Abstract
Hypocrellins and hypomycins are naturally occurring fungal perylenequinones with potential photodynamic activity against cancer and microbial diseases. This project pursued three lines of research. First, the production of perylenequinones was enhanced by investigating the effect of culture medium and light exposure on their biosynthesis. Solid-fermentation cultures on rice medium allowed for enhanced production of hypocrellins as compared to Cheerios or oatmeal medium. Alternatively, increased production of hypomycins, which are structurally related to the hypocrellins, was observed on oatmeal medium. In both cases, light exposure was an essential factor for the enhanced biosynthesis. In addition, this led to the discovery of two new perylenequinones, ent-shiraiachrome A (5) and hypomycin E (8), which were elucidated based on spectroscopic data. Finally, the photocytotoxic effects of both classes of compounds were evaluated against human skin melanoma, with EC50 values at nanomolar levels for hypocrellins and micromolar levels for hypomycins. In contrast, both classes of compounds showed reduced dark toxicity (EC50 values > 100 μM), demonstrating promising phototherapeutic indices.
Graphical Abstract
Hypocrellins are a class of fungal metabolites (perylenequinone) that were first isolated from the fruiting bodies of Hypocrella bambusae, Shiraia bambusicola, and other Shiraia-like fungi.1–4 Hypocrellin-producing fungi have been used in traditional Chinese medicine for the treatment of rheumatoid arthritis, vertigo, psoriasis, and other diseases.5 Hypocrellins are known to have an oxidized pentacyclic core, a highly conjugated ring system, and exhibit various pharmacological activities. Recently, hypocrellins have started gaining attention in photodynamic therapy (PDT) to combat cancer and bacterial, fungal, and viral infections.6–11 In photodynamic therapy, a non-toxic photosensitizer is irradiated with visible-light at a defined wavelength in the presence of oxygen to induce the production of reactive oxygen species and free radicals that are fatal to cancerous cells and microbial pathogens.12 Owing to their high quantum yields of singlet oxygen (1O2), low general toxicity in the absence of light, and light absorption properties in the red spectral region, hypocrellins are of clinical interest as naturally-derived photosensitizers.13, 14
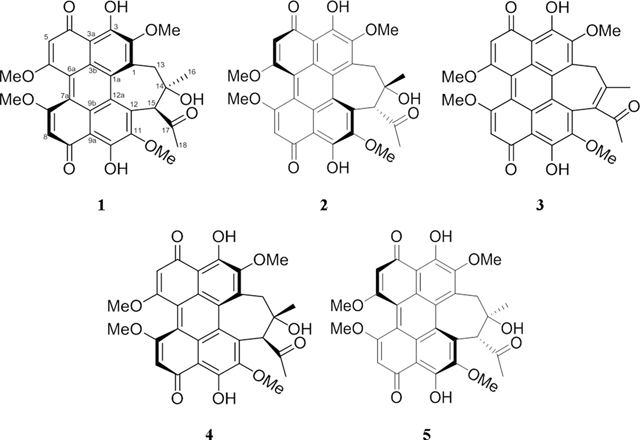
The hypocrellin family consists of four main compounds, hypocrellin (1), hypocrellin A (2), hypocrellin B (3), and shiraiachrome A (4). A great deal of confusion exists in the literature on the naming of these perylenequinones. For instance, hypocrellin (1) and hypocrellin A (2), which are enantiomers, were isolated from Hypocrella bambusae and Shiraia bambusicola, respectively.1, 2 However, several studies on the biological and photodynamic properties of these compounds do not differentiate between their structures.8, 15, 16 Moreover, hypocrellin A (2) was referred by Wu et al.2 as shiraiachrome B. Similarly, shiraiachrome A (4) was also named as hypocrellin B,17 which is the trivial name that is used most often in the literature to refer to 3.18, 19 On the other hand, hypocrellin B (3) was named as either shiraiachrome C or hypocrellin C by Wu et al.2 or Kishi et al.,17 respectively. Owing to the large number of studies that use the names hypocrellin (1), hypocrellin A (2), hypocrellin B (3), and shiraiachrome A (4), we propose that these names should take precedent; Figure S1 should be examined for an illustration of the varied nomenclature of these compounds.
There was a need to increase the production of these compounds to facilitate further investigations on their promising photodynamic properties. Moreover, the axial chirality of hypocrellins, along with the atropisomerization (inversion of axial chirality) and tautomerism exhibited by these compounds make the total synthesis of hypocrellins challenging (e.g. 19 steps with a 1.6% overall yield to generate 2).20 Fermentation constitutes a viable route to supply these compounds, and several attempts have been made to increase their production. Of these, enhanced production of hypocrellin A (2) was observed after the addition of Triton X-100 surfactant to the submerged cultures of S. bambusicola (yield ~100 mg/L).21 Co-cultivation of Shiraia sp. with Pseudomonas fulva was found to stimulate the production of various perylenequinone compounds, including hypocrellin A (2).22, 23 In another study, solid-state fermentation of Shiraia sp. using corn was found to enhance the production of 2 as compared to seven other types of grain.24 Furthermore, enhanced biosynthesis of hypocrellin A (2) along with stimulating its release to the medium was observed after treating the submerged culture of Shiraia sp. with low intensity ultrasound irradiation.25 In general, the literature suggests that hypocrellin-producing fungi are sensitive to media, growth conditions, available nutrients, and surrounding environmental factors.
Light is a major factor that regulates fungal growth, reproduction, sexual development, and production of secondary metabolites.26 Strong evidence exists on the impact of light exposure for the production of hypocrellins. Wang et al.27 investigated the effect of exposing cultures of Shiraia sp. to light at various wavelengths and found that a red light emitting diode (LED) maximized the production of 2 to >3 fold that of the dark control. Similarly, growing Shiraia sp. cultures under light and dark cycles (12:12 or 24:24 h) was found to increase the production of 2 by 1.7 fold as compared to the dark culture control.28 High intensity light (i.e. 600–800 lux) negatively affected fungal growth and decreased the production of 2, while lower intensity light (i.e. 200–400 lux) was shown to up-regulate the biosynthesis of 2.28 Although those studies suggested that light exposure might help to increase the production of 2, Gao et al.29 found that higher hypocrellin production was observed in Shiraia sp. grown under dark conditions, as compared to those grown under light of various wavelengths (i.e. white, red, yellow, green, blue, and purple). Therefore, uncertainty persists with regard to the effect of light on the biosynthesis of secondary metabolites from hypocrellin-producing fungi.
As part of ongoing studies to uncover potential anticancer leads from natural resources,30 a hypocrellin-producing fungus (i.e. strain MSX60519) was examined. This strain was grown under a suite of growth conditions to investigate the effect of culture medium and light exposure on the production of these compounds (1–8),31 resulting in the discovery of new analogues 5 and 8 and the refinement of the structures of compounds 6 and 7. In addition, the photodynamic properties were evaluated under both light and dark conditions, revealing light-induced cytotoxicity in the nanomolar range, suggestive of the pharmacological potential of these compounds.
RESULTS AND DISCUSION
As reported previously, our standard protocol is to first grow fungal cultures on a pilot scale (i.e., using ~ 10 g of rice), and then, if a biologically active sample passes dereplication (i.e., filtering out extracts that contain well studied mycotoxins),32, 33 it will be scaled for more in depth natural products studies.34 For strain MSX60519, the major compounds isolated from solid state fermentation cultures were identified as ent-shiraiachrome A (5; elucidation discussed below) and hypocrellin (1),1, 20 where the spectroscopic data matched the literature. However, the purified amounts of these two compounds were limited to a few milligrams (~2–3 mg).
To facilitate further investigations on the photodynamic properties of these compounds, larger supplies were necessary. This was initiated by first identifying the strain as a Shiraia-like sp. in the family Shiraiaceae based on maximum likelihood analysis using the ITSr_DNA region.35 Solid-state fermentations of this fungus were grown in triplicate under nine conditions to investigate the effect of culture medium and light exposure on the production of perylenequinones. Herein, we report enhanced production of hypocrellins on solid rice medium as compare to Cheerios or oatmeal media. Alternatively, it was found that oatmeal was the ideal medium to enhance the production of hypomycins, which are closely related analogues that have an additional six-membered ring. Interestingly, light exposure was an essential factor for the production of both hypocrellins and hypomycins. In addition, we report two new perylenequinones, compound 5 (which is the enantiomer of 4), and hypomycin E (8), along with identifying the absolute configuration of hypomycin A (6) and hypomycin C (7) based on analysis of their NOESY spectra and comparing their experimental ECD and VCD data with the calculated spectra.
The Effect of Growth Medium and Light on the Production of Hypocrellins.
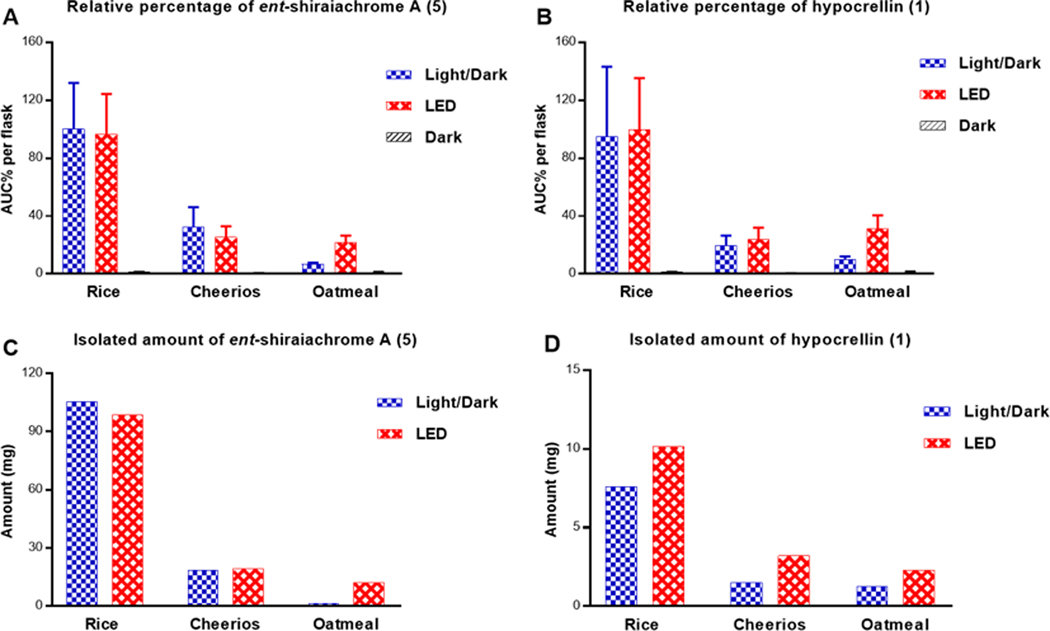
UPLC-HRMS data were collected for extracts and flash chromatography fractions of cultures of strain MSX60519 to confirm the production of ent-shiraiachrome A (5) and hypocrellin (1) across different growth conditions, to measure their relative abundance, and to guide the isolation process of these compounds (Figures S2 and S3). Cultures grown under complete darkness exhibited a low abundance of compounds 5 and 1 (Figure 1). Ent-shiraiachrome A (5) and hypocrellin (1) were optimally produced by cultures grown on rice medium and incubated under 12:12 h light:dark cycles, or continuous LED light exposure (Figure 1), supporting that light facilitates the biosynthesis of these compounds, as suggested by Wang et al.27, 28 Furthermore, the organic extracts were tested for photocytotoxic activity, and cultures grown under complete darkness exhibited significantly reduced photocytotoxic activity as compared to those grown under 12:12 h light:dark cycles or continuous LED light (Figure S4). This further supported a higher production of hypocrellins in cultures that were exposed to light.
Figure 1.
Panels A and B show the relative percentages of 5 and 1, respectively, across cultures grown on rice, Cheerios, and oatmeal media under 12:12 h light:dark cycles, continuous LED, or in darkness. The relative percentages were measured by LC-HRESIMS in three biological replicates and multiplied by the extract weight, and then normalized according to the extract with highest abundance. Panels C and D show the amounts isolated of 5 and 1, respectively, from cultures shown in panels A and B and grown on rice, Cheerios, and oatmeal media under either 12:12 h light:dark cycles or LED conditions.
Perylenequinones are produced by a variety of fungal species, particularly by plant pathogens that cause damage to plant tissues.36 Light-activated hypocrellins exhibited a broad-range of toxicities against bacteria, fungi, as well as human tumor cells.9, 10, 37, 38 As such, obvious questions pertain to how fungi that biosynthesize hypocrellins protect themselves against their own toxins or how light exposure facilitates the biosynthesis of these compounds without a noticeable decrease in fungal growth and development. Although the mechanisms of protection against autotoxicity have not been reported for these fungi, Cercospora spp. were suggested to defend themselves against the cercosporin photosensitizer by a transient reduction and detoxification of cercosporin molecules.39 Reduced cercosporin absorbs less light, and has been shown to be less toxic, than cercosporin itself.36 Similar mechanisms may be adapted by fungi that biosynthesize hypocrellins, where hypocrellins in contact with fungal hyphae are maintained in a reduced (i.e., non-photoactive) form. Aeration by diffusion out of fungal hyphae (or by extraction) results in oxidation. Furthermore, light exposure was reported to be essential for the biosynthesis of cercosporin, and the spectrum of light required for the induction of cercosporin biosynthesis resembled the absorption spectrum of cercosporin itself.40, 41 While more studies are needed, the structural similarity between hypocrellins and cercosporin suggests that analogous mechanisms for autoprotection and the stimulation of biosynthesis are at least plausible.
Based on HRESIMS data, the second fraction from flash chromatography was identified as the hypocrellins-containing fraction. Accordingly, ent-shiraiachrome A (5) and hypocrellin (1) were isolated from this fraction from all cultures grown under either light:dark cycles or continuous LED light. Interestingly, the isolated amounts of 5 and 1 from these cultures nearly matched their relative abundance ratios (Figure 1). The isolated amounts of 5 and 1 from rice cultures that were grown under light:dark cycles were ~100 mg (5) and 8–10 mg (1); nearly identical results were observed from rice cultures grown under continuous LED light. However, the purification process of these compounds using preparative HPLC was more straightforward from cultures grown under continuous LED light (Figure S5). This was due to the production of other non-hypocrellin compounds by cultures grown under light:dark cycles. In addition, < 1 mg of hypocrellin B (3) was isolated from cultures grown on rice exposed to light:dark cycles. To obtain more of this compound, 3 was produced by a dehydration reaction of 5 and 1, as described below.
The Structure Elucidation of 5 and 1 and Generation of 3.
Compound 5 was isolated as a deep red amorphous powder. Using HRESIMS data (Figure S6), the molecular formula was determined as C30H26O10, corresponding to an index of hydrogen deficiency of 18. The 1H and 13C NMR data (Table 1) matched those reported for shiraiachrome A (4),2, 17 and 2D NMR data confirmed that 5 and 4 shared the same 2D structure (Figures S8–S12). However, the ECD spectrum of 5 (Figure S7) was the opposite of the spectrum reported for 4,2, 17 indicating that 5 was the enantiomer of shiraiachrome A (4), thereby suggestive of the trivial name, ent-shiraiachrome A (5). As noted earlier (Figure S1), there are some inconsistencies in the literature regarding the nomenclature of this class of compounds, and indeed, 4 was reported differently in 1989 and 1991, where those authors referred to this compound as shiraiachrome A and hypocrellin B, respectively.2, 17 Moreover, the absolute configuration of 4 was later revised to M(R), 14S, 15S;42 therefore, the absolute configuration of ent-shiraiachrome A (5) was P(S), 14R, 15R.
Table 1.
NMR Spectroscopic Data for 5 and 8 in CDCl3 (500 MHz for 1H and 125 MHz for 13C, δ in ppm)
| 5 | 8 | |||||
|---|---|---|---|---|---|---|
| position | δC, type | δH (J, Hz) | δC, type | δH (J, Hz) | ||
| 1 | 135.2, C | 52.8, C | ||||
| 1a | 127.9, C | 141.4, C | ||||
| 2 | 151.9, C | 39.5, CH2 | 2.91, s | |||
| 3 | 171.7, C | 199.9, C | ||||
| 3a | 106.8, C | 104.3, C | ||||
| 3b | 124.5 or 125.4, C | 129.2, C | ||||
| 4 | 180.1, C | 165.0, C | ||||
| 5 | 102.2, CH | 6.56, s | 100.4, CH | 6.73, s | ||
| 6 | 167.3 or 167.7, C | 164.6, C | ||||
| 6a | 118.0, C | 115.8, C | ||||
| 7 | 167.3 or 167.7, C | 165.6, C | ||||
| 7a | 117.6, C | 112.4, C | ||||
| 8 | 101.8, CH | 6.57, s | 99.4, CH | 6.76, s | ||
| 9 | 179.7, C | 169.9, C | ||||
| 9a | 107.1, C | 107.0, C | ||||
| 9b | 124.5 or 125.4, C | 124.7, C | ||||
| 10 | 171.8, C | 181.4, C | ||||
| 11 | 149.2, C | 149.6, C | ||||
| 12 | 131.3, C | 138.6, C | ||||
| 12a | 128.3, C | 122.5, C | ||||
| 13 | 42.6, CH2 | Ha | 2.34, dd (13.8, 2.0) | 52.9, CH2 | Ha | 1.97, d (13.2) |
| Hb | 3.67, d (13.8) | Hb | 2.61, d (13.2) | |||
| 14 | 78.7, C | 79.2, C | ||||
| 15 | 64.3, CH | 3.72, s | 59.4, CH | 3.86, s | ||
| 16 | 24.9, CH3 | 1.78, s | 32.3, CH3 | 1.77, s | ||
| 17 | 206.8, C | 84.1, C | ||||
| 18 | 28.6, CH3 | 1.83, s | 21.7, CH3 | 1.18, s | ||
| CH3O-2 | 62.0 | 4.18, s | -- | |||
| CH3O-6 | 56.6 or 56.7 | 4.07, s | 56.4 | 4.05, s | ||
| CH3O-7 | 56.6 or 56.7 | 4.07, s | 56.4 | 4.09, s | ||
| CH3O-11 | 61.1 | 4.27, s | 61.2 | 4.05, s | ||
| HO-3 | 15.97, s | |||||
| HO-4 | 13.57, s | |||||
| HO-9 | 15.23, s | |||||
| HO-10 | 16.07, s | |||||
| HO-14 | 3.80, d (2.0) | |||||
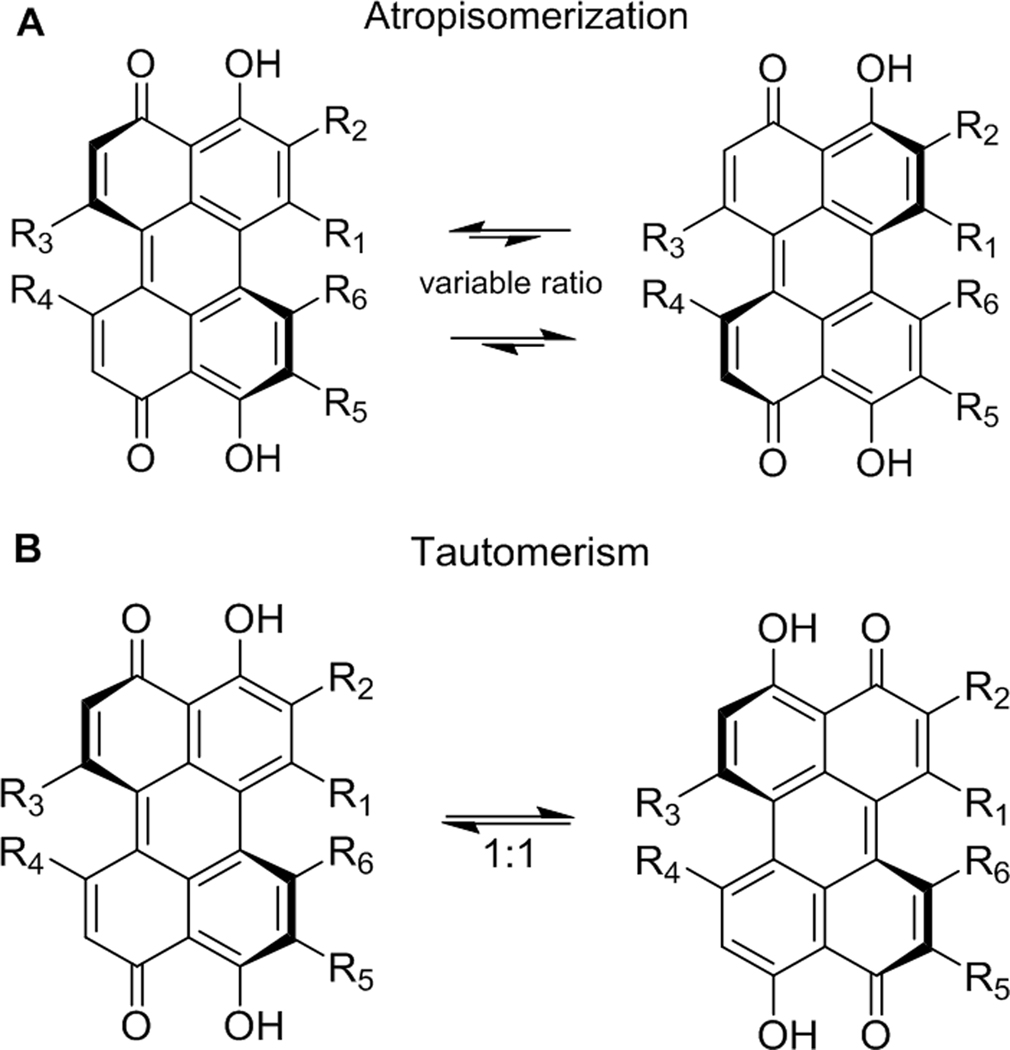
Like other hypocrellins, the fused pentacyclic core of ent-shiraiachrome A (5) is twisted out of plane, thereby establishing axial chirality. The stability of this axial chirality varies among different perylenequinones. In compounds like 5, the additional seven-membered ring, attached to the pentacyclic core, lowers the barrier of atropisomerization for these compounds, such that it can be observed at room temperature.42 For some perylenequinones, this process can be observed on the NMR timescale and involves interconversion between the compound and its atropisomer (Figure 2A).42 Unlike hypocrellin (1) and hypocrellin A (2), which each exist as an equilibrium mixture of two atropisomers in a ratio of 4:1 as recognized by their 1H NMR spectra, the atropisomerization of 4 and its enantiomer (5) occurs at a slower rate and, hence, it takes longer before the atropoisomers are observed by NMR spectroscopy. The 13C NMR spectrum of 5 (Figure S8), collected shortly after its purification, showed a total of 30 carbons, consistent with its molecular formula. However, the number of carbon signals doubled when 5 was stored for ~two months at room temperature (data not shown), suggesting that 5 existed in equilibrium with its atropisomer.
Figure 2.
The atropisomerization process (A) and the tautomerism process (B) of hypocrellins.
Another characteristic of perylenequinones is keto-enol tautomerism. All hypocrellins, including 5, exist in an equilibrium with a 1:1 ratio of the tautomers (Figure 2B). In this case, the tautomerism is too fast to be detected by 1H or 13C NMR spectroscopy using standard experiments.42 However, the HMBC correlations of the HO-3 proton with C-2, C-3, C-3a, C-4, and C-5 (Figure S11) suggested that this proton was in continuous tautomerism with the C-4 carbonyl. Similar HMBC correlations were observed for the HO-10 proton (Figure S11).
The HRESIMS (Figure S6) and NMR data (Table S1 and Figure S13) for 1 matched those reported for hypocrellin (1) and hypocrellin A (2),1, 20 both of which have a molecular formula of C30H26O10. However, 1 was identified as hypocrellin and differentiated from its enantiomer (i.e. 2) based on the ECD spectrum, which compared favorably to the literature (Figure S7).20 Hypocrellin was first isolated from Hypocrella bambusae in 1981, and the absolute configuration was determined by X-ray crystallographic analysis to be M(R), 14R, 15S.1 Hypocrellin (1) and hypocrellin A (2) are known to exist as an equilibrium mixture of two atropisomers in a ratio of 4:1, favoring the more stable atropisomer with M(R) axial chirality for 1 and P(S) for 2.42, 43 The two hypocrellin (1) atropisomers can be recognized by examining the 1H NMR spectrum collected immediately after purification (Figure S13). Moreover, each atropisomer is present in equilibrium with its tautomer in a 1:1 ratio.20, 42
The molecular formula of 3 was deduced as C30H24O9 by HRESIMS data, and the structure was identified as hypocrellin B based on favorable comparisons with the reported 1H and 13C NMR data (Table S1 and Figure S14).17 Interestingly, analysis of 3 by ECD spectroscopy produced a baseline spectrum, which has been attributed by Kozlowski et al.20 to the presence of a racemic mixture of the two atropisomeric enantiomers of 3. Effectively, compound 3 has a lower barrier of atropisomerization, and as such, the ECD Cotton effects of the M(R) and P(S) atropisomers cancel each other. The ability to produce 3 via dehydration of either hypocrellin A (2) or shiraiachrome A (4) has been reported.2, 17 By analogy, 6.5 mg of 3 were generated by a dehydration reaction of a mixture of 1 and 5.
The Effect of Growth Medium and Light on the Production of Hypomycins.
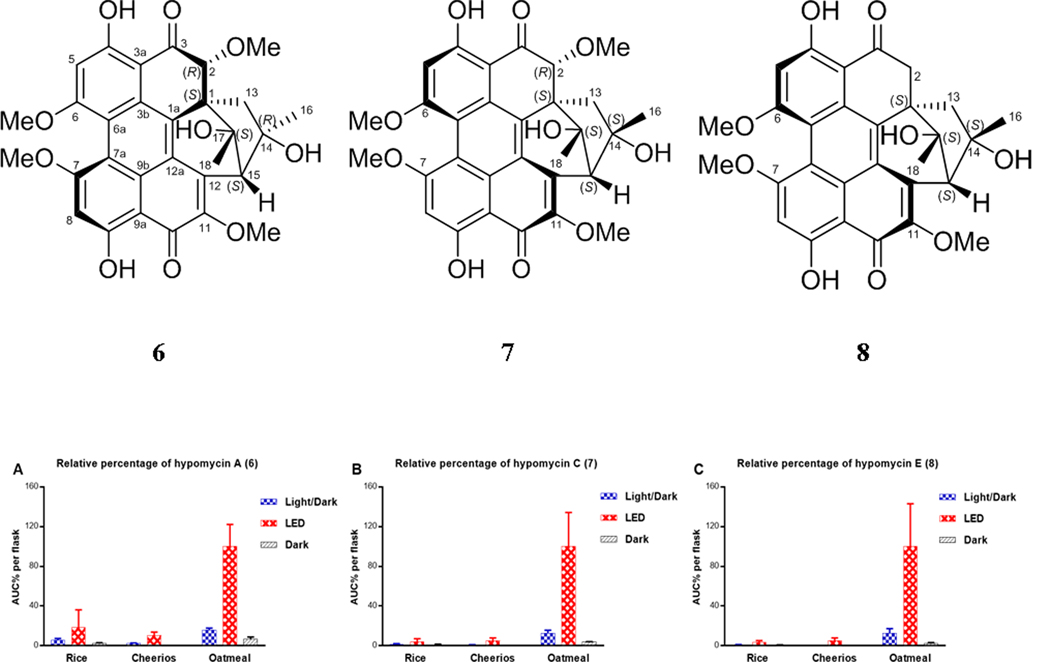
Upon examining the LC-HRESIMS data of the extracts obtained from cultures grown under LED light using oatmeal medium, there were signals for three compounds, other than 1 or 5, that exhibited the characteristic UV spectrum of perylenequinones (Figure S3). These were identified as hypomycin A (6), hypomycin C (7), and hypomycin E (8), as discussed below. Hypomycins have the same pentacyclic core of perylenequinone; however, the typical conjugated system is disrupted by the absence of the Δ1(2) double bond. In addition, the acetyl group observed in hypocrellins has been rearranged in hypomycins to form an additional six-membered ring. Based on the relative abundance of hypomycins across various growths of strain MSX60519 (Figure 3), it was obvious that both oatmeal medium and light were beneficial for the biosynthesis of these compounds. The isolated amounts of 6–8 were the highest from cultures that were grown in oatmeal medium and exposed to continuous LED light (Figure S15). This is the first report of the production of hypocrellins and hypomycins from the same fungus and represents an additional example of how compound production can be tuned based on varying fermentation conditions, including varying the light source.44–46
Figure 3.
Panels A-C show the relative percentages of 6–8, respectively, across cultures grown on rice, Cheerios, and oatmeal media under 12:12 h light:dark cycles, continuous LED, or in darkness. The relative percentages were measured by LC-HRMS in three biological replicates and multiplied by the extract weight, and then normalized according to the extract with highest abundance.
The Structure Elucidation of 6–8.
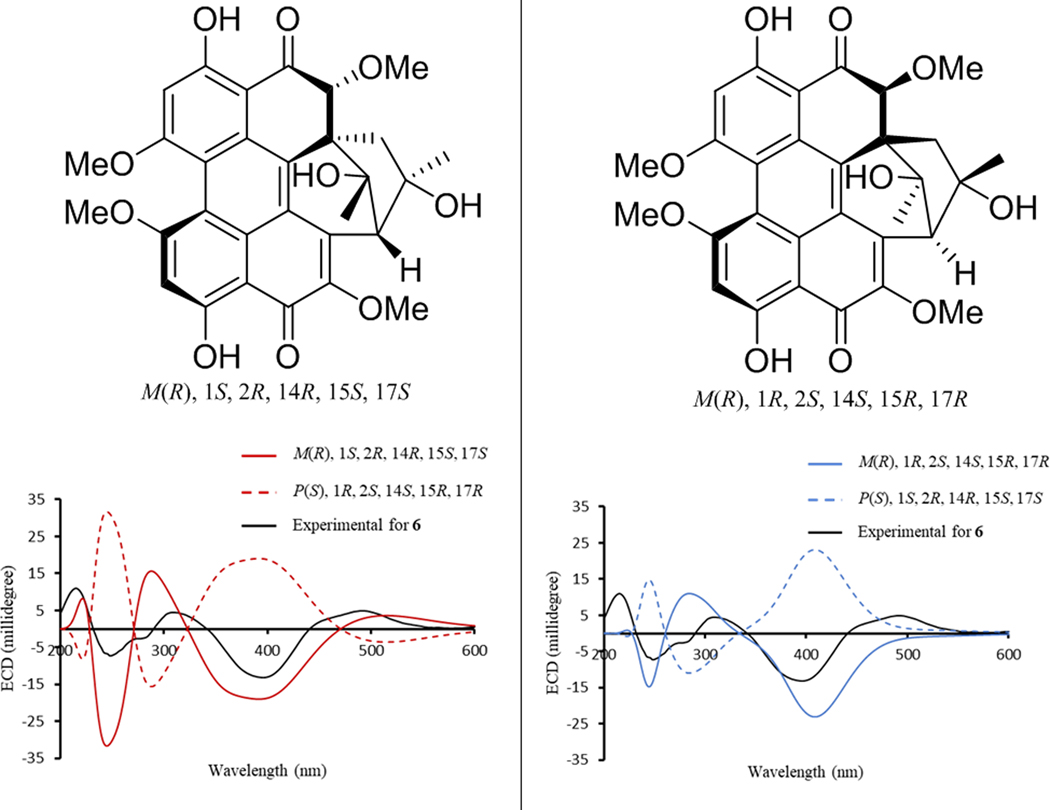
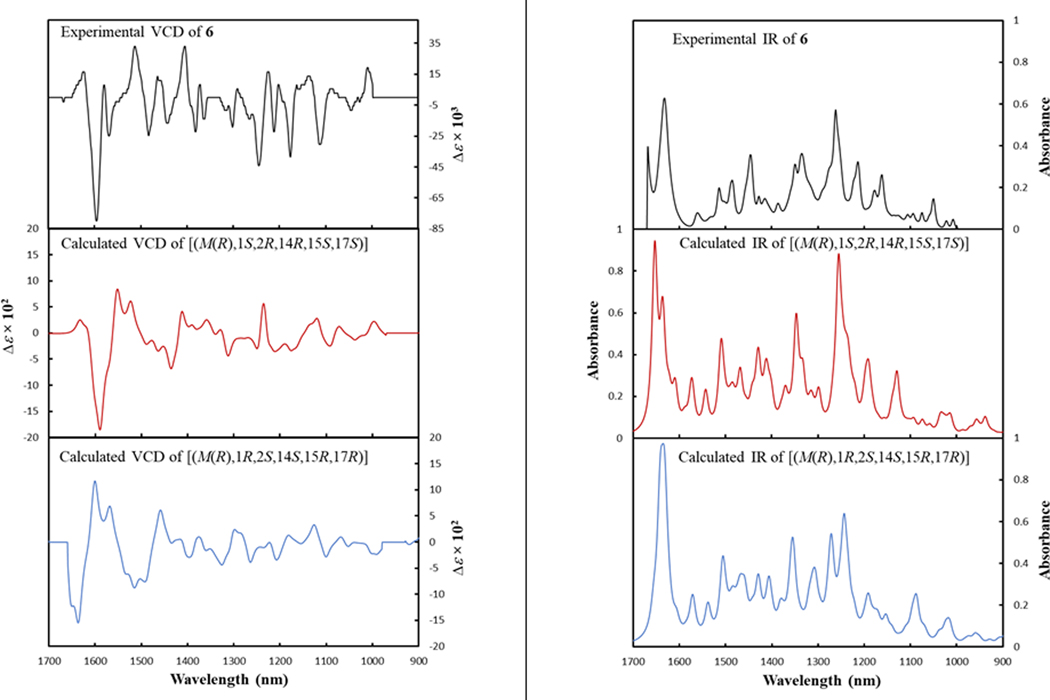
Hypomycins A (6) and C (7) had the same molecular formula, as defined by HRESIMS data (Figure S6), and these were identified by 1H and 13C NMR data (Table S1 and Figures S16–S17), which were congruent with the literature.47, 48 Both compounds were reported previously from the mycelia of Hypomyces sp. (Hypocreales, Ascomycota),47, 48 but only their relative configurations were reported. Updated chemical shift assignments of C-4, C-6, and C-7, for both compounds, based on the HMBC correlations for HO-4, H-5, and H-8 are given in Table S1. Unlike hypocrellins, the axial chirality of hypomycins has not been reported, and little is known about their biological effects or light-induced activities. Compounds 6 and 7 exhibited opposite ECD spectra (Figure S7). Moreover, the ECD spectrum of 6 was similar to that exhibited by 1, while the spectrum of 7 was similar to that of 5. Since axial chirality is the major factor that generates the Cotton effects observed for perylenequinones,49 the axial chirality would be M(R) for 6 and P(S) for 7; these assignments were confirmed via calculation of the ECD spectrum of 6. The calculated ECD spectra for the two possible configurations of 6 with M(R) axial chirality [(M(R), 1S, 2R, 14R, 15S, 17S) and (M(R), 1R, 2S, 14S, 15R, 17R)] showed good agreement with the experimental ECD spectrum of 6 (Figure 4). However, only the [M(R), 1S, 2R, 14R, 15S, 17S]- configuration reproduced the positive Cotton effect at 500 nm (Figure 4). Accordingly, the experimental ECD Cotton effects of hypomycins A (6) and C (7) were mainly generated by their axial chirality.
Figure 4.
Experimental ECD spectrum of 6 compared with calculated ECD spectra of [M(R), 1S, 2R, 14R, 15S, 17S]- configuration and its enantiomer (left frame), and [M(R), 1R, 2S, 14S, 15R, 17R]- configuration and its enantiomer (right frame).
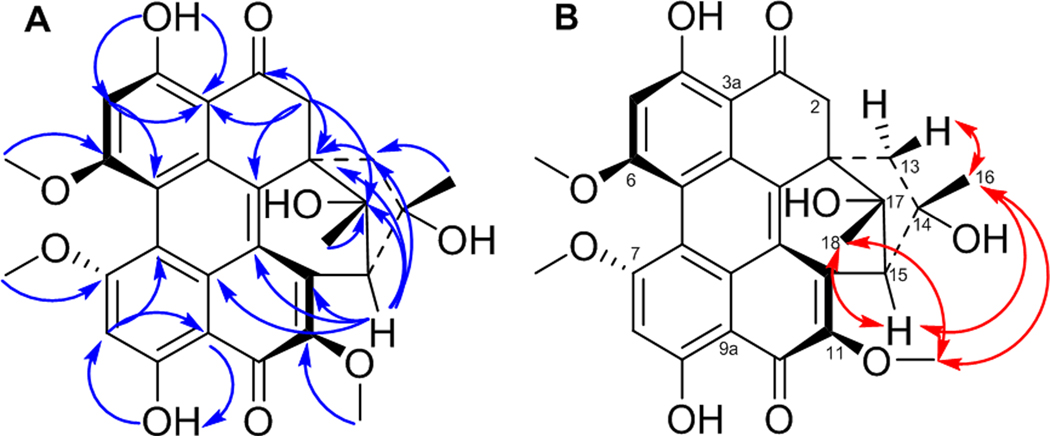
A series of NOESY correlations were used to differentiate between compounds 6 and 7 (Figures S18 and S19). The NOESY spectrum of 6 showed strong correlations between CH3-16/2-OCH3, CH3-16/11-OCH3, and CH3-16/Ha-13, suggesting the same orientation for CH3-16, 2-OCH3, and Ha-13 (Figure S20). In addition, CH3-18 and H-2 were cofacial, opposite to the correlations noted above, also based on NOESY correlations. The orientation of the six-membered ring was suggested by the NOESY correlations of Hb-13 with both the HO-14 and HO-17 protons (Figure S20). Alternatively, in hypomycin C (7), the NOESY correlations of CH3O-11/CH3-16, CH3-16/Hb-13, CH3O-11/CH3-18, CH3-18/H-2 suggested the same orientation for CH3-16, CH-18, Hb-13, and H-2 (Figure S20). The correlation of Hb-13 with CH3-16 and HO-17 supported the same orientation of the six-membered ring as in 6. Therefore, compounds 6 and 7 differed in the configuration of the C-14 stereogenic center and their axial chirality. The absolute configuration of 6 was suggested by comparing the experimental VCD spectrum with the calculated VCD spectra for the two possible configurations of 6, [M(R), 1S, 2R, 14R, 15S, 17S], or [M(R), 1R, 2S, 14S, 15R, 17R]. The experimental VCD of 6 showed the best match with the calculated VCD spectrum of [M(R), 1S, 2R, 14R, 15S, 17S] configuration (Figure 5). Accordingly, the absolute configurations of 6 and 7 were defined as [M(R), 1S, 2R, 14R, 15S, 17S] and [P(S), 1S, 2R, 14S, 15S, 17S], respectively.
Figure 5.
Experimental VCD (left frame) and IR (right frame) spectra observed for hypomycin A (6) compared with calculated VCD and IR spectra of [M(R), 1S, 2R, 14R, 15S, 17S]-configuration (0.65 similarity factor to 6) and [M(R), 1R, 2S, 14S, 15R, 17R]- configuration (0.11 similarity factor to 6). Specdis software was utilized to calculate the similarity factors.50
The molecular formula of 8 was deduced as C29H26O9 by HRESIMS data (Figure S6), suggesting a loss of a methoxy group relative to 6 and 7. The 1D and 2D NMR spectra of 8 (Figures S21–S25) confirmed the presence of methoxy groups attached to C-6, C-7, and C-11, but the absence of the methoxy group at C-2. HMBC correlations of the C-2 methylene protons with C-1, C-1a, C-3, C-3a, and C-17 supported this assignment (Figure 6A). Comparing the NOESY correlations of 8 with those observed for 6 and 7 suggested that 8 exhibited the same relative configuration as hypomycin C (7). For example, key NOESY interactions of CH3O-11/CH3-16, CH3O-11/CH3-18, and CH3-16/Hb-13 indicated the same orientation for CH3-16 and CH3-18 (Figure 6B). In addition, H-15 showed strong NOESY correlations with both CH3-16 and CH3-18. The ECD spectrum of 8 was highly similar to 7 (Figure S7), confirming the P(S) axial chirality. Accordingly, the absolute configuration of 8 was defined as [P(S), 1S, 14S, 15S, 17S], and this compound was ascribed the trivial name hypomycin E.
Figure 6.
Key HMBC (A) and NOESY (B) correlations for hypomycin E (8).
Cytotoxicity and Photocytotoxicity Toward Human Melanoma Cancer Cells (SK-MEL-28).
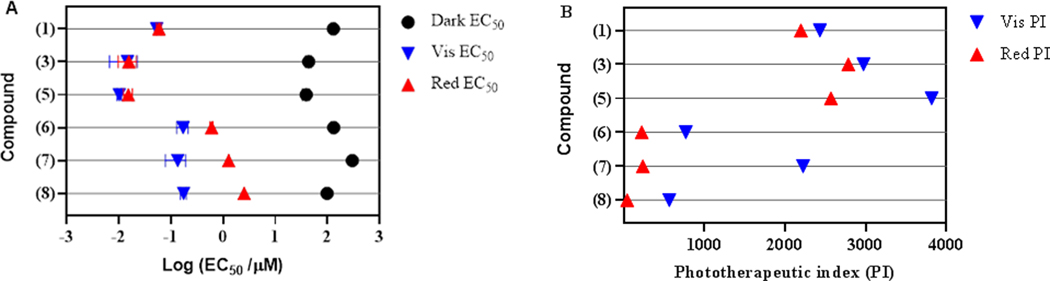
Perylenequinones are generally known for their promising light-induced cytotoxicity.51 The photocytotoxic activity of hypocrellins A (2) and B (3) was reported previously.16, 52, 53 However, the limited supply of hypomycins hindered more in depth investigations of their photoactivated cytotoxic properties. As such, the activities of hypocrellins 1, 3, and 5 and hypomycins 6–8 were evaluated against a human skin melanoma cancer cell line (SK-MEL-28) for photocytotoxicity. Briefly, melanoma cells were treated with the compound of interest at a concentration range of 1 nM to 300 μM and incubated at 37 °C (5% CO2) for 16 h before evaluation under dark, broadband visible-light (100 J/cm2), or monochromatic red-light (100 J/cm2, 625 nm) treatments, resulting in both EC50 values and phototherapeutic indices (PIs) (Table 3). The phototherapeutic index represents the ratio of the dark to light EC50 values and indicates the amplification of cytotoxicity with the light treatment. Hypocrellins 1, 3, and 5 exhibited potent photocytotoxic activities at the nanomolar level, with ent-shiraiachrome A (5) and hypocrellin B (3) being the most active. Moreover, comparable light-induced cytotoxicity of these compounds was observed with visible- and red-light treatments (Figures 7 and S26). On the other hand, hypomycins 6–8 were less potent photosensitizers with EC50 values at the micromolar level. This might be attributed to the absence of the Δ1(2) double bond in 6–8, resulting in reduced conjugation. In contrast to hypocrellins, hypomycins exhibited reduced cytotoxicity with red-light treatment as compared to visible-light treatment (Figure 7). In both classes of compounds, toxicity without the presence of light was much lower than light-induced cytotoxicity. This allowed for large phototherapeutic index (PI) values for hypocrellins that ranged between 2200–3800 for both visible- and red-light treatments. The low dark toxicity of hypomycin C (7) resulted in a large PI with visible-light treatment (> 2200). In summary, hypocrellins, and potentially hypomycins, represent promising naturally-derived photosensitizers that compare well with the photodynamic activity of the FDA approved photosensitizer for cancer treatment, photofrin.54 The compounds in this family with PIs >103 are under further investigation to test the robustness of this response in other cell lines and models, including in vivo tumors.
Table 3.
EC50 Values of Compounds 1, 3, and 5–8Against SK-MEL-28 Cancer Cells
| compound | Dark (μM) | Visa (nM) | PIvisb | Redc (nM) | PIredd |
|---|---|---|---|---|---|
| 1 | 130.0 ± 12.0 | 53.5 ± 1.0 | 2430 | 59.2 ± 6.0 | 2196 |
| 3 | 43.7 ± 5.9 | 14.7 ± 8.0 | 2973 | 15.7 ± 6.0 | 2783 |
| 5 | 39.3 ± 6.2 | 10.3 ± 1.0 | 3816 | 15.3 ± 3.0 | 2569 |
| 6 | 132.0 ± 13.0 | 172.0 ± 43.0 | 767 | 603.0 ± 41.0 | 219 |
| 7 | > 300 | 135.0 ± 56.0 | > 2222 | 1280 ± 20.0 | > 234 |
| 8 | 98.7 ± 1.7 | 176.0 ± 23.0 | 561 | 2550 ± 60.0 | 39 |
Vis: 16 hours drug-to-light interval followed by 100 J/cm2 broadband visible-light irradiation
PIvis = phototherapeutic index (ratio of dark EC50 to visible-light EC50)
Red: 16 hours drug-to-light interval followed by 100 J/cm2 light irradiation with 625 nm LEDs
PIred = phototherapeutic index (ratio of dark EC50 to red-light EC50).
Figure 7.
Panel A: photocytotoxic activity plot for compounds 1, 3, and 5–8 against SK-MEL-28 melanoma cells without (black) or with visible- (blue) or red- (red) light irradiation. Panel B: phototherapeutic index (PI) activity plot for compounds 1, 3, 5–8 against SK-MEL-28 melanoma cells treated with visible- (blue) or red- light (red).
In conclusion, the present study was carried out to enhance the production of two different classes of perylenequinones, hypocrellins and hypomycins, via fermentation. Interestingly, the biosynthesis of these fungal metabolites could be tuned based on media and light exposure. Ent-shiraiachrome A (5) and hypomycin E (8) were reported for the first time, and additional studies were carried out to refine the structure elucidation of 6 and 7, including the combined use of both ECD and VCD data for the former. Both hypocrellins and hypomycins exhibited significant photodynamic activity. Hypocrellins, in particular, showed potent light-induced cytotoxicity at the nanomolar level. Equipotent photodynamic activities with both visible- and red-light treatment renders hypocrellins as suitable candidates for further in vivo photodynamic therapy (PDT) studies.
EXPERIMENTAL SECTION
General Experimental Procedures.
Ultraviolet (UV) and electronic circular dichroism (ECD) data were measured using a Varian Cary 100 Bio UV‒Vis spectrophotometer (Varian Inc.) and an Olis DSM 17 ECD spectrophotometer (Olis, Inc.), respectively. The IR and VCD measurements were performed on a BioTools ChiralIR FT-VCD spectrometer equipped with dual photoelastic modulation (PEM). A sample of 3.0 mg of 6 was dissolved in 150 μL CDCl3 and placed in a BaF2 IR cell with a path length of 100 μm, and data were acquired at 4 cm−1 resolution with total measurement time of 12 h and 1400 cm−1 PEM setting. 1D and 2D NMR data were obtained using a JEOL ECA-500 spectrometer operating at 500 MHz or a JEOL ECS-400 spectrometer operating at 400 MHz that is equipped with a high sensitivity JEOL Royal probe and a 24-slot autosampler (both from JEOL Ltd.). Residual solvent signals were utilized for referencing. UPLC-HRMS data were collected via Thermo Fisher Scientifc Q Exactive Plus mass spectrometer equipped with an electrospray ionization source (ESI) and connected to a Waters Acquity UPLC system. A BEH Shield RP18 column (Waters, 1.7 μm; 50 × 2.1 mm) was used and heated to 40°C. The flow rate of the mobile phase was 0.3 mL/min and consisted of a gradient system of 15:85 to 100:0 of CH3CN-H2O (1% formic acid) over 10 min. MS data were collected from m/z 150 to 2000 while alternating between positive and negative modes. The same UPLC system (Waters Corp.) utilizing the same column was used to measure the purity of compounds 1, 3, and 5–8 with data collected and analyzed using Empower 3 software (Figure S30). A Varian Prostar HPLC system, equipped with ProStar 210 pumps and a Prostar 335 photodiode array detector (PDA), was used to conduct all analytical and preparative HPLC experiments, with data collected and analyzed using Galaxie Chromatography Workstation software (version 1.9.3.2, Varian Inc.). Flash chromatography was performed on a Teledyne ISCO CombiFlash Rf 200 using Silica Gold columns (from Teledyne Isco) and monitored by UV and evaporative light‒scattering detectors.
Identification of Fungal Strain.
Fungal strain MSX60519 was isolated from dry leaf litter by A. Abraham on 2/15/1992. Based on growth on potato dextrose agar and malt extract agar, Difco, the fungal strain did not show any characteristics that were useful for morphological identification. The culture only grew as sterile mycelium. Therefore, to identify this strain, genomic DNA extraction, PCR, and sequencing were performed for the nuclear ribosomal internal transcribed spacers and 5.8s gene (ITS rDNA region) using methods outlined previously.35 The PCR experiment was performed using primer combination ITS1F and ITS4.55, 56 The PCR reaction was cleaned up using a Wizard SV Gel and PCR Clean-up System (Promega) followed by Sanger sequencing using the same primer combination ITS1F and ITS4 at Eurofins Scientific, USA. The contig was assembled in Sequencher 5.2.3 (Gene Codes), optimized by eye and BLAST searched in NCBI GenBank to estimate the placement of strain MSX60519. Since hypocrellins-producing strains are produced by fungal genera such as Hypocrella bambusae (Sordariomycetes, Ascomycota), Shiraia bambusicola, and Rubroshiraia bambusae (Dothideomycetes, Ascomycota), we hypothesized that MSX60519 would likely show homology and phylogenetic affinities towards one of these classes of fungi. A BLAST search in NCBI showed that strain MSX60519 shared homology with members of the Pleosporales, Dothideomycetes, Ascomycota with one strain, Pleosporales sp. MX286 (JQ905814), which was isolated as an endophyte from Hevea brasiliensis (rubber tree) in Peru showing 99% sequence similarity, while all other strains in the Pleosporales showed ≥ 90–95% sequence similarity. Since the results of the BLAST search were equivocal, further placement of MSX60519 was evaluated by Maximum Likelihood (RAxML) phylogenetic analysis of the ITS region using methods outlined previously.35 The choice of taxon sampling was based on phylogenetic studies of Pleosporalean taxa, which were known to produce hypocrellins.3, 4 Interestingly, based on the RAxML analysis using the ITS region, strain MSX60519 was placed basal to Shiraia bambusicola, and Rubroshiraia bambusae as well as endophytic Shiraia-like fungal clade (group A) as defined by Morakatkarn et al.3 Strain MSX60519 was placed in the family Shiraiaceae (Pleosporales, Dothideomycetes, Ascomycota) with 89% RAxML bootstrap support (Figure S27). Strain MSX60519 showed 100% RAxML bootstrap support with rubber tree endophyte from Peru stain MX286 (JQ905814)57 (Figure S27). The RAxML analysis indicated that all hypocrellins-producing strains in the Shiraiaceae, Pleosporales are monophyletic (Figure S27). Furthermore, all strains in endophytic Shiraia-like fungal clade group A produced pinkish to reddish pigment in potato dextrose media,3 which was similar to the morphology of strain MSX60519 on both potato dextrose and malt extract agar, including solid-state rice fermentation (Figures S27 and S28), which was used to isolate hypocrellins and hypomycins in the present study. Therefore, based on RAxML analysis, strain MSX60159 was identified as a Shiraia-like fungus in the family Shiraiaceae, Pleosporales, Dothideomycetes, Ascomycota. The ITS sequences have been deposited in GenBank under the accession number: MN970609 and MN970610. A live culture of this strain is accessioned at Mycosynthetix, Inc. (Hillsborough, NC, USA).
Media and Fermentations.
The culture of fungal strain MSX60519 was maintained on potato dextrose agar (PDA; Difco) and was transferred periodically to fresh PDA Petri plates. An agar plug from the leading edge of the PDA culture was transferred to a sterile tube with 10 mL of YESD (2% soy peptone, 2% dextrose, and 1% yeast extract). The YESD culture was grown for 7 days on an orbital shaker (100 rpm) at room temperature (~23˚C) and then used to inoculate three types of solid fermentation media.
Nine cultures of strain MSX60519 were grown over three different grain-based media in triplicate: rice, Cheerios breakfast cereal,58 and breakfast oatmeal (old fashioned Quaker oats) for a total of 27 cultures. Solid-state fermentations were carried out in 250 mL Erlenmeyer flasks. To prepare rice medium, 10 g of rice were added to each flask with 20 mL of deionized water (DI H2O). For the oatmeal medium, the same amount was used in each flask with 17 mL of DI H2O. For Cheerios medium, 7 g of Cheerios were used in each flask without water. After autoclaving these samples at 120° C for 20 min, the flasks were inoculated with YESD seed cultures (described above) and incubated at room temperature for 15 days under three different conditions.
Out of the nine cultures that were grown on rice medium, three were incubated under usual room light with 12:12 h light:dark cycles to simulate daily light:dark cycles. Three were placed inside a light box (33 cm × 24 cm × 15 cm) that was equipped with light-emitting diode (LED) lamps (Ustellar, flexible LED strip lights; 24 watt) to incubate these cultures under continuous light exposure for 15 days. Finally, three cultures were incubated in complete darkness by covering the flasks with aluminum foil and placing them inside a dark area for 15 days. The same growth parameters were employed for the nine cultures that were grown on Cheerios and oatmeal medium, respectively (Figure S29). Over the incubation period, the fungal cultures grew normally with no sign of growth retardation, except for the cultures growing on oatmeal medium under 12:12 h light:dark cycles, which were dried out by the time of extraction (Figure S28).
Extraction, Fractionation, and Isolation.
Each flask of solid culture was extracted with 90 mL of 1:2 CH3OH-CHCl3. Each culture was chopped and shaken for 20 h at ~125 rpm. The extract was vacuum filtered, and the remaining residues were washed with 30 mL of CHCl3. To the filtrate, 90 mL of CHCl3 and 100 mL of DI H2O were added, and the mixture was stirred for 30 min and then transferred into a separatory funnel. The bottom layer was drawn off and evaporated to dryness. The dried organic extract was re-constituted in 100 mL of 1:1 CH3OH-CH3CN and 100 mL of hexanes. The biphasic solution was shaken vigorously and transferred into a separatory funnel. The CH3OH/CH3CN layer was drawn off and evaporated to dryness under vacuum. The extract amounts that were produced by each culture were not significantly different across cultures grown under the different conditions (Figure S2).
Before performing further purifications, quantitative UPLC-HRMS data were collected. The organic extracts collected from the triplicate cultures, which were grown under the same growth conditions, were combined and dissolved in CHCl3, adsorbed onto Celite 545, and subdivided into four fractions via normal-phase flash chromatography using a gradient solvent system of hexanes-CHCl3-CH3OH at a 30 mL/min flow rate and 75 column volumes over 42 min. The second flash chromatography fraction of each extract was subjected to preparative HPLC over a Phenomenex Synergi C12 preparative column using an isocratic system of 60:40 of CH3CN-H2O (0.1% formic acid) over 45 min at a flow rate of 21.24 mL/min to yield ent-shiraiachrome A (5), hypocrellin (1), and hypomycin A (6). The third flash chromatography fraction of each extract was subjected to preparative HPLC using a Phenomenex Synergi preparative column on an isocratic system of 50:50 of CH3CN-H2O (0.1% formic acid) over 30 min to give hypomycin C (7) and hypomycin E (8). The isolated amounts of 1, 5 and 6–8, based on the suite of media and light conditions, are provided in Figures 1 and S15. The purity of 1, 3, and 5–8 were evaluated using an Acquity UPLC system, which showed > 97% purity (Figure S30).
Hypocrellin (1):
Dark red amorphous powder; UV (CH3OH) λmax (log ε) 581 (3.96), 540 (3.94), 464 (4.25), 341 (3.62), 285 (4.31), 265 (4.38), 213 (4.56) nm; ECD (c 3.7 × 10−4 M, CH3OH) λmax (Δε) 232 (5.22), 268 (−4.48), 294 (−4.02), 346 (2.66), 446 (−2.72), 532 (2.52), 576 (2.33) nm; 1H NMR (CDCl3, 400 MHz) and 13C NMR (CDCl3, 100 MHz) (see Table 1); HRESIMS m/z 547.1596 [M + H]+ (calcd. for C30H27O10, 547.1604).
Hypocrellin B (3):
Dark red amorphous powder; UV (CH3OH) λmax (log ε) 590 (3.92), 548 (4.12), 460 (4.40), 335 (3.92), 223 (4.71) nm; 1H NMR (CDCl3, 500 MHz) and 13C NMR (CDCl3, 125 MHz) (see Table 1); HRESIMS m/z 529.1491 [M + H]+ (calcd. for C30H25O9, 529.1498).
Ent-shiraiachrome A (5):
Dark red amorphous powder; UV (CH3OH) λmax (log ε) 581 (4.07), 540 (4.05), 467 (4.34), 343 (3.72), 286 (4.38), 265 (4.48), 214 (4.65) nm; ECD (c 3.7 × 10−4 M, CH3OH) λmax (Δε) 202 (−11.43), 270 (8.79), 352 (−7.00), 460 (9.47), 582 (−4.54) nm; 1H NMR (CDCl3, 500 MHz) and 13C NMR (CDCl3, 125 MHz) (see Table 1); HRESIMS m/z 547.1590 [M + H]+ (calcd. for C30H27O10, 547.1604).
Hypomycin A (6):
Orange amorphous powder; UV (CH3OH) λmax (log ε) 527 (3.68), 494 (3.03), 465 (3.99), 414 (4.39), 394 (4.03), 276 (4.55), 242 (4.53), 215 (4.58) nm; ECD (c 3.7 × 10−4 M, CH3OH) λmax (Δε) 216 (10.95), 248 (−7.33), 308 (4.45), 396 (−13.22), 490 (4.88) nm; 1H NMR (CDCl3, 400 MHz) and 13C NMR (CDCl3, 100 MHz) (see Table 1); HRESIMS m/z 549.1746 [M + H]+ (calcd. for C30H29O10, 549.1760).
Hypomycin C (7):
Orange amorphous powder; UV (CH3OH) λmax (log ε) 522 (3.46), 489 (3.82), 462 (3.80), 413 (4.24), 393 (4.16), 274 (3.40), 241 (4.36), 214 (4.44) nm; ECD (c 3.7 × 10−4 M, CH3OH) λmax (Δε) 218 (−11.66), 256 (12.37), 310 (−7.46), 394 (20.39), 492 (−5.28) nm; 1H NMR (CDCl3, 400 MHz) and 13C NMR (CDCl3, 100 MHz) (see Table 1); HRESIMS m/z 549.1749 [M + H]+ (calcd. for C30H29O10, 549.1760).
Hypomycin E (8):
Orange amorphous powder; UV (CH3OH) λmax (log ε) 489 (3.86), 461 (3.85), 412 (4.29), 394 (4.15), 336 (3.80), 273 (4.44), 243 (4.42), 215 (4.45) nm; ECD (c 3.9 × 10−4 M, CH3OH) λmax (Δε) 216 (−18.56), 252 (20.00), 310 (−6.63), 388 (23.60), 486 (−7.25) nm; 1H NMR (CDCl3, 400 MHz) and 13C NMR (CDCl3, 125 MHz) (see Table 1); HRESIMS m/z 519.1646 [M + H]+ (calcd. for C29H27O9, 519.1655).
Semi-synthesis of Hypocrellin B.
Hypocrellin B (3) was produced from a mixture of 5 and 1 (~11 mg) by a dehydration reaction, as previously described.2 In brief, the mixture was treated with 7 mL of a 3% KOH solution and left at room temperature for 5 h. The reaction mixture was neutralized with 7 mL of a 10% HCl solution and extracted with 20 mL CHCl3. The organic fraction was evaporated to dryness under vacuum and then subjected to preparative HPLC over a Phenomenex PFP preparative column using an isocratic system of 60:40 of CH3CN-H2O (0.1% formic acid) over 45 min at a flow rate of 21.24 mL/min to yield 6.5 mg of hypocrellin B (3).
Computational Methods.
Molecular Merck force field (MMFF) and density functional theory (TD-DFT/DFT) calculations were carried out with Spartan’10 (Wavefunction Inc., Irvine, CA, USA) and GaussView 06 software, respectively. The conformers were optimized using DFT calculations at the B3LYP/cc-pVTZ level in CH3OH for ECD prediction of 6, and in CDCl3 for VCD prediction of 6. The ECD and VCD spectra were generated using SpecDis 1.71 software.50
Cytotoxicity and Photocytotoxicity Assay.
The in vitro cytotoxic and photocytotoxic activities of compounds 1, 3, and 5–8 were evaluated against SK-MEL-28 melanoma cells as previously described.59, 60 In brief, SK-MEL-28 cells (~550,000–600,000 cells/mL) were transferred in 50 μL aliquots to the inner wells of a 96-well TC-treated microtiter plates, and then incubated at 37 ˚C under 5% CO2 for 3 h. Solutions of compounds 1, 3, and 5–8 were serially diluted with DPBS and aliquots of 25 μL of the appropriate dilutions were added to cells, and the plates were incubated at 37 ˚C under 5% CO2 for 16 h. Dark (control) microplates were kept in the incubator, and light-treated microplates were irradiated for approximately 48 and 60 min under one of the following conditions: visible-light (400−700 nm, 34.7 mW/cm2) using a 190 W BenQ MS 510 overhead projector or red-light (625 nm, 27.8 mW/cm2) from an LED array. Following the dark or light treatment, the microplates were incubated for 48 h followed by the addition of 10 μL of prewarmed alamarBlue reagent (Life Technologies DAL 1025) to all sample wells. The plates were subsequently incubated for another 15–16 h. Cell viability was determined by fluorescence using a Cytofluor 4000 fluorescence microplate reader (excitation = 530 ± 25 nm and emission = 620 ± 40 nm). Cisplatin was used as a positive control for cytotoxicity, yielding an EC50 value of 3.4 μM in the dark, with no significant changes upon illumination with visible or red light. [Ru(bpy)2(dppn]Cl2 was used as a negative control for cytotoxicity (dark EC50 >100 μM) and a positive control for photocytotoxicity (vis EC50=290 nM, red EC50=760 nM).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by the National Institutes of Health via the National Cancer Institute (P01 CA125066) and by the National Science and Engineering Research Council of Canada (NSERC). We thank Tyler Graf for helpful suggestions and edits and Mario Augustinović for help submitting ECD and VCD calculations.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Images of MSX60519 on different solid-state growth conditions, UPLC chromatograms of culture extracts, phototoxic activity (EC50 values) of extracts, UPLC chromatograms and (+)-HRESIMS of compounds 1, 3, 5–8, selected 1D and 2D NMR spectra for compounds 1, 3, 5–8, key NOESY correlations for 6 and 7, UV and ECD spectra, representative analytical HPLC chromatograms for the purification of 1 and 5, and phylograms of the most likely tree.
REFRENCES
- (1).Wei-Shin C; Yuan-Teng C; Xiang-Yi W; Friedrichs E; Puff H; Breitmaier E. Liebigs Ann. Chem. 1981, 1981, 1880–1885. [Google Scholar]
- (2).Wu H; Lao X-F; Wang Q-W; Lu R-R; Shen C; Zhang F; Liu M; Jia L. J. Nat. Prod. 1989, 52, 948–951. [Google Scholar]
- (3).Morakotkarn D; Kawasaki H; Seki T; Okane I; Tanaka K. Mycoscience 2008, 49, 258–265. [Google Scholar]
- (4).Dai DQ; Wijayawardene NN; Tang LZ; Liu C; Han LH; Chu HL; Wang HB; Liao CF; Yang EF; Xu RF; Li YM; Hyde KD; Bhat DJ; Cannon PF MycoKeys 2019, 58, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Diwu Z. Photochem. Photobiol. 1995, 61, 529–539. [DOI] [PubMed] [Google Scholar]
- (6).Jiang Y; Leung AW; Wang X; Zhang H; Xu C. Int. J. Radiat. Biol. 2014, 90, 575–579. [DOI] [PubMed] [Google Scholar]
- (7).Qi S; Guo L; Yan S; Lee RJ; Yu S; Chen S. Acta. Pharm. Sin. B 2019, 9, 279–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Du W; Sun C; Liang Z; Han Y; Yu J. World J. Microbiol. Biotechnol. 2012, 28, 3151–3157. [DOI] [PubMed] [Google Scholar]
- (9).Yang Y; Wang C; Zhuge Y; Zhang J; Xu K; Zhang Q; Zhang H; Chen H; Chu M; Jia C. Front. Microbiol. 2019, 10, 1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Su Y; Sun J; Rao S; Cai Y; Yang Y. J. Photochem. Photobiol. B, Biol. 2011, 103, 29–34. [DOI] [PubMed] [Google Scholar]
- (11).Hudson JB; Zhou J; Chen J; Harris L. Photochem. Photobiol. 1994, 60, 253. [DOI] [PubMed] [Google Scholar]
- (12).Donohoe C; Senge MO; Arnaut LG; Gomes-da-Silva LC Biochim. Biophys. Acta. Rev. Cancer. 2019, 1872, 188308. [DOI] [PubMed] [Google Scholar]
- (13).Zang LY; Zhang ZY; Misra HP Photochem. Photobiol. 1990, 52, 677–83. [DOI] [PubMed] [Google Scholar]
- (14).Ali S; Olivo M. Int. J. Oncol. 2002, 21, 1229–1237. [PubMed] [Google Scholar]
- (15).Ma G; Khan SI; Jacob MR; Tekwani BL; Li Z; Pasco DS; Walker LA; Khan IA Antimicrob. Agents Chemother. 2004, 48, 4450–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ma F; Huang H; Ge X; Yang X; Yang C; Han L; Zhou J; Zhou L. Bioorg. Med. Chem. Lett. 2013, 23, 1689–1692. [DOI] [PubMed] [Google Scholar]
- (17).Kishi T; Tahara S; Taniguchi N; Tsuda M; Tanaka C; Takahashi S. Planta Med. 1991, 57, 376–9. [DOI] [PubMed] [Google Scholar]
- (18).Liu X; Xie J; Zhang L; Chen H; Gu Y; Zhao JJ Photochem. Photobiol. B, Biol. 2009, 94, 171–178. [DOI] [PubMed] [Google Scholar]
- (19).Ma J-H; Zhao J-Q; Jiang L-J Free Radic. Res. 2001, 35, 607–617. [DOI] [PubMed] [Google Scholar]
- (20).O’Brien EM; Morgan BJ; Mulrooney CA; Carroll PJ; Kozlowski MC J. Org. Chem. 2010, 75, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lei XY; Zhang MY; Ma YJ; Wang JW J. Ind. Microbiol. Biotechnol. 2017, 44, 1415–1429. [DOI] [PubMed] [Google Scholar]
- (22).Ma YJ; Zheng LP; Wang JW Microb. Cell Fact. 2019, 18, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ma YJ; Zheng LP; Wang JW Front. Microbiol. 2019, 10, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Cai Y; Liang X; Liao X; Ding Y; Sun J; Li X. Appl. Biochem. Biotechnol. 2010, 160, 2275–86. [DOI] [PubMed] [Google Scholar]
- (25).Sun CX; Ma YJ; Wang JW Ultrason. Sonochem. 2017, 38, 214–224. [DOI] [PubMed] [Google Scholar]
- (26).Tisch D; Schmoll M. Appl. Biochem. Biotechnol. 2010, 85, 1259–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ma YJ; Sun CX; Wang JW Photochem. Photobiol. 2019, 95, 812–822. [DOI] [PubMed] [Google Scholar]
- (28).Sun CX; Ma YJ; Wang JW J. Photochem. Photobiol. B, Biol. 2018, 182, 100–107. [DOI] [PubMed] [Google Scholar]
- (29).Gao R; Xu Z; Deng H; Guan Z; Liao X; Zhao Y; Zheng X; Cai Y. Arch. Microbiol. 2018, 200, 1217–1225. [DOI] [PubMed] [Google Scholar]
- (30).Kinghorn AD; Carache de Blanco EJ; Lucas DM; Rakotondraibe HL; Orjala J; Soejarto DD; Oberlies NH; Pearce CJ; Wani MC; Stockwell BR; Burdette JE; Swanson SM; Fuchs JR; Phelps MA; Xu L; Zhang X; Shen YY Anticancer Res. 2016, 36, 5623–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Bills G; Martin J; Collado J; Platas G; Overy D; Ruben Tormo J; Vicente F; Verkley GJM; Crous PW SIMB News 2009, 133–147. [Google Scholar]
- (32).Paguigan ND; El-Elimat T; Kao D; Raja HA; Pearce CJ; Oberlies NH J. Antibiot. 2017, 70, 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).El-Elimat T; Figueroa M; Ehrmann BM; Cech NB; Pearce CJ; Oberlies NH J. Nat. Prod. 2013, 76, 1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Graf TN; Kao D; Rivera-Chávez J; Gallagher JM; Raja HA; Oberlies NH Planta Med. 2020, DOI: 10.1055/a-1130-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Raja HA; Miller AN; Pearce CJ; Oberlies NH J. Nat. Prod. 2017, 80, 756–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Daub ME; Ehrenshaft M. Physiol. Plant. 1993, 89, 227–236. [Google Scholar]
- (37).Leung AW; Ip M; Xu CS; Wang XN; Yung PT; Hua HY Hong Kong Med. J. 2017, 23, 36–37. [PubMed] [Google Scholar]
- (38).Wang Z-J; He Y-Y; Huang C-G; Huang J-S; Huang Y-C; An J-Y; Gu Y; Jiang L-J Photochem. Photobiol. 1999, 70, 773–780. [PubMed] [Google Scholar]
- (39).Sollod CC; Jenns AE; Daub ME Appl. Environ. Microbiol. 1992, 58, 444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lynch FJ; Geoghegan MJ Trans. Br. Mycol. Soc. 1979, 73, 311–327. [Google Scholar]
- (41).Ehrenshaft M; Upchurch RG Appl. Environ. Microbiol. 1991, 57, 2671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Mazzini S; Merlini L; Mondelli R; Scaglioni LJ Chem. Soc., Perkin Trans. 2 2001, 409–416. [Google Scholar]
- (43).Smirnov A; Fulton DB; Andreotti A; Petrich JW J. Am. Chem. Soc. 1999, 121, 7979–7988. [Google Scholar]
- (44).Elias BC; Said S; de Albuquerque S; Pupo MT Mycol. Res. 2006, 161, 273–280. [DOI] [PubMed] [Google Scholar]
- (45).Bills GF; Gloer JB Microbiol. Spectr. 2016, 4, FUNK-0009–2016. [DOI] [PubMed] [Google Scholar]
- (46).Frisvad JC Methods Mol. Biol. 2012, 944, 47–58. [DOI] [PubMed] [Google Scholar]
- (47).Liu WZ; Shen YX; Liu XF; Chen YT; Xie JL Chin. Chem. Lett. 2001, 12, 431–432. [Google Scholar]
- (48).Shen YX; Liu WZ; Rong XG; Sun YH Acta. Pharm. Sin. 2003, 38, 834–7. [PubMed] [Google Scholar]
- (49).Hu J; Sarrami F; Li H; Zhang G; Stubbs KA; Lacey E; Stewart SG; Karton A; Piggott Andrew M.; Chooi Y-H Chem. Sci. 2019, 10, 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Bruhn T; Schaumloffel A; Hemberger Y; Bringmann G. Chirality 2013, 25, 243–9. [DOI] [PubMed] [Google Scholar]
- (51).Mulrooney CA; O’Brien EM; Morgan BJ; Kozlowski MC European. J. Org. Chem. 2012, 2012, 3887–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Zhou Z; Liu Y; Qin M; Sheng W; Wang X; Li Z; Zhong R. J. Photochem. Photobiol. B, Biol. 2014, 134, 1–8. [DOI] [PubMed] [Google Scholar]
- (53).Cao EH; Xin SM; Cheng LS Int. J. Radiat. Biol. 1992, 61, 213–9. [DOI] [PubMed] [Google Scholar]
- (54).Shi R; Li C; Jiang Z; Li W; Wang A; Wei J. Molecules 2017, 22, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Gardes M; White TJ; Fortin JA; Bruns TD; Taylor JW Can. J. Bot. 1991, 69, 180–190. [Google Scholar]
- (56).Innis MA; Gelfand DH; Sninsky JJ; White TJ, PCR Protocols : a Guide to Methods and Applications. Elsevier Science: St. Louis, 2014. [Google Scholar]
- (57).Gazis R; Chaverri P. Fungal Ecol. 2010, 3, 240–254. [Google Scholar]
- (58).Du L; King JB; Cichewicz RH J. Nat. Prod. 2014, 77, 2454–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Monro S; Yin H; McFarland SA; Colon KL; Roque J; Cameron CG; McFarland SA; Konda P; Gujar S; Gujar S; McFarland SA; Gujar S; Gujar S; Thummel RP; Lilge L. Chem. Rev. 2019, 119, 797–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).McCain J; Colón KL; Barrett PC; Monro SMA; Sainuddin T; Roque Iii J; Pinto M; Yin H; Cameron CG; McFarland SA Inorg. Chem. 2019, 58, 10778–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.