Abstract
Background
Artificial intelligence (AI) is increasingly a part of daily life and offers great possibilities to enrich health care. Imaging applications of AI have been mostly developed by large, well‐funded companies and currently are inaccessible to the comparatively small market of point‐of‐care ultrasound (POCUS) programs. Given this absence of commercial solutions, we sought to create and test a do‐it‐yourself (DIY) deep learning algorithm to classify ultrasound images to enhance the quality assurance work‐flow for POCUS programs.
Methods
We created a convolutional neural network using publicly available software tools and pre‐existing convolutional neural network architecture. The convolutional neural network was subsequently trained using ultrasound images from seven ultrasound exam types: pelvis, heart, lung, abdomen, musculoskeletal, ocular, and central vascular access from 189 publicly available POCUS videos. Approximately 121,000 individual images were extracted from the videos, 80% were used for model training and 10% each for cross validation and testing. We then tested the algorithm for accuracy against a set of 160 randomly extracted ultrasound frames from ultrasound videos not previously used for training and that were performed on different ultrasound equipment. Three POCUS experts blindly categorized the 160 random images, and results were compared to the convolutional neural network algorithm. Descriptive statistics and Krippendorff alpha reliability estimates were calculated.
Results
The cross validation of the convolutional neural network approached 99% for accuracy. The algorithm accurately classified 98% of the test ultrasound images. In the new POCUS program simulation phase, the algorithm accurately classified 70% of 160 new images for moderate correlation with the ground truth, α = 0.64. The three blinded POCUS experts correctly classified 93%, 94%, and 98% of the images, respectively. There was excellent agreement among the experts with α = 0.87. Agreement between experts and algorithm was good with α = 0.74. The most common error was misclassifying musculoskeletal images for both the algorithm (40%) and POCUS experts (40.6%). The algorithm took 7 minutes 45 seconds to review and classify the new 160 images. The 3 expert reviewers took 27, 32, and 45 minutes to classify the images, respectively.
Conclusions
Our algorithm accurately classified 98% of new images, by body scan area, related to its training pool, simulating POCUS program workflow. Performance was diminished with exam images from an unrelated image pool and ultrasound equipment, suggesting additional images and convolutional neural network training are necessary for fine tuning when using across different POCUS programs. The algorithm showed theoretical potential to improve workflow for POCUS program directors, if fully implemented. The implications of our DIY AI for POCUS are scalable and further work to maximize the collaboration between AI and POCUS programs is warranted.
Keywords: artificial intelligence, deep learning, emergency medicine, emergency ultrasound, point‐of‐care ultrasound
1. INTRODUCTION
1.1. Background
Artificial intelligence (AI) is a general term applied to a range of methods through which computers complete tasks that have traditionally relied on human intelligence. Breakthroughs in 2012 in the architecture of neural networks—coupled with increasing computational power and access to large amounts of data—led to the recent explosion of applied AI technologies within a variety of domains. One of the most widely studied domains is deep learning. Deep learning refers to the application of neural networks in image analysis, as commonly seen in facial recognition and object detection in self driving car operation.
Within medicine, deep learning algorithms have shown particular promise in the machine interpretation of diagnostic imaging techniques across various organ systems. For example, the application of deep learning techniques to the interpretation of chest x‐rays and computed tomography (CT) scans of the head and chest have all been shown to yield improved diagnostic accuracy when compared to radiologists. 1 , 2 , 3 , 4 However, some of these studies and algorithms have come under justified criticism for inadequate validation in real world applications. 5
1.2. Importance
Although far less studied, deep learning may be applied to assist in the interpretation of ultrasound images as well. This is timely for the point‐of‐care ultrasound (POCUS) arena in particular as the number of POCUS users across all specialties is growing rapidly, yet educational capacity to train these users remains a significant roadblock. 6 , 7 Specifically, the amount of data (ultrasound examinations) that requires sorting, interpretation, and quality assurance is rapidly outpacing the resources of ultrasound program directors. 8 , 9 The paucity of faculty and corresponding quality assurance pressures have been widely acknowledged in emergency medicine and other POCUS communities. 10 An automated method for image and study classification based on body location could decrease POCUS director workload by allowing automatic sorting of POCUS examinations for review by faculty assigned to reviewing different examination types and for image storage.
1.3. Goals
We sought to describe a proof of concept for methods through which individual POCUS program directors and others could begin to access deep learning for their workflow needs by creating and testing a “do‐it‐yourself” (DIY) deep learning algorithm designed to help expedite physician review and categorization of POCUS studies. We also explore alternatives for those who do not have access to large restricted or highly expensive ultrasound image databases. To these ends, we created a convolutional neural network, based on an existing a convolutional neural network skeleton architecture, capable of automatically classifying ultrasound images into one of seven different point‐of‐care exam sub‐types and tested its accuracy in a real world scenario against three blinded POCUS experts. Rather than an evaluation of diagnostic accuracy, we tested the ability of an algorithm to categorize by each ultrasound image by scan type or body location.
2. METHODS
2.1. Study design
This was a study of deep learning algorithm development for automated categorization of POCUS studies into one of seven common classes. The study was exempted by the Institutional Review Board; neither patients nor patient data were used in this study, including creation and testing of the deep learning algorithm.
2.2. Data
Ultrasound image data focusing on pelvic, cardiac, lung, abdominal, musculoskeletal, central vascular access, and ocular examinations was obtained from public domain, open sources with no accompanying patient information. Compared to large public domain radiology imaging databases for chest X‐rays, head, and body CT images, no such ultrasound databases are currently available. 11 , 12 , 13 Ultrasound videos, which are commonly obtained in POCUS scanning for education, yield large numbers of individual images that can be used for convolutional neural network training. Convolutional neural networks are modeled after mammalian cortex, but have mathematical equations or nodes in place of neurons, which communicate with other nodes.
Sources used included internet‐posted ultrasound videos, open source image and video bank repositories, and open source educational multimedia materials and videos available from unrestricted ultrasound vendor tutorials. No patient identifiers were present on any of the image sources and this was manually confirmed by study authors for each video. Videos were collected from a wide variety of ultrasound machine types with varying image quality, resolution, scanning quality, and presence of pathology as might be encountered in a typical POCUS program. Researcher performed searches over a 5‐month period as time allowed, using various search methods and key terms/word related to ultrasound.
Images from the same examination type/class performed with different transducer formats (linear, curvilinear, and phased array) were included. This should ensure a more robust convolutional neural network training set that does not simply associate the transducer type with certain examinations, a potential pitfall seen in deep learning. Such an association is likely to otherwise lead to examination classification errors when applied to outside data sets. Additionally, a recent review of all radiology deep learning and medical image studies published over a 2‐year period highlighted the vulnerability of deep learning algorithms to be fine‐tuned for researchers data but not built robust enough for real world data encounters. 5
2.3. Data manipulation
Videos downloaded from websites, repositories, and those extracted from open source educational multimedia sources were checked for labels revealing examination type. When labels were identified, they were masked, or frames containing them were deleted using video editing software. The image size and aspect ratios of videos were not altered or standardized before algorithm training. A total of 189 ultrasound videos containing normal and abnormal findings were used in the initial algorithm training, validation, and testing. No videos or images were excluded. All videos were broken down into individual frames/still images using FFMPEG. FFMPEG is an open source video manipulation software, which can be encoded to perform various batch tasks on videos and images (http://ffmpeg.org).
The Bottom Line
The number of ultrasounds requiring quality assurance is growing in emergency medicine, placing a burden on those who review these studies. This study used a do‐it‐yourself deep learning algorithm to aid categorization of POCUS by body site to expedite retrieval and review.
The resultant individual images were randomly divided into three groups, 80% assigned to a training data set, 10% into a cross validation data set (for validation during convolutional neural network training), and 10% for a testing data set applied after training and validation. The training data set contained 97,822 images (14,321 pelvic, 13,971 abdominal, 12,668 musculoskeletal, 13,774 ocular, 13,234 cardiac, 16,024 lung, and 13,830 vascular access), the validation and testing data sets contained 10,873 and 12,079 images, respectively. Dividing an image data set in such a fashion and into these categories is standard in deep learning algorithm development. The validation data was used to fine tune convolutional neural network weights to optimize network performance during network training. The testing data set was used after training completion to test the algorithm's performance on images that were originally selected from the same pool as the training and validation images. This would represent new images coming from the same POCUS program, covering the same range of examinations and machines.
Finally, we sought to examine the effect of transferring this algorithm from one POCUS program to another. We hypothesized that the algorithm may encounter ultrasound images from previously unseen ultrasound machines and formats that differ from those the algorithm was trained on. Failure to perform such real‐world testing on never before seen data from different locations and from different systems from the original data pool has been cited as a weakness in many deep learning image classification studies. 5 , 14 Researchers identified and downloaded another batch of ultrasound videos covering the seven examination types. Several videos selected were of poor image quality and orientation, reflecting real‐world POCUS scan challenges. These videos were similarly broken into individual frames and examination labels that revealed examination type masked where applicable. A total of 160 frames were randomly selected from a batch of 11,210 frames covering the seven randomly ordered image sets. These frames were put through the algorithm to test image recognition and classification with the total time required tracked. The same frames were used, in random order, to populate an online survey designed with Microsoft Forms that was sent to three POCUS experts who were blinded to examination type. The expert reviewers were asked to assign an image class and a probability for the answer being correct. This matched the standard algorithm output of the top three possibilities with a probability of up to 100% assigned to each. In cases where 100% probability was assigned, that answer was treated as the only likely answer with the other two representing noise. This is similar to the results used to interpret algorithm output.
2.4. Algorithm design
Researchers used open access Anaconda software and libraries along with open access TensorFlow (a deep learning utility used for coding and convolutional neural network creation) to customize the convolutional neural network and train it. We used transfer learning, a method where a pre‐trained convolutional neural network algorithm architecture is then trained on specific image types, such as ultrasound images. All code was written in Python 3.6 programming language. VGG‐19 architecture was used because of its good excellent performance in other image classification tasks (eg, ImageNet—a public image database) as well as relative computational efficiency given that training would be performed on a home PC. VGG‐19 has a depth of 19 layers and has been pre‐trained using the ImageNet database that contains over 20,000,000 images made up of >20,000 image classes. 15 It is publicly available for use from a variety of sources. Using this existing architecture allows for more efficient training of the network with fewer ultrasound images using the network's prior training for general image recognition. All images are automatically cropped by the python code to 224 × 224 pixels as required by the VGG‐19 network architecture before image analysis. Images are also randomly rotated to increase network robustness and train it to recognize a more diverse variety of image presentations.
2.5. Algorithm training
The convolutional neural network was trained using 120,774 images. The images were kept in individual folders with randomly generated numerical file names corresponding to specific examination type that was used as ground truth (deep learning term for gold standard or actual value of the data/class of the image) for convolutional neural network training. A JSON (a text file listing examination type by number) file was created for connecting image code to specific examination types, identifying the image category type during training, validation, and testing. A desktop Microsoft PC was used for training with a NVIDIA GeForce RTX 2018 Ti 11 GB GPU graphics card required for deep learning computations. Training optimization was accomplished through incremental refinements. Figure 1 shows a flow chart of the training and evaluation process.
Figure 1.
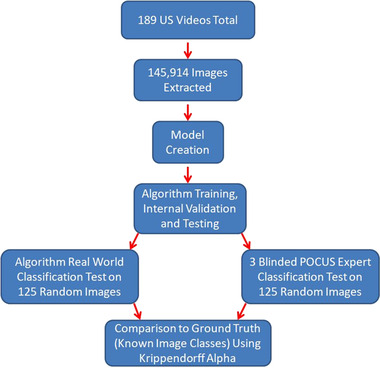
Flow diagram of the video/image acquisition, training, and testing process
2.6. Algorithm validation
We used 10,873 images that were randomly selected to serve as a training validation set and another 12,079 for testing. The validation occurs through algorithm training automatically and the testing at the end of training. This is common practice in convolutional neural network training to confirm that the algorithm is not suffering from overfitting or the capacity to very accurately predict results for the training set, but not on novel images. No overfitting was observed with the optimized training regimen. However, these steps are not enough to act as an actual real‐world use case test of an algorithm.
2.7. Additional testing representing algorithm application at different POCUS program
To test the robustness of the algorithm, AI researchers have recommend using images from alternative institutions and not used in the training or cross validation phase. This is seen in only 6% out of nearly 500 recently reviewed radiology AI studies, but is felt to be critical before widespread use. 5 We used novel images, formats, and previously unused ultrasound machines by downloading additional new ultrasound video and images from public internet sources. Videos were screened for labels specifying examination type and any labels were masked. These videos, representing the selected seven ultrasound examination types, were similarly broken down into individual images. A variety of transducer formats for each examination class were obtained to test real‐world variation seen in practice. A total of 160 images were randomly selected from the seven examination classes.
2.8. Expert classification of real‐world image set
The same randomly selected 160 images were inserted into an online survey, at original resolution rather than the reduced resolution trained on by the algorithm, and sent to three blinded POCUS experts. Reviewers were given no information regarding the scan, only single images to rate the probability (0% to 100%) of examination type, mirroring the process followed by the algorithm. Image order was randomized. Algorithm and POCUS expert answers were compared to the known image class for each image, or ground truth as it is known in deep learning design. Reviewer answers were recorded in an Excel database for later analysis. As with the training images obtained from videos, real‐world test images from video were susceptible to having low heterogeneity if operators scanned the same anatomic area with little transducer movement for a prolonged period of time.
2.9. Statistical analysis
Regression analysis and descriptive statistics available with the Anaconda packages were used to evaluate algorithm accuracy during training, validation, and testing. Descriptive statistics and Krippendorff α reliability estimates were calculated for the real‐world testing portion. Krippendorff α reliability estimate was chosen for comparison of >2 raters.
3. RESULTS
Training was first attempted using batches of 50 images and 20 total epochs. An epoch is one training cycle through an entire data set; multiple epochs are used in deep learning training. Accuracy reached 99% on cross validation early and the total training took ≈29 hours. The code was then progressively changed and we found best performance with batches of 100 images; this increased speed but did not degrade accuracy on algorithm validation. Eight epochs were used instead of the original 20. Training took 12 hours to complete and an accuracy of 99% was reached on cross validation and 98% on the test image batch. Table 1 shows POCUS experts and algorithm results for the 160 real‐world image classification test. There was excellent agreement among the experts with α = 0.87, agreement between experts and algorithm was good with α = 0.74; the algorithm showed moderate correlation with ground truth/gold standard with α = 0.64. The most common error was misclassifying musculoskeletal images for both the algorithm (40%) and POCUS experts (40.6%).
Table 1.
Data for algorithm and POCUS expert performance on 160 real‐world image test classification
| Image classifier | Review time | % Classified correctly | Most common classification error |
|---|---|---|---|
| POCUS 1 | 27 min | 93 | Musculoskeletal |
| POCUS 2 | 32 min | 94 | Musculoskeletal |
| POCUS 3 | 45 min | 98 | Musculoskeletal |
| Convolutional neural network (VGG19) | 7 min 45 s | 70 | Musculoskeletal |
Analysis of the most common errors for both algorithm and POCUS experts revealed that most originated from a lengthy musculoskeletal video showing a large knee effusion that dominated the image (Figure 2). These musculoskeletal images were misclassified as lung, abdominal, and pelvic images. The video largely consisted of a similar angle image and changed little for a prolonged time, resulting in a disproportionately large representation of these images (that were difficult to interpret because of few obvious anatomic landmarks that caused confusion with other class types repeatedly). Similarly, the next most common error, misclassification of abdominal images, was related to zoomed‐in video of an abdominal aortic aneurysm (Figure 3). This portion of the video generated numerous still images that were confused with other classes such as pelvic and lung both by algorithm and POCUS expert reviewer.
Figure 2.
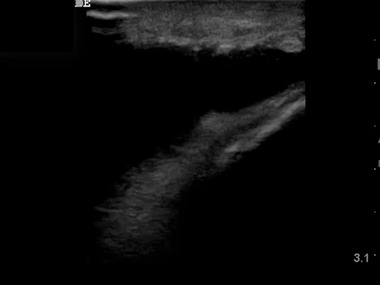
Image of a knee effusion from a lengthy video of an aspiration attempt by a point‐of‐care ultrasound (POCUS) user. Random selection of frames for testing resulted in several representative frames from this video that lacks identifiable landmarks and caused confusion for both POCUS experts and the algorithm with lung and abdominal images
Figure 3.

An image of a zoomed view of an abdominal aortic aneurysm. This image came from a lengthy explorative video of a particularly large and well‐defined abdominal aortic aneurysm. The length of the video resulted in multiple random image samples with a zoomed‐in view potentially suggesting other scan types being included in the test data set. The image was most commonly confused with a pelvic scan by POCUS experts and deep learning algorithm
4. DISCUSSION
The recent advances of AI have been described as the “fourth industrial revolution” and are predicted to have a dramatic influence on all aspects of life. In medicine, imaging analysis is among the most prominent and exciting AI applications. This technology is being driven largely by industry rather than by frontline clinicians. 16 , 17 Given this, the vast majority of deep learning activity by corporations are focused on radiology AI use and POCUS uses have been largely overlooked. 18
Although the pace of AI development accelerates, so too has POCUS use with multiple new clinical specialties beginning to adopt POCUS in clinical practice and education. 19 , 20 , 21 Additionally, ultrasound education into medical school curricula has significantly increased. 22 , 23 , 24 , 25 All of these developments mean the generation of increasing amount of ultrasound image data, not only from the mandated emergency medicine ultrasound in residency training, but throughout the house of medicine. 9 , 26 , 27 As many hospitals and medical systems attempt to promote and ensure safe use of POCUS, an increasing number of emergency medicine, critical care, and internal medicine physicians are taking on roles as system ultrasound directors. 8 The massive amounts of ultrasound video and images generated by faculty, residents, medical students, and others frequently overwhelm quality assurance and educational directors responsible for reviewing and assessing these studies. 28 , 29 Automated classification of images and videos into specific categories can enable distribution of studies to specific faculty focusing on different ultrasound applications without initial review by the ultrasound director, decreasing their manual work burden. We were unable to identify any publications describing internal creation of similar automated classification described for POCUS settings.
Although a faculty‐created neural network may seem ambitious, our work highlights the ability to create customized, DIY AI deep learning algorithms using freely available images, software, and algorithms. Our convolutional neural network focused on image identification and classification into multiple categories of examinations as would be present in many POCUS programs. We chose this task because labeling POCUS images by their exam type is the first step and common workflow bottleneck in the image review and quality assurance process. However, such categorization and labeling is also essential for program management and any informatics or data‐driven research in the future. Further, we saw this as an easily digested starting point as correct image classification is a necessary achievement before proceeding with more subtle AI image interpretation tasks. This is a familiar application of AI, akin to facial recognition in snapshots on one's smartphone or social media.
The ability to use open source ultrasound video for generation of training data may be unique to POCUS applications because of the history and growth of POCUS throughout the world. As opposed to traditional imaging, such as radiology and cardiology, which had their start with ultrasound before the digital age, POCUS clinicians honed the craft in the digital video and YouTube era, resulting in large volumes of ultrasound data being readily available online. POCUS clinicians also scan themselves rather than having technologists scan and provide example still images to be read. POCUS directors with large and available ultrasound image and video databanks may choose to use them for training of deep learning algorithms. Our study indicates very high accuracy can be attained in image classification, but also points out that this accuracy may decrease when applied to different ultrasound machines and scanning styles, such as with the introduction of new users and purchase of new equipment. This means additional training of an algorithm may be required when applying in a new hospital or when new machines are purchases by a POCUS program.
Some ultrasound review programs will sort images into exam types based on a combination of the transducer and exam type selected. For instance, cardiac preset on a phased array probe will lead to categorization as a cardiac exam. Although correct a portion of the time, users commonly cross over exam types (intentionally or unintentionally) and the errors produced through this “classify‐by‐probe‐type” logic mandate manual, exam‐by‐exam verification by ultrasound program directors. By eliminating this tedious process, our algorithm could further improve the workflow of POCUS programs.
When created for a specific POCUS program and trained on its own inherent range of ultrasound examinations, this algorithm produced high accuracy (98%). This reliability will allow POCUS directors to rely on algorithms to categorize submitted videos and images that are not labeled automatically. In our results, the algorithm reviewed and classified images greater than four times faster than expert human reviews. When operating 24 hours per day, automatically when an examination is submitted to a cloud or other central storage system or off of existing stored studies, a computer can run through unsorted studies without regard to hours spent on the task.
A limitation of our algorithm was that when taking on ultrasound images mimicking multiple different POCUS programs, its accuracy declined. This shows that our algorithm would require additional training on new sample images when applied at a different POCUS program. When faced with ultrasound images from previously unseen videos from randomly selected ultrasound examinations of the same seven categories and using different ultrasound equipment, the algorithm correctly classified 70% of examinations. This occurred secondary to algorithm exposure to ultrasound images that were unrelated to the original training data set, including one generated on ultrasound machines not included in the training data. The algorithm had particular difficulty with test musculoskeletal ultrasound images, which was the most common error for both algorithm and expert reviewers. Because of random selection, many of the test frames came from a long musculoskeletal procedure video showing a highly magnified view of a knee with an effusion and its drainage. The video contributed a large number of frames to the total real‐world test pool and happened to yield challenging individual frames to interpret as seen in Figure 2.
Another problematic image type came from a length transverse trace down a large abdominal aortic aneurysm in a very thin patient. The sonologist used a long slow pass through the abdominal aorta, zooming in on the area of interest. This again created an appearance that overlapped with other examination types leaving both the POCUS experts and algorithm few distinct anatomic landmarks to use in classification (Figure 3). These results suggest that images from a wider array of sources would result in optimal results. Yet, this requires access to larger ultrasound image databases.
While performing worse than the POCUS experts, the algorithm had good agreement with the ground truth and the experts alike. Had it simply picked classes for images by random, the expected accuracy would be ≈14%. The drop in algorithm accuracy from classifying images from the same general data set it was trained on to the real world test of previously unseen image sets and equipment support the criticism leveled by Kim et al, 5 who noted most radiology AI algorithms were not properly tested in institutions outside of their creation and in different environments. Clearly, our data suggest this is important and challenging, likely requiring larger and broader data sets. Our data set size was still suboptimal, despite the considerable breadth of data sources utilized.
A limitation of this paper is that this approach requires some baseline fluency in convolutional neural networks and Python programming language. Additionally, we did not prospectively enroll patients and test their data. However, although this was not a prospective human data study, and our algorithm could not be tested on prospectively acquitted images, the use of new real‐world ultrasound video simulated important deep learning study design criteria suggested in a recent radiology deep learning article. 5 The degree of computer and AI understanding provides a barrier to entry for some clinicians; however, readily accessible online programs on AI can bridge this gap more readily for a clinician than any online clinical coursework could for a trained non‐clinical AI engineer. However, this results in a high percentage of images that are relatively similar, especially if the video is recorded with the transducer being held in one location and when the ultrasound scenery is not changing, rather than sweeping through an area of interest. Similar images can lead to poorer generalizability of the trained convolutional neural network.
5. CONCLUSIONS
Our algorithm accurately classified 98% of new images, by body scan area, related to its training pool, simulating POCUS program workflow. Performance was diminished with exam images from an unrelated image pool and ultrasound equipment, suggesting additional images and convolutional neural network training are necessary for fine tuning when using across different POCUS programs. The algorithm showed theoretical potential to improve workflow for POCUS program directors, if fully implemented. The implications of our DIY AI for POCUS are scalable and further work to maximize the collaboration between AI and POCUS programs is warranted.
CONFLICT OF INTEREST
MB consults with EchoNous Inc and 410Medical. Neither company had influence or contribution to this study and manuscript, nor knowledge of its performance.
AUTHOR CONTRIBUTIONS
MB, RA, and MW conceived the study concept, gathered data, performed image classification, and wrote the manuscript. MB created algorithmic approach and performed algorithm training and testing.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the help of Laura Blaivas, BS, in algorithm development, programming, and execution.
Biography
Michael Blaivas is an affiliate faculty member at the University of South Carolina School of Medicine and practices clinically at St. Francis Hospital in Columbus, GA.

Blaivas M, Arntfield R, White M. DIY AI, deep learning network development for automated image classification in a point‐of‐care ultrasound quality assurance program. JACEP Open 2020;1:124–131. 10.1002/emp2.12018
Supervising Editor: Steven G. Rothrock, MD.
Abstract Presented at 2019 ACEP Scientific Assembly at #344.
Funding and support: By JACEP Open policy, all authors are required to disclose any and all commercial, financial, and other relationships in any way related to the subject of this article as per ICMJE conflict of interest guidelines (see www.icmje.org). The authors have stated that no such relationships exist.
REFERENCES
- 1. Chilamkurthy S, Ghosh R, Tanamala S, et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet. 2018;392(10162):2388‐2396. [DOI] [PubMed] [Google Scholar]
- 2. Singh R, Kalra MK, Nitiwarangkul C, et al. Deep learning in chest radiography: detection of findings and presence of change. PLoS One. 2018;13(10):e0204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cha MJ, Chung MJ, Lee JH, Lee KS. Performance of deep learning model in detecting operable lung cancer with chest radiographs. J Thorac Imaging. 2019;34(2):86‐91. [DOI] [PubMed] [Google Scholar]
- 4. Abiyev RH, Ma'aitah MKS. Deep convolutional neural networks for chest diseases detection. J Healthc Eng. 2018;2018:4168538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim DW, Jang HY, Kim KW, Shin Y, Park SH. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol. 2019;20(3):405‐410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arntfield RT. The utility of remote supervision with feedback as a method to deliver high‐volume critical care ultrasound training. J Crit Care. 2015;30(2):441.e1‐6. [DOI] [PubMed] [Google Scholar]
- 7. LoPresti CM, Boyd JS, Schott C, et al. A national needs assessment of point‐of‐care ultrasound training for hospitalists. Mayo Clin Proc. 2019;94(9):1910‐1912. [DOI] [PubMed] [Google Scholar]
- 8. Emergency ultrasound imaging criteria compendium. Ann Emerg Med. 2016;68(1):e11‐e48. [DOI] [PubMed] [Google Scholar]
- 9. American College of Emergency Physicians . Emergency ultrasound imaging criteria compendium. American College of Emergency Physicians. Ann Emerg Med. 2006;48(4):487‐510. [DOI] [PubMed] [Google Scholar]
- 10. Strony R, Marin JR, Bailitz J, et al. Systemwide clinical ultrasound program development: an expert consensus model. West J Emerg Med. 2018;19(4):649‐653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim TK, Yi PH, Wei J, et al. Deep learning method for automated classification of anteroposterior and posteroanterior chest radiographs. J Digit Imaging. 2019;32(6):925‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuh EL, Gean AD, Manley GT, Callen AL, Wintermark M. Computer‐aided assessment of head computed tomography (CT) studies in patients with suspected traumatic brain injury. J Neurotrauma. 2008;25(10):1163‐1172. [DOI] [PubMed] [Google Scholar]
- 13. Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574‐582. [DOI] [PubMed] [Google Scholar]
- 14. Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross‐sectional study. PLoS Med. 2018;15:e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simonyan K, Zisserman A. Very deep convolutional networks for large‐scale image recognition. ICLR. 2015;1409:1‐15. [Google Scholar]
- 16. Ahmad T, Lund LH, Rao P, et al. Machine learning methods improve prognostication, identify clinically distinct phenotypes, and detect heterogeneity in response to therapy in a large cohort of heart failure patients. J Am Heart Assoc. 2018;7(8): e008081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MaxQ AI to add ICH software to Philips CT systems. Available at: https://www.auntminnie.com/index.aspx?Sec=sup&Sub=aic&Pag=dis&ItemId=126840
- 18. NVIDIA and American College of Radiology AI‐LAB Team to Accelerate Adoption of AI in Diagnostic Radiology Across Thousands of Hospitals. Available at: https://www.globenewswire.com/news-release/2019/04/08/1798842/0/en/NVIDIA-and-American-College-of-Radiology-AI-LAB-Team-to-Accelerate-Adoption-of-AI-in-Diagnostic-Radiology-Across-Thousands-of-Hospitals.html
- 19. Soni NJ, Schnobrich D, Matthews BK, et al. Point‐of‐care ultrasound for hospitalists: a position statement of the Society of Hospital Medicine. J Hosp Med. 2019;14:E1‐E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. ACP Statement in Support of Point‐of‐Care Ultrasound in Internal Medicine. Available at: https://www.acponline.org/meetings-courses/focused-topics/point-of-care-ultrasound-pocus-for-internal-medicine/acp-statement-in-support-of-point-of-care-ultrasound-in-internal-medicine
- 21. Bornemann P, Barreto T. Point‐of‐care ultrasonography in family medicine. Am Fam Physician. 2018;98(4):200‐202. [PubMed] [Google Scholar]
- 22. Dietrich CF, Goudie A, Chiorean L, et al. Point‐of‐care ultrasound: a WFUMB position paper. Ultrasound Med Biol. 2017;43(1):49‐58. [DOI] [PubMed] [Google Scholar]
- 23. AIUM practice parameter for the performance of point‐of‐care ultrasound examinations. J Ultrasound Med. 2019;38(4):833‐849. [DOI] [PubMed] [Google Scholar]
- 24. Society of Ultrasound in Medical Education home page . Available at: https://www.susme.org/
- 25. Hoppmann RA, Rao VV, Bell F, e al. The evolution of an integrated ultrasound curriculum (iUSC) for medical students: 9‐year experience. Crit Ultrasound J. 2015;7(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akhtar S, Theodoro D, Gaspari R, et al. Resident training in emergency ultrasound: consensus recommendations from the 2008 Council of Emergency Medicine Residency Directors Conference. Acad Emerg Med. 2009;16 (Suppl 2):S32‐S36. [DOI] [PubMed] [Google Scholar]
- 27. Point‐of‐Care Ultrasonography by Pediatric Emergency Medicine Physicians statement by ACEP. Available at: https://www.acep.org/globalassets/uploads/uploaded-files/acep/clinical-and-practice-management/policy-statements/point-of-care-ultrasonography.pdf
- 28. Thomas‐Mohtat R, Breslin K, Cohen JS. Quality assurance for point‐of‐care ultrasound in North American pediatric emergency medicine fellowships. Pediatr Emerg Care. 2019. [DOI] [PubMed] [Google Scholar]
- 29. Amini R, Wyman MT, Hernandez NC, Guisto JA, Adhikari S. Use of emergency ultrasound in Arizona community emergency departments. J Ultrasound Med. 2017;36(5):913‐921. [DOI] [PubMed] [Google Scholar]


