontaneous foot lifting (SFL) was used as a measure of spontaneous pain as described previously.25 Rats were pre-treated with capsaicin (n = 5) or vehicle (n = 6) for 30 min prior to surgical incision. Frequency and duration of SFL were obtained longitudinally at 3 h and at 1, 2, 3, 4, 5, 7, 9, and 16 days after surgery. In brief, animals were placed in individual Plexiglass chambers on a mesh floor above an angled mirror and habituated for 30 min to minimize exploratory behavior during the assessment period. Spontaneous, rapid lifting of the incised hind paw was recorded with a hand tally counter and the duration of prolonged (>1 s) paw elevation was recorded with a stopwatch. Each rat was tested over four 5-min periods with 5 to 10 min resting intervals between each assessment. The sum frequency of SFL and sum duration of paw lifts were determined for each rat.
Measurement of hyperalgesia to heat
Sensitivity to heat was determined by measuring paw withdrawal latencies to heat (PWL). The same rats tested for SFL were used. PWL were determined 3, 2, and 1 day before surgery and at 3 h and 1, 2, 3, 4, 5, 7, 9, and 16 days after surgery. Rats were placed under Plexiglass boxes (23.6 cm × 13.8 cm × 13.7 cm) on a tempered glass floor (3-mm thickness) that was maintained at 30 °C and allowed to acclimate for 20 min. A focused radiant heat source (50 W projector lamp with an aperture diameter of 6 mm) underneath the glass floor was aimed at the plantar surface of the injured paw and withdrawal latencies were determined. PWLs were measured to the nearest 0.01 s by an automated system. The intensity of the heat was adjusted to produce withdrawal latencies in normal, naive rats of 10 to 12 s. Mean PWL for each rat was obtained from three trials with a 5-min inter-stimulus interval between trials. The experimenter collecting all behavioral data was blinded to treatment condition of rats; however, blinding was limited due to distinct changes in PWL due to capsaicin pretreatment.
Measurement of mechanical hyperalgesia
The same rats tested for SFL were used to test for mechanical paw withdrawal (PWT) 3, 2, and 1 day before surgery and at 3 h and 1, 2, 3, 4, 5, 7, 9, and 16 days after surgery. Rats were placed in individual Plexiglass chambers on a mesh floor and allowed to habituate for 30 min. Mechanical PWT was determined using an electronic von Frey aesthesiometer (IITC Life Science, Woodland Hills, CA). This consists of a handheld force transducer with a series of rigidity-graded, attachable 0.8 mm polypropylene tips. Starting with the least rigid tip, force was transversely applied for to the mid-plantar surface of the hind paw (adjacent to the site of surgical injury) until the occurrence of a rapid paw withdrawal response. Stimuli were applied for 2 to 3 s with an inter-stimulus interval of 5 min. The second and third trials were conducted using the tip that was two grades lower than the tip which elicited the first withdrawal response. Withdrawal thresholds (g) were expressed as the mean from three trials.
Immunohistochemistry and confocal laser scanning microscopy
The epidermal and dermal nerve fibers were assessed by immunohistochemistry as previously described by our group.28,29 In brief, rats were anesthetized with isoflurane as described above, and treated with 1% capsaicin (n = 3) or vehicle (n = 3) 30 min prior to the surgical incision. After 24 h, rats were euthanized with euthasol (390 mg/mL Pentobarbital), and after no righting reflex, 200 µL of Zamboni’s fixative was injected into the hind paw. The hind paw was removed and placed in Zamboni’s fixative at 4 °C for 24 h, then 20% sucrose at 4 °C for 24 h. A 3-mm punch biopsy tool was used to remove tissue from the edge of the incision. For sectioning, tissues were immersed in OCT mounting medium (Electron Microscopy Sciences, Hatfield, PA) frozen on a cryo-microtome and cut into 50 µm thick sections. Free floating tissue sections were incubated 1 h in blocking buffer (10% normal donkey serum/PBS/0.1% Triton X-100), then overnight in antibody buffer (3% normal donkey serum/PBS/0.1% Triton X-100) containing DAPI, rabbit anti-advillin, and goat anti-collagen type IV. Sections were rinsed with antibody buffer three times, 1 h each time, and then incubated for an additional day in Cy5-conjugated donkey anti-goat and Cy3-conjugated donkey anti-rabbit secondary antibodies. Sections were rinsed in antibody buffer three times, 1 h each time, and then mounted in hot liquid noble agar onto 22 mm2 coverslips that were dehydrated in 95%, 100% ethanol, and cleared with 100% methyl salicylate for 30 min each before placing in DPX mounting medium on slides. Fluorescent images were captured using a Nikon Ti2 laser scanning confocal microscope equipped with an oil immersion objective using Nikon Elements software. Digitized images were collected in successive frames of 0.5-µm serial optical sections (z-series) throughout the thickness of the sections and flattened into a single image. The immunostaining of type IV collagen was used to localize the dermo-epidermal boundary.
RNA sequencing
Thirty min prior to incisional surgery, rats were treated either with (n = 4) or without (n = 4) capsaicin (see above). The L4–L6 DRGs collected from control (sham) rats (n = 4) and rats at 24 h after plantar incision. Surgery was carried out in the afternoon between 3 and 5 pm. Total RNA isolated from DRGs (RNAqueous RNA isolation kit, Life Technologies Inc., Carlsbad, CA) were quantified and qualified by RiboGreen RNA quantification (Invitrogen, Carlsbad, CA) and an Agilent 2100 Bioanalyzer (Agilent Inc., Santa Clara, CA). Median RNA integrity score in samples was 7.9 (interquartile range, 7.825–8.275). The median mass input was 5698 ng (Supplemental Figure 3). A total of 100 ng of RNA samples with RIN values ≥8.0 were used for library construction using a TruSeq RNA v2 kit (Illumina, San Diego, CA). Libraries were size-selected ∼200-bp inserts and sequenced as a 50-bp pair end using the HiSeq2000 (Agilent Inc.).
Bioinformatics
2 × 50 bp FastQ paired end reads (n = 18.8 Million average per sample) were trimmed using Trimmomatic (v 0.33) enabled with the optional “-q” option; 3 bp sliding-window trimming from 3’ end requiring minimum Q30. Quality control on raw sequence data for each sample were performed with FastQC. Read mapping was performed via Hisat230 (v2.1.0) using the rat genome (rn6) as reference. Gene quantification was done via Feature Counts for raw read counts. Differentially expressed genes were identified using the edgeR31,32 (negative binomial) feature in CLCGWB (Qiagen, Redwood city, CA) using raw read counts. We filtered the generated list based on a minimum 1.5× Absolute Fold Change and false discovery rate corrected p < 0.05.
Gene networks mapping by Ingenuity Pathway Analysis
The differentially expressed transcripts were uploaded to Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA) to identify regulatory networks operant in rat DRGs. IPA provides an extensive description of biological processes, molecular functions, and cellular components to test for enrichment of associated gene.33 Analysis was restricted to 126 genes, whose expression was changed in the incised rats. Moreover, p value is calculated by determining the probability that a disease or biological function assigned to the data. To show the connectivity between genes, and to classify them according to the molecular mechanisms, differentially expressed genes are categorized by Medical Subject headings terms and proprietary ontology. The significance of the association is measured by the ratio of the number of genes from the dataset that map to the canonical pathway divided by the total number of known genes in that pathway.
Quantitative real-time PCR
The quantitative real-time PCR (qPCR) experiments were carried out with biological replicates (n = 3–5/group). In brief, 1 µg of total RNA was used to generate complementary DNA (cDNA; High Capacity cDNA reverse transcription kit; Life Technologies). Gene expressions were quantified using TaqMan Gene Expression Assay probes (Applied Biosystems, Foster City, CA) and a DNA analyzer (Quant Studio, Applied Biosystems Inc.). Samples were analyzed in technical duplicates. Fold changes are means of duplicates, normalized to β-actin (internal loading control). Taqman gene expression assays include Rn00565371_m1 (IGFBP6), Rn01454518_m1 (IGF2), and Rn00667869_m1 (Actb).
Statistical analysis
Two-way analyses of variance (ANOVAs) with repeated measures were used to compare differences in the frequency and duration of SFL, PWT, and PWL. Post hoc comparisons (Bonferroni) were used to determine differences between vehicle and capsaicin-treated groups at specific time points. For qPCR validation, interactions were analyzed using ANOVA or unpaired t-test for differences between groups. α was set at <0.05. For IPA comparison analysis, Fisher’s exact test was used to calculate a p-value for the association among genes in the data sets, pathways, and functions. The generated diseases or functional annotations or upstream regulators were ranked by the activation z score. An absolute z score of more than 2 was considered as significant. All values are expressed as means ± SDs
Results
Intra-plantar capsaicin infiltration attenuated pain behaviors after plantar incision and reduced innervation in the dermis and epidermis
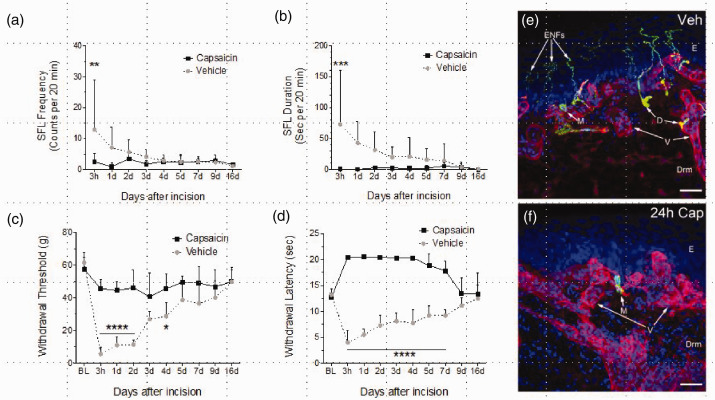
Following plantar incision, rats exhibited spontaneous pain-like behaviors and hypersensitivity to mechanical and heat stimuli that persisted for three to nine days (Figure 1(a) to (d)). Compared to vehicle (n = 6), intra-plantar injection with 1% capsaicin (n = 5) 30 min before surgical incision significantly attenuated SFL frequency (F2,11 = 2.0, p = 0.059); duration (F2,11 = 2.20, p = 0.037) and prevented the development of both mechanical (F2,11 = 11.2, p < 0.0001) and heat hyperalgesia (F2,11 = 28.9, p < 0.0001). Epidermal and dermal innervation following vehicle or 1% capsaicin infiltration was examined in rats 24 h following plantar incision. Representative confocal images of immune-stained skin from each of these groups are shown in Figure 1(e) and (f). A complete loss of epidermal and dermal nerve fibers were observed in hind paw skin pretreated with capsaicin (Figure 1(f), arrow).
Figure 1.
Capsaicin pretreatment attenuates the development of pain-like behaviors 24 h after plantar incision. The frequency (a) and duration (b) of plantar incision-induced spontaneous foot lifting behaviors (see methods) as well as mechanical (c) and heat (d) hyperalgesia were decreased in capsaicin-treated groups compared to vehicle-treated rats. The capsaicin pretreatment completely abolished epidermal and dermal nerves (f) compared with vehicle treated (e) rats 24 h after incision. In immunohistochemistry experiments, the tissue cryosections were immune-stained with anti-advillin antibody to label nerve fibers (green) and with anti-collagen type IV antibody to label basement membranes (red). Yellow indicates colocalization of the two antibodies. Cell nuclei were labeled with DAPI (blue). Each image in this figure represents a maximum projection of an image stack captured as 0.5 µm optical slices through the immune-stained tissue. Merkel cells (M) were unchanged with capsaicin treatment. No changes were evident in blood vessels (V) in the dermis. E indicates the location of epidermis and Drm indicates dermis. Scale bar = 50 microns.
Identification of differentially expressed genes in DRGs after plantar incision
To determine the transcriptomic changes in the primary afferent neurons that occur following plantar incision, we sequenced DRG transcripts using NGS. Three groups of rats were used such as control (sham) rats, rats with plantar incision, and rats treated with intra-plantar capsaicin 30 min prior to plantar incision. This approach allowed us to identify genes that are potentially involved in incisional pain behaviors because intra-plantar capsaicin completely denervated epidermal and dermal nerves around the incision and abolished hyperalgesia and SFL behaviors (Figure 1).
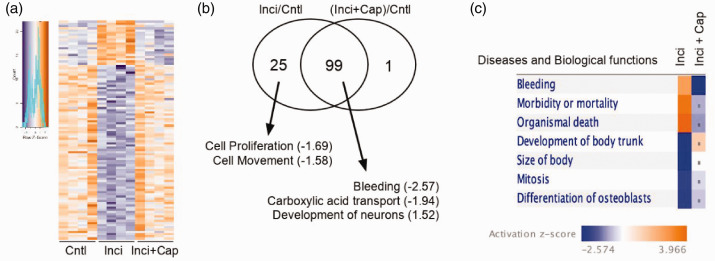
Using selection criteria that include an absolute fold change ≥1.5×, p < 0.05, and a false discovery rate <0.05, 126 genes were identified in the incised compared to sham group, including 36 that were up-regulated (incision vs. control; Table 1) and 90 that were down-regulated (incision vs. control; Table 2). Capsaicin pretreatment prevented transcriptional changes in 99 of 126 genes, including 20 of the upregulated and 79 of the down-regulated genes (Figure 2(a) and (b), Inci vs. Inci + Cap). These genes are displayed in the heat map (Figure 2(a)) to illustrate the degree of reproducibility among biological replicates. IPA mapped these genes onto specific diseases and biological functions (Figure 2(c)). Compared to sham control (Cntl) group, incised groups showed changes in the expression of genes associated with increased morbidity, organismal death, and decreased growth and differentiation (Figure 2(c), Inci). These changes in gene expression were abrogated with capsaicin pretreatment (Figure 2(c), Inci+Cap).
Table 1.
List of upregulated genes in the dorsal root ganglia after plantar incision.
| Gene Name | Gene symbol | log2(Fold change) | Location | Type(s) |
|---|---|---|---|---|
| Germ cell-less homolog 1 (Drosophila)-like | LOC302576 | 53.20 | Other | Other |
| Interferon-induced protein with tetratricopeptide repeats 1B | Ifit1 | 50.10 | Cytoplasm | Other |
| Inhibin subunit beta A | Inhba | 42.46 | Extracellular space | Growth factor |
| Interleukin 2 receptor subunit alpha | Il2ra | 34.32 | Plasma membrane | Transmembrane receptor |
| Chitinase domain containing 1 | Chid1 | 27.50 | Extracellular space | Other |
| Aquaporin 4 | Aqp4 | 13.53 | Plasma membrane | Transporter |
| Exocyst complex component 7 | Exoc7 | 9.69 | Cytoplasm | Transporter |
| Urocortin | Ucn | 9.19 | Extracellular space | Other |
| CD300c molecule | RGD1559482 | 6.24 | Plasma membrane | Transmembrane receptor |
| Solute carrier family 22 member 6 | Slc22a6 | 4.11 | Plasma membrane | Transporter |
| ALK and LTK Ligand 2 | Fam150b | 4.06 | Extracellular space | Other |
| Coiled-coil domain containing 153 | Ccdc153 | 3.70 | Other | Other |
| C-X-C motif chemokine ligand 13 | Cxcl13 | 3.56 | Extracellular space | Cytokine |
| Prostaglandin D2 synthase | Ptgds | 3.27 | Cytoplasm | Enzyme |
| Troponin T2, cardiac type | Tnnt2 | 3.09 | Cytoplasm | Other |
| Insulin-like growth factor 2 | Igf2 | 2.97 | Extracellular space | Growth factor |
| Aldehyde dehydrogenase 1 family member A2 | Aldh1a2 | 2.92 | Cytoplasm | Enzyme |
| Hematopoietic SH2 domain containing | Hsh2d | 2.88 | Cytoplasm | Other |
| Solute carrier family 22 member 8 | Slc22a8 | 2.69 | Plasma membrane | Transporter |
| Prepronociceptin | Pnoc | 2.58 | Extracellular space | Other |
| Mitochondria localized glutamic acid-rich protein | Mgarp | 2.52 | Cytoplasm | Other |
| Solute carrier family 6 member 13 | Slc6a13 | 2.52 | Plasma membrane | Transporter |
| Ankyrin repeat domain 33B | Ankrd33b | 2.27 | Other | Other |
| Gap junction protein beta 6 | Gjb6 | 2.14 | Plasma membrane | Transporter |
| Astacin-like metalloendopeptidase | Astl | 2.09 | Cytoplasm | Peptidase |
| Solute carrier family 16 member 11 | Slc16a11 | 1.99 | Cytoplasm | Transporter |
| Tripartite motif containing 45 | Trim45 | 1.95 | Cytoplasm | Other |
| Myomesin 2 | Myom2 | 1.90 | Cytoplasm | Other |
| Activating transcription factor 3 | Atf3 | 1.83 | Nucleus | Transcription regulator |
| Sulfotransferase family 1A, phenol-preferring, member 1 | Sult1a1 | 1.70 | Cytoplasm | Enzyme |
| CD274 molecule | Cd274 | 1.66 | Plasma membrane | Enzyme |
| Insulin-like growth factor binding protein 6 | Igfbp6 | 1.65 | Extracellular space | Other |
| Serine peptidase inhibitor, Kunitz type 2 | Spint2 | 1.59 | Extracellular space | Other |
| Cholinergic receptor nicotinic alpha 6 subunit | Chrna6 | 1.55 | Plasma membrane | Transmembrane receptor |
| RNA binding motif protein 3 | Rbm3 | 1.52 | Cytoplasm | Other |
Table 2.
List of downregulated genes in the dorsal root ganglia after plantar incision.
| Gene Name | Gene symbol | log2(Fold change) | Location | Type(s) |
|---|---|---|---|---|
| Myosin light chain 1 | Myl1 | −6.00 | Cytoplasm | Other |
| H19 imprinted maternally expressed transcript | H19 | −4.32 | Cytoplasm | Other |
| DNA topoisomerase II alpha | Top2a | −4.05 | Nucleus | Enzyme |
| Collagen type II alpha 1 chain | Col2a1 | −3.76 | Extracellular space | Other |
| Centromere protein T | Cenpt | −3.59 | Cytoplasm | Transcription regulator |
| Ribonucleotide reductase regulatory subunit M2 | Rrm2 | −3.46 | Nucleus | Enzyme |
| BUB1 mitotic checkpoint serine/threonine kinase B | Bub1b | −3.42 | Nucleus | Kinase |
| Family with sequence similarity 84 member A | Fam84a | −3.37 | Other | Other |
| Integrin subunit beta 3 | Itgb3 | −3.33 | Plasma membrane | Transmembrane receptor |
| Syntrophin gamma 2 | Sntg2 | −2.98 | Plasma membrane | Other |
| Kinesin family member 18B | Kif18b | −2.95 | Cytoplasm | Other |
| Beta-1,4-N-acetyl-galactosaminyltransferase 3 | B4galnt3 | −2.85 | Cytoplasm | Enzyme |
| C1q and TNF-related 6 | C1qtnf6 | −2.81 | Extracellular space | Other |
| Cyclin B2 | Ccnb2 | −2.76 | Cytoplasm | Other |
| Centromere protein F | Cenpf | −2.76 | Nucleus | Other |
| Protein regulator of cytokinesis 1 | Prc1 | −2.71 | Nucleus | Other |
| CD93 molecule | Cd93 | −2.56 | Plasma membrane | Other |
| Grainyhead like transcription factor 3 | Grhl3 | −2.36 | Nucleus | Transcription regulator |
| Insulin-like growth factor 1 | Igf1 | −2.33 | Extracellular space | Growth factor |
| Collagen type III alpha 1 chain | Col3a1 | −2.31 | Extracellular space | Other |
| Collagen type XI alpha 1 chain | Col11a1 | −2.30 | Extracellular space | Other |
| Apelin receptor | Aplnr | −2.23 | Plasma membrane | G-protein-coupled receptor |
| Ewing sarcoma breakpoint region 1 | Ewsr1 | −2.21 | Nucleus | Other |
| Periostin | Postn | −2.21 | Extracellular space | Other |
| Nucleolar and spindle-associated protein 1 | Nusap1 | −2.19 | Nucleus | Other |
| Collagen type VIII alpha 1 chain | Col8a1 | −2.18 | Extracellular space | Other |
| Chromosome 2 open reading frame 40 | RGD1305645 | −2.15 | Extracellular space | Other |
| Collagen type I alpha 1 chain | Col1a1 | −2.13 | Extracellular space | Other |
| Tenascin C | Tnc | −2.11 | Extracellular space | Other |
| NUF2 component of NDC80 kinetochore complex | Nuf2 | −2.08 | Nucleus | Other |
| Collagen type XIV alpha 1 chain | Col14a1 | −2.07 | Extracellular space | Other |
| Fibrillin 2 | Fbn2 | −2.05 | Extracellular space | Other |
| Transmembrane protein 26 | Tmem26 | −2.04 | Other | Other |
| Myosin heavy chain 6 | Myh6 | −2.01 | Cytoplasm | Enzyme |
| RAB7B, member RAS oncogene family | Rab7b | −1.99 | Cytoplasm | Peptidase |
| EMI domain containing 1 | Emid1 | −1.97 | Extracellular space | Other |
| Secreted frizzled-related protein 4 | Sfrp4 | −1.96 | Plasma membrane | Transmembrane receptor |
| Kirre-like nephrin family adhesion molecule 1 | Kirrel | −1.91 | Plasma membrane | Other |
| Minichromosome maintenance complex component 6 | Mcm6 | −1.88 | Nucleus | Enzyme |
| Feline leukemia virus subgroup C cellular receptor family member 2 | Flvcr2 | −1.84 | Plasma membrane | Transporter |
| Peripheral myelin protein 2 | Pmp2 | −1.80 | Cytoplasm | Other |
| Niban apoptosis regulator 1 | Fam129a | −1.77 | Cytoplasm | Other |
| Acyl-CoA thioesterase 1 | Acot1 | −1.75 | Cytoplasm | Enzyme |
| Collagen type XXVII alpha 1 chain | Col27a1 | −1.73 | Extracellular space | Other |
| Collagen type IV alpha 1 chain | Col4a1 | −1.73 | Extracellular space | Other |
| ETS transcription factor ELK1 | Elk1 | −1.72 | Nucleus | Transcription regulator |
| Collagen type V alpha 2 chain | Col5a2 | −1.68 | Extracellular space | Other |
| POU class 3 homeobox 2 | Pou3f2 | −1.68 | Nucleus | Transcription regulator |
| Leucine-rich repeat containing G protein-coupled receptor 5 | Lgr5 | −1.67 | Plasma membrane | Transmembrane receptor |
| Fibrillin 1 | Fbn1 | −1.67 | Extracellular space | Other |
| Nidogen 2 | Nid2 | −1.66 | Extracellular space | Other |
| Pyridine nucleotide-disulphide oxidoreductase domain 2 | Pyroxd2 | −1.66 | Other | Other |
| TLR4 interactor with leucine-rich repeats | Tril | −1.65 | Other | Other |
| Dedicator of cytokinesis 1 | Dock1 | −1.64 | Cytoplasm | Other |
| Transmembrane serine protease 5 | Tmprss5 | −1.64 | Plasma membrane | Peptidase |
| Fatty acid desaturase 2 | Fads2 | −1.63 | Plasma membrane | Enzyme |
| Bone morphogenetic protein 1 | Bmp1 | −1.61 | Extracellular space | Peptidase |
| Protein phosphatase 1 regulatory inhibitor subunit 14C | Ppp1r14c | −1.60 | Cytoplasm | Other |
| KDEL motif containing 1 | Kdelc1 | −1.60 | Cytoplasm | Enzyme |
| Myelin protein zero | Mpz | −1.60 | Plasma membrane | Other |
| Sclerostin domain containing 1 | Sostdc1 | −1.59 | Extracellular space | Growth factor |
| Thrombospondin 2 | Thbs2 | −1.59 | Extracellular space | Other |
| Laminin subunit alpha 4 | Lama4 | −1.59 | Extracellular space | Enzyme |
| C-X-C motif chemokine receptor 4 | Cxcr4 | −1.59 | Plasma membrane | G-protein-coupled receptor |
| Protein tyrosine phosphatase receptor type Z1 | Ptprz1 | −1.58 | Plasma membrane | Phosphatase |
| Transforming growth factor beta receptor 3 | Tgfbr3 | −1.58 | Plasma membrane | Kinase |
| Cysteine and tyrosine rich 1 | Cyyr1 | −1.58 | Other | Other |
| Glycerophosphodiester phosphodiesterase domain containing 2 | Gdpd2 | −1.58 | Plasma membrane | Enzyme |
| Fms-related tyrosine kinase 1 | Flt1 | −1.58 | Plasma membrane | Kinase |
| Uncharacterized LOC100912041 | LOC100912041 | −1.58 | Plasma membrane | Transmembrane receptor |
| Sortilin-related VPS10 domain containing receptor 2 | Sorcs2 | −1.58 | Plasma membrane | Transporter |
| Endothelin receptor type B | Ednrb | −1.58 | Plasma membrane | G-protein-coupled receptor |
| UDP glycosyltransferase 8 | Ugt8 | −1.57 | Cytoplasm | Enzyme |
| ADAM metallopeptidase with thrombospondin type 1 motif 5 | Adamts5 | −1.57 | Extracellular space | Peptidase |
| Collagen type V alpha 1 chain | Col5a1 | −1.57 | Extracellular space | Other |
| Chrondromodulin | Lect1 | −1.57 | Extracellular space | Other |
| Plexin B3 | Plxnb3 | −1.56 | Plasma membrane | Transmembrane receptor |
| Eadherin EGF LAG seven-pass G-type receptor 2 | Celsr2 | −1.56 | Plasma membrane | G-protein-coupled receptor |
| ANTXR cell adhesion molecule 1 | Antxr1 | −1.56 | Plasma membrane | Transmembrane receptor |
| SLIT- and NTRK-like family member 2 | Slitrk2 | −1.54 | Plasma membrane | Other |
| Myelin-associated glycoprotein | Mag | −1.54 | Plasma membrane | Other |
| Epithelial membrane protein 2 | Emp2 | −1.54 | Plasma membrane | Other |
| nudE neurodevelopment protein 1 | Nde1 | −1.53 | Nucleus | Other |
| Sphingosine kinase 1 | Sphk1 | −1.52 | Cytoplasm | Kinase |
| Growth arrest specific 7 | Gas7 | −1.52 | Cytoplasm | Transcription regulator |
| Insulin-like growth factor binding protein 3 | Igfbp3 | −1.52 | Extracellular space | Other |
| PDZ domain containing 2 | Pdzd2 | −1.52 | Plasma membrane | Other |
| Leucine-rich repeat transmembrane neuronal 1 | LRRTM1 | −1.51 | Plasma membrane | Other |
| Erb-b2 receptor tyrosine kinase 3 | Erbb3 | −1.51 | Plasma membrane | Kinase |
| Podoplanin | Pdpn | −1.50 | Plasma membrane | Other |
| Peroxidasin | Pxdn | −1.50 | Extracellular space | Enzyme |
Figure 2.
The global transcriptome analysis on rat dorsal root ganglia (DRG) identifies the genes that are significantly altered in one day after plantar incision. The infiltration of capsaicin 30 min prior to surgical incision completely prevented incision-induced transcriptional changes in 99 of 126 (79%) genes; shown in a heat map (a). Each line represents a gene across three groups (n = 4/group). Shades of blue and orange indicate negative and positive z-scores, respectively. (b) Venn diagram showing altered functions associated with two distinct groups of genes. Both groups underwent plantar incision but one was pretreated with capsaicin (inci + cap) and the other was not (Inci). Values in parentheses indicate z-score where negative and positive values predict inhibition and activation, respectively. (c) The differentially expressed genes from Inci + cap and Inci were compared by IPA. The top diseases and biological functions were related to “mortality morbidity” and “organismal death”. Dots indicate z-score <2.0.
Regulatory networks involved in plantar incision-induced pain behaviors
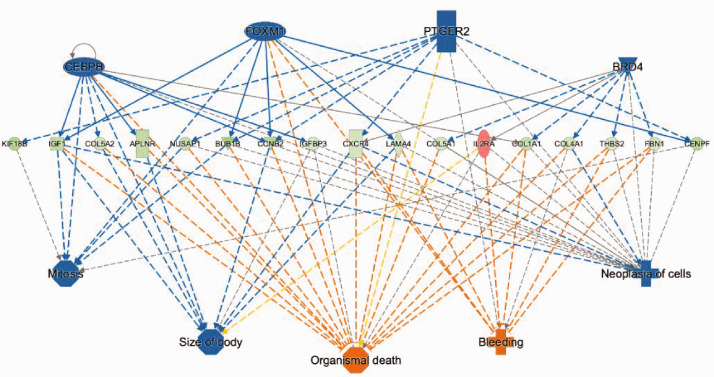
To identify the regulatory networks involved in the development of incisional pain, IPA was used to annotate these genes onto known functional gene networks. Specific analysis of DRG genes, which are altered 24 h after plantar incision but unchanged when rats were pretreated with 1% capsaicin in skin, predicted lower activity of CCAAT/enhancer-binding protein beta (CEBPB), forkhead box M1 (FOXM1), prostaglandin E(2) receptor (PTGER2), and BRD4 (Figure 3). These changes are associated with reduced cell proliferation, increased bleeding, and organismal death (Figure 3).
Figure 3.
Altered regulatory networks and associated downstream effects in DRG following plantar incision. Analysis of differentially expressed genes indicated reduced activity of CEBPB, FOXM1, PTGER2, and BRD4 regulatory factors (z-score < −2.0), leading to increased risk organismal mortality accompanied by decreased mitosis and tissue size. Legends: Upregulation: red; downregulation: green; predicted inhibition: blue; predicted activation: orange.
Changes in inflammatory genes following plantar incision
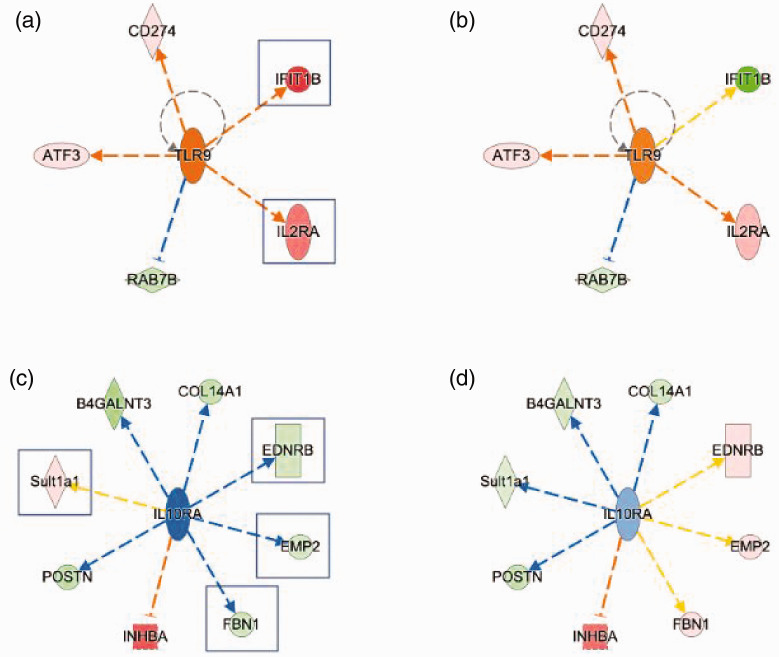
We have identified that plantar incision induces the expression of DRG genes that regulates inflammatory processes. In particular, markedly increased expression of IFIT1B (∼50-fold), IL2Ra, CD274 were predictive of increased toll-like receptor 9 (TLR9) signaling (Figure 4(a)), which was attenuated with capsaicin pretreatment (Figure 4(b)). Conversely, reduced expression of EDNRB, EMP2, and FBN1 coupled with increased expression of INHBA and Sult1a1 were indicative of reduced IL10Ra signaling (Figure 4(c)), which was attenuated by capsaicin pretreatment (Figure 4(d)). These results suggest plantar incision produces a change in genes leading to increased pro-inflammatory responses and decreased anti-inflammatory responses in the DRG.
Figure 4.
Changes in gene expression leading to increased pro-inflammatory and decreased anti-inflammatory responses in DRG following plantar incision (a, b). IPA predicted increased toll-like receptor 9 (TLR9) activity due to increased expression of IFIT1B and IL2RA (a, boxed). Capsaicin pretreatment (b) diminished these transcriptional responses. (c–d) Altered gene expression suggests a decreased anti-inflammation in incision group (c), which were not altered with capsaicin pretreatment (d).
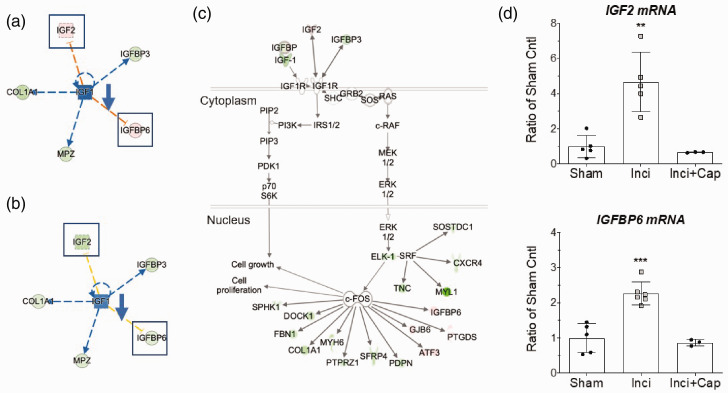
Insulih-like growth factor signaling pathway is involved in incisional pain
The analysis of differentially expressed DRG genes showed evidence of reduced insulin-like growth factor (IGF) signaling pathway with downregulation of IGF1, IGFBP3, MPZ, and COL1A1 accompanied by upregulation of IGF2 and IGFBP6 (Figure 5(a)). Capsaicin pretreatment prevented the upregulation of IGF2 and IGFBP6 (Figure 5(b)). Further, IPA linked these transcriptional changes to reduced PI3K and ERK signaling, leading to altered expression of ERK target genes (Figure 5(c)). To independently validate NGS data, expression of IGF2 and IGFBP6 were quantified by qPCR using a different set of biological replicates. The significant upregulation of IGF2 and IGFBP6 were confirmed in the DRGs from incised rats, which were prevented with capsaicin pretreatment (Figure 5(d); IGF2: F2,10 = 16.99, p = 0.0006; IGFBP6: F2,11 = 26.22, p < 0.0001).
Figure 5.
Altered IGF1 signaling in DRG of rats after plantar incision. (a, b) IGF1 gene networks showing altered expression level in incision group (a) and incision with capsaicin pretreated group (b). Boxed genes indicate those that responded to capsaicin treatment. (c) IPA mapped differentially expressed genes onto IGF1 canonical pathway suggesting reduced cell growth and proliferation. (d) qPCR validation of IGF2 and IGFBP6 in DRG. Values are mean SD, n = 3–6/group, t-test, **p < 0.01, ***p < 0.001.
Plantar incision-induced changes in genes not related to pain
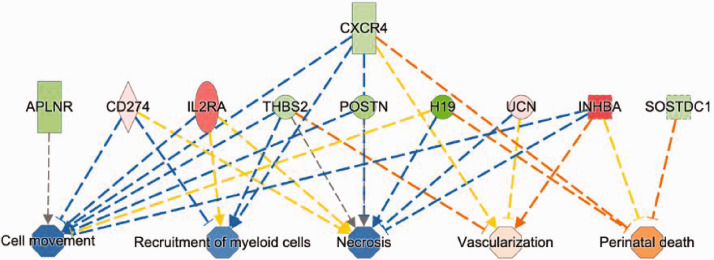
IPA annotation of 27 DRG genes that are not influenced by capsaicin pretreatment in plantar skin (Figure 2(b)) revealed changes in cellular responses including decreased recruitment of myeloid cells, cell movement, and cellular necrotic death. These effects were accompanied by increased vascularization and organismal death (Figure 6). These responses may have a role in surgical injury and repair and unlikely to be involved in the pain processing after incision.
Figure 6.
Functional annotation of DRG genes that were regulated by plantar incision but were not influenced by the capsaicin pretreatment. IPA mapped functions of these 25 genes indicating reduced cell movement, recruitment of myeloid cells, and necrosis as well as increased vascularization and risk of perinatal death.
Discussion
Our study is the first genome-wide transcriptional profiling of DRG tissues in a rat model of incisional pain. We identified changes in gene expression in the DRG at one day after plantar incision, a time point at which pain behaviors were most prominent. The skin infiltration of capsaicin 30 min prior to surgical incision attenuated incision-induced pain behaviors, completely denervated the epidermis and dermis around the incision, and prevented incision-induced transcriptional changes in 99 of 126 DRG genes (79%), suggesting the majority of these transcriptional changes were related to post-operative pain development. These findings highlight novel gene networks associated post-operative pain including neuro-inflammation and the IGF signaling.
Global analysis of transcriptome in rat DRG following plantar incision
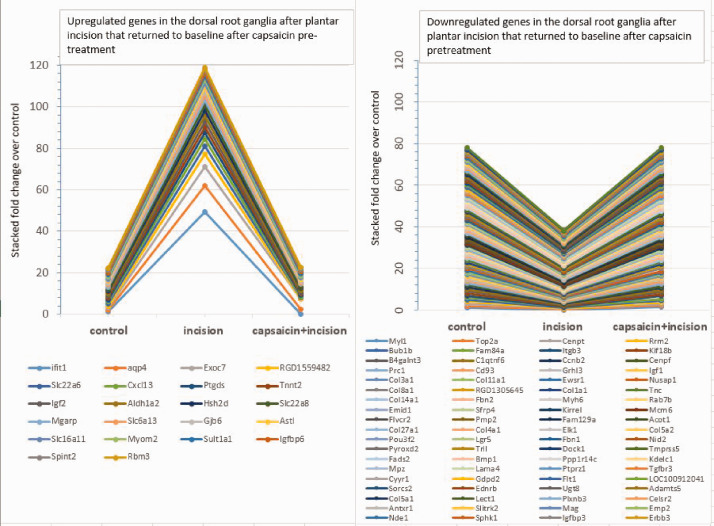
Plantar incision alone induced 36 upregulated and 90 downregulated genes in L4–L6 DRGs. These results are significant and robust compared to a previous polymerase chain reaction array study by Spofford and Brennan,34 who showed few changes in gene expression in DRGs using similar incision model.34 The disparate results were likely due to the use of different techniques. The qPCR approach by Spofford and Brennan34 is limited to measuring single transcripts and requires priori of known gene targets. In contrast, RNA Seq, used in the present study, is an agnostic and more comprehensive approach that covers the entire transcriptome (>26,000 gene loci) present in DRGs. Moreover, this approach allows a better resolution of complex nature of the transcriptome including a detailed and quantitative measure of gene expression. Notably, the number of downregulated genes much greater than the number of upregulated genes; however, the fold change in expression for the upregulated genes was markedly higher than that for the downregulated genes (Figure 7, Tables 1 and 2). The higher number of downregulated versus upregulated transcripts in DRG is consistent with previous studies that used lumbar disc herniation models35 but it contrasts with studies that used neuropathic pain models.36,37 These findings suggest that altered gene expression in sensory neurons are specific to the types of tissue injury. Interestingly, our findings showed a strong similarity to a transcriptomic analysis of the spinal cord in a similar model of post-operative pain by Raithel et al. by identifying increased inflammation as a possible contributing factor for pain development at day 2 after plantar incision.38 Together, these two studies showed a prolonged common physiological response following plantar incision at the DRG and spinal cord. However, our findings showed an opposite effect from Raithel et al.38 in cell proliferation. This differential effect could stem from differences in the timing (24 h vs. 48 h) or tissue type (DRG vs. spinal cord) of analysis following plantar incision. Given a positive effects of the CEBPB and the PTGER2 in the genesis of neuropathic pain,39 the reduced activity of these factors in the DRG of the incision model could have a role in wound repair. The role of the cell cycle regulator FOXM1 in post-operative pain is unknown. Due to its high expression level in macrophages, reduced FOXM1 activity could have a role in inflammatory response and wound repair following plantar incision.40
Figure 7.
Data presented in figure 2 are shown as stacked percentage chart showing degree of expression changes relative to the total changes in gene expression in dorsal root ganglia (DRG) following plantar incision. Left: upregulated genes; Right: downregulated genes.
Plantar incision induces Toll-like receptor 9 mediated pro-inflammatory response
Gene expression changes in the DRG suggest TLR9 as a significant regulator pain development following plantar incision (Figure 4(a) and (b)). Among the DRG genes, IFIT1b and ILR2a were profoundly upregulated by 50- and 34-fold, respectively. TLRs signaling are thought to protect and defend the host organism by initiating inflammatory responses during tissue injury. Activation of TLRs in glia, sensory neurons, and other cell types has been shown to alter nociceptive processing leading to pain.41 Though our findings pinpoint TLR9 activation in the DRG as a contributing factor in a model of post-operative pain, this signaling mechanism has been postulated in neuropathic and inflammatory pain models.42,43 Thus, TLR9 activity is likely a common mechanism in DRG that modulates pain signaling in multiple pain models. Stimulation of TLR9 can exacerbate neuronal injury through excessive release of Tumor necrosis factor and nitric oxide from glial cells.44 Conversely, inhibition of TLR9 signaling can reduce neuropathic pain45 and thermal hyperalgesia following spinal cord injury.46 TLR9 signals through MyD88, which activates the transcription factor NF-κB for production of pro-inflammatory cytokines.41 Further studies are needed to determine the mechanisms by which TLR9 contributes to pain hypersensitivity following plantar incision.
Plantar incision inhibits IL10RA-mediated anti-inflammatory responses
The reduced DRG expression of EDNRB, EMP2, FBN1 predicted the inhibition of upstream regulator interleukin 10 receptor (IL10RA) activity after plantar incision (Figure 4(c) and (d)). Emerging evidence indicates that IL-10 is a potent anti-inflammatory molecule that acts to dampen initial pro-inflammatory responses through a poorly defined feedback mechanism.47 In addition, IL-10 is a positive regulator of neuropathic pain, where intrathecal administration of IL-10 attenuates mechanical allodynia and thermal hyperalgesia after chronic constriction injury of the sciatic nerves.48 Moreover, IL-10 can be induced by electropuncture to produce analgesic effects in the plantar incision model.49 Our finding of reduced IL10RA activity suggests a role in modulating pain behaviors following plantar incision. More studies are needed to delineate the mechanism by which IL10 signaling contributes to the development of incisional pain.
Activation of IGF signaling pathway following plantar incision
We identified insulin-like growth factor signaling (IGF1, IGF-2, and IGF binding proteins) as a major gene network in the development of pain behaviors (Figure 5(a) to (c)). The upregulation of IGF2 and IGFBP6 after incision (Figure 5(d)) was ameliorated by capsaicin pretreatment, suggesting active roles of IGF2 and IGFBP6 in the development of incisional pain (Figure 7). IGF2 is a neurotrophic factor critical for cell proliferation, neuronal survival, and nerve regeneration. Dysregulated IGF2 expression is implicated in a growing number of diseases.50–54 IGF2 binds with IGF1 receptor (IGF1R), IGF2R, and insulin receptors. Activation of IGF1-receptors engages multiple signaling pathways, including the PI3 kinase/AKT and MAPK, which modulate neuronal function and plasticity.55–58 Activation of IGF1R can stimulate voltage-gated T-type Ca2+ (CaV3) channels in mouse DRG through a mechanism dependent on heterotrimeric G protein signaling.59,60 The inhibition of IGF-1R signaling or knock down of CaV3 in DRG neurons abolished the increased mechanical and thermal sensitivity after chronic inflammation.60 In addition, IGF-receptor activation enhanced TrpV1-mediated membrane currents in heterologous expression systems and cultured DRG neurons.61 We have previously shown that TrpV1 contributes to spontaneous activity and sensitization to heat of primary afferent nociceptors following skin incision in rats.22 Taken together, these findings suggest that IGF signaling is a novel pharmacologic target with therapeutic potential to manage pain following surgical procedures.
Conclusions
We highlight three signaling pathways in the DRG that are potentially involved in the processing of incisional pain. These pathways may modulate nociceptor activation and sensitization, tissue repair, nerve regeneration, and inflammatory responses. The present study is the first to employ NGS, which is a genome-wide approach to identify molecular factors, in the DRG of incisional pain model. Our data suggest that patterns of transcriptome changes after incision share similar processes in the development of pain associated with diabetes and inflammation12–16; but distinct from neuropathic pain37 and sciatic nerve transection36 pain model, which include process for neuronal cell death and regeneration. This study is limited in its pathway predictions, which are based on a knowledge-based database, and a lack of validation at the protein level. While the transcriptomic changes that were attenuated by capsaicin pretreatment are likely associated with postoperative pain development, we cannot rule out the possibility of other non-pain-related processes contributed to these changes. Future studies are warranted to validate our results. Nevertheless, investigation of this process on a molecular level by examining whole-genome transcriptional activity provides initial insights into the mechanisms of incisional pain, which may ultimately guide efforts to identify novel therapeutics and biomarkers for incisional pain in the future.
Acknowledgments
The work is dedicated to Professor Timothy J Brennan, MD, PhD, Emeritus Professor of the Department of Anesthesiology, University of Iowa, on the eve of his retirement after many years of invaluable contribution to acute postoperative pain model and mechanism.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a startup fund to RKB from the Department of Anesthesiology, University of Minnesota and Fairview Medical Center and in part by NIH grants HL135895 and CA241627 to DS.
ORCID iD
Ratan K Banik https://orcid.org/0000-0001-7126-5768
References
- 1.Weiser TG, Regenbogen SE, Thompson KD, Haynes AB, Lipsitz SR, Berry WR, Gawande AA. An estimation of the global volume of surgery: a modelling strategy based on available data. Lancet 2008; 372: 139–144. [DOI] [PubMed] [Google Scholar]
- 2.DeFrances CJ, Podgornik MN. National hospital discharge survey. Adv Data 2004; 2006: 1–19. http://www.ncbi.nlm.nih.gov/pubmed/16703980. [PubMed] [Google Scholar]
- 3.Kehlet H, Jensen TS, Woolf CJ. Persistent postsurgical pain: risk factors and prevention. Lancet (London, England) 2006; 367: 1618–1625. [DOI] [PubMed] [Google Scholar]
- 4.Polomano RC, Dunwoody CJ, Krenzischek DA, Rathmell JP. Perspective on pain management in the 21st century. Pain Manag Nurs 2008; 9: 3–10. [DOI] [PubMed] [Google Scholar]
- 5.Imam MZ, Kuo A, Ghassabian S, Smith MT. Progress in understanding mechanisms of opioid-induced gastrointestinal adverse effects and respiratory depression. Neuropharmacology 2018; 131: 238–255. [DOI] [PubMed] [Google Scholar]
- 6.Dumas EO, Pollack GM. Opioid tolerance development: a pharmacokinetic/pharmacodynamic perspective. Aaps J 2008; 10: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fletcher D, Martinez V. Opioid-induced hyperalgesia in patients after surgery: a systematic review and a meta-analysis. Br J Anaesth 2014; 112: 991–1004. [DOI] [PubMed] [Google Scholar]
- 8.de Boer HD, Detriche O, Forget P. Opioid-related side effects: postoperative ileus, urinary retention, nausea and vomiting, and shivering. A review of the literature. Best Pract Res Clin Anaesthesiol 2017; 31: 499–504. [DOI] [PubMed] [Google Scholar]
- 9.Boyer EW. Management of opioid analgesic overdose. N Engl J Med 2012; 367: 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alam A, Gomes T, Zheng H, Mamdani MM, Juurlink DN, Bell CM. Long-term analgesic use after low-risk surgery: a retrospective cohort study. Arch Intern Med 2012; 172: 425–430. [DOI] [PubMed] [Google Scholar]
- 11.Bates C, Laciak R, Southwick A, Bishoff J. Overprescription of postoperative narcotics: a look at postoperative pain medication delivery, consumption and disposal in urological practice. J Urol 2011; 185: 551–555. [DOI] [PubMed] [Google Scholar]
- 12.Athie MCP, Vieira AS, Teixeira JM, dos Santos GG, Dias EV, Tambeli CH, Sartori CR, Parada CA. Transcriptome analysis of dorsal root ganglia’s diabetic neuropathy reveals mechanisms involved in pain and regeneration. Life Sci 2018; 205: 54–62. [DOI] [PubMed] [Google Scholar]
- 13.Pande M, Hur J, Hong Y, Backus C, Hayes JM, Oh SS, Kretzler M, Feldman EL. Transcriptional profiling of diabetic neuropathy in the BKS db/db mouse: a model of type 2 diabetes. Diabetes 2011; 60: 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perkins JR, Antunes-Martins A, Calvo M, Grist J, Rust W, Schmid R, Hildebrandt T, Kohl M, Orengo C, McMahon SB, Bennett DLH. A comparison of RNA-seq and exon arrays for whole genome transcription profiling of the L5 spinal nerve transection model of neuropathic pain in the rat. Mol Pain 2014; 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wangzhou A, McIlvried LA, Paige C, Barragan-Iglesias P, Guzman CA, Dussor G, Ray PR, Gereau RW, Price TJ. Pharmacological target-focused transcriptomic analysis of native vs cultured human and mouse dorsal root ganglia. Pain 2020; 161(7): 1497–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin C, Hu Q, Liu B, Tai Y, Zheng X, Li Y, Xiang X, Wang P, Liu B. Transcriptome profiling of dorsal root ganglia in a rat model of complex regional pain syndrome type-I reveals potential mechanisms involved in pain. J Pain Res 2019; 12: 1201–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haroutiunian S, Nikolajsen L, Finnerup NB, Jensen TS. The neuropathic component in persistent postsurgical pain: a systematic literature review. Pain 2013; 154: 95–102. [DOI] [PubMed] [Google Scholar]
- 18.Richebé P, Capdevila X, Rivat C. Persistent postsurgical pain: pathophysiology and preventative pharmacologic considerations. Anesthesiology 2018; 129: 590–607. [DOI] [PubMed] [Google Scholar]
- 19.Xu Q, Yaksh TL. A brief comparison of the pathophysiology of inflammatory versus neuropathic pain. Curr Opin Anaesthesiol 2011; 24: 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banik RK, Brennan TJ. Sensitization of primary afferents to mechanical and heat stimuli after incision in a novel in vitro mouse glabrous skin-nerve preparation. Pain 2008; 138: 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banik RK, Brennan TJ. Spontaneous discharge and increased heat sensitivity of rat C-fiber nociceptors are present in vitro after plantar incision. Pain 2004; 112: 204–213. [DOI] [PubMed] [Google Scholar]
- 22.Banik RK, Brennan TJ. Trpv1 mediates spontaneous firing and heat sensitization of cutaneous primary afferents after plantar incision. Pain 2009; 141: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banik RK, Kabadi RA. A modified Hargreaves’ method for assessing threshold temperatures for heat nociception. J Neurosci Methods 2013; 219: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banik RK, Subieta AR, Wu C, Brennan TJ. Increased nerve growth factor after rat plantar incision contributes to guarding behavior and heat hyperalgesia. Pain 2005; 117: 68–76. [DOI] [PubMed] [Google Scholar]
- 25.Kabadi R, Kouya F, Cohen HW, Banik RK. Spontaneous pain-like behaviors are more sensitive to morphine and buprenorphine than mechanically evoked behaviors in a rat model of acute postoperative pain. Anesth Analg 2015; 120: 472–478. [DOI] [PubMed] [Google Scholar]
- 26.Kang S, Wu C, Banik RK, Brennan TJ. Effect of capsaicin treatment on nociceptors in rat glabrous skin one day after plantar incision. Pain 2010; 148: 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhelski ML, McAdams B, Johns ME, Kabadi RA, Simone DA, Banik RK. Lack of relationship between epidermal denervation by capsaicin and incisional pain behaviors: a laser scanning confocal microscopy study in rats. Eur J Pain 2020; 24: 1197–1208. [DOI] [PubMed] [Google Scholar]
- 28.Kaliappan S, Simone DA, Banik RK. Nonlinear inverted-U shaped relationship between aging and epidermal innervation in the rat plantar hind paw: a laser scanning confocal microscopy study. J Pain 2018; 19: 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy WR, Wendelschafer-Crabb G, Johnson T. Quantitation of epidermal nerves in diabetic neuropathy. Neurology 1996; 47: 1042–1048. [DOI] [PubMed] [Google Scholar]
- 30.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012; 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson M, McCarthy D, Bioinformatics GS. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. academic.oup.com n.d, https://academic.oup.com/bioinformatics/article-abstract/26/1/139/182458 (2010, undefined.). [DOI] [PMC free article] [PubMed]
- 33.Tran PV, Kennedy BC, Pisansky MT, Won K-J, Gewirtz JC, Simmons RA, Georgieff MK. Prenatal choline supplementation diminishes early-life iron deficiency–induced reprogramming of molecular networks associated with behavioral abnormalities in the adult rat hippocampus. J Nutr 2016; 146: 484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spofford CM, Brennan TJ. Gene expression in skin, muscle, and dorsal root ganglion after plantar incision in the rat. Anesthesiology 2012; 117: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Ai H, Liu J, Xu M, Zhou Z, Qian C, Xie Y, Yan J. Characterization of novel lnc RNAs in the spinal cord of rats with lumbar disc herniation. J Pain Res 2019; 12: 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, Liu Q, Wang Y, Gu Y, Liu D, Wang C, Ding G, Chen J, Liu J, Gu X. Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PLoS One 2013; 8: e57000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uttam S, Wong C, Amorim IS, Jafarnejad SM, Tansley SN, Yang J, Prager-Khoutorsky M, Mogil JS, Gkogkas CG, Khoutorsky A. Translational profiling of dorsal root ganglia and spinal cord in a mouse model of neuropathic pain. Neurobiol Pain 2018; 4: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raithel SJ., Sapio MR, LaPaglia DM, Iadarola MJ, Mannes AJ. Transcriptional changes in dorsal spinal cord persist after surgical incision despite preemptive analgesia with peripheral resiniferatoxin. Anesthesiology 2018; 128: 620–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki M, Hashimoto S, Sawa T, Amaya F. Tumor necrosis factor-alpha induces expression of C/EBP-beta in primary afferent neurons following nerve injury. Neuroscience 2014; 279: 1–9. [DOI] [PubMed] [Google Scholar]
- 40.Ren X, Zhang Y, Snyder J, Cross ER, Shah Ta Kalin TV, Kalinichenko VV. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol Cell Biol 2010; 30: 5381–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacagnina MJ, Watkins LR, Grace PM. Toll-like receptors and their role in persistent pain. Pharmacol Ther 2018; 184: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David BT, Ratnayake A, Amarante Ma Reddy Np Dong W, Sampath S, Heary RF, Elkabes SA. Toll-like receptor 9 antagonist reduces pain hypersensitivity and the inflammatory response in spinal cord injury. Neurobiol Dis 2013; 54: 194–205. [DOI] [PubMed] [Google Scholar]
- 43.Luo X, Huh Y, Bang S, He Q, Zhang L, Matsuda M, Ji RR. Macrophage toll-like receptor 9 contributes to chemotherapy-induced neuropathic pain in male mice. J Neurosci 2019; 39: 6848–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iliev AI, Stringaris AK, Nau R, Neumann H. Neuronal injury mediated via stimulation of microglial toll-like receptor-9 (TLR9). Faseb J 2004; 18: 412–414. [DOI] [PubMed] [Google Scholar]
- 45.Liu F, Wang Z, Qiu Y, Wei M, Li C, Xie Y, Shen L, Huang Y, Ma C. Suppression of MyD88-dependent signaling alleviates neuropathic pain induced by peripheral nerve injury in the rat. J Neuroinflammation 2017; 14: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.David BT, Ratnayake A, Amarante MA, Reddy NP, Dong W, Sampath S, Heary RF, Elkabes S. A toll-like receptor 9 antagonist reduces pain hypersensitivity and the inflammatory response in spinal cord injury. Neurobiol Dis 2013; 54: 194–205. [DOI] [PubMed] [Google Scholar]
- 47.Iyer SS, Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol 2012; 32: 23–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milligan ED, Langer SJ, Sloane EM, He L, Wieseler-Frank J, O'Connor K, Martin D, Forsayeth JR, Maier SF, Johnson K, Chavez RA, Leinwand LA, Watkins LR. Controlling pathological pain by adenovirally driven spinal production of the anti-inflammatory cytokine, interleukin-10. Eur J Neurosci 2005; 21: 2136–2148. [DOI] [PubMed] [Google Scholar]
- 49.Dai W, Sun J-L, Li C, Mao W, Huang Y-K, Zhao Z-Q, Zhang Y-Q, Lu N. Involvement of interleukin-10 in analgesia of electroacupuncture on incision pain. Evidence-Based Complement Altern Med 2019; 2019: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvino CL, Ong SC, McNeil KA, Delaine C, Booker GW, Wallace JC, Forbes BE. Understanding the mechanism of insulin and insulin-like growth factor (IGF) receptor activation by IGF-II. PLoS One 2011; 6: e27488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergman D, Halje M, Nordin M, Engström W. Insulin-like growth factor 2 in development and disease: a mini-review. Gerontology 2013; 59: 240–249. [DOI] [PubMed] [Google Scholar]
- 52.Chao W, D'Amore P A. IGF2: epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev 2008; 19: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clemmons DR. Insulin-like growth factor binding proteins and their role in controlling IGF actions. Cytokine Growth Factor Rev 1997; 8: 45–62. [DOI] [PubMed] [Google Scholar]
- 54.Engström W, Shokrai A, Otte K, Granérus M, Gessbo Å, Bierke P, Madej A, Sjölund M, Ward A. Transcriptional regulation and biological significance of the insulin like growth factor II gene. Cell Prolif 1998; 31: 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foncea R, Andersson M, Ketterman A, Blakesley V, Sapag-Hagar M, Sugden PH, LeRoith D, Lavandero S. Insulin-like growth factor-I rapidly activates multiple signal transduction pathways in cultured rat cardiac myocytes. J Biol Chem 1997; 272: 19115–19124. [DOI] [PubMed] [Google Scholar]
- 56.Monnier D, Boutillier AL, Giraud P, Chiu R, Aunis D, Feltz P, Zwiller J, Loeffler JP. Insulin-like growth factor-I stimulates c-fos and c-jun transcription in PC12 cells. Mol Cell Endocrinol 1994; 104: 139–145. [DOI] [PubMed] [Google Scholar]
- 57.Pugazhenthi S, Boras T, O’Connor D, Meintzer MK, Heidenreich KA, Reusch JEB. Insulin-like growth factor I-mediated activation of the transcription factor camp response element-binding protein in pc12 cells: involvement of p38 mitogen-activated protein kinase-mediated pathway. J Biol Chem 1999; 274: 2829–2837. [DOI] [PubMed] [Google Scholar]
- 58.Zheng WH, Quirion R. Insulin-like growth factor-1 (IGF-1) induces the activation/phosphorylation of Akt kinase and cAMP response element-binding protein (CREB) by activating different signaling pathways in PC12 cells. BMC Neurosci 2006; 7: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stemkowski PL, Zamponi GW. The tao of IGF-1: insulin-like growth factor receptor activation increases pain by enhancing T-type calcium channel activity. Sci Signal 2014; 7: pe23. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Qin W, Qian Z, Liu X, Wang H, Gong S, Sun YG, Snutch TP, Jiang X, Tao J. Peripheral pain is enhanced by insulin-like growth factor 1 through a G protein-mediated stimulation of T-type calcium channels. Sci Signal 2014; 7: ra94. [DOI] [PubMed] [Google Scholar]
- 61.Van Buren JJ, Bhat S, Rotello R, Pauza ME, Premkumar LS. Sensitization and translocation of TRPVI by insulin and IGF-I. Mol Pain 2005; 1: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]