Background:
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused severe pneumonia at December 2019. Since then, it has been wildly spread from Wuhan, China, to Asia, European, and United States to become the pandemic worldwide. Now coronavirus disease 2019 were globally diagnosed over 3 084 740 cases with mortality of 212 561 toll. Current reports variants are found in SARS-CoV-2, majoring in functional ribonucleic acid (RNA) to transcribe into structural proteins as transmembrane spike (S) glycoprotein and the nucleocapsid (N) protein holds the virus RNA genome; the envelope (E) and membrane (M) alone with spike protein form viral envelope. The nonstructural RNA genome includes ORF1ab, ORF3, ORF6, 7a, 8, and ORF10 with highly conserved information for genome synthesis and replication in ORF1ab.
Methods:
We apply genomic alignment analysis to observe SARS-CoV-2 sequences from GenBank (http://www.ncbi.nim.nih.gov/genebank/): MN 908947 (China, C1); MN985325 (United States: WA, UW); MN996527 (China, C2); MT007544 (Australia: Victoria, A1); MT027064 (United States: CA, UC); MT039890 (South Korea, K1); MT066175 (Taiwan, T1); MT066176 (Taiwan, T2); LC528232 (Japan, J1); and LC528233 (Japan, J2) and Global Initiative on Sharing All Influenza Data database (https://www.gisaid.org). We adopt Multiple Sequence Alignments web from Clustalw (https://www.genome.jp/tools-bin/clustalw) and Geneious web (https://www.geneious.com.
Results:
We analyze database by genome alignment search for nonstructural ORFs and structural E, M, N, and S proteins. Mutations in ORF1ab, ORF3, and ORF6 are observed; specific variants in spike region are detected.
Conclusion:
We perform genomic analysis and comparative multiple sequence of SARS-CoV-2. Large scaling sequence alignments trace to localize and catch different mutant strains in United possibly to transmit severe deadly threat to humans. Studies about the biological symptom of SARS-CoV-2 in clinic animal and humans will be applied and manipulated to find mechanisms and shield the light for understanding the origin of pandemic crisis.
Keywords: Genomic analysis, Multiple sequence, Severe acute respiratory syndrome coronavirus 2
1. INTRODUCTION
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused severe pneumonia at December 2019.1 Since then, it has been wildly spread from Wuhan, China, to Asia, European, and United States to become pandemic worldwide.2 Severe cases beginning from Huanan Seafood Wholesale market in China which confirmed human pneumonia with the infection of a novel coronavirus (2019-nCoV),3 and named as SARS-CoV-2 by International Committee on Taxonomy of Viruses.4,5 Now coronavirus disease 2019 were globally diagnosed over 3 084 740 cases with mortality of 212 516 toll.6
Current reports single nucleotide variants are found in many patients with SARS-CoV-2, which belongs to beta-coronavirus species. SARS-CoV-2 contains functional genomic ribonucleic acid (RNA) to transcribe into structural proteins as transmembrane spike (S) glycoprotein for mediating the virus to entry the host cell by utilizing host’s cellular angiotensin-converting enzyme 2 (ACE2), and the nucleocapsid (N) protein holds the major nuclear viral RNA genome; the envelope (E) and membrane (M) alone with spike protein form viral envelope.7 The nonstructural RNA genome including ORF1ab, ORF3, ORF6, 7a, 8, and ORF10 contains highly conserved information for genome RNA synthesis and replication in ORF1ab and unclear-verified function in other ORF proteins.8
The transmission mechanisms with the start of SARS-CoV attaches host cell membrane receptor and then induce the membrane endocytosis to entry host cells. ORF1 of virus genome leads its replication and synthesize the subgenomic RNAs afterward. Meanwhile, N protein and new genomic RNA assemble to form helical nucleocapsids with M protein inserted in endoplasmic reticulum (ER) and anchored Golgi of host cells. E and M proteins then begin to trigger budding processes. S together with helical N on membrane-bound ER triggers the translation-required viral structure proteins and transport to Golgi. During the final cycle, virions are released by exocytosis to finish the life cycle and replication of the virus.9
Previous SARS-CoV-1 in 2003 transmits possibly through Bat and Civet as its intermediate hosts, and finally to human with the symptoms of severe respiratory impacts in a 10% mortality rate. However, Wuhan SARS-CoV-2 is suspected to be transmitted from bat (RaTG13) to pangolin as intermediate hosts before transmitted to humans by some unknown mechanisms with symptoms of severe respiratory impacts with highest mortality now.10 The genomic sequence of RaTG13 cited the 96% similarity with Wuhan coronavirus.11 Although intermediate host is not clear at present, genomic sequence comparison obviously points out spike receptor-binding domain (RBD) of Wuhan SARS-CoV-2 with the similarity in 90% homolog of pangolin. Thus, the possibility that pangolin might contribute the spike protein region to cross-transmitted to RATG13 forms a new recombinant mutant Wuhan SARS-CoV-2 to transmit onto human finally.12
The S protein of SARS-CoV-1 and SARS-CoV-2 responsible for viral entry mediates the binding to host cell membrane of ACE2 through its RBD.13 The surface S spike protein of SARS-CoV comprises two components (S1 and S2). The S protein of SARS-CoV-2 binds to the host receptor ACE2 through its S1 subunit, which contains RBD, and follows by fusing the viral and host membranes through the S2 subunit, which contains the fusion peptide primed by host protease.
Major six ORFs exist in SARS-CoV-2. ORF1ab occupies the two-thirds length of the whole genome and subgenome RNA to play roles in viral pathogenesis excluding its replication function as well as involving in cellular signaling and modification of cellular gene expression.14
There is no clue for antiviral therapy and treatment for SARS-CoV-2 at present. Further study approaches the molecular genomic variants for selection and packaging is critical for developing antiviral strategies. We will verify and compare various SARS-CoV-2 sequences from different countries by analyzing the possible genomic networks of disease from its origin to evolution, providing the moving development of strategy against the worldwide SARS-CoV-2 pandemic threat.
2. METHODS
2.1. Sequence resource
Studies focus on evolutionary and phylogenetic analysis have applied in disease progression for Wuhan lung pneumonia treatment. Herein, we apply genomic analysis to observe SARS-CoV-2 sequences from GenBank (http://www.ncbi.nim.nih.gov/genebank/): MN 908947 (China, C1); MN985325 (United States: WA, UW); MN996527 (China, C2); MT007544 (Australia: Victoria, A1); MT027064 (United States: CA, UC); MT039890 (South Korea, K1); MT066175 (Taiwan, T1); MT066176 (Taiwan, T2); MT192759 (Taiwan, T3); MT198652 (Spain, SP); LC528232 (Japan, J1); LC528233 (Japan, J2); MT093571 (Sweden, SW); MT066156 (Italy, IT); and MT050493 (India, In) for genomic sequence alignment analysis.
2.2. Method applied
Multiple Sequence Alignment by Clustalw (https://www.genome.jp/tools-bin/clustalw) web is applied as our alignment tool. Phylogenetic analysis platform performs at Geneious website (https://www.geneious.com).
3. RESULTS
3.1. ORF1ab
ORF1ab joins 16 proteins together to perform viral genomic replication and synthesis. From the data analysis, it reveals eight mutations from a different country: During this long 6796 amino acids protein, we observe eight mutations located in different regions from various countries; position T609I mutation in California/United States sequence, G818S in Sweden and India, M902I in Korea, F3071Y in Spain, S3120L China, L3606X in Italy and L3606F in Japan, F4321L in Sweden and India, and T6891M in Korea.
3.2. ORF3a
ORF3a functions as accessory protein to help new viral synthesis and escape from the host cell. We find four position mutations; M128L in Korea, K136X in Spain, G196V in Spain, and G251V in Italy, Korea, and Sweden.
3.3. ORF6, ORF7a, ORF8, and ORF10
There are no mutations in ORF6, ORF7a, and ORF10, but we do find one mutation in ORF8 located at L84S from Spain, India, and China.
3.4. E protein
E protein has a short and hydrophilic N-terminus consisting of 7-12 amino acids, followed by a large hydrophobic transmembrane domain of 25 amino acids, and ends with a long, hydrophilic C-carboxyl terminus (C-terminal), which comprises the majority of the E protein. Analyzing of E protein alignment, we find one amino acid mutation at L37H from Korea.
3.5. M and N protein
The M protein abundantly defines the shape of the viral envelope. N protein functions primarily to bind to RNA genome of SARS-CoV, making up the nucleocapsid.15 Although N is most involved in processes viral genome signaling, it is also involved RNA replication cycle with host cellular response to viral infection. Although many differences between SARS-CoV-1 and SARS-CoV-2 within in M and N protein, there is no variant observed in M protein but we find a point mutation S197L from Spain.
3.6. S protein
S protein mediates the attachment of SARS-CoV-1 to the host cell surface receptors and subsequently fuse them to facilitate viral entry into the host cell.15 The expression of S protein at the cell membrane can mediate cell-cell fusion. This formation offers a strategy to spread the virus between cells to subvert function of virus-neutralizing antibodies mechanisms, which play major controlling of protein interaction. By analysis of S protein, we find four mutations from 10 countries; S221W in Korea, S247R in Australia, F737C in Sweden, and A870V in India (Figs. 3–6).
Fig. 3.

Genomic analysis of ORF6, ORF7a, ORF8, and ORF10 protein amino acid sequence. There are not any mutations in ORF6, ORF7a, and ORF10, but we find one mutation in ORF8 located at L84S in Taiwan, United States, Spain, India, and China.
Fig. 6.

Genomic analysis of S protein amino acid sequence. During analysis of S protein, we find four mutations from 10 countries; S221W in Korea, S247R in Australia, F737C in Sweden, and A870V in India.
4. DISCUSSION
4.1. Point mutation
Six ORFs in SARS-CoV-2 function variously. ORF1ab joins 16 proteins together to perform viral genomic replication and synthesis. Our first finding reveals eight mutations in different countries. Eight mutation in different regions from various countries are; position T609I mutation in California/United States sequence, G818S in Sweden and India, M902I in Korea, F3071Y in Spain, S3120L China, L3606X in Italy and L3606F in Japan, F4321L in Sweden and India, and T6891M in Korea. No direct evidence proves if each mutant will enhance or decrease viral RNA polymerase and replication (Fig. 1).
Fig. 1.

Genomic analysis of ORF1ab protein amino acid sequence. We detect eight mutations in different regions from various countries, T609I mutation in United States, G818S in Sweden and India, M902I in Korea, F3071Y in Spain, S3120L in China, L3606X in Italy and L3606F in Japan, F4321L in Sweden and India, and T6891M in Korea.
ORF3a functions as accessory protein to help new viral synthesis and escape from the host cell. We find four position mutation; M128L in Korea, K136X in Spain, G196V in Spain, and G251V in Italy, Korea, and Sweden (Fig. 2). We do not observe any mutations in ORF6, ORF7a, and ORF10 proteins, but we find one mutation in ORF8, which located at L84S from Spain, India, and China. No inclusion can explain the mutations happened at present (Fig. 3).
Fig. 2.
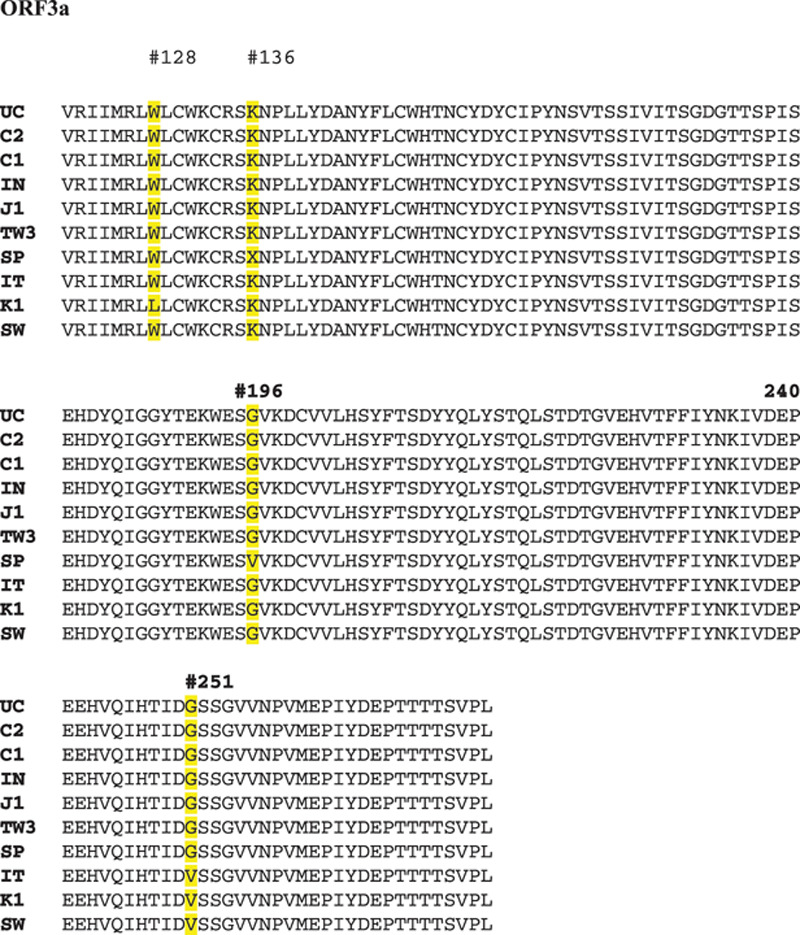
Genomic analysis of ORF3a protein amino acid sequence. We find four position mutations; M128L in Korea, K136X in Spain, G196V in Spain, and G251V in Italy, Korea, and Sweden.
In comparison of 10 strains from different countries, one mutation of E protein is observed at L37H in Korea (Fig. 4). Inside the envelope, there is the nucleocapsid, which is formed from multiple copies of the nucleocapsid (N) protein, which are bound to the positive-sense single stranded RNA genome in a continuous beads-on-a-string type conformation.16 The lipid bilayer envelope, membrane proteins, and nucleocapsid protect the virus when it is outside the host cell.17
Fig. 4.

Genomic analysis of E protein amino acid sequence. We found one amino acid mutation at position 37th L37H as “H” from South Korea comparing the “L” from other nine sequences. Yellow line indicates the difference in 10 sequence alignment.
Although the N protein holds the viral RNA, and M protein joins with E and S proteins together to create the viral envelope for protection when it is outside the host cell, we do not find point mutation of M protein.
We do find a point mutation S197L of N protein in Spain. The binding of M to N stability the nucleocapsid (N protein-RNA complex), as well as the internal core of virions, and, ultimately, promotes completion of viral assembly.18 No evidence demonstrates if S197L will abolish function of N protein (Fig. 5).
Fig. 5.

. Genomic analysis of M and N protein amino acid sequences. We do not observe any mutation in 10 sequences of M protein region but detect one mutation in Spain at S197L of N protein.
By analysis of S protein, we find four mutations from 10 countries; S221W in Korea, S247R in Australia, F737C in Sweden, and A870V in India (Fig. 6). Report19 mentioned a single amino acid reversion (L294Q) in the S protein is sufficient to abrogate the phenotype and grows well at and below 32oC.
4.2. Large scaling alignment of spike protein mutations and phylogenetic analysis
Although SARS-CoV-1 and SARS-CoV-2 share the sequence similarity with 80% homolog. After performing the alignment, they reveal their 75% similarity in spike protein. The S protein mediates viral entry into host cells by first binding to a host receptor through the RBD in the S1 subunit and then fusing the viral and host membranes through the S2 subunit priming by host cell proteases.20–23 Unraveling which cellular factors are used by SARS-CoV-2 for entry might provide insights into viral transmission and reveal therapeutic targets. SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) RBDs recognize different receptors. SARS-CoV recognizes ACE2 as its receptor, whereas MERS-CoV recognizes dipeptidyl peptidase 4 as its receptor.14,24 Since SARS-CoV-2 recognizes ACE2 as its host receptor binding to viral S protein.25 Therefore, it is critical to define the RBD in SARS-CoV-2 S protein as the most likely target for the mechanism of virus attachment such as new developing inhibitors, neutralizing antibodies, and vaccines.
Authors from the group of Tai et al26 demonstrate by characterizing of SARS-CoV-2 RBD to display a multiple sequence alignment of RBDs of SARS-CoV-2, SARS-CoV, and MERS-CoV spike (S) proteins.
They identified the RBD in SARS-CoV-2 S protein and found that the RBD protein bound strongly to human and bat ACE2 receptors. SARS-CoV-2 RBD displayed significantly higher binding affinity to ACE2 receptor than SARS-CoV RBD. Subsequently, SARS-CoV RBD-specific antibodies could cross-react with SARS-CoV-2 RBD protein. Meanwhile, SARS-CoV RBD-induced antisera could cross-neutralize SARS-CoV-2 which suggested the potentials to develop SARS-CoV RBD-based vaccines for prevention of SARS-CoV-2 and SARS-CoV infection.26
Hoffmann group mentions SARS-CoV-1 and SARS-CoV-2 share 76% amino acid identity in spike protein region. By the amino acid alignment, they observe the receptor-binding motif of SARS-CoV-1 corresponding to the sequences of bat-associated beta-coronavirus S proteins. Demonstration of high or low similarity by taking advantage of ACE2 as cellular receptor reveals SARS-CoV-2 possesses crucial amino acid residues for ACE2 binding.
They also find similarity signal to points out between SARS-CoV-2 and SARS-CoV-1 during transmitting host cells stage and then identify a potential target for antiviral intervention. Inspecting conserved amino acids within ACE2 domain, Hoffmann group perform SARS-CoV-2 to transmit cell entry depends on ACE2 and transmembrane serine protease 2 two proteins and is blocked by applied clinically proven protease inhibitor.27,28
By deep and large scaling analysis of spike protein from many countries, we do have variants found in US case including specimen from east coast United States. We do find variants in United States comparing with China origin (Fig. 7). Mutant-1 expresses a “G” amino acid at 614 instead of China “D” (D614G). Mutant-2 strain displays the position at 614 same as China strain with “D” but other mutations found in different regions (Fig. 8A). Mutant 2-2 with same position of 614 “D” but only display one mutation same as China pointed as QIS60546 strain (Fig. 8B). Studies suggest various viral strains originally spread from China to Europe which one strain should be deadly mutations as observed and then they spread to New York finally. The other milder strains also spread to west coast in United States from China.29 Since this report cites SARS-CoV2 acquired mutations capable of substantially changing its pathogenicity. Will this observation be matched with our finding that three variants found in New York become more severe transmitted to humans than west coast in the United States?
Fig. 7.
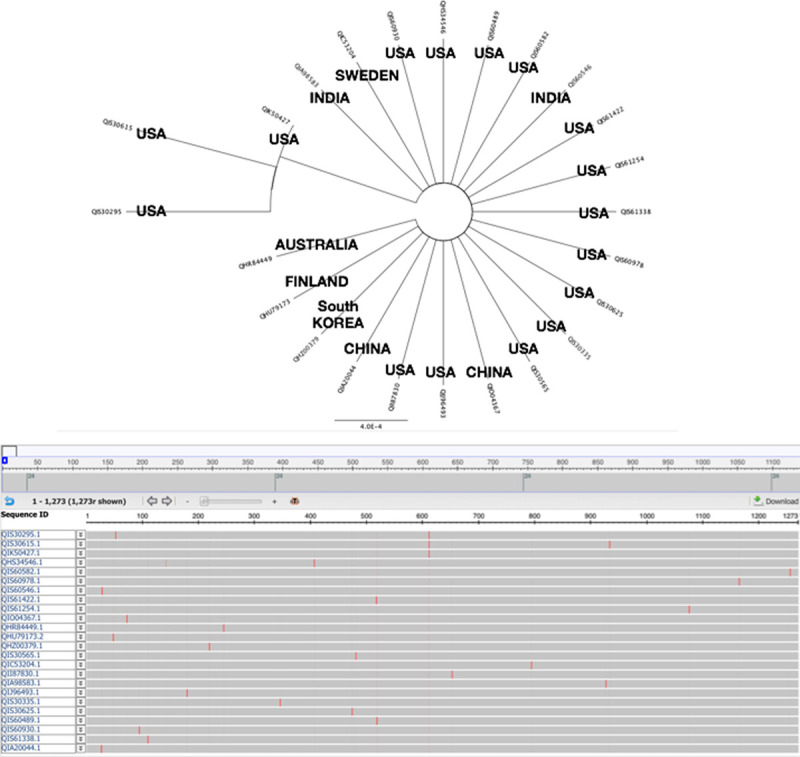
Spike protein reveals variants in the world. We find many variants in spike protein by alignment and phylogenetic analysis.
Fig. 8.
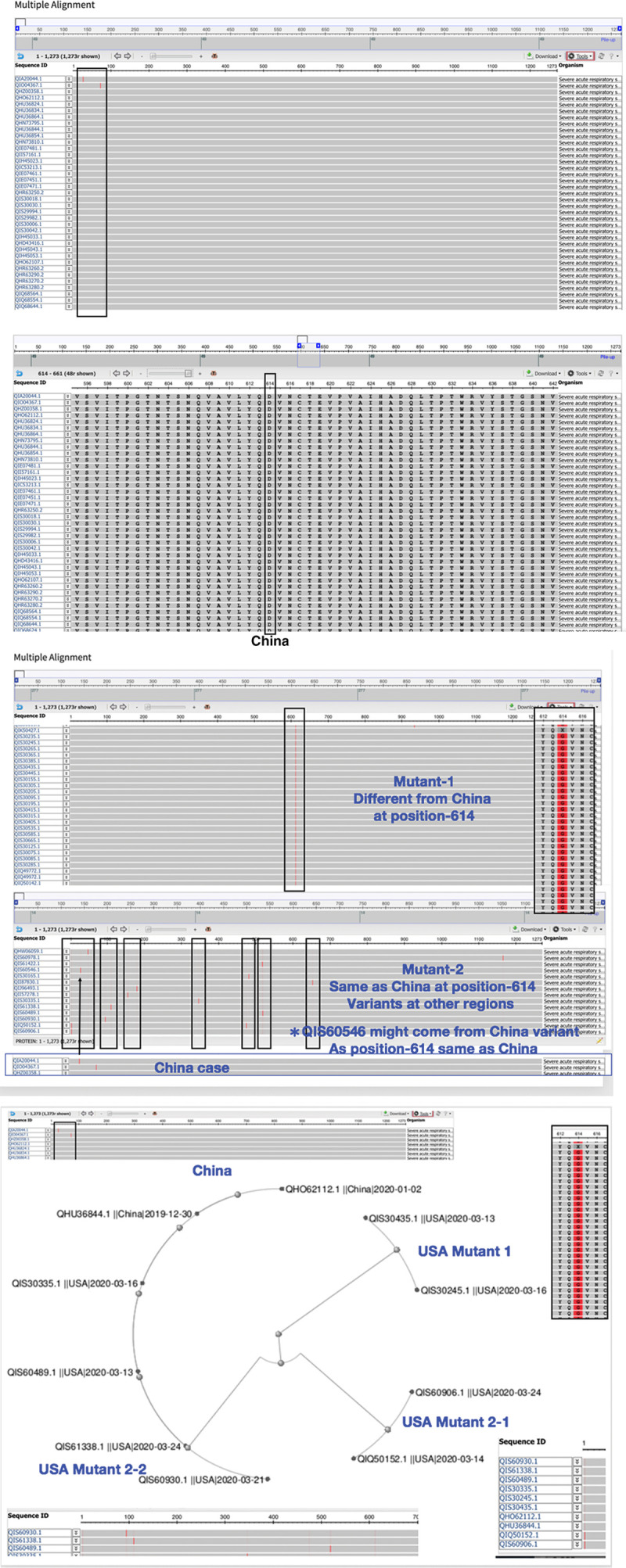
A, Spike protein in China sequences exhibit a conserved amino acid. We found a conserved amino acid “D” at position 614 of Spike protein in most China sequences. B, Analysis indicates three variants of spike protein in the United States. We observe three variants in the analysis of United States sequences; mutant-1 found with different amino acid “G” at position 614, mutant 2-1 with same “D” at position 614 same as China but various variants at other regions. Mutant 2-2 same as 2-1 at 614 but same as China in one region as QIS60546 indicated. (I) US case. (II) Phylogenetic analysis to map three mutants in United States and China.
Limitedly in the study, we perform our study either data mining by alignment and phylogenetic analysis from public domains such as Global Initiative on Sharing All Influenza Data and National Center for Biotechnology Information. There will be interesting to demonstrate biological approaches with specimens in hands to observe the correlation from clinical to lab analysis directly.
In conclusion, we analyze database by genome alignment search for nonstructural ORFs and structural E, M, N, and S proteins. Large scaling performance to catch different mutant strains in American possibly induce severe deadly threat to humans. More studies about the biological symptom of SARS-CoV-2 in clinic animal and humans will manipulate and shield the light for understanding the origin of pandemic crisis.
ACKNOWLEDGMENTS
This research was funded by Taipei Veterans General Hospital (grant number V107E-002-2, V108D46-004-MY2-1, V108E-006-4, 108E-006-5, and 109VACS-003).
Footnotes
Conflicts of interest: The authors declare that they have no conflicts of interest related to the subject matter or materials discussed in this article.
REFERENCES
- 1.Fehr AR, Channappanavar R, Perlman S. Middle East respiratory syndrome: emergence of a pathogenic human coronavirus. Annu Rev Med 2017;68:387–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 2016;14:523–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Novel coronavirus (2019-nCoV) situation report 23. 2020Available at https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 4.World Health Organization. Coronavirus disease 2019. Retrieved March 15, 2020. Available at https://covid19.who.int.
- 5.ICTV website. The International Committee on Taxonomy of Viruses (ICTV).February 5, 2020Available at https://talk.ictvonline.org.
- 6.nCoV2019.Live. Available at https://ncov2019.live/data.
- 7.Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, et al. The genome sequence of the SARS-associated coronavirus. Science 2003;300:1399–404. [DOI] [PubMed] [Google Scholar]
- 8.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003;300:1394–9. [DOI] [PubMed] [Google Scholar]
- 9.Anderson KG, Rambaut A, Lipkin WI, Holes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med 2020;17:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 2015;1282:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020;395:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 2020;9:221–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003;426:450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham RL, Sparks JS, Eckerle LD, Sims AC, Denison MR. SARS coronavirus replicase proteins in pathogenesis. Virus Res 2008;133:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J 2019;16:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang CK, Hou MH, Chang CF, Hsiao CD, Huang TH. The SARS coronavirus nucleocapsid protein–forms and functions. Antiviral Res 2014;103:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, et al. A structural analysis of M protein in coronavirus assemblyand morphology. Journal Struct Biol 2011;174:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escors D, Ortego J, Laude H, Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J Virol 2001;75:1312–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen S, Law YC, Liu DX. A single amino acid mutation in the spike protein of coronavirus infectious bronchitis virus hampers its maturation and incorporation into virions at the nonpermissive temperature. Virology 2004;326:288–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, Xiao G, Chen Y, He Y, Niu J, Escalante CR, et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 2004;363:938–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Wong G, Lu G, Yan J, Gao GF. MERS-CoV spike protein: targets for vaccines and therapeutics. Antiviral Res 2016;133:165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005;309:1864–8. [DOI] [PubMed] [Google Scholar]
- 24.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013;500:227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013;495:251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell mole Immunol 2020;17:613–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao H, Lu X, Chen Q, Xu K, Chen Y, Cheng L, et al. Patient-derived mutations impact pathogenicity of SARS-CoV-2. medRxiv 2020 10.1101/2020.04.14.20060160. [DOI] [Google Scholar]


