Highlights
-
•
The value of swab types on the detection of SARS-CoV-2 from patients during infection late stage is studied.
-
•
The effect of specimen collection time on the detection rate of novel coronavirus was explored.
-
•
Nasopharyngeal /nasal swabs collected before washing in the morning are more suitable for screening of large-scale specimens.
Keywords: SARS-Cov-2, Specimen types, Collection timepoint, RT-PCR, Positive detection rate
Abstract
Objective
Low viral load from patients infected with SARS-CoV-2 during infection late stage easily lead to false negative nucleic acid testing results, thus having great challenges to the prevention and control of the current pandemic. In present study, we mainly aimed to evaluate specimen types and specimen collection timepoint on the positive detection of 2019 novel coronavirus from patients at infection late stage based on RT-PCR testing.
Methods
Paired nasopharyngeal swabs, nasal swabs, oropharyngeal swabs and anal swabs were collected from patients infected with SARS-CoV-2 during infection late stage before washing in the morning and afternoon on the same day. Then virus RNA was extracted and tested for 2019-nCoV identification by RT-PCR within 24 h.
Results
Viral load was low at late infection stage. Specimens collected before washing in the morning would increase the detection ratio of 2019-nCoV. Detection ratio of nasopharyngeal swab [65 (95 % CI: 49.51–77.87) vs 42.5(95 % CI: 28.51–57.8)] or nasal swab [57.5 (95 % CI: 42.2–71.49) vs 35 (95 % CI: 22.13–50.49)] is higher not only than oropharyngeal swab[22.5 (95 % CI: 12.32–37.5) vs 7.5 (95 % CI: 2.58–19.86)], but also anal swab[2.5 (95 % CI: 0.44–12.88) vs 5 (95 % CI: 1.38–16.5)].
Conclusions
In summary, our research discovers that nasopharyngeal or nasal swab collected before washing in the morning might be more suitable for detecting of large-scale specimens from patients infected with low SARS-CoV-2 load during infection late stage. Those results could facilitate other laboratories in collecting appropriate specimens for improving detection of SARS-CoV-2 from patients during infection late stage as well as initially screening.
1. Introduction
The pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) with high morbidity and mortality has become the most serious health crisis in modern times, which makes everyone at risk of significant harm (Zhu and Zhang, 2019; Zheng, 2020). Nucleic acid test is a simple and rapid novel coronavirus identification method, which has become the gold standard in the diagnosis of novel coronavirus infected pneumonia (Li et al., 2020). Low viral load from patients infected with SARS-CoV-2 during infection late stage easily lead to false negative nucleic acid testing results, thus having great challenges to the prevention and control of the current pandemic.
Nucleic acid amplification tests (NAAT) based on detection of unique sequences of virus, such as whole genome sequencing analysis and real-time reverse-transcriptase polymerase-chain-reaction (RT-PCR) assay, could achieve early and high-throughput detection of low viral load and play an important role in the detection of COVID-19. Whole genome sequencing was the method that SARS−COV-2 was initially identified in the laboratory (Chen et al., 2020a). This method is not suitable for large-scale specimen detection under the current pandemic because of its longer turnaround time, higher cost and professional requirements for interpretation of results. RT-PCR recommended in the New Coronavirus Pneumonia Prevention and Control Program (6th edition) published by the National Health Commission of China was still used as one of the vital criteria for initial screen and discharge diagnostic (Corman et al., 2020; Lo et al., 2020). At present, reports of false negative nucleic acid testing results and discharged patients that turn positive again are increasing (Ai et al., 2020; Zhang et al., 2020). Appropriate specimen selection is important for improving the detection of SARS-Cov-2 through RT-PCR method and reducing current false negative detection. Lower respiratory tract samples, such as bronchoalveolar lavage fluid (BALF), is most accurate for laboratory diagnosis of COVID-19 based on some reports (Xiong et al., 2020). However, BALF is not feasible for the routine rapid laboratory diagnosis of the SARS-Cov-2 because collection of BALF requires both a suction device and a skilled operator, is also painful for the patients. Instead, swabs collected from upper respiratory tract or anal is more suitable for large-scale specimen detection under the current pandemic because of its rapid, simple and safe (Lambert et al., 2008; Lin et al., 2020a; Peng et al., 2019). Some studies have investigated the effects of swab type on SARS-Cov-2 detection (Lin et al., 2020a). The value of swab collection timepoint on the detection of SARS-Cov-2 is ignored.
Accordingly, in this study, we mainly aimed to evaluate specimen types and specimen collection timepoint on the positive detection of SARS-CoV-2 for patients during late infection using RT-PCR testing. We hope to assist other laboratories in selecting appropriate specimen for the fast and accurate detection of SARS-CoV-2 from COVID-19 infected patients that are not only in late stage but also in initial screening.
2. Material and methods
2.1. Patient information
48 confirmed or highly suspected 2019-nCoV infected patients who were hospitalized in Yichang Central people's Hospital from Jan. 31 to Mar. 16, 2020 were included. Patient inclusion criteria were based on the following points. Firstly, these patients were diagnosed based on the National recommendations for diagnosis and treatment of pneumonia caused by 2019-nCoV through epidemiology survey, imaging features, laboratory examinations, clinical characteristics and nucleic acid testing results (Chen et al., 2020b; Jin et al., 2020). If the nucleic acid test result is positive, the patient is judged as a confirmed case. If the nucleic acid test result is negative but the patient has relative epidemiological history, clinical characteristics or imaging features, the patient is judged as a suspected case. Secondly, the patient should have the ability to understand and agree to participate in the study. Patients who lack the ability to understand and agree, object to participate and could not collect specimen needed in this study because of physical reasons would be excluded. Patients in this study were divided into three groups based on initial results of chest computerized tomography (CT) test. Common type: CT shows signs of pneumonia with multiple small plaques and interstitial changes, especially in the outer lung. Heavy type: Both lungs showed typical multiple ground glass shadow and infiltration shadow. Severe type: Lung consolidation appeared besides the characteristics of heavy type. The recovery index was used to represent the stage of the patient. The recovery index = the patient's hospitalization time on the day of testing /the patient's whole hospitalization time. The closer the recovery index is to 1, the closer the patient is to discharge criteria. Discharge time was determined by experts based on national standards. First, the body temperature returned to normal for more than 3 days. Second, respiratory symptoms significantly improved. Third, pulmonary imaging showed that acute exudative lesions were significantly improved. Fourth, the nucleic acid test of respiratory tract specimens were negative for two consecutive times (the sampling time was at least 1 day).
2.2. Study plan
The study plan is described as follows. Ⅰ. Patients wrote informed consent. Ⅱ. Querying medical history data and grouping. Ⅲ. Paired nasopharyngeal, nasal, oropharyngeal and anal swabs were collected by nurses based on a standard protocol from the same patient before washing in the morning and after on the same day (Li et al., 2013; Lin et al., 2020b). Ⅳ. Virus RNA was extracted and tested for 2019-nCoV identification by RT-PCR within 24 h.Ⅴ. Data analysis. Quality controls including three negative and one positive were used for monitoring the whole detection process. Two consecutively negative RT-PCR test results in respiratory tract specimens is required for the evaluation of discharge from hospital in china. If the test results of all swabs from the same patient are negative, the swabs from the patient should be collected and detected again according to the negative criteria depicted as above. The study was approved by the Ethics Committees from Yichang Central people's Hospital.
2.3. Quantitative reverse transcription polymerase chain reaction
Viral RNA was extracted with a High Pure Viral Nucleic Acid Kit, as described by the manufacturer (Sansure Biotech). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using a China Food and Drug Administration (CFDA) approved commercial kit specific for 2019-nCoV detection (Sansure biotechnology co., Ltd, China) in accordance with manufacturer instructions. RT-PCR assay was used to detect viral RNA by targeting a consensus open reading frame (ORF 1ab) and nucleoprotein (N) gene region that recommended by the China CDC. A housekeeper gene is used to monitor the reaction. The limit of detection (LoD) of the kit could reach 200 copies/mL. The qPCR reaction mixture (40 μL each) were prepared by mixing 30 μL of Master Mix and 10 μL of virus RNA. The qPCR amplification was performed in the ABI DX real-time PCR system with reverse transcription at 50 °C for 30 min, followed by denaturation at 95 °C for 1 min and then 45 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 30 s. The positive controls consisted of viral RNA plasmids. Interpretation based on kit instructions shows that the specimen is judged as positive if the Ct value was ≤ 40.0, and negative if the result is undetermined. Specimen with a Ct value during 40 to 45 is in a grey area and the specimen should be tested again. The specimen is judged as positive if the repeat result is still during 40–45. If the repeat Ct is undetectable, the specimen is judged as negative.
2.4. Statistics analyses
All 95 % confidence intervals were used to determine whether differences in specimens and timepoint were statistically significant. Statistical analyses were based on McNemar's Test for Correlated Proportions in the Marginals of a 2 × 2 Contingency Table. A p-value of <0.05 was considered statistically significant.
3. Results
3.1. The characteristics of the patients in this study
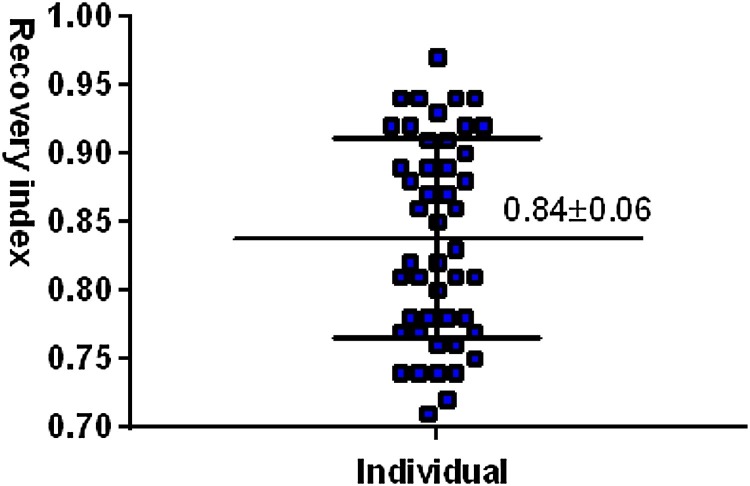
A total of 48 volunteers were recruited in this study. One patient (No. 22) was excluded because this patient failure to meet the inclusion criteria based on the comprehensive analysis from nucleic acid testing and other inspection. Patient information and test results are shown in Table 1 (Detailed information shown in Table S1). The whole average age of all patients was 53.45 ± 12.08 old. The average age and number of ordinary type patients, heavy patients and serve patients were 52 ± 11.44 and 32[68.09 % (95 % CI: 53.84–79.61)], 51.25 ± 11.08 and 12[25.53 % (95 % CI: 15.25–39.51)], 77.67 ± 7.11 and 3[6.38 % (95 % CI: 2.19–17.16)], respectively. The number of female patients was 25/47, 25 % [53.19 % (95 % CI: 39.23–66.67)]. The average age of female patients was 52.68 ± 11.40 old. The average age and number of ordinary type patients, heavy patients and serve patients were 50.53 ± 10.79 and 17[68 % (95 % CI: 48.41–82.79)], 51.5 ± 10.5 and 6[24 % (95 % CI: 11.5–43.43)], 74.5 ± 7.5 and 2[8% (95 % CI: 2.22–24.97)], respectively. The number of male patients was 22/47, 22 [46.81 % (95 % CI: 33.33–60.77)]. The average age of male patients was 54.32 ± 12.86 old. The average age and number of ordinary type patients, heavy patients and serve patients were 53.67 ± 12.22 and 15 [68.18 % (95 % CI: 47.32–83.63)], 51 ± 11.67 and 6 [27.27 % (95 % CI: 13.15–48.15)], 84 ± 0 and 1 [4.55 % (95 % CI: 0.81–21.8)], respectively. The average recovery index of the patients is 0.84 ± 0.06 (Fig. 1 ), showing all patient were at late infection. The average hospitalization time (Table 1) for all patients was 36.81 ± 3.16. The average hospitalization time for three type patients were 36 ± 3, 39.08 ± 2.60 and 36.33 ± 1.78, respectively. The average hospitalization time for female patients was 37.36 ± 2.72. The average hospitalization time for three type patients were 36.88 ± 2.57, 38.67 ± 2.78 and 37.5 ± 1.5, respectively. The average hospitalization time for male patients was 36.18 ± 3.65. The average hospitalization time for three type patients were 35 ± 3.47, 39.5 ± 3.25 and 34 ± 0, respectively. It should be noted that for sever patients, the data lack statistical significance due to the too few scale specimens.
Table 1.
The clinical dada of patients in this study.
| Group | Number [% (95 % CI)] | Age + SD | Hospitalization time + SD |
|---|---|---|---|
| Total | 47 | 53.45 ± 12.08 | 36.81 ± 3.16 |
| Common | 32 [68.09 (53.84−79.61)] | 52 ± 11.44 | 36 ± 3 |
| Heavy | 12 [25.53 (15.25−39.51)] | 51.25 ± 11.08 | 39.08 ± 2.60 |
| Sever | 3 [6.38 (2.19−17.16)] | 77.67 ± 7.11 | 36.33 ± 1.78 |
| Male | 22 [46.81 (33.33−60.77)] | 54.32 ± 12.86 | 36.18 ± 3.65 |
| Common | 15 [68.18 (47.32−83.63)] | 53.67 ± 12.22 | 35 ± 3.47 |
| Heavy | 6 [27.27 (13.15−48.15)] | 51 ± 11.67 | 39.5 ± 3.25 |
| Sever | 1 [4.55 (0.81−21.8)] | 84 ± 0 | 34 ± 0 |
| Female | 25 [53.19 (39.23−66.67)] | 52.68 ± 11.40 | 37.36 ± 2.72 |
| Common | 17[68 (48.41−82.79)] | 50.53 ± 10.79 | 36.88 ± 2.57 |
| Heavy | 6[24 (11.5−43.43)] | 51.5 ± 10.5 | 38.67 ± 2.78 |
| Sever | 2[8 (2.22−24.97)] | 74.5 ± 7.5 | 37.5 ± 1.5 |
Fig. 1.
Recovery index of patients in this study.
Note: The recovery index = The patient's hospitalization time on the day of testing /the patient's whole hospitalization time.
3.2. Detection of 2019-nCoV virus from nasopharyngeal swabs, nasal swabs, oropharyngeal swabs and anal swabs collected before washing in the morning and afternoon
Nucleic acid testing results of patients including No.2, No.4, No.14, No.18, No.24, No.33, and No.38 (shown in Table S1) were final judged as negative based on nucleic acid testing discharge criteria. Results shown that the specificity for four different swab types were 100 %. Forty enrolled patients available were used for further methodological analysis. As shown in Table 2 , the detection number of nasopharyngeal swabs, nasal swabs, oropharyngeal swabs and anal swabs collected at before washing in the morning were 26/47[65 %(95 % CI: 49.51–77.87)], 23/47[57.5 %(95 % CI: 42.2–71.49)], 9/47[22.5 %(95 % CI: 12.32–37.5)] and 1/47[2.5 %(95 % CI: 0.44–12.88)], respectively. The detection number of nasopharyngeal swabs, nasal swabs, oropharyngeal swabs and anal swabs collected at afternoon were 17/47[42.5(95 %CI:28.51–57.8)], 14/47[35(95 %CI:22.13–50.49)], 3/47[7.5(95 %CI:2.58–19.86)] and 2/47[5(95 %CI:1.38–16.5)], respectively.
Table 2.
Comparison of detection ratio based on different swab type collected between before washing in the morning and afternoon.
| Swab type | Before washing in the morning |
p-value | Afternoon |
||||
|---|---|---|---|---|---|---|---|
| Detection |
p-value | Detection |
p-value | ||||
| No. | % Ratio (95 % CI:) | No. | % Ratio (95 % CI:) | ||||
| Nasopharyngeal | 26 | 65 (49.51−77.87) | 0.035 | 17 | 42.5 (28.51−57.8) | ||
| Nasal | 23 | 57.5(42.2−71.49) | 0.25 | 0.064 | 14 | 35(22.13−50.49) | 0.375 |
| Oropharyngeal | 9 | 22.5(12.32−37.5) | <0.001 | 0.031 | 3 | 7.5(2.58−19.86) | <0.001 |
| Anal | 1 | 2.5(0.44−12.88) | <0.001 | 1 | 2 | 5(1.38−16.5) | <0.001 |
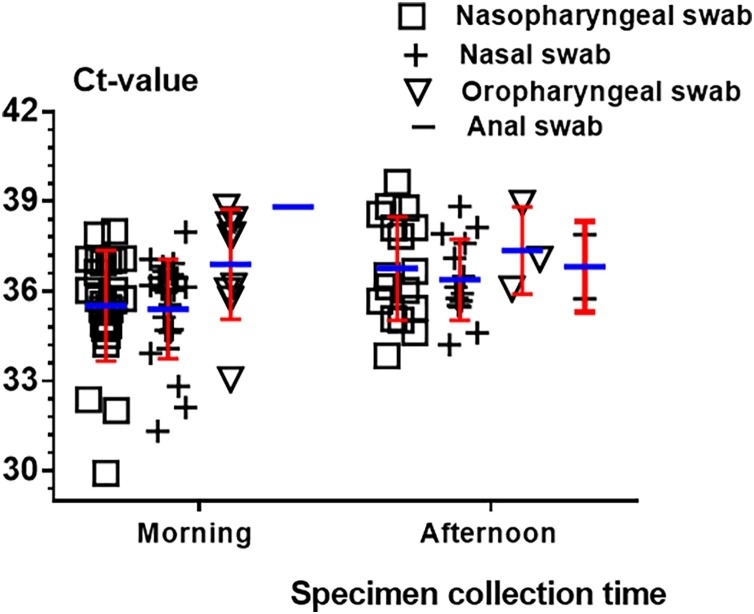
Further statistical analysis results from the Fig. 2 demonstrated that there is no statistical significance between nasopharyngeal swabs and nasal swabs before washing in the morning or afternoon (p = 0.25 and 0.375), respectively. To No.10, No.39 and No.48, the virus was only detected in nasopharyngeal swab before washing in the morning. To No.9, No.21, No.25 and No.48, the virus was only detected in nasopharyngeal swab afternoon. There is statistical significance among nasopharyngeal swabs, oropharyngeal swabs and anal swabs before washing in the morning or afternoon (p < 0.001/p < 0.001, p < 0.001/p < 0.001), respectively. Statistical analysis performed on the same swab type collected before washing in the morning and afternoon show that there is statistical significance for nasopharyngeal swab and oropharyngeal swab (p = 0.035, p = 0.031) and there is no statistical significance for nasal swab and anal swab (p = 0.064, p = 1).To No.16, the virus was only detected in nasal swab. Ct value for nasopharyngeal / nasal swab, oropharyngeal swab and anal swab were 35.51 ± 1.29 vs 36.76 ± 1.48, 35.39 ± 1.33 vs 36.38 ± 1.02, 36.90 ± 1.47 vs 37.36 ± 1.06 and 37.89 vs 37.03 ± 1.29 before washing in the morning and afternoon. Overall, data demonstrated that viral load in these patients was low at late infection stage.
Fig. 2.
Ct value for different swab specimens collected before washing in the morning and afternoon.
Note: Morning and afternoon show swab specimen were collected before washing in the morning and afternoon.
4. Discussion
SARS-CoV-2 viral load in novel coronavirus patients gradual declines since onset of symptoms (Ai et al., 2020). Low SARS-CoV-2 viral load in novel coronavirus patients at late infection stage, especially, easily lead to false negative nucleic acid testing results, thus having great challenges to current detection method. Appropriate specimens would be one key factor for improving current detection ratio of SARS-CoV-2.
In this study, we first statistically analyzed the age distribution of those patients. Results showed that the average age of 47 patients was 53.45 ± 12.08, which was consistent with most of the current reports (Wang et al., 2020), indicating that the middle-aged and elderly were mainly targets attacked by novel coronavirus. The infected population of female patients occupy 53.19 % (95 % CI: 39.23–66.67), which show that female might be more likely to be infected. The average hospitalization time is basically the same as the disease condition (shown in Table 1 and Table S1). There are several points to be explained that some patients rejected to discharge from hospital because of worrying about prejudice and other reasons, which increase the hospitalization time. This may be a new social problem facing the treatment of novel coronavirus patients at present. To sever patients, the scale of population is too small, which affects the accuracy of the results of this part of patients.
Two consecutively negative RT-PCR test results in respiratory tract specimens are required for the evaluation of discharge from hospital. However, reports of false negative of RT-PCR detection in COVID-19 and discharged patient turning positive are increasing (Zhang et al., 2020; Winichakoon et al., 2020). Low SARS-CoV-2 viral load might be main reason (Zou et al., 2020). In addition, the specimen type selection have a significant impact on the accuracy of nucleic acid detection. Although it is reported that sputum specimens or bronchoalveolar lavage fluid specimens have a very high detection rate for novel coronavirus (Yang et al., 2020; Han et al., 2020), some patients are unable to carry out bronchoalveolar lavage or expectoration difficulties. At present, nasal swabs and oropharyngeal swabs have been widely used in clinical novel coronavirus detection (Lin et al., 2020a; Yang et al., 2020). Anal swab had also been used as specimen for detection of virus (Chen et al., 2020c). Few reports have systematically compared the detection rate of nasopharyngeal swabs, nasal swabs, oropharyngeal swab and anal swabs for novel coronavirus patients in late infection stage. Based on our results (Fig. 2), the detection ratio of nasopharyngeal is higher than nasal swab but there is no significant difference between them, oropharyngeal swab is the second, and the anal swab is the lowest. These are consistent with the current literature reports (Lin et al., 2020a). Based on our clinical testing experience, for initial screening or monitoring of diagnosed patients, we prefer nasopharyngeal swab or solo nasal swab. For patients who could not be diagnosed by multiple times or waiting for discharge after a negative nucleic acid test, the collection of different specimen types including oropharyngeal swab and anal swab might prevent missed detection and reduce the probability that patients turn positive again after discharge.
Few studies pay attention to the effect of specimen collection time on the detection rate of novel coronavirus. Through our study, we found the detection rate of nasopharyngeal swab, nasal swab and oropharyngeal swab were the higher before washing in the morning. This may be because the load of virus for patients at late infection stage was low. Meanwhile, the human body is in a resting state at night, and the virus propagates or falls off in the nasal cavity or oral mucosa, so the detection rate is higher in the morning. During the day, individual activities may affect the local accumulation of the virus, so the virus detection rate of patients in the afternoon is relatively low. Especially for oropharyngeal swab, the related inhibitory components are accumulating, so the detection rate of oropharyngeal swab collected in the afternoon is lower obviously than other specimens. This may be one of the reasons why there is a significant difference in the detection rate of nasopharyngeal swabs between before washing in the morning and afternoon. As mentioned above, many reports pointed out that nasal or oropharyngeal swab testing lead to false negative (Ai et al., 2020; Zhang et al., 2020). The false negative results of virus detection might be reduced through changing collection time based on our results.
There are some shortcomings in this study. Firstly, initial patients were not included. Then the number of patients in this study is too small, which needs to be verified by increasing specimen scales. Otherwise, whether the detection of an RT-PCR sample with CT value above 35 implies the individual is infectious at this stage should be tested in future.
5. Conclusions
In summary, our research discovers that the detection rate of virus before washing in the morning is higher. Nasopharyngeal swabs and nasal swabs are more suitable for general screening of large-scale specimens. Our results might be of significance for improving the positive detection rate of COVID-19 infection patients at late infection stage as well as in initial screening.
CRediT authorship contribution statement
Min Liu and Qianyuan Li carried out nucleic acid detection and data analysis. Rong Guo, Xiaoyin Li, Xiandi Wu, Haiying Xu, Ling Jiang, Huaqin Zhang, Jing Chen and Lili Tian collected samples and coordinate patients to sign informed consent form. Wen Ai, Xiaoling Zheng, Jingjing Zeng, Yuwen Liu and Xiying Xiang carried out nucleic acid detection and data filling. Jun Luo carried nucleic acid detection and paper writing. Jun Zhou revised this paper and gave clinical guidance. Chunhua Luo designed projects and provided guidance.
Ethical approval
The study was approved by the Ethics Committees from Yichang Central people's Hospital.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The work was financially supported by the National Natural Science Foundation of China No.81902168. We thank Gong Feng, Xie Wenjuan, Wang Xi for their assistance.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2020.113974.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Ai J.-W., Zhang H.-C., Xu T. Optimizing diagnostic strategy for novel coronavirus pneumonia, a multi-center study in Eastern China. medRxiv. 2020 [Google Scholar]
- Chen Liangjun, Liu Weiyong, Zhang Qi. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 2020;9(1):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Fu J., Shu Q. Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World J. pediatr. WJP. 2020;16(3):240–246. doi: 10.1007/s12519-020-00345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Weilie, Lan Yun, Yuan Xiaozhen. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg. Microbes Infect. 2020;9(1):469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman Victor M., Landt Olfert, Kaiser Marco. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2020;25(3) doi: 10.2807/1560-7917.es.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Huanqin, Luo Qingfeng, Mo Fan, Long Lieming, Zheng Weiqiang. SARS-CoV-2 RNA more readily detected in induced sputum than in throat swabs of convalescent COVID-19 patients. Lancet Infect. Dis. 2020 doi: 10.1016/s1473-3099(20)30174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Cai L., Cheng Z. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil. Med. Res. 2020;7(1):4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert Stephen B., Whiley David M., O’Neill Nicholas T. Comparing nose-throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real-time polymerase chain reaction. Pediatrics. 2008;122(3):e615–620. doi: 10.1542/peds.2008-0691. [DOI] [PubMed] [Google Scholar]
- Li L., Chen Q.Y., Li Y.Y., Wang Y.F., Zhong N.S. Comparison among nasopharyngeal swab, nasal wash, and oropharyngeal swab for respiratory virus detection in adults with acute pharyngitis. BMC Infect. Dis. 2013;13(1):281. doi: 10.1186/1471-2334-13-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Wang D., Dong J. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-Based CT diagnosis and insights from two cases. Korean J. Radiol. 2020;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Xiang J., Yan M., Li H., Huang S., Shen C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2) infected pneumonia (COVID-19) medRxiv. 2020 doi: 10.1515/cclm-2020-0187. [DOI] [PubMed] [Google Scholar]
- Lin W., Xie Z., Li Y. Association between detectable SARS-COV-2 RNA in anal swabs and disease severity in patients with Coronavirus Disease 2019. J. Med. Virol. 2020 doi: 10.1002/jmv.26307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Iek Long, Lio Chon Fu, Cheong Hou Hon. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16(10):1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Liu J., Xu W. Novel Coronavirus can be detected in urine, blood, anal swabs and oropharyngeal swabs samples. medRxiv. 2019:2020. doi: 10.1002/jmv.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C.M. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winichakoon P., Chaiwarith R., Liwsrisakun C. Negative nasopharyngeal and oropharyngeal swab does not rule out COVID-19. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Yong, Liu Yuan, Cao Liu. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang M., Shen C. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv. 2020 doi: 10.1101/2020.02.11.20021493. 2020.02.11.20021493. [DOI] [Google Scholar]
- Zhang Jing-Feng, Yan Kun, Ye Hong-Hua, Lin Jie, Zheng Jian-Jun, Cai Ting. SARS-CoV-2 turned positive in a discharged patient with COVID-19 arouses concern regarding the present standard for discharge. Int. J. infect. diseases IJID: off. Pub. Int. Soc. Infect. Diseases. 2020 doi: 10.1016/j.ijid.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Jun. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16(10):1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D. 2019. A Novel Coronavirus from Patients with Pneumonia in China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Lirong, Ruan Feng, Huang Mingxing. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.