Abstract
Genetically engineered mouse models through gene deletion are useful tools for analyzing gene function. To delete a gene in a certain tissue temporally, tissue-specific and tamoxifen-inducible Cre transgenic mice are generally used. Here, we generated transgenic mouse with cardiac-specific expression of Cre recombinase fused to a mutant estrogen ligand-binding domain (ERT2) on both N-terminal and C-terminal under the regulatory region of human vasoactive intestinal peptide receptor 2 (VIPR2) intron and Hsp68 promoter (VIPR2-ERT2CreERT2). In VIPR2-ERT2CreERT2 transgenic mice, mRNA for Cre gene was highly expressed in the heart. To further reveal heart-specific Cre expression, VIPR2-ERT2CreERT2 mice mated with ROSA26-lacZ reporter mice were examined by X-gal staining. Results of X-gal staining revealed that Cre-dependent recombination occurred only in the heart after treatment with tamoxifen. Taken together, these results demonstrate that VIPR2-ERT2CreERT2 transgenic mouse is a useful model to unveil a specific gene function in the heart.
Keywords: VIPR2 intron, ERT2CreERT2, Heart, Tamoxifen-inducible Cre transgenic mouse
Introduction
The heart is the first organ to develop. It is also the most important organ for survival during embryonic development [1]. Gene knockout experiments have shown that abnormal development of the heart is the main cause of embryonic lethality [2], making it difficult to study specific gene function in the adult heart. To resolve this limitation, Cre/loxP recombination system has been developed to examine gene functions through gene deletion using adult mice [3, 4]. Such Cre/loxP-mediated conditional gene targeting can be used to study tissue-specific gene function, including genes expressed in the heart [5]. To activate Cre in a spatiotemporal manner, tamoxifen-inducible Cre/loxP recombination system requires Cre recombinase fused with a mutant form of estrogen receptor (CreERT2) which can migrate to the nucleus and induce site-specific recombination after treatment with tamoxifen [6]. Leaky Cre recombinase activity irrelevant to tamoxifen treatment can be more tightly controlled by placing the mutant estrogen receptor to both ends of Cre protein such as MerCreMer [7] and ERT2CreERT2 [8].
Regulatory DNA sequences are essential to achieve specific Cre expression. However, most Cre mouse lines express Cre in more than one specific tissue, leading to misinterpretation of data due to indirect effect of Cre-mediated gene deletion in other tissues on overall phenotype. In the present study, we selected an enhancer for heart-specific gene expression using VISTA enhancer browser and experimentally tested enhancer activity in transgenic mice [9]. Among these enhancers, hs1753 located in intron 4 of human VIPR2 gene exhibited LacZ expression only in the heart, not in other region of embryonic day 11.5 (E11.5) mouse embryos. We generated a tamoxifen-inducible Cre transgenic mice using VIPR2-ERT2CreERT2 expression vector consisting of hs1753 regulatory sequence, Hsp68 promoter, ERT2CreERT2 cDNA, and the polyadenylation site from the SV40 early region. We investigated whether Cre-dependent recombination occurs only in the heart of the embryo or adult after tamoxifen treatment on mice born by further crossing the VIPR2-ERT2CreERT2 transgenic mouse with the ROSA26-lacZ reporter mouse.
Materials and methods
Construction of VIPR2-ERT2CreERT2 expression vector
Plasmid pCAG-ERT2CreERT2 was a gift from Connie Cepko (Addgene plasmid # 13777). pHsp68-LacZ-Gateway was a gift from Nadav Ahituv (Addgene plasmid # 37843) and the plasmid pCre-ERT2 was a gift from Pierre Chambon. All restriction enzymes and Phusion® high-fidelity DNA polymerase for PCR amplification were purchased from NEB (Ipswich, MA, USA). EcoR1-SacI fragment (2.0 kb) and SacI-SalI (0.17 kb) fragment of pCre-ERT2 were cloned to EcoR1 and SalI sites of pBluescript II SK(+) to generate pCre-ERT2pA vector. A SmaI-ClaI fragment (1.8 kb) of pCAG-ERT2CreERT2 was further cloned to SmaI and ClaI sites of pCre-ERT2pA to generate pERT2CreERT2pA vector without promoter or enhancer. Finally, VIPR2-driven ERT2CreERT2 expression vector (pVIPR2-ERT2CreERT2pA) was constructed as follows. Human VIPR2 genomic DNA located in intron 4 was amplified by PCR using a BAC clone (RP11-645 K21, BACPAC Resources, Oakland, CA, USA) with sense primer (5′-AAGCGGCCGCTGGGAGGAGAAGGGCTCTGC-3′, NotI site underlined) and antisense primer (5′-TTGACGCGTGAGAACAGGAGTGTCACCGG-3′; MluI site underlined). The PCR product (3.0 kb) was digested with NotI and MluI followed by purification with QIAquick PCR purification kit (Qiagen, Germany). The minimal promoter sequence of Hsp68 was amplified by PCR using a pHsp68-LacZ-Gateway (Ref: PubMed 17086198) with sense primer (5′- TTGACGCGTGAGCTTCCAGGAACATCCAAA-3′, MluI site underlined) and antisense primer (5′-AATCTAGACGCTCTGCTTCTGGAAGGCT-3′, XbaI site underlined). The PCR product (0.9 kb) was digested with MluI and XbaI followed by purification with QIAquick PCR purification kit. Both fragments were cloned to NotI and SpeI sites of pERT2CreERT2pA to obtain pVIPR2-ERT2CreERT2pA. 5′ VIPR2 sequence of pVIPR2-ERT2CreERT2pA was verified by DNA sequencing. NotI-KpnI fragment of VIPR2-ERT2CreERT2 expression vector was purified with QIAquick gel extraction kit (Qiagen) and dissolved in TE buffer (10 mM Tris-HCl, pH 7.4, 0.25 mM EDTA) to 1 μg/ml for microinjection.
Generation and maintenance of transgenic mice
Three-week-old female FVB/N mice were injected intraperitoneally with pregnant mare serum gonadotropin (Merck KGaA, Germany) and human chorionic gonadotropin (hCG, Merck KGaA) 48 h later. After administration of hCG, female mice were mated with FVB/N male mice. The ampulla region of oviduct from plugged females was teared with a fine needle in 1x PBS containing 0.1% hyaluronidase (Merck KGaA) and 0.1% polyvinylpyrrolidone (M.W. 40 kDa, Merck KGaA). Fertilized eggs were collected using mouth-controlled pipet. These eggs were washed several times with M2 medium (Merck KGaA) and cultured in the drop of KSOM medium (Merck KGaA) under mineral oil (Merck KGaA) until microinjection. After pronuclear injection of DNA, eggs survived microinjection were then transferred into oviducts of pseudopregnant ICR females. Founder mice containing injection gene were identified with PCR genotyping. Subsequent generation of transgenic mice were maintained congenic on C57BL/6 J genetic background. All mice experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Ewha Womans University (Permit number: 2015–01-072).
PCR genotyping
Toe clips from 7 to 10 days old pups were used to extract DNA for PCR genotyping with 4 primers. PCR primers Hsp68-S1 (5′-CAGGTCACCAGACGCTGACA-3′) and ERT2-AS1 (5′-TCATGTCTCCAGCCATGGTG-3′) produced a 276-bp PCR product specific for transgene while PCR primers Ereg-S3 and Ereg-AS1 primers produced a 159-bp PCR product from Ereg gene for endogenous control [10]. PCR product was resolved with 1.2% agarose gel electrophoresis and visualized with ethidium bromide staining.
RNA preparation, reverse-transcription (RT), and quantitative RT-PCR (qRT-PCR)
After euthanasia of mouse with CO2, tissues were isolated and immediately frozen in liquid nitrogen. Total RNA was prepared using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol. To remove any residual DNA in RNA preparation, RNA solution was treated with DNaseI (Thermo Fisher Scientific) at 37 °C for 30 min and followed by RNA clean up with RNeasy MinElute Cleanup Kit (Qiagen, Germany). Then cDNA was generated from RNA using Superscript III reverse transcriptase (Thermo Fisher Scientific) and random hexamers as described previously [10]. Reverse transcription products were amplified by real-time PCR with Cre-S (5′-GGCATGGTGCAAGTTGAAT-3′) and Cre-AS (5′-AGCATTGCTGTCACTTGGTC-3′); Il6-S (5′-ATGAGAAAAGAGTTGTGCAATGGC-3′) and Il6-AS (5′-CCAGGTAGCTATGGTACTCCAGAA-3′); Col3a1-S (5′-GATGAGCTTTGTGCAAAGTGG-3′) and Col3a1-AS (5′-CGCAAAGGACAGATCCTGA-3′) primers. 18S rRNA-S (5′-TCAACTTTCGATGGTAGTCGCC-3′) and 18S rRNA-AS (5′-GGCCTCGAAAGAGTCCTGTATTGT-3′) primers were used to determine expression level of 18S rRNA as endogenous control. Real-time PCR was performed with KAPA SYBR FAST ABI Prism qPCR Kit (KAPA Biosystems, Wilmington, MA, USA) using ABI Prism 7300 (Thermo Fisher Scientific) machine. The relative amount of transcript was quantified as described previously [11].
Tamoxifen treatment
Tamoxifen (Merck KGaA) was dissolved in ethanol to 100 mg/ml or 200 mg/ml and further diluted with sunflower seed oil (Merck KGaA) to 10 mg/ml and 20 mg/ml, respectively [12]. For collection of embryos, pregnant mice at 9 days post coitum were injected intraperitoneally with 0.1 ml of 10 mg/ml tamoxifen solution for three consecutive days. Embryos were harvested at 1 day or 3 days after the last injection. For collection of adult tissues, mice (3 to 5 months old) were injected intraperitoneally with 0.1 ml of 20 mg/ml tamoxifen for five consecutive days and tissues were harvested at 7 days after the final injection. Ethanol in sunflower seed oil at identical concentration was injected intraperitoneally as a negative control.
LacZ staining
Embryos were collected from pregnant Gtrosa26tm1Sor/tm1Sor females mated with VIPR2-ERT2CreERT2 males and stained with X-gal as described previously [13]. Whole-mount embryos were imaged using a SMZ 1000 dissecting microscope (Nikon). Tissues from adult mice derived from breeding Gtrosa26tm1Sor/tm1Sor females with VIPR2-ERT2CreERT2Tg/+ males were collected, fixed, and stained with X-gal as described previously [10]. Slides were analyzed using a Nikon Eclipse 80i microscope and images were captured with a DS-Ri1 camera (Nikon).
Statistical analysis
Experimental groups were compared with one-way ANOVA with Turkey’s multiple comparison test using GraphPad Prism program (GraphPad Software, San Diego, CA, USA).
Results and discussion
Tamoxifen-dependent Cre activity in embryonic heart of transgenic mice
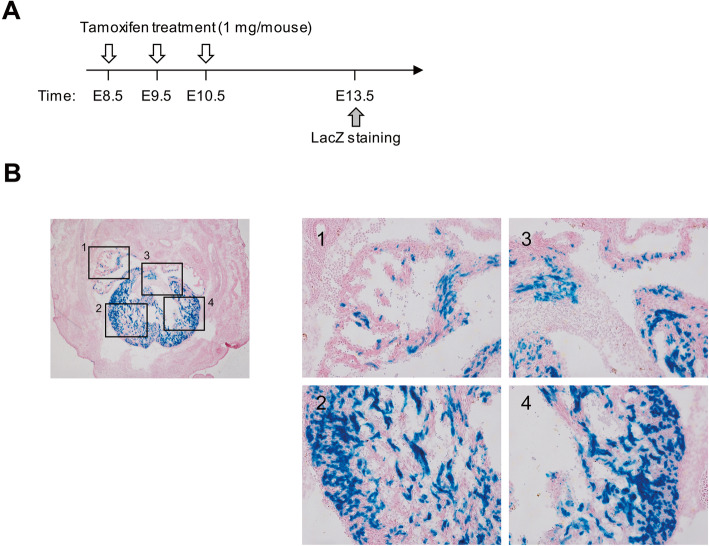
VISTA enhancer browser was used to identify enhancer for heart-specific gene expression [9]. Based on LacZ staining of E11.5 embryos (https://enhancer.lbl.gov), hs1753 (chr7:158,888,320-158,891,362) located in intron 4 of human VIPR2 gene was subcloned into Hsp68 promoter fused with ERT2CreERT2-pA to generate VIPR2-ERT2CreERT2 expression vector (Fig. 1a). A total of nine transgenic founders were obtained out of 22 newborn pups. Subsequent generations of transgenic mice were identified by PCR genotyping (Fig. 1b). To determine Cre expression, transgenic males mated with homozygous Gtrosa26tm1Sor [14] females were examined for the expression of LacZ due to excision of Gtrosa26tm1Sor by tamoxifen-dependent Cre recombinase. After three consecutive injections of tamoxifen, whole embryos were stained with X-gal (Fig. 1c). All transgenic lines showed heart-specific LacZ staining only in VIPR2-ERT2CreERT2Tg/+ and Gtrosa26tm1Sor/+ compound heterozygous (VIPR2:R26) embryos (Fig. 1d). Because VIPR2-ERT2CreERT2–001 line exhibited the strongest expression of LacZ, further studies were performed with this line as a representative. To confirm the specificity of Cre recombination in the heart, we analyzed VIPR2:R26 E13.5 embryo sections after treating pregnant mice with tamoxifen (Fig. 2a). Heart-specific LacZ expression was detected in the whole region of the heart with punctate pattern (Fig. 2b). These results indicate that the 3.04-kb of VIPR2 intron 4 harbors cis-regulatory elements for heart-specific expression during embryonic development.
Fig. 1.
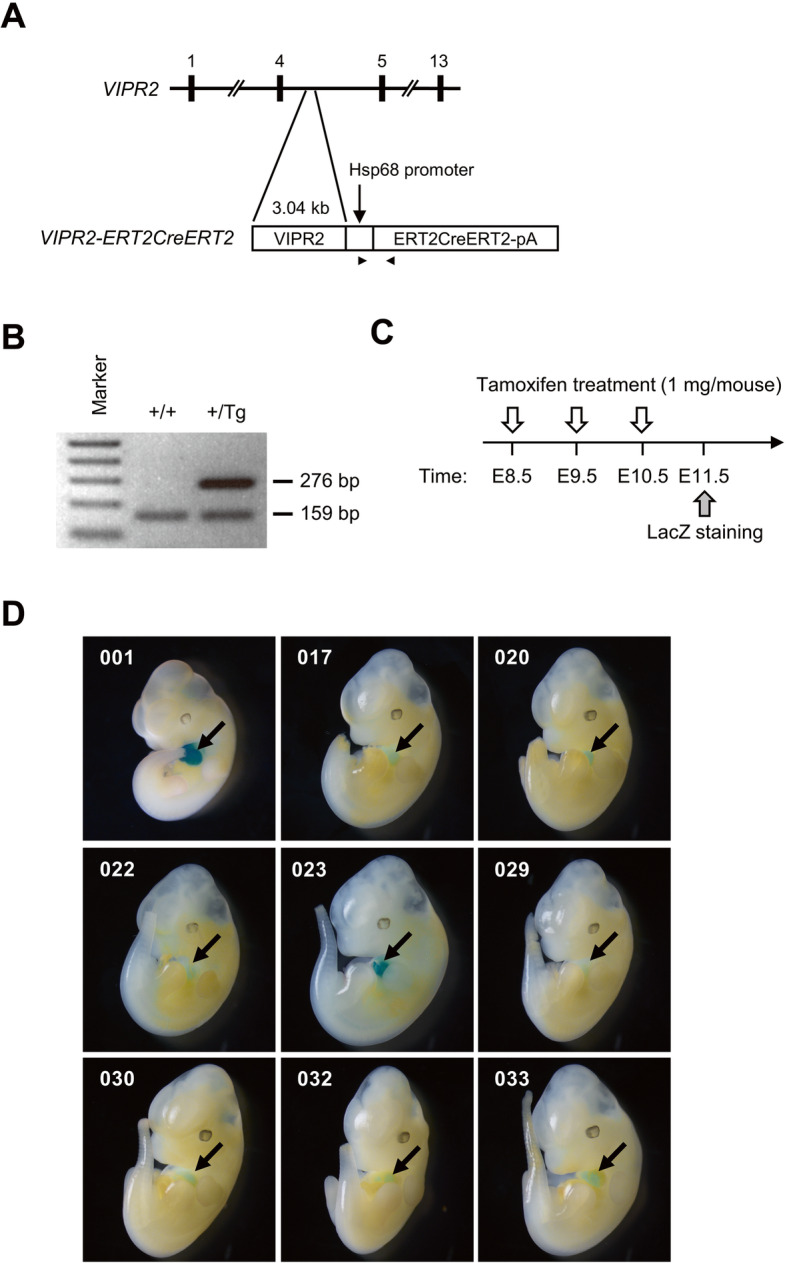
Generation of VIPR2-ERT2CreERT2 transgenic mice and analysis of transgene expression. a Schematic structure of VIPR2-ERT2CreERT2 transgene. Numbers indicate exons of human VIPR2 gene. Arrowheads indicate primers for genotyping. b PCR genotyping of representative founder mice showing a 276-bp PCR product specific for transgene and a 159-bp product for endogenous gene (+/+, wild-type; +/Tg, heterozygous transgenic mouse). c Strategies for treating pregnant mice with tamoxifen followed by LacZ staining. d Whole-mount LacZ staining of VIPR2:R26 compound heterozygotes E11.5 embryos. Each number indicates VIPR2-ERT2CreERT2 transgenic mouse line. Arrows indicate heart-specific X-gal staining. Original magnification, 20 ×
Fig. 2.
LacZ staining of E13.5 embryos. a Strategies for treating pregnant mice (Gtrosa26tm1Sor/tm1Sor mated with VIPR2-ERT2CreERT2Tg/+) with tamoxifen followed by LacZ staining. b Frozen section was stained with X-gal followed by counterstaining with hematoxylin and eosin Y (H&E) briefly. Left, original magnification, 40×; Right, 100× magnification of rectangle 1 to 4. Scale bar, 50 μm
Heart-specific Cre activity in adult VIPR2-ERT2CreERT2 transgenic mice
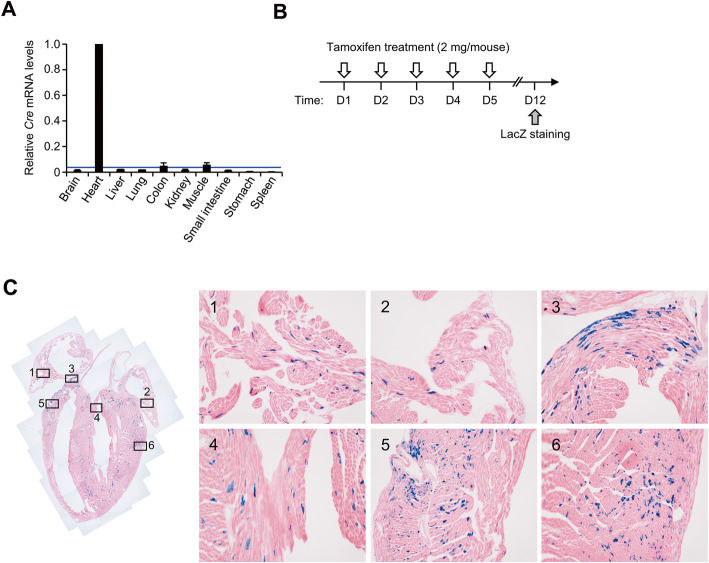
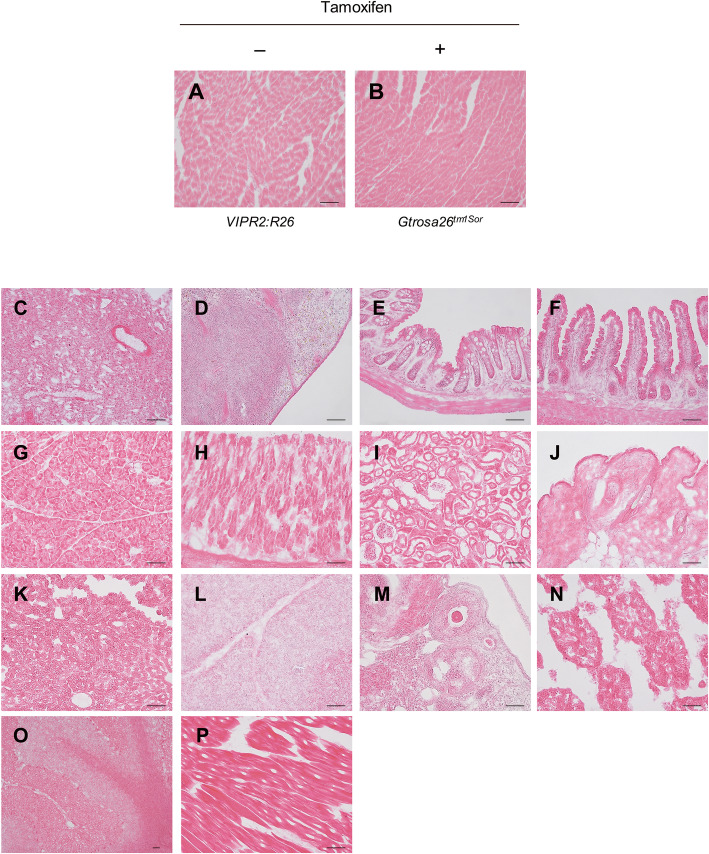
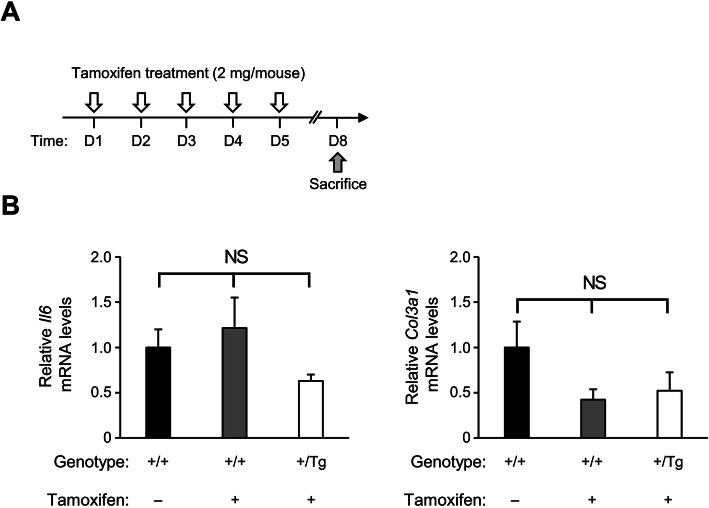
Before analyzing LacZ activity in adult VIPR2:R26 mouse, we determined Cre mRNA levels in various organs of VIPR2-ERT2CreERT2–001 line. Relative expression levels of Cre mRNA were basal in most organs compared to those in the heart (Fig. 3a). To further confirm tamoxifen induced Cre recombination, VIPR2:R26 adult mice were treated with either 2 mg of tamoxifen or the same volume of ethanol with sunflower oil mixture (1:9, v/v) as control injection for five consecutive days. Organs were harvested 7 days after the last injection (Fig. 3b). LacZ staining was detected mainly in cardiomyocytes of all regions with punctate pattern similar to that in the embryo (Fig. 3c). In contrast, LacZ staining was not observed in the heart of VIPR2:R26 adult mouse treated with solvent alone or Gtrosa26tm1Sor/+ mouse treated with tamoxifen solution (Fig. 4a, b). The heart was the only organ stained with X-gal while all other organs were negative for the X-gal staining (Fig. 4c to p). The degree of X-gal staining of the heart did not differ by gender. Moreover, consecutive treatment of VIPR2-ERT2CreERT2 transgenic mouse with tamoxifen did not show harmful effects such as inflammatory response or heart reorganization (Fig. 5a to c). Overall, these results indicate that VIPR2-ERT2CreERT2 transgenic mouse is useful for heart-specific gene knockout experiment using adult mice without leaky expression or adverse effect.
Fig. 3.
Analysis of tissue-specific Cre activity. a qRT-PCR analysis of relative Cre mRNA levels in various tissues. Total RNA prepared from VIPR2-ERT2CreERT2Tg/+ adult mice was analyzed by real-time PCR and the relative Cre mRNA level in each tissue was compared to the level of the heart (n = 2). The blue line indicates the non-specific Cre mRNA level of’ the wild-type heart. Data are presented as mean ± SD. Only the level of the heart is statistically significant (p < 0.01). b Strategies for treating adult VIPR2:R26 compound heterozygotes with tamoxifen followed by LacZ staining. c Frozen sections of the heart were stained with X-gal followed by counterstaining with H&E briefly. 1, right atrium; 2 and 3, left atrium; 4, aortic valve; 5, right ventricle; 6, left ventricle. Original magnification, 40×; Right, 100× magnification of rectangles 1 to 6
Fig. 4.
Analysis of tissue-specific Cre activity by LacZ staining. Frozen sections were stained with X-gal followed by counterstaining with H&E briefly. a Heart section of adult VIPR2:R26 compound heterozygote mouse without tamoxifen treatment. b Heart section of adult Gtrosa26tm1Sor/+ treated with tamoxifen; c-p Adult VIPR2:R26 compound heterozygote mouse treated with tamoxifen: c Lung; d Spleen; e Colon; f Small intestine; g Pancreas; h Stomach; i Kidney; j Skin; k Liver; l Thymus; m Ovary; n Testis; o Brain; p Muscle. Scale bar, 50 μm
Fig. 5.
Analysis of short-term treatment of tamoxifen on heart. a Strategies for treating adult VIPR2-ERT2CreERT2Tg/+ and wild-type mice with tamoxifen. b and c Total RNA prepared from each group of mice (n = 4) was analyzed by qRT-PCR. Relative Il-6 mRNA and Col3a1 mRNA levels in the heart were then compared. Data are presented as mean ± SEM. NS, statistically insignificant, one-way ANOVA
The adult heart is composed of several cell types, including cardiomyocytes, cardiac fibroblasts, vascular smooth muscle cells, and endothelial cells. Fluorescence-activated cell sorting (FACS) analysis has revealed that cardiomyocytes, the majority of cell types, account for 56% of heart cell types while the rest of heart cells are cardiac fibroblasts (27%), vascular smooth muscle cells (10%), and endothelial cells (7%) [15]. In the present study, cardiomyocytes were the major cell types for expressing LacZ while vascular smooth muscle cells and endothelial cells were rare in the heart of transgenic mice. The most widely used gene for driving heart-specific Cre expression is α-myosin heavy chain (Myh6) [16]. Inducible Cre recombinase using Myh6 promoter has also been established [17, 18]. However, adverse effect of tamoxifen treatment on normal heart function in Myh6-MerCreMer mice requires appropriate control mice to avoid misinterpretation of conditional knockout mice phenotype [19, 20]. Recently developed Cre knock-in mouse by targeting a MerCreMer cassette into the start codon of Myh6 seems to be able to replace Myh6-MerCreMer mice since these knock-in mice exhibit normal heart function regardless of tamoxifen treatment [21]. In contrast to robust recombination in cardiomyocytes of Myh6 promoter-driven Cre lines, VIPR2-ERT2CreERT2 transgenic mouse exhibited far less recombination in the heart, suggesting that this transgenic mouse could be useful for analyzing genes whose cardiomyocytes-wide deletion might lead to heart failure.
Conclusions
LacZ staining after crossing with Rosa26-lacZ reporter mouse revealed that VIPR2-ERT2CreERT2 transgenic mice showed tamoxifen-dependent Cre activity only in the heart. This transgenic mouse can be used as a tool to temporally and spatially target genes in the heart and then, making it a useful model for revealing specific gene functions in the heart.
Acknowledgments
We thank Daekee’s lab member for animal care and helpful discussion.
Abbreviations
- CreERT2
Cre recombinase fused with a mutant form of estrogen receptor
- E11.5
Embryonic day 11.5
- ERT2
Mutant form of estrogen receptor
- FACS
Fluorescence-activated cell sorting
- H&E
Hematoxylin and eosin Y
- hCG
Human chorionic gonadotropin.
- IACUC
Institutional Animal Care and Use Committee
- Myh6
α-myosin heavy chain
- qRT-PCR
Quantitative RT-PCR
- RT
Reverse-transcription
- VIPR2
Vasoactive intestinal peptide receptor 2
Authors’ contributions
HJC, S-yL and DL designed research and performed experiments; HJC and DL analyzed data and wrote the manuscript. The author(s) read and approved the final manuscript.
Funding
This research was supported by a 2014 grant (14182MFDS978) from the Ministry of Food and Drug Safety, Republic of Korea.
Availability of data and materials
VIPR2-ERT2CreERT2–001 transgenic mouse was backcrossed with C57BL/6 J mice for 10 generations, and then deposited in the Korea National Institute of Food and Drug Safety Evaluation for distribution [Stock number, 18-NIFDS-M-ET-001; Stock name, B6.Cg-Tg (VIPR2-ERT2CreERT2)Dkl/Korl].
Competing interests
The authors declare that there are no conflicts of interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gittenberger-de Groot AC, Bartelings MM, Deruiter MC, Poelmann RE. Basics of cardiac development for the understanding of congenital heart malformations. Pediatr Res. 2005;57(2):169–176. doi: 10.1203/01.PDR.0000148710.69159.61. [DOI] [PubMed] [Google Scholar]
- 2.Copp AJ. Death before birth: clues from gene knockouts and mutations. Trends Genet. 1995;11(3):87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- 3.Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6(1):7–28. doi: 10.1016/S1534-5807(03)00399-X. [DOI] [PubMed] [Google Scholar]
- 4.Kim H, Kim M, Im SK, Fang S. Mouse Cre-LoxP system: general principles to determine tissue-specific roles of target genes. Lab Anim Res. 2018;34(4):147–159. doi: 10.5625/lar.2018.34.4.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doetschman T, Azhar M. Cardiac-specific inducible and conditional gene targeting in mice. Circ Res. 2012;110(11):1498–1512. doi: 10.1161/CIRCRESAHA.112.265066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237(3):752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Riesterer C, Ayrall AM, Sablitzky F, Littlewood TD, Reth M. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 1996;24(4):543–548. doi: 10.1093/nar/24.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casanova E, Fehsenfeld S, Lemberger T, Shimshek DR, Sprengel R, Mantamadiotis T. ER-based double iCre fusion protein allows partial recombination in forebrain. Genesis. 2002;34(3):208–214. doi: 10.1002/gene.10153. [DOI] [PubMed] [Google Scholar]
- 9.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA enhancer browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35(Database issue):D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee D, Pearsall RS, Das S, Dey SK, Godfrey VL, Threadgill DW. Epiregulin is not essential for development of intestinal tumors but is required for protection from intestinal damage. Mol Cell Biol. 2004;24(20):8907–8916. doi: 10.1128/MCB.24.20.8907-8916.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K, Lee H, Threadgill DW, Lee D. Epiregulin-dependent amphiregulin expression and ERBB2 signaling are involved in luteinizing hormone-induced paracrine signaling pathways in mouse ovary. Biochem Biophys Res Commun. 2011;405(2):319–324. doi: 10.1016/j.bbrc.2011.01.039. [DOI] [PubMed] [Google Scholar]
- 12.Chen M, Lichtler AC, Sheu TJ, Xie C, Zhang X, O'Keefe RJ, et al. Generation of a transgenic mouse model with chondrocyte-specific and tamoxifen-inducible expression of Cre recombinase. Genesis. 2007;45(1):44–50. doi: 10.1002/dvg.20261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K, Kim H, Lee D. Site-specific modification of genome with cell-permeable Cre fusion protein in preimplantation mouse embryo. Biochem Biophys Res Commun. 2009;388(1):122–126. doi: 10.1016/j.bbrc.2009.07.132. [DOI] [PubMed] [Google Scholar]
- 14.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol. 2007;293(3):H1883–H1891. doi: 10.1152/ajpheart.00514.2007. [DOI] [PubMed] [Google Scholar]
- 16.Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100(1):169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minamino T, Gaussin V, DeMayo FJ, Schneider MD. Inducible gene targeting in postnatal myocardium by cardiac-specific expression of a hormone-activated Cre fusion protein. Circ Res. 2001;88(6):587–592. doi: 10.1161/01.RES.88.6.587. [DOI] [PubMed] [Google Scholar]
- 18.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89(1):20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 19.Bersell K, Choudhury S, Mollova M, Polizzotti BD, Ganapathy B, Walsh S, et al. Moderate and high amounts of tamoxifen in alphaMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis Model Mech. 2013;6(6):1459–1469. doi: 10.1242/dmm.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lexow J, Poggioli T, Sarathchandra P, Santini MP, Rosenthal N. Cardiac fibrosis in mice expressing an inducible myocardial-specific Cre driver. Dis Model Mech. 2013;6(6):1470–1476. doi: 10.1242/dmm.010470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J, Zhang L, Sultana N, Park DS, Shekhar A, Bu L, et al. A murine Myh6MerCreMer Knock-in allele specifically mediates temporal genetic deletion in Cardiomyocytes after Tamoxifen induction. PLoS One. 2015;10(7):e0133472. doi: 10.1371/journal.pone.0133472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
VIPR2-ERT2CreERT2–001 transgenic mouse was backcrossed with C57BL/6 J mice for 10 generations, and then deposited in the Korea National Institute of Food and Drug Safety Evaluation for distribution [Stock number, 18-NIFDS-M-ET-001; Stock name, B6.Cg-Tg (VIPR2-ERT2CreERT2)Dkl/Korl].