Abstract
Regorafenib confers the benefit of longer survival in metastatic colorectal cancer patients. The CCL5/CCR5 pathway modulates endothelial progenitor cell migration and vascular endothelial growth factor A production. Genetic variants of CCL4 and CCL3 may predict outcomes, and the different frequencies of CCL5 homozygote may explain ethnic differences in the development of severe hand–foot skin reactions.
Background:
The C-C motif chemokine ligand 5/C-C motif chemokine receptor 5 (CCL5/CCR5) pathway has been shown to induce endothelial progenitor cell migration, resulting in increased vascular endothelial growth factor A expression. We hypothesized that genetic polymorphisms in the CCL5/CCR5 pathway predict efficacy and toxicity in patients with metastatic colorectal cancer (mCRC) treated with regorafenib.
Patients and Methods:
We analyzed genomic DNA extracted from 229 tumor samples from 2 different cohorts of patients who received regorafenib: an evaluation cohort of 79 Japanese patients and a validation cohort of 150 Italian patients. Single nucleotide polymorphisms of CCL5/CCR5 pathway-related genes were analyzed by PCR-based direct sequencing.
Results:
CCL4 rs1634517 and CCL3 rs1130371 were associated with progression-free survival in the evaluation cohort (hazard ratio [HR] 1.54, P = .043; HR 1.48, P = .064), and progression-free survival (HR 1.74, P < .001; HR 1.66, P = .002) and overall survival (HR 1.65, P = .004; HR 1.65, P = .004) in the validation cohort. The allelic frequencies of CCL5 single nucleotide polymorphisms varied between the evaluation and validation cohorts (G/G variant in rs2280789, 21.5% vs. 1.3%, P < .001; T/T variant in rs3817655, 22.8% vs. 2.7%, P < .001). In the evaluation cohort, patients with the G/G variant in rs2280789 had a higher incidence of grade 3+ hand–foot skin reaction compared to any A allele (53% vs. 27%, P = .078), and similarly to the T/T variant in rs3817655 compared to any A allele (56% vs. 26%, P = .026).
Conclusion:
Genetic variants in the CCL5/CCR5 pathway may serve as prognostic markers and may predict severe hand–foot skin reaction in mCRC patients receiving regorafenib therapy.
Keywords: CCL5/CCR5 signaling, Colorectal cancer, Ethnic difference, Hand-foot skin reaction, Regorafenib
Introduction
Regorafenib, an oral multikinase inhibitor, confers the benefit of longer survival to patients with refractory metastatic colorectal cancer (mCRC).1,2 Tumor mutation status, plasma DNA concentration, and plasma protein concentration, including its target protein kinases, have been examined by a retrospective exploration of the CORRECT study to identify predictive markers of this agent, while real-time circulating DNA analysis has shown potential prognostic markers for clinical outcomes.3 However, no validated predictive markers of efficacy and/or toxicity have been identified. Hand–foot skin reaction (HFSR) is a well-known toxicity of regorafenib that obliges patients to interrupt treatment, and an ethnic difference in the frequency of HFSR has been reported between Japanese and non-Japanese patients in the CORRECT study.4
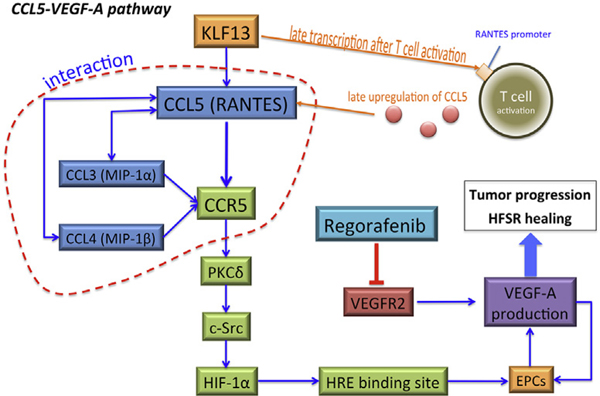
A recent study that investigated whether serum cytokine levels are associated with clinical outcomes in mCRC patients receiving regorafenib reported that baseline serum C-C motif chemokine ligand 5 (CCL5) levels and decrease of serum vascular endothelial growth factor (VEGF) A levels after start of treatment predicted the efficacy of regorafenib in refractory mCRC. Furthermore, low CCL5 levels were associated with the onset of HFSR.5 C-C motif chemokine receptor 5 (CCR5) is a receptor of CCL5, and CCL5 can promote endothelial progenitor cell (EPC) migration in a CCR5-dependent manner. The CCL5/CCR5 pathway is involved in VEGF-A production via EPC migration.6 CCL5 is characterized as late expression after T-cell activation, and it localizes with tumor-infiltrating leukocytes.7 It is also known as regulated on activation, normal T-cell expressed and secreted (RANTES). Krüppel-like transcription factor (KLF) 13 is a transcription factor that regulates RANTES expression in T lymphocytes; it is known as RANTES factor of late activated T lymphocytes 1 (RFLAT-1).8 Other CCR5 ligands–C-C motif chemokine ligand-3 (CCL3) and −4 (CCL4)—also participate in EPC migration via binding to CCR5; however, a recent in vitro study showed that CCL5 is the most potent chemoattractant of EPCs.9 The CCL5/CCR5 signaling pathway positively activates protein kinase Cδ (PKCδ), c-Src, and hypoxia-inducible factor 1α (HIF1A) in activating VEGF-A expression (Figure 1).6
Figure 1. Illustration of CCL5-CCR5 Signaling Pathway for VEGF-A production in Regorafenib Treatment.
Abbreviations: EPC = endothelial progenitor cell; HFSR = hand–foot skin reaction; HRE = hypoxia-response element; MIP-1 = macrophage inflammatory protein 1; RANTES = regulated on activation, normal T-cell expressed and secreted; VEGF-A = vascular endothelial growth factor A.
We therefore tested whether genetic polymorphisms in the CCL5/CCR5 pathway are associated with clinical outcomes and toxicity, particularly HFSR, in patients with refractory mCRC treated with regorafenib.
Patients and Methods
Study Design and Patients
This study investigated 2 independent cohorts composed of patients with refractory, histologically confirmed mCRC: an evaluation cohort of 79 patients treated with regorafenib at the Cancer Institute Hospital in Japan between May 2013 and December 2015, and a validation cohort of 150 patients treated with regorafenib at Azienda Ospedaliero–Universitaria Pisana (Pisa, Italy) and Istituto Oncologico Veneto (Padua, Italy) between August 2010 and November 2015. All patients met the eligibility criteria: history of standard chemotherapy including 5-fluorouracil, oxaliplatin, irinotecan, bevacizumab, and cetuximab or panitumumab for KRAS or RAS wild type; measurable or evaluable disease according to the Response Evaluation Criteria in Solid Tumors v1.1; and signed informed consent. Adverse events were graded according to the Common Terminology Criteria for Adverse Events, version 4.0.
In the evaluation and validation cohorts, patients received 160 mg regorafenib (Bayer, Leverkusen, Germany) once daily from day 1 to day 21 every 4 weeks. Doses were adjusted on the basis of adverse events at a physician’s discretion, following the manufacturer’s recommendations. We were fully compliant with the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) guidelines. The analyses were approved by the institutional review board of each institute, and they were conducted at the University of Southern California/Norris Comprehensive Cancer Center and in accordance with the Declaration of Helsinki and good clinical practice guidelines.
Selection of Candidate Single Nucleotide Polymorphisms
The 9 candidate single nucleotide polymorphisms (SNPs) in this study inhabited 7 genes—CCL3, CCL4, CCL5, CCR5, PRKCD, KLF13, and HIF1A—and were selected on the basis of the following criteria: (1) SNP with biological significance according to published literature review; (2) tagging SNPs selected using the HapMap genotype data with r2 threshold = 0.8 (https://snpinfo.niehs.nih.gov/snpinfo/snptag.html); or (3) minor allele frequency with a cutoff of ≥ 10% in both whites and East Asians (in the Ensembl Genome Browser, http://uswest.ensembl.org/index.html). Functional significance was predicted on the basis of the Functional Single Nucleotide Polymorphism database (http://compbio.cs.queensu.ca/F-SNP/) (Supplemental Table 1 in the online version). Details of DNA extraction and genotyping are provided in the Supplemental Methods in the online version.
Analysis of Serum VEGF-A and CCL5 Levels
Blood samples were obtained from 57 Japanese patients enrolled onto the evaluation cohort, at baseline before the first dose of regorafenib, and at day 21 in the first cycle (Supplemental Methods in the online version).
Statistical Analysis
The primary end point of the current study was progression-free survival (PFS), and the secondary end points were overall survival (OS) and disease control rate. All analyses were performed by SAS 9.4 (SAS Institute, Cary, NC). All tests were 2 sided at a significance level of .05. P values were not adjusted for multiple testing (Supplemental Methods in the online version).
Results
Patient and Tumor Baseline Characteristics
In the evaluation cohort, the median follow-up time was 15.3 months, and median PFS and OS were 2.0 and 8.7 months, respectively. In the validation cohort, the median follow-up time was 36.4 months, and median PFS and OS were 2.1 and 6.0 months, respectively. The baseline characteristics of the evaluation and validation are summarized in Supplemental Table 2 in the online version. The associations between baseline characteristics and clinical outcomes are summarized in Supplemental Tables 3 and 4 in the online version for evaluation and validation, respectively. All candidate SNPs were within the Hardy-Weinberg equilibrium when tested using HaploView 4.2. CCL5 rs2280789 and CCL5 rs3817655 showed high linkage disequilibrium in both evaluation and validation cohorts (evaluation cohort: D′ = 0.97, r2 = 0.92; validation cohort: D′ = 0.97, r2 = 0.73).
Associations Between Candidate SNPs and Clinical Outcomes in Evaluation Cohort
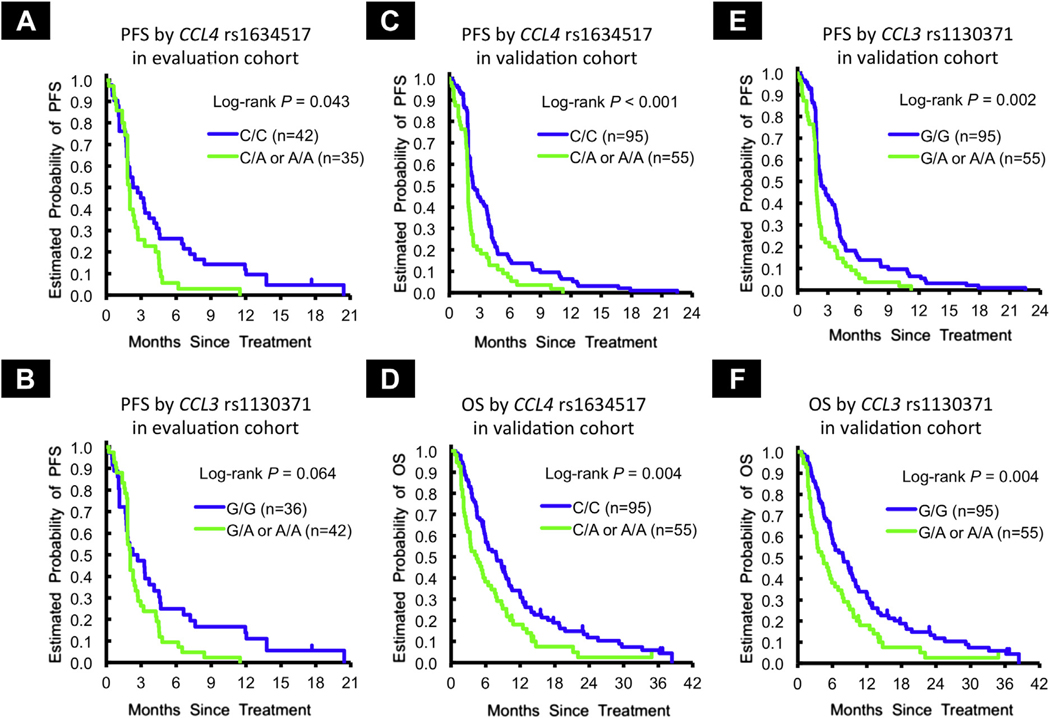
Patients carrying the G/G variant in CCL5 rs2280789 showed a significant benefit in OS compared to those with any A allele per the multivariable analysis (12.9 vs. 7.9 months; hazard ratio [HR] 0.45, P = .032). Similarly, patients carrying the T/T variant in CCL5 rs3817655 also showed longer OS (12.9 vs. 7.9 months, HR 0.50, P = .055). In the univariate analysis, patients with any A allele in CCL4 rs1634517 had significantly shorter PFS compared to those with the C/C variant (2.0 vs. 2.5 months, HR 1.54, 95% confidence interval [CI] 0.96–2.50, P = .043) (Figure 2A). The effect remained in the multivariable analysis (P = .058). Patients carrying any A allele in CCL3 rs1130371 had shorter PFS than those with the G/G variant (2.0 vs. 2.5 months, HR 1.48, 95% CI, 0.91–2.39, P = .064) (Figure 2B, Table 1, Supplemental Table 5 in the online version).
Figure 2.
PFS and OS in Evaluation and Validation Cohorts
Table 1.
Association Between Gene Polymorphism and Clinical Outcome
| N | Disease Control | Progression-Free Survival | Overall Survival | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PR + SD | PD | P value* | Median (95% CI), mo | Univariate HR (95% CI)† | P value* | Multivariable HR (95% CI)‡ | P value* | Median (95% CI), mo | Univariate HR (95% CI)† | P value* | Multivariable HR (95% CI)‡ | P value* | ||
| CCL5 rs2280789 | .42 | .56 | .21 | .36 | .039 | |||||||||
| A/A | 27 | 13 (59%) | 9 (41%) | 2.3 (1.8, 3.3) | 1 (Reference) | 1 (Reference) | 7.9 (4.0, 29.9) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 35 | 12 (46%) | 14 (54%) | 2.0 (1.5, 2.5) | 1.01 (0.61, 1.69) | 1.20 (0.72, 2.02) | 8.1 (4.6, 12.6) | 1.19 (0.64, 2.22) | 1.53 (0.81, 2.91) | |||||
| G/G | 17 | 6 (38%) | 10 (63%) | 2.0 (1.7, 4.7) | 0.76 (0.41, 1.43) | 0.68 (0.36, 1.30) | 12.9 (5.0, 27.7) | 0.71 (0.32, 1.57) | 0.56 (0.25, 1.27) | |||||
| .39 | .28 | .11 | .19 | .032 | ||||||||||
| Any A | 62 | 25 (52%) | 23 (48%) | 2.0 (1.8, 2.7) | 1 (Reference) | 1 (Reference) | 7.9 (5.8, 11.8) | 1 (Reference) | 1 (Reference) | |||||
| G/G | 17 | 6 (38%) | 10 (63%) | 2.0 (1.7, 4.7) | 0.76 (0.43, 1.32) | 0.62 (0.35, 1.11) | 12.9 (5.0, 27.7) | 0.64 (0.32, 1.28) | 0.45 (0.21, 0.93) | |||||
| CCL5 rs3817655 | .74 | .60 | .40 | .52 | .12 | |||||||||
| A/A | 27 | 12 (55%) | 10 (45%) | 2.0 (1.8, 3.2) | 1 (Reference) | 1 (Reference) | 7.1 (4.0, 29.9) | 1 (Reference) | 1 (Reference) | |||||
| A/T | 34 | 12 (48%) | 13 (52%) | 2.0 (1.6, 3.7) | 0.87 (0.52, 1.46) | 0.98 (0.58, 1.67) | 8.7 (4.6, 12.6) | 1.00 (0.54, 1.86) | 1.28 (0.67, 2.44) | |||||
| T/T | 18 | 7 (41%) | 10 (59%) | 1.9 (1.7, 4.5) | 0.75 (0.40, 1.38) | 0.67 (0.35, 1.26) | 12.9 (5.0, 27.7) | 0.68 (0.32, 1.47) | 0.57 (0.26, 1.24) | |||||
| .58 | .40 | .18 | .25 | .055 | ||||||||||
| Any A | 61 | 24 (51%) | 23 (49%) | 2.0 (1.8, 2.7) | 1 (Reference) | 1 (Reference) | 7.9 (5.8, 11.8) | 1 (Reference) | 1 (Reference) | |||||
| T/T | 18 | 7 (41%) | 10 (59%) | 1.9 (1.7, 4.5) | 0.81 (0.47, 1.39) | 0.67 (0.38, 1.19) | 12.9 (5.0, 27.7) | 0.68 (0.35, 1.34) | 0.50 (0.25, 1.02) | |||||
| CCR5 rs1799988 | .019 | .25 | .50 | .43 | .55 | |||||||||
| T/T | 25 | 5 (25%) | 15 (75%) | 1.8 (1.1, 2.3) | 1 (Reference) | 1 (Reference) | 6.3 (4.0, 27.2) | 1 (Reference) | 1 (Reference) | |||||
| T/C | 29 | 12 (50%) | 12 (50%) | 2.3 (1.8, 3.3) | 0.65 (0.37, 1.14) | 0.71 (0.40, 1.26) | 9.6 (5.0, 13.6) | 0.88 (0.45, 1.70) | 0.88 (0.45, 1.73) | |||||
| C/C | 24 | 14 (70%) | 6 (30%) | 2.7 (1.6, 4.5) | 0.74 (0.42, 1.30) | 0.80 (0.45, 1.42) | 12.6 (6.5, 15.5) | 0.64 (0.32, 1.30) | 0.68 (0.33, 1.39) | |||||
| .016 | .11 | .26 | .35 | .43 | ||||||||||
| T/T | 25 | 5 (25%) | 15 (75%) | 1.8 (1.1, 2.3) | 1 (Reference) | 1 (Reference) | 6.3 (4.0, 27.2) | 1 (Reference) | 1 (Reference) | |||||
| Any C | 53 | 26 (59%) | 18 (41%) | 2.5 (1.8, 3.3) | 0.69 (0.42, 1.12) | 0.75 (0.46, 1.23) | 10.3 (6.7, 12.9) | 0.76 (0.42, 1.38) | 0.78 (0.42, 1.44) | |||||
| .031 | .81 | .86 | .22 | .30 | ||||||||||
| Any T | 54 | 17 (39%) | 27 (61%) | 2.0 (1.8, 2.3) | 1 (Reference) | 1 (Reference) | 7.8 (5.0, 10.8) | 1 (Reference) | 1 (Reference) | |||||
| C/C | 24 | 14 (70%) | 6 (30%) | 2.7 (1.6, 4.5) | 0.94 (0.58, 1.54) | 0.96 (0.58, 1.58) | 12.6 (6.5, 15.5) | 0.69 (0.38, 1.26) | 0.73 (0.40, 1.33) | |||||
| CCL3 rs1130371 | .31 | .064 | .41 | .76 | .23 | |||||||||
| G/G | 36 | 16 (57%) | 12 (43%) | 2.5 (1.7, 4.1) | 1 (Reference) | 1 (Reference) | 8.1 (4.6, 12.9) | 1 (Reference) | 1 (Reference) | |||||
| Any A | 42 | 15 (42%) | 21 (58%) | 2.0 (1.8, 2.4) | 1.48 (0.91, 2.39) | 1.23 (0.75, 2.01) | 9.6 (6.1, 12.9) | 1.09 (0.62, 1.90) | 0.70 (0.39, 1.26) | |||||
| CCL4 rs1634517 | .12 | .12 | .14 | .13 | .17 | |||||||||
| C/C | 42 | 20 (57%) | 15 (43%) | 2.5 (1.7, 3.7) | 1 (Reference) | 1 (Reference) | 8.1 (5.1, 13.6) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 26 | 6 (30%) | 14 (70%) | 1.9 (1.6, 2.4) | 1.60 (0.95, 2.69) | 1.67 (1.00, 2.78) | 11.8 (7.1, 15.3) | 0.89 (0.48, 1.68) | 0.79 (0.41, 1.50) | |||||
| A/A | 9 | 5 (63%) | 3 (38%) | 2.0 (0.6, 4.5) | 1.40 (0.66, 2.97) | 1.35 (0.64, 2.85) | 6.1 (1.5, 13.9) | 1.96 (0.91, 4.20) | 1.76 (0.80, 3.86) | |||||
| .21 | .043 | .058 | .71 | .96 | ||||||||||
| C/C | 42 | 20 (57%) | 15 (43%) | 2.5 (1.7, 3.7) | 1 (Reference) | 1 (Reference) | 8.1 (5.1, 13.6) | 1 (Reference) | 1 (Reference) | |||||
| Any A | 35 | 11 (39%) | 17 (61%) | 2.0 (1.8, 2.4) | 1.54 (0.96, 2.50) | 1.58 (0.98, 2.53) | 9.6 (6.1, 12.9) | 1.11 (0.64, 1.94) | 0.99 (0.56, 1.74) | |||||
| KLF13 rs2241779 | .62 | .21 | .019 | .57 | .20 | |||||||||
| C/C | 30 | 14 (56%) | 11 (44%) | 3.0 (1.8, 4.6) | 1 (Reference) | 1 (Reference) | 10.3 (6.1, 15.5) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 33 | 12 (46%) | 14 (54%) | 1.8 (1.5, 2.3) | 1.48 (0.89, 2.45) | 2.13 (1.24, 3.65) | 8.1 (5.1, 12.0) | 1.37 (0.74, 2.55) | 1.78 (0.94, 3.40) | |||||
| A/A | 15 | 5 (38%) | 8 (62%) | 2.0 (1.6, 2.7) | 1.49 (0.79, 2.80) | 1.83 (0.95, 3.50) | 7.9 (3.1, 29.9) | 1.32 (0.61, 2.85) | 1.54 (0.71, 3.37) | |||||
| .44 | .079 | .006 | .29 | .081 | ||||||||||
| C/C | 30 | 14 (56%) | 11 (44%) | 3.0 (1.8, 4.6) | 1 (Reference) | 1 (Reference) | 10.3 (6.1, 15.5) | 1 (Reference) | 1 (Reference) | |||||
| Any A | 48 | 17 (44%) | 22 (56%) | 1.9 (1.7, 2.3) | 1.48 (0.93, 2.36) | 2.02 (1.23, 3.33) | 7.9 (5.2, 11.8) | 1.35 (0.76, 2.40) | 1.70 (0.94, 3.07) | |||||
| CCL5 rs2280789 | .55 | .27 | .17 | .29 | .42 | |||||||||
| A/A | 106 | 38 (37%) | 64 (63%) | 2.0 (1.8, 2.3) | 1 (Reference) | 1 (Reference) | 5.9 (4.7, 7.8) | 1 (Reference) | 1 (Reference) | |||||
| A/Ga | 41 | 12 (32%) | 26 (68%) | 2.1 (1.9, 2.8) | 0.82 (0.57, 1.19) | 0.76 (0.52, 1.12) | 8.0 (4.3, 10.1) | 0.82 (0.56, 1.20) | 0.85 (0.57, 1.26) | |||||
| G/Ga | 2 | 0 (0%) | 2 (100%) | |||||||||||
| CCL5 rs3817655 | .42 | .42 | .10 | .51 | .34 | |||||||||
| A/A | 98 | 35 (37%) | 60 (63%) | 2.0 (1.8, 2.3) | 1 (Reference) | 1 (Reference) | 6.3 (5.0, 7.9) | 1 (Reference) | 1 (Reference) | |||||
| A/Ta | 47 | 15 (32%) | 32 (68%) | 2.2 (1.9, 2.5) | 0.87 (0.62, 1.23) | 0.74 (0.51, 1.06) | 6.0 (4.3, 9.7) | 0.89 (0.62, 1.27) | 0.83 (0.57, 1.21) | |||||
| T/Ta | 4 | 0 | 4 (100%) | |||||||||||
| CCR5 rs1799988 | .32 | .40 | .076 | .66 | .21 | |||||||||
| T/T | 52 | 13 (27%) | 36 (73%) | 2.1 (1.8, 2.3) | 1 (Reference) | 1 (Reference) | 5.5 (4.0, 7.8) | 1 (Reference) | 1 (Reference) | |||||
| T/C | 64 | 24 (39%) | 37 (61%) | 2.1 (1.8, 3.7) | 0.78 (0.54, 1.13) | 0.64 (0.43, 0.94) | 8.9 (5.7, 10.5) | 0.84 (0.57, 1.24) | 0.70 (0.47, 1.04) | |||||
| C/C | 34 | 13 (39%) | 20 (61%) | 1.9 (1.8, 3.4) | 0.89 (0.57, 1.37) | 0.76 (0.49, 1.18) | 5.6 (3.4, 8.7) | 0.93 (0.59, 1.47) | 0.87 (0.54, 1.40) | |||||
| .14 | .22 | .032 | .44 | .12 | ||||||||||
| T/T | 52 | 13 (27%) | 36 (73%) | 2.1 (1.8, 2.3) | 1 (Reference) | 1 (Reference) | 5.5 (4.0, 7.8) | 1 (Reference) | 1 (Reference) | |||||
| Any C | 98 | 37 (39%) | 57 (61%) | 2.1 (1.8, 2.7) | 0.81 (0.58, 1.15) | 0.68 (0.48, 0.97) | 7.1 (5.1, 9.5) | 0.87 (0.61, 1.24) | 0.75 (0.52, 1.08) | |||||
| CCL3 rs1130371 | .10 | .002 | .027 | .004 | .047 | |||||||||
| G/G | 95 | 37 (40%) | 55 (60%) | 2.3 (2.1, 3.4) | 1 (Reference) | 1 (Reference) | 7.9 (5.9, 9.6) | 1 (Reference) | 1 (Reference) | |||||
| Any A | 55 | 13 (25%) | 38 (75%) | 1.8 (1.7, 2.0) | 1.66 (1.18, 2.33) | 1.50 (1.05, 2.15) | 4.4 (2.7, 6.4) | 1.65 (1.16, 2.34) | 1.44 (1.01, 2.07) | |||||
| CCL4 rs1634517 | .077 | .003 | .040 | .004 | .034 | |||||||||
| C/C | 95 | 37 (41%) | 54 (59%) | 2.3 (2.1, 3.6) | 1 (Reference) | 1 (Reference) | 7.9 (5.9, 9.6) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 48 | 10 (22%) | 35 (78%) | 1.8 (1.7, 2.0) | 1.73 (1.21, 2.48) | 1.58 (1.08, 2.30) | 4.9 (2.7, 7.6) | 1.56 (1.08, 2.24) | 1.37 (0.94, 1.99) | |||||
| A/A | 7 | 3 (43%) | 4 (57%) | 1.7 (0.9, 3.1) | 1.78 (0.82, 3.89) | 1.72 (0.78, 3.78) | 3.4 (1.0, 5.9) | 2.67 (1.22, 5.86) | 2.53 (1.15, 5.56) | |||||
| .070 | <.001 | .012 | .004 | .041 | ||||||||||
| C/C | 95 | 37 (41%) | 54 (59%) | 2.3 (2.1, 3.6) | 1 (Reference) | 1 (Reference) | 7.9 (5.9, 9.6) | 1 (Reference) | 1 (Reference) | |||||
| Any A | 55 | 13 (25%) | 39 (75%) | 1.8 (1.7, 2.0) | 1.74 (1.24, 2.45) | 1.59 (1.11, 2.29) | 4.4 (2.7, 6.4) | 1.65 (1.16, 2.34) | 1.46 (1.01, 2.09) | |||||
| KLF13 rs2241779 | .21 | .28 | .17 | .88 | .51 | |||||||||
| C/C | 33 | 13 (43%) | 17 (57%) | 2.3 (1.8, 4.1) | 1 (Reference) | 1 (Reference) | 8.0 (3.6, 10.0) | 1 (Reference) | 1 (Reference) | |||||
| A/C | 72 | 19 (28%) | 50 (72%) | 1.9 (1.8, 2.1) | 1.38 (0.91, 2.09) | 1.37 (0.90, 2.09) | 5.9 (4.5, 7.8) | 1.06 (0.69, 1.63) | 1.05 (0.68, 1.62) | |||||
| A/A | 44 | 17 (40%) | 26 (60%) | 2.3 (1.9, 2.8) | 1.15 (0.73, 1.82) | 0.99 (0.62,1.58) | 5.7 (4.4, 9.5) | 0.96 (0.60, 1.53) | 0.83 (0.51, 1.34) | |||||
| .28 | .21 | .38 | .93 | .83 | ||||||||||
| C/C | 33 | 13 (43%) | 17 (57%) | 2.3 (1.8, 4.1) | 1 (Reference) | 1 (Reference) | 8.0 (3.6, 10.0) | 1 (Reference) | 1 (Reference) | |||||
| Any A | 116 | 36 (32%) | 76 (68%) | 2.0 (1.8, 2.3) | 1.28 (0.86, 1.90) | 1.20 (0.80, 1.78) | 5.9 (4.7, 7.8) | 1.02 (0.68, 1.52) | 0.96 (0.64, 1.44) | |||||
Abbreviations: PD = progressive disease; PR = partial response; SD = stable disease.
P values<.05 were shown in bold.
P value was based on the Fisher’s exact test for response, log-rank test in the univariate analysis
() and Wald test in the multivariable analysis within Cox regression model
() adjusted for liver metastasis and LN metastasis in the evaluation cohort; time to REGO started (<18 vs ≥18 months), ECOG performance status (0 vs 1 or 2), primary tumor resection (yes vs no), and Kohne score (low-intermediate vs high) in the validation cohort.
Grouped together for estimate of HR.
Association Between Candidate SNPs and Clinical Outcomes in Validation Cohort
In the univariate analysis, patients with any A allele in CCL4 rs1634517 had significantly shorter PFS (1.8 vs. 2.3 months, HR 1.74, 95% CI, 1.24–2.45, P < .001) and OS (4.4 vs. 7.9 months; HR 1.65, 1.16–2.34, P = .004) compared to those with the C/C variant (Figure 2C and D). This remained significant in the multivariable analysis for PFS and OS (HR 1.59, P = .012; HR 1.46, P = .041, respectively). Patients carrying any A allele in CCL3 rs1130371 had significantly shorter PFS and OS (PFS: 1.8 vs. 2.3 months, HR 1.66, 95% CI, 1.18–2.33, P = .002; OS: 4.4 vs. 7.9 months, HR 1.65, 95% CI, 1.16–2.34, P = .004) compared to those with the G/G variant (Figure 2E and F); these effects remained significant in the multivariable model (PFS: HR 1.50, P = .027; OS: HR 1.44, P = .047). Uni- and multivariate analyses using recessive models in each CCL5 SNP were not available for analysis owing to the low frequency of the homozygote: G/G variant in rs2280789, 2 (1.3%) of 149; and T/T variant in rs3817655, 4 (2.7%) of 149 (Table 1).
Association Between Candidate SNPs and Toxicity in Both Cohorts
Grade 3 or higher adverse events were analyzed to investigate their associations with clinical outcomes and candidate SNPs. In the evaluation and validation cohorts, patients with grade 3 or higher hypertension and rash showed longer PFS and OS, respectively (Supplemental Table 6 in the online version).
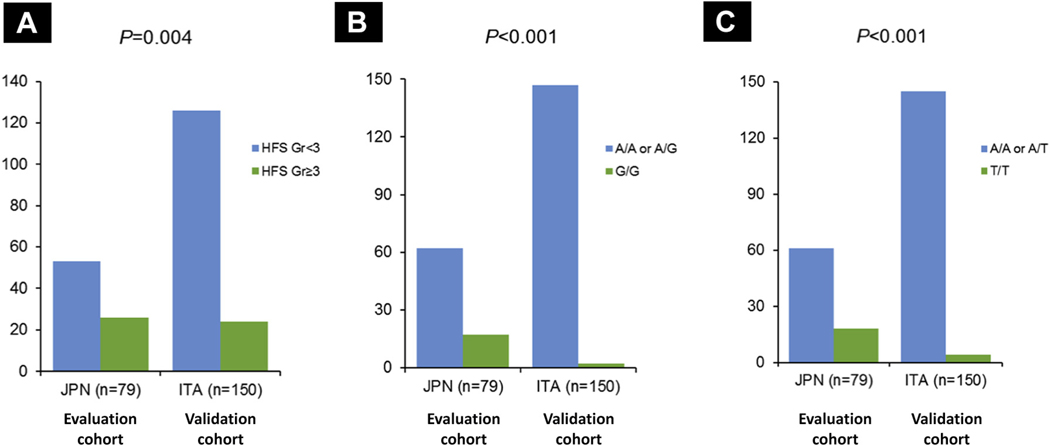
Allelic distribution of SNPs was compared between the evaluation and validation cohorts. The frequency of homozygotes in CCL5 SNPs varied between Japanese and Italian patients (G/G variant in rs2280789, 21.5% vs. 1.3%, P < .001; T/T variant in rs3817655, 22.8% vs. 2.7%, P < .001). Grade 3 or higher HFSR was more frequent in the evaluation cohort than in the validation cohort (32.9% vs. 16.0%, P = .004) (Figure 3 and Table 2).
Figure 3. Different in Frequency of Grade 3 + HFSR (A), and Allelic Distribution of CCL5 rs2280789 (B) and CCL5 rs3817655 (C) in Evaluation and Validation Cohorts.
Abbreviations: ITA = Italian; JPN = Japanese.
Table 2.
Association Between SNPs and Toxicities
| SNP | Evaluation Cohort | Validation Cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| N | Grade 3 or Higher | Grade 2 or Lower | P | N | Grade 3 or Higher | Grade 2 or Lower | P | |
| Hand–Foot Skin Reaction | ||||||||
| CCL5 rs2280789 | .16 | .24 | ||||||
| A/A | 27 | 8 (30%) | 19 (70%) | 106 | 15 (14%) | 91 (86%) | ||
| A/G | 35 | 9 (26%) | 26 (74%) | 41 | 8 (20%) | 33 (80%) | ||
| G/G | 17 | 9 (53%) | 8 (47%) | 2 | 1 (50%) | 1 (50%) | ||
| .078 | .30 | |||||||
| Any A | 62 | 17 (27%) | 45 (73%) | 147 | 23 (16%) | 124 (84%) | ||
| G/G | 17 | 9 (53%) | 8 (47%) | 2 | 1 (50%) | 1 (50%) | ||
| CCL5 rs3817655 | .084 | .67 | ||||||
| A/A | 27 | 7 (26%) | 20 (74%) | 98 | 15 (15%) | 83 (85%) | ||
| A/T | 34 | 9 (26%) | 25 (74%) | 47 | 8 (17%) | 39 (83%) | ||
| T/T | 18 | 10 (56%) | 8 (44%) | 4 | 1 (25%) | 3 (75%) | ||
| .026a | .51 | |||||||
| Any A | 61 | 16 (26%) | 45 (74%) | 145 | 23 (16%) | 122 (84%) | ||
| T/T | 18 | 10 (56%) | 8 (44%) | 4 | 1 (25%) | 3 (75%) | ||
| Hypertension | ||||||||
| CCL5 rs2280789 | .037a | .81 | ||||||
| A/A | 27 | 0 | 27 (100%) | 106 | 25 (24%) | 81 (76%) | ||
| A/G | 35 | 6 (17%) | 29 (83%) | 41 | 11 (27%) | 30 (73%) | ||
| G/G | 17 | 1 (6%) | 16 (94%) | 2 | 0 | 2 (100%) | ||
| .089 | .83 | |||||||
| A/A | 27 | 0 | 27 (100%) | 106 | 25 (24%) | 81 (76%) | ||
| Any G | 52 | 7 (13%) | 45 (87%) | 43 | 11 (26%) | 32 (74%) | ||
| CCL5 rs3817655 | .040a | .75 | ||||||
| A/A | 27 | 0 | 27 (100%) | 98 | 24 (24%) | 74 (76%) | ||
| A/T | 34 | 6 (18%) | 28 (82%) | 47 | 12 (26%) | 35 (74%) | ||
| T/T | 18 | 1 (6%) | 17 (94%) | 4 | 0 | 4 (100%) | ||
| .089 | 1.00 | |||||||
| A/A | 27 | 0 | 27 (100%) | 98 | 24 (24%) | 74 (76%) | ||
| Any T | 52 | 7 (13%) | 45 (87%) | 51 | 12 (24%) | 39 (76%) | ||
| CCR5 rs1799988 | .026a | .37 | ||||||
| Any T | 54 | 2 (4%) | 52 (96%) | 116 | 30 (26%) | 86 (74%) | ||
| C/C | 24 | 5 (21%) | 19 (79%) | 34 | 6 (18%) | 28 (82%) | ||
| Diarrhea | ||||||||
| CCL5 rs2280789 | .52 | .034a | ||||||
| A/A | 27 | 0 | 27 (100%) | 106 | 11 (10%) | 95 (90%) | ||
| Any G | 52 | 3 (6%) | 49 (94%) | 43 | 0 | 43 (100%) | ||
| Rash | ||||||||
| KLF13 rs2241779 | .50 | .010a | ||||||
| C/C | 30 | 5 (17%) | 25 (83%) | 33 | 10 (30%) | 23 (70%) | ||
| Any A | 48 | 5 (10%) | 43 (90%) | 116 | 12 (10%) | 104 (90%) | ||
| AST/ALT | ||||||||
| CCL3 rs1130371 | .65 | .004a | ||||||
| G/G | 36 | 2 (6%) | 34 (94%) | 95 | 7 (7%) | 88 (93%) | ||
| G/A | 39 | 1 (3%) | 38 (97%) | 52 | 1 (2%) | 51 (98%) | ||
| A/A | 3 | 0 | 3 (100%) | 3 | 2 (67%) | 1 (33%) | ||
| .59 | .75 | |||||||
| G/G | 36 | 2 (6%) | 34 (94%) | 95 | 7 (7%) | 88 (93%) | ||
| Any A | 42 | 1 (2%) | 41 (98%) | 55 | 3 (5%) | 52 (95%) | ||
| CCL4 rs1634517 | .22 | .052 | ||||||
| C/C | 42 | 2 (5%) | 40 (95%) | 95 | 7 (7%) | 88 (93%) | ||
| C/A | 26 | 0 | 26 (100%) | 48 | 1 (2%) | 47 (98%) | ||
| A/A | 9 | 1 (11%) | 8 (89%) | 7 | 2 (29%) | 5 (71%) | ||
| 1.00 | .75 | |||||||
| C/C | 42 | 2 (5%) | 40 (95%) | 95 | 7 (7%) | 88 (93%) | ||
| Any A | 35 | 1 (3%) | 34 (97%) | 55 | 3 (5%) | 52 (95%) | ||
Abbreviations: ALT = alanine aminotransferase; AST = aspartate aminotransferase; SNP = single nucleotide polymorphism.
Statistically significant (P < .05) by Fisher’s exact test.
Serum CCL5 and VEGF-A Levels by SNPs in Evaluation Cohort
Associations between SNPs and cytokine levels are summarized in Table 3. The CCL5 rs2280789 G/G variant was significantly associated with lower CCL5 levels compared to any A allele at baseline and day 21 (P = .003; P = .009). Serum VEGF-A levels at baseline appeared to be lower in the CCL5 rs2280789 G/G variant than those in any A allele, although no statistical significance was observed; meanwhile, it was significantly lower at day 21 (P = .024). Similarly, the CCL5 rs3817655 T/T variant was associated with lower serum CCL5 levels and VEGF-A levels at baseline and day 21 compared to any A allele (CCL5: P = .015 and P = .006; VEGF-A: P = .086 and P = .013). In the detection of changes between baseline and day 21, increased CCL5 levels at day 21 were highly expressed in patients with the CCL3 rs1130371 G/G variant (63.0 vs. 34.5%, P = .060), CCL4 rs1634517 C/C variant (61.3 vs. 32.0%, P = .035), and the CCR5 rs1799988 T/T variant (70.6 vs. 38.5%, P = .042). However, no significant difference for serum VEGF-A levels was observed.
Table 3.
Association Between Serum Cytokine Levels and SNPs in Evaluation Cohort
| SNP | N | Mean Serum CCL5 Levels (pg/mL) | Mean Serum VEGF-A Levels (pg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | Day 21 | Quantitative Change | Increase (%) | Baseline | Day 21 | Quantitative Change | Increase (%) | ||
| CCL5 rs2280789 | |||||||||
| A/A | 20 | 67,125.2 | 66,218.1 | −1167.7 | 47.4 | 404.0 | 621.0 | 205.2 | 68.4 |
| A/G | 22 | 63,290.4 | 64,017.6 | 727.2 | 50.0 | 369.4 | 509.0 | 139.6 | 54.5 |
| G/G | 15 | 44,844.5 | 43,344.6 | −1499.9 | 46.7 | 251.4 | 324.1 | 72.7 | 66.7 |
| P | .010a | .008a | .97 | 1.00 | .14 | .062 | .34 | .65 | |
| Any A | 42 | 65,116.5 | 65,037.4 | −150.9 | 48.8 | 385.8 | 560.9 | 170.0 | 61.0 |
| G/G | 15 | 44,844.5 | 43,344.6 | −1499.9 | 46.7 | 251.4 | 324.1 | 72.7 | 66.7 |
| P | .003a | .009a | .82 | 1.00 | .13 | .024a | .23 | .76 | |
| CCL5 rs3817655 | |||||||||
| A/A | 20 | 67,125.2 | 66,218.1 | −1167.7 | 47.4 | 404.0 | 621.0 | 205.2 | 68.4 |
| A/T | 21 | 61,796.6 | 64,922.4 | 3125.8 | 52.4 | 381.9 | 525.8 | 143.9 | 52.4 |
| T/T | 16 | 47,958.0 | 43,449.1 | −4508.8 | 43.8 | 242.3 | 313.5 | 71.2 | 68.8 |
| P | .039a | .005a | .68 | .94 | .45 | .041a | .32 | .58 | |
| Any A | 41 | 64,396.0 | 65,537.9 | 1086.4 | 50.0 | 392.7 | 571.0 | 173.0 | 60.0 |
| T/T | 16 | 47,958.0 | 43,449.1 | −4508.8 | 43.8 | 242.3 | 313.5 | 71.2 | 68.8 |
| P | .015a | .006a | .39 | .77 | .086 | .013a | .21 | .76 | |
| CCL3 rs1130371 | |||||||||
| G/G | 28 | 58,307.0 | 59,695.6 | 1531.8 | 63.0 | 325.3 | 423.0 | 92.3 | 66.7 |
| G/A | 27 | 60,243.5 | 57,254.6 | −2988.9 | 33.3 | 369.6 | 539.1 | 169.5 | 55.6 |
| A/A | 2 | 74,195.5 | 79,523.5 | 5328.0 | 50.0 | 444.4 | 940.8 | 496.5 | 100 |
| P | .65 | .56 | .81 | .078 | .87 | .64 | .29 | .55 | |
| G/G | 28 | 58,307.0 | 59,695.6 | 1531.8 | 63.0 | 325.3 | 423.0 | 92.3 | 66.7 |
| Any A | 29 | 61,205.7 | 58,790.3 | −2415.3 | 34.5 | 374.8 | 566.8 | 192.0 | 58.6 |
| P | .64 | .91 | .60 | .060 | .65 | .23 | .32 | .59 | |
| Any G | 55 | 59,257.6 | 58,475.1 | −728.6 | 48.1 | 347.0 | 481.0 | 130.9 | 61.1 |
| A/A | 2 | 74,195.5 | 79,523.5 | 5328.0 | 50 | 444.4 | 940.8 | 496.5 | 100 |
| P | .38 | .30 | .77 | 1.00 | .74 | .70 | .17 | .52 | |
| CCL4 rs1634517 | |||||||||
| C/C | 32 | 56,542.2 | 60,138.3 | 3777.7 | 61.3 | 318.8 | 415.0 | 91.7 | 61.3 |
| C/A | 17 | 58,021.9 | 56,411.3 | −1610.6 | 41.2 | 392.7 | 623.8 | 231.1 | 70.6 |
| A/A | 8 | 76,479.8 | 61,677.9 | −14,801.9 | 12.5 | 387.4 | 548.3 | 160.9 | 50.0 |
| P | .085 | .88 | .24 | .036a | .80 | .29 | .46 | .64 | |
| C/C | 32 | 56,542.2 | 60,138.3 | 3777.7 | 61.3 | 318.8 | 415.0 | 91.7 | 61.3 |
| Any A | 25 | 63,928.4 | 58,096.6 | −5831.8 | 32.0 | 391.0 | 599.7 | 208.6 | 64.0 |
| P | .24 | .79 | .20 | .035a | .51 | .14 | .24 | 1.00 | |
| Any C | 49 | 57,055.6 | 58,818.3 | 1869.4 | 54.2 | 344.4 | 489.0 | 141.1 | 64.6 |
| A/A | 8 | 76,479.8 | 61,677.9 | −14,801.9 | 12.5 | 387.4 | 548.3 | 160.9 | 50.0 |
| P | .026a | .79 | .12 | .052 | .78 | .73 | .89 | .46 | |
| CCR5 rs1799988 | |||||||||
| T/T | 17 | 55,005.2 | 62,594.8 | 7589.5 | 70.6 | 288.4 | 436.2 | 147.8 | 70.6 |
| T/C | 21 | 60,885.6 | 56,854.4 | −3966.9 | 45 | 386.8 | 608.0 | 210.8 | 65.0 |
| C/C | 19 | 62,835.4 | 58,710.6 | −4124.8 | 31.6 | 365.7 | 435.9 | 70.2 | 52.6 |
| P | .59 | .82 | .36 | .071 | .75 | .39 | .48 | .54 | |
| T/T | 17 | 55,005.2 | 62,594.8 | 7589.5 | 70.6 | 288.4 | 436.2 | 147.8 | 70.6 |
| Any C | 40 | 61,811.8 | 57,758.7 | −4043.8 | 38.5 | 376.8 | 524.2 | 142.3 | 59.0 |
| P | .31 | .56 | .15 | .042a | .45 | .50 | .96 | .55 | |
| Any T | 38 | 58,254.9 | 59,491.9 | 1342.8 | 56.8 | 342.8147 | 529.0511 | 181.8 | 67.6 |
| C/C | 19 | 62,835.4 | 58,710.6 | −4124.8 | 31.6 | 365.7442 | 435.9084 | 70.2 | 52.6 |
| P | .49 | .92 | .49 | .095 | .84 | .40 | .29 | .38 | |
Abbreviations: SNP = single nucleotide polymorphism; VEGF-A = vascular endothelial growth factor A.
Statistically significant (P < .05). P values were calculated by Student’s unpaired t test and 1-way ANOVA for means of continuous measurements and Fisher’s exact test for change patterns in each cytokines.
Discussion
Our data provide the first evidence that SNPs of genes in the CCL5/CCR5 signaling pathway are associated with not only clinical outcomes of but also HFSR caused by regorafenib in mCRC patients.
The CCL5/CCR5 axis is involved in the immune microenvironment and is exploited for network-enabling tumor progression.10 CCL5 is expressed and localized within CD8+ T cells and CXCL10 in tumor cells and macrophages within the invasive margin. CCL3 and CCL4, macrophage inflammatory protein 1 proteins, are produced particularly by macrophages, dendritic cells, and lymphocytes activating CCR5 downstream. RNA expression analysis in colorectal cancer showed that CCL4 was the most strongly expressed in cancer tissues compared to those expressed in nonneoplastic mucosal tissues. CCL3 was also highly expressed in cancer tissue. In contrast, CCL5 was widely expressed not only in cancer tissue but also in nonneoplastic mucosal tissues.11
Our approach was based on preliminary data obtained from a previous translational study that identified both low serum CCL5 levels at baseline and decreased serum VEGF-A levels under treatment with regorafenib,5 indicating CCL5 as potential regulator of VEGF-A production. Recent studies demonstrated that both CCL5 and CCR5 are the key players in activating the signaling.6,12 The clinical significance of gene polymorphisms in the CCL5/CCR5 signaling pathway in carcinogenesis and their predictive and prognostic value with regard to chemotherapeutic agents remains unclear. In our study, patients with the homozygous G/G variant in CCL5 rs2280789 or T/T variant in CCL5 rs3817655 had a trend toward longer OS in the evaluation cohorts. However, these findings were not confirmed in the validation cohort owing to the quite low frequency of these homozygotes in the validation cohort compared to those in the evaluation cohort (approximately 1% vs. 10%). Intriguingly, SNPs of other CCR5 ligands, CCL4 and CCL3, were associated with PFS and OS in both evaluation and validation cohorts. In addition, allelic distributions of these SNPs were similar between the evaluation and validation cohorts, unlike CCL5. Our data are also consistent with findings that mRNA expressions of CCL4 and CCL3 were more specific in cancer tissue than in normal tissue, while CCL5 expression was not limited to cancer tissue.11 In addition, an in vivo study revealed that only CCL5 could induce EPC migration in a dose-dependent manner at a wound site with CCL3, CCL4, CCL5, and CCR5 expression, while CCL3 and CCL4 lacked this activity.9
Regarding the genetic functionality of SNPs, An et al13 demonstrated that transcriptional regulation of CCL5 was primarily governed by CCL5 rs2280789 in the promoter region, to which the G allele corresponded with a strong decrease in transcriptional activity of RANTES. In our study, CCL5 SNPs were the only ones showing a significant relationship with CCL5 and VEGF-A, suggesting that the homozygote might have low productivity of CCL5 leading to lower VEGF-A production. We speculate that the demand for VEGF-A increased in response to regorafenib, which was supported by a phase I study.14 showing that plasma VEGF-A concentration increased over 21 days of multiple doses of regorafenib followed by a decrease to baseline levels during a 7-day treatment rest. By contrast, the plasma soluble VEGF receptor (VEGFR)-2 concentration as a molecular target of regorafenib showed a dose-dependent decrease in each treatment cycle.15 VEGF-A is known to increase vascular permeability and promote angiogenesis in tumor progression, particularly through VEGFR-2 activation.16,17 Meanwhile, another VEGF receptor, VEGFR-1, also acts as a mediator for vascular permeability, and unique cross talk between VEGFR-1 and VEGFR-2 corresponding to vascular permeability and angiogenesis was suggested.18 Altogether, the above assumptions may help to explain the mechanism of action of CCL5 in VEGF-A production triggered by inhibiting VEGFR-1 and VEGFR-2 in response to regorafenib.
Another interesting result of our study is the relationship between CCL5 SNPs and the onset of severe HFSR. Assuming that recovery from HFSR mainly depends on wound-healing ability, the individual capacity of VEGF-A production could be a critical factor in the likelihood or severity of HFSR in addition to the pathologic findings of HFSR in patients treated with multiple kinase inhibitors such as hyperkeratosis, keratinocyte necrosis, and dermal inflammation.19 This idea corresponds to our findings that the CCL5 rs2280789 G/G variant and CCL5 rs3817655 T/T variant were associated with grade 3 or higher HFSR showing lower serum CCL5 levels compared to those with the other variants. These differences can consequently explain the ethnic difference: the high incidence of severe HFSR in the evaluation cohort of Japanese patients compared to the validation cohort of Italian patients; that is, the homozygote of the CCL5 SNPs was extremely rare in the validation cohort compared to the evaluation cohort (1%−3% vs. 21%−23%). Considering that most of the circulating CCL5 derives from the host and not tumors,20 CCL5 genotyping is suggested as a solid resource for precision medicine in managing HFSR due to regorafenib.
Our study has some limitations. It has a retrospective study design; it lacks preclinical data regarding the function of the SNPs; and all cytokine data came from a Japanese population with limited cytokine testing. In addition, other different angiogenic signaling that might affect VEGF-A production could not be excluded. Ideally, a population receiving best supportive care with refractory mCRC should be tested. Further validation research is thus warranted to confirm our findings. A strength of our study is the presence of a validation group of patients with comparable clinical characteristics receiving the same treatment. Furthermore, we first clarified the relationship between serum cytokine levels and SNPs for regorafenib on the basis of data from the previous translational study.
In conclusion, CCL5/CCR5 signaling for VEGF-A production may affect both clinical outcome and HFSR in refractory mCRC patients receiving regorafenib. CCL4 rs1634517 and CCL3 rs1130371 may serve as prognostic markers, and the different percentage of homozygotes in CCL5 SNPs lead to ethnic differences in developing severe HFSR between Italian and Japanese patients.
Supplementary Material
Clinical Practice Points.
Regorafenib improves survival in mCRC patients.
CCL5/CCR5 signaling pathway modulates VEGF-A production.
Genetic variants of CCL4 and CCL3 are associated with clinical outcomes.
CCL5 homozygote is associated with severe HFSR.
Frequencies of CCL5 homozygote accounts for the ethnic differences of severe HFSR.
Acknowledgments
Supported in part by the National Cancer Institute (P30CA014089), the Gloria Borges Wunderglo Project, the Dhont Family Foundation, the Dave Butler Research Fund, and the Call to Cure Research Fund. M.S. is the recipient of Takashi Tsuruo Memorial Fund and JSPS KAKENHI grant 15K06860. M.D.B. received a grant from the Swiss Cancer League (BIL KLS-3334–022014) and the Werner and Hedy Berger-Janser Foundation for cancer research. Y.M. received a grant from Japan Society for the Promotion of Science (S2606).
Footnotes
Disclosure
The authors have stated that they have no conflict of interest.
Supplemental Data
Supplemental methods and tables accompanying this article can be found in the online version https://doi.org/10.1016/j.clcc.2018.02.010.
References
- 1.Grothey A, Van Cutsem E, Sobrero A, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 2013; 381: 303–12. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Qin S, Xu R, et al. Regorafenib plus best supportive care versus placebo plus best supportive care in Asian patients with previously treated metastatic colorectal cancer (CONCUR): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2015; 16:619–29. [DOI] [PubMed] [Google Scholar]
- 3.Tabernero J, Lenz HJ, Siena S, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol 2015; 16:937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshino T, Komatsu Y, Yamada Y, et al. Randomized phase III trial of regorafenib in metastatic colorectal cancer: analysis of the CORRECT Japanese and non-Japanese subpopulations. Invest New Drugs 2015; 33:740–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suenaga M, Mashima T, Kawata N, et al. Serum VEGF-A and CCL5 levels as candidate biomarkers for efficacy and toxicity of regorafenib in patients with metastatic colorectal cancer. Oncotarget 2016; 7:34811–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang SW, Liu SC, Sun HL, et al. CCL5/CCR5 axis induces vascular endothelial growth factor mediated tumor angiogenesis in human osteosarcoma microenvironment. Carcinogenesis 2015; 36:104–14. [DOI] [PubMed] [Google Scholar]
- 7.Song A, Nikolcheva T, Krensky AM. Transcriptional regulation of RANTES expression in T lymphocytes. Immunol Rev 2000; 177:236–45. [DOI] [PubMed] [Google Scholar]
- 8.Song A, Patel A, Thamatrakoln K, et al. Functional domains and DNA-binding sequences of RFLAT-1/KLF13, a Krüppel-like transcription factor of activated T lymphocytes. J Biol Chem 2002; 277:30055–65. [DOI] [PubMed] [Google Scholar]
- 9.Ishida Y, Kimura A, Kuninaka Y, et al. Pivotal role of the CCL5/CCR5 interaction for recruitment of endothelial progenitor cells in mouse wound healing. J Clin Invest 2012; 122:711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Balkwill F. and the chemokine network. Nat Rev Cancer 2004; 4:540–50. [DOI] [PubMed] [Google Scholar]
- 11.Musha H, Ohtani H, Mizoi T, et al. Selective infiltration of CCR5(+) CXCR3(+) T lymphocytes in human colorectal carcinoma. Int J Cancer 2005; 116:949–56. [DOI] [PubMed] [Google Scholar]
- 12.Liu GT, Huang YL, Tzeng HE, Tsai CH, Wang SW, Tang CH. CCL5 promotes vascular endothelial growth factor expression and induces angiogenesis by downregulating miR-199a in human chondrosarcoma cells. Cancer Lett 2015; 357: 476–87. [DOI] [PubMed] [Google Scholar]
- 13.An P, Nelson GW, Wang L, et al. Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci U S A 2002; 99: 10002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mross K, Frost A, Steinbild S, et al. A phase I dose-escalation study of regorafenib (BAY 73–4506), an inhibitor of oncogenic, angiogenic, and stromal kinases, in patients with advanced solid tumors. Clin Cancer Res 2012; 18: 2658–67. [DOI] [PubMed] [Google Scholar]
- 15.Wilhelm SM, Dumas J, Adnane L, et al. Regorafenib (BAY 73–4506): a new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int J Cancer 2011; 129:245–55. [DOI] [PubMed] [Google Scholar]
- 16.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011; 473:298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson N, Powner MB, Sarker MH, et al. Differential apicobasal VEGF signaling at vascular bloodeneural barriers. Dev Cell 2014; 30:541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi H, Hattori S, Iwamatsu A, Takizawa H, Shibuya M. A novel snake venom vascular endothelial growth factor (VEGF) predominantly induces vascular permeability through preferential signaling via VEGF receptor-1. J Biol Chem 2004; 279:46304–14. [DOI] [PubMed] [Google Scholar]
- 19.Lacouture ME, Reilly LM, Gerami P, Guitart J. Hand foot skin reaction in cancer patients treated with the multikinase inhibitors sorafenib and sunitinib. Ann Oncol 2008; 19:1955–61. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Lv D, Kim HJ, et al. A novel role of hematopoietic CCL5 in promoting triple-negative mammary tumor progression by regulating generation of myeloidderived suppressor cells. Cell Res 2013; 23:394–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.