Abstract
Single-cell gene expression (transcriptomics) data are becoming robust and abundant, and are increasingly used to track organisms along their life-course. This allows investigation into how aging affects cellular transcriptomes, and how changes in transcriptomes may underlie aging, including chronic inflammation (inflammaging), immunosenescence and cellular senescence. We compiled and tabulated aging-related single-cell datasets published to date, collected and discussed relevant findings, and inspected some of these datasets ourselves. We specifically note insights that cannot (or not easily) be based on bulk data. For example, in some datasets, the fraction of cells expressing p16 (CDKN2A), one of the most prominent markers of cellular senescence, was reported to increase, in addition to its upregulated mean expression over all cells. Moreover, we found evidence for inflammatory processes in most datasets, some of these driven by specific cells of the immune system. Further, single-cell data are specifically useful to investigate whether transcriptional heterogeneity (also called noise or variability) increases with age, and many (but not all) studies in our review report an increase in such heterogeneity. Finally, we demonstrate some stability of marker gene expression patterns across closely similar studies and suggest that single-cell experiments may hold the key to provide detailed insights whenever interventions (countering aging, inflammation, senescence, disease, etc.) are affecting cells depending on cell type.
Keywords: Single-cell sequencing, Aging, Inflammaging, Cellular senescence, Transcriptional heterogeneity, Biomarkers
1. Introduction
Aging research offers exciting prospects. For example, in recent years, research on cellular senescence, one of the hallmarks of aging (Lopez-Otin et al., 2013), gained momentum with the finding that the elimination of senescent cells can improve health and survival in mice (Baar et al., 2017; Baker et al., 2016; Xu et al., 2018). Around the same time, single-cell analyses gained momentum (Svensson et al., 2018; Zappia et al., 2018). Now, both fields are increasingly coming together. As we will see, aging, inflammation and senescence have different effects on different cell-types, providing a direct justification for single-cell approaches. In turn, the processes underlying aging are often subtle, and will provide interesting challenges for the development of single-cell methods. And indeed, the development of molecular clocks, that is biomarker signatures predicting age, or aging, based on single-cell data requires an interdisciplinary approach. We start with a brief review of the development of the two fields as separate areas of research. In the main part, we will summarize, compare and contrast the results of recently published investigations of aging, aging-related chronic inflammation (inflammaging) and senescence (that is, cellular senescence and immunosenescence) based on single-cell RNA-sequencing (scRNA-seq). We will specifically focus on markers of cellular senescence. We do not consider other omics data, as these are still few and far between. We will describe multi-tissue studies and studies of whole blood, as well as of specific organ tissues, such as pancreas, brain, lung, skin and muscle. We will also describe some single-cell studies based on in-vitro material.
Investigating single-cell data with a focus on aging processes, or, more generally, along a time axis, all transcriptomics data are necessarily cross-sectional on the cell level: no single cell can be investigated twice. On the individual level, it is possible in principle to repeatedly take blood or tissue samples from the same individual. Longitudinal (multi-)omics studies have been done using bulk transcriptomics, e.g., (Schussler-Fiorenza Rose et al., 2019), but longitudinal in vivo studies at the single cell level, with relevance to aging processes, are still lacking; longitudinal samples of cancer biopsies were subjected to single-cell sequencing, however (Hamza et al., 2019; Maynard et al., 2019). Thus, the insights reported here regarding changes across time are cross-sectional, based on different human or animal donors for different time-points, who come with their inter-individual variability. This consideration is important regarding the signal in the data, referring to patterns of expression changes associated with aging processes; we would expect that this signal would increase if longitudinal data were investigated. Cross-sectionally at least, an increase in average heterogeneity of gene expression with age can be found in most of the single-cell data that are specifically investigated regarding this aspect; it is also known as transcriptional noise or variability and estimated in some single-cell studies as the correlation of gene expression patterns among cells and similar observations are long known from bulk data (Bahar et al., 2006; Işıldak et al., 2019; Martinez-Jimenez et al., 2017).
In some single-cell analyses, specific sets of marker genes are explicitly established to define cells, though in most cases, cell type definitions are done based on a priori published marker gene data. In that case, individual marker genes, or sets of these, can still be used to characterize the existing cells further, as well as new-found clusters of cells. Whether such further descriptions necessitate to define “sub-cell-types” or not may be debated; we just note that “cell types” are man-made constructs based on bona-fide boundaries or thresholds with an eye on utility and applicability, not scientific rigor. In our case, our default assumption is that aging processes do not imply the change of one cell type to another; rather we speak about a specific cell type in various states due to age- or aging-related change, which usually amounts to a deterioration of function. For example, we consider that cells of a certain cell type can be in “early” or “late” as well as “partial” or “full” states of, e.g., cellular senescence, depending on their expression of marker genes associated with cellular senescence.
Moreover, combining omics data with endpoint data, we can aim to identify biomarkers that predict these endpoints. Standard aging-related endpoints are related to mortality or morbidity, where the latter can be defined as reflecting disease and/or dysfunction. Biomarker candidates can then be some specific single-cell omics measurement, or sets (signatures) of such measurements, or sum scores and the like. A biomarker of aging then is expected to predict an endpoint better than chronological age (see also Fuellen et al. (2019)). Transferring these ideas to the single-cell world will require appropriate longitudinal single-cell datasets, including the clinical characterization of the donor animals or humans in terms of mortality or morbidity, and there will be exciting opportunities for biomarker discovery and validation once these data will become available (Rajewsky et al. (2020)).
2. Cellular senescence, inflammation and aging
While we review all aging-related single-cell datasets that we could identify, we will pay specific attention to chronic inflammation (inflammaging), and, most specifically, we will focus on the inflammaging-related hallmark of aging that is known as cellular senescence. Cellular senescence was first described almost 60 years ago when it was discovered that human diploid fibroblasts have a finite replicative potential in culture, after which the cells enter a state of irreversible replicative arrest (Hayflick and Moorhead, 1961). This phenomenon, later called replicative senescence, is linked to the constant loss of telomeric DNA that is associated with each cell division (Harley et al., 1990). Because of the 5′->3′ directionality of DNA-polymerases, the replication machinery is not able to copy the very ends of linear chromosomes. The ensuing loss of DNA, as time progresses, eventually triggers cell cycle checkpoints that preclude further cell divisions. This type of replicative senescence, also known as the Hayflick limit, exists in many species and correlates with maximum lifespan (Röhme, 1981). Consequently, it has given rise to the telomere theory of aging (Harley, 1991).
However, in recent years it became clear that telomere attrition is only one of several mechanisms that can induce cellular senescence. Various types of stress, such as reactive oxygen species, radiation, DNA damaging agents and the unfolded protein response are among such mechanisms (Campisi, 2013; Coppe et al., 2010). Thus, cellular senescence appears to be a special stress response state of the cell that is accompanied by broad morphological and biochemical changes (McHugh and Gil, 2018). Transcriptional changes include an up-regulation of tumor suppressor genes and a down-regulation of cell-cycle promoting genes (Chatsirisupachai et al., 2019; Wiley et al., 2017). Importantly, senescent cells frequently secrete an inflammatory mix of cytokines, growth factors and matrix metalloproteinases, which form the so-called senescence-associated secretory phenotype (SASP) (Coppe et al., 2010; de Keizer, 2017; Gorgoulis et al., 2019). This paracrine signaling has a range of negative effects involving tissue remodeling and also tumorigenesis, as well as chronic inflammation (inflammaging) (Coppe et al., 2010; Franceschi et al., 2000, 2018; Sun et al., 2018). In a nutshell, senescent cells are non-dividing but secretory, damaged, and metabolically altered cells, which frequently escape apoptosis. Molecular identification of senescent cells is not trivial, since the senescent state induced by different triggers in different tissues displays a certain degree of heterogeneity (Hernandez-Segura et al., 2017). However, key markers used for identification are a senescence-associated form of beta-(β)-galactosidase (Dimri et al., 1995) and p16 (CDKN2A), which is expressed by a majority of senescent cells (Campisi and d’Adda di Fagagna, 2007; Collins and Sedivy, 2003).
Clear support that senescent cells are not only correlated with aging and diseases, but instead are causally involved, as already suggested by Yanai and Fraifeld (2018), comes for instance from recent studies which transplanted senescent cells from old into young mice (Xu et al., 2018). This resulted in persistent functional impairment as well as spread of cellular senescence to host tissues. Another strong line of evidence comes from experiments that actually removed senescent cells from aged mice (Baar et al., 2017; Baker et al., 2016; Xu et al., 2018). In each case, an increase in lifespan and a delay of typical age related diseases has been observed. A good summary of the involvement of senescent cells in human diseases can be found in McHugh and Gil (2018), and a good yet brief account of the state of the art is van Deursen (2019). Cellular senescence is a frequently cited hallmark of aging (Lopez-Otin et al., 2013), and it is no surprise that it is implicated in the morbidity and co-morbidity of many (if not most or even all) age-associated diseases.
3. Single-cell analyses
Many fundamental questions in biology and medicine require the understanding of molecular processes at single-cell resolution. How does a zygote change and multiply to become a full-fledged organism made up of trillions of cells including many types of cells, tissues, and organs? Which cell within a tumor is the first ancestral cell that developed into a cancerous state and what molecular changes caused that cell to gain uncontrollable growth and proliferation? What kind of immune cells start accumulating at a damaged blood vessel and eventually cause the rupture of an arterial plaque that causes stroke or acute myocardial infarction? How close is a cell to reaching the Hayflick limit - is it still functional or is it in a senescent state? These and many other questions have been traditionally interrogated by studying single cells, but only few genes or few cells at a time, because, technologically it was not possible until recently to have an unbiased view of the whole genomes, transcriptomes, or epigenomes of single cells at an affordable manner at scale.
Whole transcriptome sequencing of a single cell was first achieved in 2009 (Tang et al., 2009). The single-cell sequencing technologies have since evolved quite rapidly from sequencing merely a manually picked individual cell (Tang et al., 2009), to massively multiplexed sequencing of hundreds of thousands of cells (Svensson et al., 2018). Even more recent protocols have achieved sequencing more than 2 million cells in one experiment at a cost of $0.01 per cell (Cao et al., 2019), which reflects the rapid decline in sequencing costs at single cell resolution.
Currently, a variety of competing single-cell RNA-seq protocols exist, each with its own advantages and trade-offs (Ding et al., 2019; Mereu et al., 2020; Svensson et al., 2017). Moreover, along with the sequencing protocols, single-cell sequencing data analysis methods have also seen a rapid rise. Multiple tools have been developed for almost every aspect of the data analysis. A resource (Zappia et al., 2018) that tracks the available single-cell sequencing data analysis tools from the scientific community, as of writing this manuscript, has tracked more than 630 tools; a new tool is added to the compendium almost every other day. Having many alternatives for each data analysis step is on one hand a luxury, on the other hand a challenge. Fortunately, there have been benchmarking studies carried out for each step of the workflow. Thus, one can make an informed decision about which tool to use depending on the requirements of the project at hand.
At least one benchmarking study exists for each step of the single-cell sequencing and data analysis workflow: sample preparation and sequencing (Mereu et al., 2020), alignment of raw reads (Baruzzo et al., 2017) or alignment-free quantification of read counts (Wu et al., 2018), count matrix pre-processing and normalization (Cole et al., 2019) -- this may include imputation for drop-outs (Zhang and Zhang, 2020), or batch correction (Tran et al., 2020) --, clustering (Duo et al., 2018), differential expression analysis to determine cluster-specific markers (Soneson and Robinson, 2018), cell type annotation (Abdelaal et al., 2019), dimension reduction (Sun et al., 2019), visualization, and downstream analyses, e.g., based on biological network data (Gawel et al., 2019; Mohammadi et al., 2019). As decisions made at each step of this workflow are likely to affect the downstream options and possible outcomes, combinatorial assessment of each component of this workflow has been assessed by various studies (Germain et al., 2020; Tian et al., 2019; Vieth et al., 2019). In some cases, a pre-defined set of tools and parameters can boost analysis speed and reproducibility (Wurmus et al., 2018). However, given the amount of tools and computational approaches and the fast development of the field, the analysis of single cell data is a moving target.
The common denominator of the benchmark studies could be summarized as that there is no one single combination of experimental protocols and computational tools that clearly dominate other options in all possible scenarios; the choices are context dependent, they need to be made according to the biological question at hand, and there must be multiple quality control checkpoints along the workflow. However, most of the single-cell sequencing data analysis challenges have been addressed, and the scientific community is converging onto a set of best practices for data analysis (Luecken and Theis, 2019). Thus, the last decade witnessed single-cell RNA-seq (scRNA-seq) becoming a mature, affordable, accessible technology suited to address the fundamental questions in biology. Finally, data from single-cell transcriptomics can be used to de-convolute bulk transcriptomics data (Dong et al., 2020), i.e., it is also a good companion to earlier technology and furthers the understanding of data already available.
4. Single-cell analyses of aging and senescence
Most publications on single-cell analyses of aging and senescence were published in the last two years. A first type of study is based on in-vivo data, investigating the effects of aging mostly in mice and humans, for a specific tissue and sometimes for a set of tissues (“multi-tissue”). In such studies, the data may be inspected specifically for markers of cellular senescence such as p16 (CDKN2A) or p21 (CDKN1A). A second type of study specifically investigates collections of senescent cells, in-vitro, identifying or inspecting specific biological mechanisms. We will first present the in-vivo studies.
4.1. Multi-tissue studies
At time of writing, two multi-tissue in-vivo mouse studies are available: the Tabula Muris Senis (Almanzar et al., 2020) and the Calico Study (Kimmel et al., 2019), see Table 1 for details.
Table 1.
List of single-cell publications and data sets related to aging and senescence.
| In-vivo datasets | |
|---|---|
| Tabula Muris Senis (Almanzar et al., 2020) | |
| Tissues: | bladder, bone marrow, brain (cerebellum, cortex, hippocampus and striatum), fat (brown, gonadal, mesenteric and subcutaneous), heart and aorta, kidney, large intestine, limb muscle and diaphragm, liver, lung, mammary gland, pancreas, skin, spleen, thymus, tongue and trachea |
| Animals/Cells: | ∼530 000 cells from 30 male & female C57BL/6 J N mice: 6 age groups: 1 mo, 3 mo, 18 mo, 21 mo, 24 mo, 30 mo |
| Heterogeneity: | Not investigated |
| Exp. setup: | 10x Genomics, also FACS-based for the 3 mo & 24 mo age groups; Nova-seq 6000 |
| Remarks: | The number of p16-expressing cells doubled in old mice, and in the 10x Genomics data, p16 expression doubled as well. |
| Calico Study (Kimmel et al., 2019) | |
| Tissues: | kidney, lung, spleen |
| Animals/Cells: | ∼55 000 cells from 7 male C57Bl/6 mice: 7 mo, and 22/23 mo |
| Heterogeneity: | Not investigated |
| Exp. setup: | 10X Genomics; HiSeq 4000 |
| Remarks: | No change in p16 expression, nor of any specific senescence-related gene signature. |
| Immune Aging Atlas (Martinez-Jimenez et al., 2017) | |
| Tissues: | blood (CD4+ T-cells) |
| Animals/Cells: | ∼1 500 cells from 3 mo and 21 mo old male mice (C57/BL6 and CAST/EiJ strains; number of animals not stated) |
| Heterogeneity: | increasing cell-to-cell transcriptional variability, based on a Mann-Whitney-Wilcoxon test of distributional differences |
| Exp. setup: | Fluidigm protocol; Illumina HiSeq2500 |
| Remarks: | Recording of immunological activation response, but no specific investigation of marker genes of aging or senescence. |
| Aging Hematopoietic Stem Cell Atlas (Grover et al., 2016) | |
| Tissues: | blood: hematopoietic stem cells |
| Animals/Cells: | 135 cells from male and female CD45.2 C57BL/6 mice: 2/3 mo and 20–25 mo |
| Heterogeneity: | Not investigated |
| Exp. setup: | Fluidigm C1 System, HiSeq 2000 |
| Remarks: | HSC aging shows upregulation of platelet lineage gene numbers and expression |
| Aging T-cell Atlas (Elyahu et al., 2019) | |
| Tissues: | blood (CD4+ T-cells) |
| Animals/Cells: | ∼24 000 cells from 8 C57BL/6 mice (of unspecified gender), 2–3 mo and 22–24 mo |
| Heterogeneity: | Not investigated |
| Exp. setup: | 10x Genomics GemCode Chromium, Illumina NextSeq 500 |
| Remarks: | Investigation of subsets of cells associated with chronic inflammation and immunity decline, but no specific investigation of marker genes of aging or senescence. |
| Aging Artery Atlas (Zhang et al., 2020) | |
| Tissues: | aorta and coronary arteries |
| Animals/Cells: | ∼8 000 cells from 16 male & female cynomolgus monkeys, 4–6 y and 18–21 y |
| Heterogeneity: | increasing transcriptional noise, following the criterion of Enge et al. (2017) |
| Exp. setup: | 10x Genomics, HiSeq4000 |
| Remarks: | FOXO3A may be a key regulator of arterial aging. |
| Aging Pancreas Atlas (Enge et al., 2017) | |
| Tissues: | pancreas |
| Animals/Cells: | 2 544 cells from 8 human donors (both genders), 1 mo to 56 y old |
| Heterogeneity: | increasing transcriptional noise, following their criterion, but no correlation to mutational load |
| Exp. setup: | Illumina NextSeq, Smartseq2, also FACS |
| Remarks: | p16 was found upregulated in a higher fraction of cells. |
| Pancreatic Cancer Atlas (Elyada et al., 2019) | |
| Tissues: | pancreatic cancer tissue + adjacent tissue |
| Animals/Cells: | 21 200 cells from 6 patients, 64–87 y old (and 11 000 cells from 4 KPC model mice) |
| Heterogeneity: | Not investigated |
| Exp. setup: | 10X Genomics Chromium; Illumina HiSeq 4000 |
| Remarks: | Inflammatory CAFs were found, expressing interleukins and cytokines that are also characteristic for the SASP. |
| Aging/T1D Beta Cell Atlas (Thompson et al., 2019) | |
| Tissues: | pancreas |
| Animals/Cells: | ∼21 000 cells from 9 female NOD mice, 8 w, 14 w & 16 w |
| Heterogeneity: | Not investigated |
| Exp. setup: | 10X Genomics Chromium; Illumina NovaSeq 6000 |
| Remarks: | Islet cells become senescent and express SASP proteins (marked in particular by upregulation of p21 and Ccxl10). |
| Aging Brain Atlas (Ximerakis et al., 2019) | |
| Tissues: | brain |
| Animals/Cells: | ∼50 000 cells from 16 male C57BL/6 J mice, 2/3 mo and 21–23 mo |
| Heterogeneity: | no broadly increasing transcriptional variability, based on coefficient of variation of expression for selected sets of transcripts |
| Exp. setup: | 10X Genomics Chromium, Illumina NextSeq500 |
| Remarks: | Senescence pathway activation was found in a cluster of endothelial cells. |
| Aging Cerebromicrovascular Atlas (Kiss et al., 2020) | |
| Tissues: | brain |
| Animals/Cells: | ∼9000 cells from 6 male C57BL/6 mice, 3 mo and 28 mo |
| Heterogeneity: | Not investigated |
| Exp. setup: | 10X Genomics Chromium; Illumina NovaSeq 6000 |
| Remarks: | ∼10 % of cells display cellular senescence, based on a precompiled list of markers. |
| Aging Microglia Atlas (Hammond et al., 2019) | |
| Tissues: | brain |
| Animals/Cells: | ∼76 000 microglia cells from male & female C57BL/6 J mice, embryonic, and at 4 d, 5 d, 30 d, 100 d and 540 d. |
| Heterogeneity: | increasing cellular diversity, potentially due to increasing transcriptional heterogeneity |
| Exp. setup: | 10X Genomics Chromium, Illumina NextSeq500 |
| Remarks: | Recording of the upregulation of genes linked to disease and inflammaging, but no specific investigation of marker genes of cellular senescence. |
| Aging Lung Atlas (Angelidis et al., 2019) | |
| Tissues: | lung |
| Animals/Cells: | ∼15 000 cells from 15 C57BL/6 mice, 3 mo and 24 mo |
| Heterogeneity: | increasing transcriptional noise, similar to Enge et al. (2017) |
| Exp. setup: | DropSeq; Illumina HiSeq4000 |
| Remarks: | Increased cholesterol biosynthesis in type-2 pneumocytes and lipofibroblasts and altered relative frequency of airway epithelial cells were observed. |
| Lung Aging/Fibrosis Atlas (Reyfman et al., 2019) | |
| Tissues: | lung |
| Animals/Cells: | ∼76 000 cells from 16 humans (both genders), 8 lung tissue donors (21–63 y) and 8 patients (37–72 y) with pulmonary fibrosis |
| Heterogeneity: | Not investigated |
| Exp. setup: | Illumina HiSeq 4000; also bulk RNA-sequencing of flow-sorted cells |
| Remarks: | Senescent cells were identified as a rare cell population by a 1,311-gene-signature (but not by a 4-gene signature including p16). |
| Lung Fibrosis Mouse Model (Yao et al., 2019) | |
| Tissues: | lung |
| Animals/Cells: | 11 686 cells from 8 transgenic mice (both genders), 10–12 w |
| Heterogeneity: | Not investigated |
| Exp. setup | Illumina Novaseq 6000, FACS sorted cells, 10X Genomics Chromium |
| Remarks: | Alveolar type 2 (AT2) stem cells isolated from IPF lung tissue exhibit characteristic transcriptomic features of cellular senescence. |
| Aging Skin Atlas (Solé-Boldo et al., 2019) | |
| Tissues: | skin |
| Animals/Cells: | ∼15 000 cells from 5 male humans, 25–27 y and 53–70 y |
| Heterogeneity: | less clarity in GO annotations, potentially due to increasing transcriptional heterogeneity |
| Exp. setup: | 10X Genomics Chromium, Illumina HiSeq 4000 |
| Remarks: | An upregulation of “skin aging-associated secreted proteins” (SAASP) was observed. |
| Aging Muscle Stem Cell Atlas (Hernando-Herraez et al., 2019) | |
| Tissues: | muscle |
| Animals/Cells: | 377 cells from 4 male mice (transgenic Pax7-nGFP17 strain, on a (C57BL/6;DBA2 F1/JRj) genetic background), 1.5-2.1 mo and 23.3–27 mo |
| Heterogeneity: | increasing variability, based on the pairwise correlation of gene expression of cells, both within and between individual mice, correlated with methylation |
| Exp. setup: | FACS-sorting for muscle stem marker; bead transcription & amplification using Smartseq2 |
| Remarks: | Quiescent cells were investigated, and accordingly, no change in expression of senescence markers Cdkn2a (p16) & Cdkn1b (p27) was observed. |
| In-vitrodatasets | |
| HCA2 Fibroblast Atlas (Tang et al., 2019) | |
| Tissues: | HCA2 fibroblasts, after replicative senescence after 38, 48 and 71 population doublings |
| Animals/Cells: | 1 200 + 400 human cells |
| Heterogeneity: | Not investigated |
| Exp. setup: | Drop-seq; Library prep: Illumina Nextera XT |
| Remarks: | Investigation of senescence (beta-galactosidase, rate of EdU incorporation), role of telomeres. |
| Senescence Entry Cell Atlas (Zirkel et al., 2018) | |
| Tissues: | HUVECs (mesodermal) IMR90 s (endodermal), and MSCs (multipotent) |
| Animals/Cells: | 5 200 cells from 8 human donors & 2 cell lines |
| Heterogeneity: | Not investigated |
| Exp. setup: | 10x Genomics Chromium; Illumina HiSeq 4000 |
| Remarks: | Investigation of senescence (beta-galactosidase, reduced proliferation, 6 specific CpGs, HMGB2 and CTCF). |
| mTOR shRNA Atlas (Aarts et al., 2017) | |
| Tissues: | primary human fibroblasts (IMR90) expressing reprogramming factors |
| Animals/Cells: | in-vitro shRNA screen |
| Heterogeneity: | Not investigated |
| Exp. setup: | Illumina Nextera XT; Illumina HiSeq 2500 |
| Remarks: | Investigation of senescence (p16, p21, p19, SASP) & reprogramming and of the role of mTOR. |
The tissues/animals/cells investigated are described as well as the experimental setup. Statements with respect to transcriptional heterogeneity, and observations regarding aging/inflammaging and cellular senescence are also provided. Abbreviations: d, day; w, week; mo, month; y, year; Exp, experimental.
The Tabula Muris Senis is an extension of the Tabula Muris study (Schaum et al., 2018) beyond the three-month (3 mo) time-point. It features 18 tissues at a maximum of 6 time points, from 1 month to 30 months, reporting gene expression for ∼530 000 cells. Of note, the fraction of cells expressing the senescence marker p16 (Cdkn2a) was found to double in older mice. A subsequent analysis of these data (Zhang et al., 2019) noted that many “global aging genes” (whose expression changes significantly with age in more than half of the cell types) are related to cellular senescence, citing App, Ctnnb1, Mapk1, Rac1, Arf1, and Junb in particular. On the pathway level, the (senescence-related) mTOR pathway is downregulated; this insight matches the overall trend of transcriptional downregulation, but it leads to some discussion in the paper regarding the plausibility of this finding, possibly as being adaptive. Also, Ubq is the only well-characterized gene among the top 20 global aging genes, suggesting that proteostasis plays a prominent role overall in mouse aging. We took the Tabula Muris Senis as the starting point to demonstrate that this data set is, like most others, a treasure trove that can (still) be mined in various ways, generating further insights of relevance to inflammaging and cellular senescence, see Box 1 .
Box 1. Inflammaging and cellular senescence in the Tabula Muris Senis.
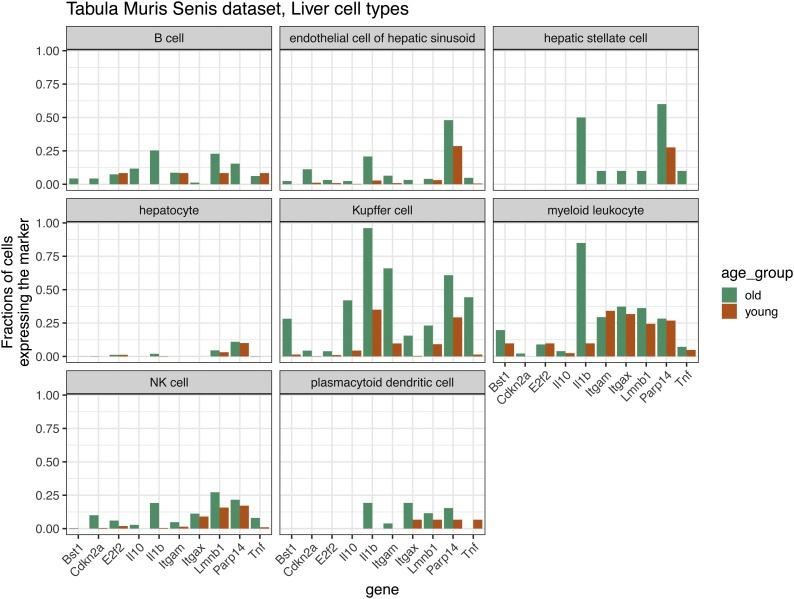
We processed the Tabula Muris Senis data (Almanzar et al., 2020) (see Supplement) and investigated 10 senescence-associated genes that are expressed in more cells from old mice, as compared to young mice (see also Box 2). Curiously, the by far largest differences in the fraction of cells expressing these markers can be found in the liver, and among the cells of the liver, in the Kupffer cells (see Fig. 1 ). In particular, Interleukin 1b (IL1b), which is one of the cytokines involved in inflammaging (Franceschi et al., 2017) and a component of the senescence-associated secretory phenotype (SASP), can be found in almost 100 % of Kupffer cells in old mice, but only in ∼37 % of these cells in young mice. We find that IL1b is also upregulated in other liver cells, e.g., hepatic stellate cells, but only in 50 % of these. Kupffer cells are resident liver macrophages (Hunt et al., 2019). Aged Kupffer cells have a reduced capacity for phagocytosis and autophagy, and display an increased inflammatory phenotype contributing to inflammaging (Hunt et al., 2019). Further, IL1b was shown to be involved in inflammation and aging in the liver context, at least in case of alcoholic steato-hepatitis (Morsiani et al., 2019). As a small caveat, we acknowledge that merely looking into the differences in terms of fraction of cells expressing a marker between the young and old animals might not accurately reflect the actual amount of cellular senescence. For that purpose, one needs to additionally account for inter-individual and inter-generational cell type compositional differences, which is beyond the scope of this review.
Fig. 1.
Fraction of cells expressing 10 aging-related marker genes in liver cells of the Tabula Muris Senis.
Alt-text: Box 1
The Calico Study (Kimmel et al., 2019) analyzed ∼55 000 single cells from lung, kidney and spleen of young (7 mo) and old (21 mo) mice. They find that age-related changes of gene expression are largely cell type specific and depend less on the type of tissue. However, they also found some 200 genes that are differentially expressed in more than five cell types. These genes are preferentially involved in transport of proteins to the endoplasmic reticulum as well as in inflammation. Inflammation pathways were found upregulated in immune as well as non-immune cells, in agreement with inflammaging. Interestingly, this study did not find an age related increase of the fraction of cells that express p16, nor of p16-expression on average.
4.2. Single-tissue studies
Further studies up to now focused on the blood (Elyahu et al., 2019; Grover et al., 2016; Martinez-Jimenez et al., 2017), the pancreas (Elyada et al., 2019; Enge et al., 2017; Thompson et al., 2019), the brain (Hammond et al., 2019; Kiss et al., 2020; Ximerakis et al., 2019) and the lung (Angelidis et al., 2019; Reyfman et al., 2019), see Table 1 and the next sections. Of note, comparing data of the Tabula Muris Senis and some of the single-tissue studies, we note high consistency among studies when looking at aging/senescence-related markers, see Box 2 .
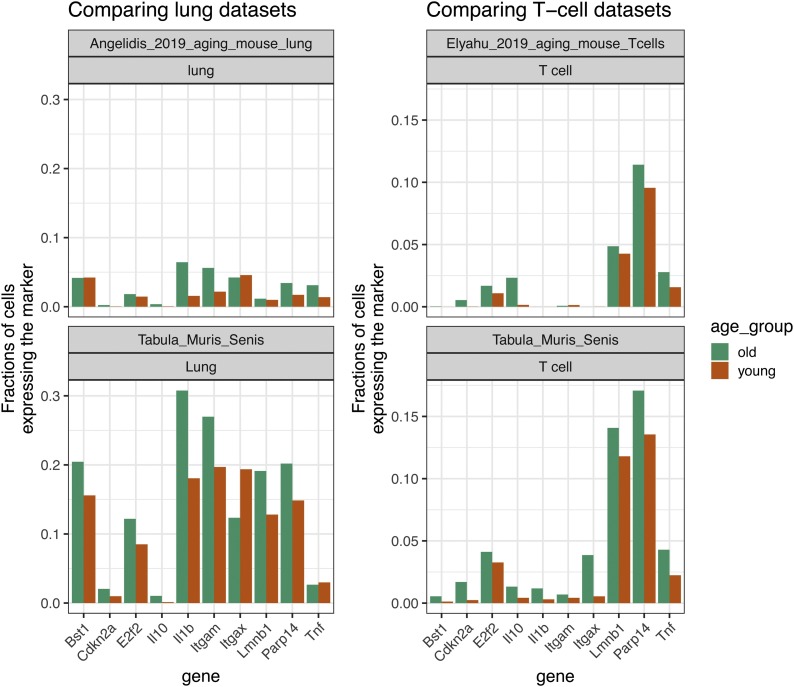
Box 2. Between-study consistency of expression patterns of markers of aging and senescence.
For this review, we checked the consistency of marker gene expression across studies. In particular, we compared the Tabula Muris Senis lung cell data to the Aging Lung Atlas (Angelidis et al., 2019), and the Tabula Muris Senis T cell data (bone marrow, spleen, and thymus) to the Aging T-cell Atlas (Elyahu et al., 2019), for 10 senescence-associated genes that are expressed in more cells from old mice, as compared to young mice (see also Box 1). As can be seen in the Fig. 2 , the cell fractions, as well as the increases in cell fraction in old mice, are roughly the same in case of the T cells, and the increases are mostly comparable in case of the lung cells. In case of lung, the cell fractions featuring a marker are, however, much lower overall than in the Tabula Muris Senis, which can be explained by the differences in the depth of sequencing between the two studies (an average of 2200 genes per cell were detected in the Tabula Muris Senis data, while only 446 genes per cell were detected in the Aging Lung Atlas).
Fig. 2.
Fraction of cells expressing 10 markers in lung cells and T cells of the Tabula Muris Senis, compared to the Aging Lung Atlas and the Aging T-cell Atlas.
Alt-text: Box 2
4.3. Blood & vascular system
Due to its easy accessibility, among the first “tissues” inspected in a single-cell fashion was blood.
In what we call an “Immune Aging Atlas”, (Martinez-Jimenez et al., 2017) studied naive CD4+ T cells in young (3 mo) and old (21 mo) mice by investigating the transcriptional pattern of ∼1500 cells before and after in-vitro immune stimulation. In old mice, most genes involved in this activation process responded to the stimulus, but with a reduced magnitude compared to young mice, reflecting immunosenescence. The authors also found that old mice showed a larger cell-to-cell transcriptional variability. Moreover, ∼10 % of the genes were differentially expressed between young and old, and these genes were often only expressed in a small subset of cells. This may lead to the observed variability, which the authors see as a hallmark of the aging process. The data of this paper was recently re-analyzed by (Hao et al., 2019), who proposed an “optimal gene filtering algorithm for scRNAseq data” (OGFSC). Using their algorithm, the originally reported reduction of the transcriptional response in old mice could no longer be established. This highlights the influence that the choice of a specific bioinformatics method can have. Thus, not only differences of the experimental conditions, but also differences in the analysis pipeline can affect the reproducibility of results.
An “Aging Hematopoietic Stem Cell (HSC) Atlas” (Grover et al., 2016) of young (2–3 month) and old (20–25 month) mice investigated age dependent transcriptional signatures, showing that the previously observed age associated decrease of lymphoid cells and erythrocytes is accompanied by the accumulation of a specific “old” HSC subgroup of cells showing an upregulation of platelet lineage gene expression. The respective HSCs predominantly generate platelets but recover their ability to increase lymphoid cell generation when the key platelet programming factor FOG-1 is compromised. The authors discovered an aging-related subclass of HSC that explains the apparent decline in engrafting ability, exploiting the single-cell resolution of their data.
The “Aging T-Cell Atlas” (Elyahu et al., 2019) investigated the transcriptomes of ∼24 000 CD4+ T cells in young and old mice and reports the drop of naive CD4+ cells typical for immunosenescence, but also a surprising accumulation of three CD4+ subtypes that occur only rarely in young animals, namely cytotoxic, exhausted and activated regulatory T cells, characterized in part by their expression of inflammatory cytokines (primarily IL-27, IFNb, and IL-6), possibly reflecting inflammaging in the old mice.
In what we refer to as an “Aging Artery Atlas”, Zhang et al. (2020) studied the vascular system of cynomolgus monkeys at the single-cell level to investigate differences between aortas and coronary arteries in young (∼6 year-old) and old (∼20 year-old) animals. Using almost 8000 endothelial cells, they identified on average 3100 genes per cell, and among these they found 515 downregulated vs. 259 upregulated genes that were differentially expressed in at least one type of vascular cell. Bioinformatics analysis indicated that FOXO3A is a key regulator of arterial aging and inactivation of this transcription factor in human endothelial cells in vitro reproduced many of the phenotypic changes observed in aged monkey arteries. Also in this study, an increase of transcriptional noise associated with age was found. Following Enge et al. (2017), transcriptional noise was defined by the ratio of biological to technical variation, based on correlating gene expression in individual cells with the average of the corresponding cell type; technical variation was in turn based on a spike-in approach.
4.4. Pancreas
The human pancreas is a focus of surprisingly many single-cell studies, and at the same time, cellular senescence is implicated in many aspects of development and therapy of pancreatic cancer (specifically PDAC, pancreatic ductal adenocarcinoma) and diabetes (both type 1 and type 2) (Elyada et al., 2019; Enge et al., 2017; Thompson et al., 2019).
An “Aging Pancreas Atlas” was thus developed (Enge et al., 2017) by performing single cell transcriptomics of ∼2500 human pancreas cells from donors spanning six decades of age. The authors found an increasing transcriptional noise (defined as described above, see the “Aging Artery Atlas”) and an increasing mutational load with age, but they concluded that the number of mutations per cell is insufficient to explain the observed transcriptional noise, though epigenetic changes may do so. The senescence marker p16 (CDKN2A) was investigated as a well-known senescence marker and was found in a higher fraction of cells in old compared to young donors, though its expression was not found to increase on average (over all cells). We may speculate that, even though the fraction of senescent cells increases in the aging pancreas, the cells surrounding these are not affected due to large amounts of extracellular matrix, which may block any paracrine effects. The Aging Pancreas Atlas data were further analyzed by (Chen et al., 2019), but without any consideration of age, with the aim of learning classification rules for six different pancreatic cell types. For this purpose they used various machine learning techniques such as Monte Carlo feature selection and support vector machines. Using only the best 7 features (biomarkers) they reached a classification accuracy of 0.966. It would be of interest to use the same or similar methods to learn and test classification rules for predicting donor age, compare their performance, and to understand which genes underlie such classification.
As a more specialized resource, the “Pancreatic Cancer Atlas” is a detailed study of various populations of cancer-associated fibroblasts (CAFs) implicated in PDAC (Elyada et al., 2019). Often, CAFs promote tumor growth by modifying collagen cross-linking, secreting diverse cytokines, chemokines and growth factors and recruiting immune-suppressive cells, while other studies indicated that CAFs actually restrict tumor growth (Ozdemir et al., 2014; Rhim et al., 2014). The single-cell studies by (Elyada et al., 2019), investigating human and mouse cells, help to come closer to an explanation, by showing that CAFs are actually a heterogeneous cell population consisting of different subtypes such as myofibroblastic CAFs (myCAF), inflammatory CAFs (iCAF) and antigen presenting CAFs (apCAF). Regarding inflammaging and cellular senescence, iCAFs are worth noting, since they express interleukins and cytokines that are also characteristic for the SASP.
Finally, the “Aging/T1D Beta Cell Atlas” (Thompson et al., 2019) describes how cellular senescence of islet cells is involved in the development of type 1 diabetes (T1D) in humans and in the non-obese diabetic (NOD) mouse model. For this purpose the authors performed scRNA-seq of ∼21 000 cells of 8, 14 and 16 week old NOD mice; specifically they collected CD45+ immune-cell-depleted but insulin-expressing cells from the pancreas islets. Remarkably, the authors find that removal of such cells via senolytic compounds (inhibiting BCL-2) was able to prevent the appearance of diabetes in these mice, pointing to a failure of the immune system to remove such senescent cells in case of T1D and to senolysis as a remedy. In the single-cell data, the senescence marker p21 (Cdkn1a) was found enriched in a cluster of cells in the diseased state at 14/16 weeks, whose overall transcriptional profile was “reminiscent of a senescence stress response”. In turn, the beta cell maturation marker Ucn3 was found depleted in that cluster.
4.5. Brain
Single cell studies are especially useful for studying the brain, since this complex organ contains a multitude of different cell types that are difficult to separate. However, brain omics of human tissue are almost always and necessarily post-mortem, which may pose problems caused by artifact patterns (Ferreira et al., 2018).
In the “Aging Brain Atlas”, brain tissue of young and old mice was studied by single-cell transcriptomics of ∼50 000 cells (Ximerakis et al., 2019). By differential gene expression analysis, the authors found that expression patterns do not change uniformly across cell types. The coefficient of variation of expression for selected sets of transcribed genes differed between young and old cells in many cell types, but the direction of change was not consistent among cell types, suggesting no broad increase of transcriptional variability. The latter observation makes this study the only one in this review that does not report a general increase in transcriptional heterogeneity. Another noteworthy result is that in most brain cell types the transcription of ribosomal proteins appears to be upregulated. In their study the authors also found that aging- and senescence-related pathways were upregulated, specifically in a cluster of cells identified as endothelial cells. A web interface is available, at https://portals.broadinstitute.org/single_cell/study/aging-mouse-brain.
Another study investigating the brain is the “Aging Cerebromicrovascular Atlas” (Kiss et al., 2020). It describes the age-induced dysregulation of microvascular endothelial function in the mouse brain, which contributes to diseases such as vascular cognitive impairment and Alzheimer’s disease, based on a new approach to identify senescent cerebromicrovascular endothelial cells. Extracted and digested brain tissues isolated from mice were separated by centrifugation and filtering, and senescent cerebromicrovascular endothelial cells were identified through clustering by specific expression patterns and by a precompiled list of senescence markers such as Cdkn2a and Bmi1. ∼10 % of the cerebromicrovascular endothelial cells experience cellular senescence at a biological age equivalent to a 75-year-old human; microglia are similarly affected.
Finally, the “Aging Microglia Atlas” (Hammond et al., 2019) describes microglia, the macrophages of the brain, that have been historically classified as resting, M1 proinflammatory, or M2 anti-inflammatory, based on in vitro stimulation methods. As the resting-M1-M2 classification system might be oversimplifying the real phenotypic heterogeneity of these cells in vivo, Hammond et al. (2019) carried out single-cell transcriptomics of mouse microglia ranging from embryogenesis to old age. They found that microglia subpopulations became less heterogeneous in adulthood, but heterogeneity increased again during aging. Some aged microglia expressed the Ccl4 chemokine, and based on the authors observations, we suggest that this cytokine contributes to inflammaging.
4.6. Lung
The lung has become a key tissue of recent single-cell investigations due to the Covid-19 pandemic. For example, we recently described (Fuellen et al., 2020) a single-cell dataset investigating the lung immune microenvironment, based on bronchoalveolar lavage fluid of Covid-19 patients (Liao et al., 2020). The arrival of longitudinal single-cell data helping to investigate the interconnections between Covid-19 disease susceptibility & sequelae, in particular inflammaging, is only a question of time.
Investigating lung aging in mice (“Aging Lung Atlas”), scRNA-seq was combined with in-depth proteomics by (Angelidis et al., 2019). They sequenced ∼15 000 cells of young (3 month) and old (24 month) animals and identified 30 different cell types. Their mass spectrometry based proteomics is based on bulk tissue, but it could still be related to their scRNA-seq data, finding a significant correlation between fold changes at the transcriptome and proteome level. However, there were notable exceptions such as collagen IV, which showed an age-related decline at the mRNA level, but an increase at the protein level. As in (Enge et al., 2017), increased transcriptional noise was noted. Enrichment analyses pointed to increased cholesterol biosynthesis in some cell types, and an altered relative frequency of airway epithelial cells was found as a hallmark of lung aging. Finally, chronic inflammation / inflammaging was observed, putatively related to the deregulation of lipid homeostasis and epithelial cellular senescence. The “Aging Lung Atlas” is available online and can be searched via a webtool (see https://theislab.github.io/LungAgingAtlas/).
A human “Lung Aging/Fibrosis Atlas” is presented by (Reyfman et al., 2019) based on the analysis of ∼76 000 single cells from eight healthy donors and eight patients with pulmonary fibrosis, of various ages. In their clustering analysis (by t-Distributed Stochastic Neighbor Embedding, t-SNE), the authors did not find a cluster of cells that specifically express senescence markers (they inspected p16, IL-6, Serpine1 and GLB), but a 1311-gene signature of replicative senescence scored highest in a population of alveolar type II stem cells that originated almost exclusively from the fibrotic lung tissue. Furthermore, this gene signature could be seen in a small fraction of practically all cell types present in the lung. This is consistent with the finding that many cell types can develop a senescent phenotype, but that this phenotype is very heterogeneous depending on the original cell type (Hernandez-Segura et al., 2017). Notably, a recent study using a transgenic mouse model investigated the role of alveolar type II stem cells for lung fibrosis further by artificially inducing cellular senescence in this cell type (Yao et al., 2019), initiating progressive lung fibrosis, which could be blocked by a senolytic treatment with dasatinib and quercetin. In this loss-of-function study, the authors performed scRNA-seq of ∼10 000 epithelial cells collected by fluorescence-activated cell sorting (FACS).
4.7. Skin
Studies that investigate the aging process using skin have to take special care, since “photo-aging” due to the exposure of humans to sunlight can strongly influence aging-related processes, acting as a major confounder. Thus, Solé-Boldo et al. (2019) collected cells from the sun protected inguinoiliac skin region of male human donors for their “Skin Aging Atlas”, comprising more than 15 000 cells. They found 14 known cell types in their sample and among these, they could identify 4 subtypes of fibroblasts based on clustering (t-SNE) plots and analysis of GO term enrichments, which they called secretory-papillary fibroblasts, secretory-reticular fibroblasts, mesenchymal fibroblasts and pro-inflammatory fibroblasts. Furthermore, they compared the samples from young (25 & 27 years) to old (53, 69, 70 years) donors to investigate age-dependent changes, revealing that fibroblasts feature a subtype-specific set of “skin-aging associated secreted proteins” (SAASP), as a set of proteins secreted by the fibroblasts from old human donors and already described previously (Waldera Lupa et al., 2015). The functional identity of cells appeared to become blurred, since fewer GO terms with less significant p-values were found in case of the old samples; higher transcriptional heterogeneity may be an explanation for this observation.
4.8. Muscle
In their “Muscle Aging Atlas”, Hernando-Herraez et al. (2019) used 377 single muscle stem cells from young (∼2 months) and old (∼25 months) mice to measure the transcriptome as well as DNA methylation, in the same cells at the same time. Muscle stem cells were specifically selected as the subject of the study, also motivated by their assumed “deep” quiescence; a transgene was used to sort out these cells based on the marker Pax7. Based on the pairwise correlation of gene expression of cells, both within and between individual mice, the authors found an increased variability at the transcriptome level comparing young versus old mice; some immunostaining was done and an increased heterogeneity on the protein level was also observed. Further, the correlation between the degree of methylation of promoter regions, and gene expression, at the single-cell level was negative, supporting the idea that age related changes in the methylome may be triggering the age related changes in the transcriptome. Assessing epigenetic age using adult muscle stem cells yielded surprising results, though. While the estimated age in the young mice was around 9 weeks, the age prediction for the muscle stem cells of the old mice was 10–11 weeks. Thus, epigenetic clocks based on bulk tissue may not confer the full picture, if some cell types (e.g. muscle stem cells) maintain a young epigenetic age, but dwindle in number in old individuals. Further studies involving single-cell methylation data from different (stem) cell populations are needed to deeper investigate single-cell-based epigenetic clocks and their ability to predict chronological or biological age.
4.9. In-vitro studies
Telomere erosion was investigated by the in-vitro study of senescent cells reported in (Tang et al., 2019). They cultured HCA2 fibroblasts until replicative senescence was reached and performed single cell transcriptomics on a total of 1200 cells from early, middle and late passage. They also irradiated early passage cells to induce SIPS (stress induced premature senescence) and analyzed 400 of those cells. Visualization via t-SNE clearly separated cells from different passages and especially late passage senescent cells appear to be more heterogeneous than irradiation-induced senescent cells. This is a further confirmation that, to a certain degree, cellular senescence is not only different in different cell types, but also depends on the stressor that caused the senescent state. Nevertheless, (Tang et al., 2019) identify a senescence core signature of up-regulated genes that include p21 (CDKN1A) and members of the SASP.
The entry into senescence was also investigated in another in-vitro study using umbilical vein endothelial cells (HUVECs), fetal lung fibroblasts (IMR90 s) and mesenchymal stromal cells (MSCs) (Zirkel et al., 2018). The levels of HMGB2, which is involved in the maintenance of the 3D chromatin structure, diminish in the nuclei of old and senescent tissues and the authors hypothesized that this decline triggers an entry into cellular senescence. They demonstrate the decline of HMGB2 before the appearance of established senescence markers, by scRNA-seq on 8300 proliferating and 5200 senescent HUVECS. Cellular senescence can also be caused by transcription-factor-induced reprogramming stress, (Banito et al., 2009), which can lower the efficiency of iPSC generation, and a combination of an shRNA library intervention approach with single-cell sequencing was presented by (Aarts et al., 2017), based on primary human fibroblasts (IMR90) expressing reprogramming factors. This allowed to identify those shRNAs that are most suitable to suppress this type of cellular senescence. The four top scoring target genes were CDKN1A, MTOR, MYOT and UBE2E1. While CDKN1A (p21), and MTOR are known to influence senescence, the other two, being part of sarcomeres and being an E2 ubiquitin-conjugating enzyme, were new candidates.
Overall, it is an open yet relevant question how faithfully the in-vitro models of cellular senescence reflect the all-important in-vivo situation, and single-cell analyses may enable a more fine-grained comparison. At least, the markers of cellular senescence first identified in-vitro are often, but not always, following the same or similar plausible patterns of age-related change in gene expression in-vivo, for bulk and single-cell data alike.
5. Conclusions
Overall, an increasing frequency of cells expressing aging/senescence markers, and/or an overall increasing expression of aging/senescence markers (albeit often at low absolute levels in the cells in which they are expressed), and an increase in transcriptional heterogeneity are the most frequent patterns revealed by the aging-related single-cell studies we reviewed. A potential corollary of the latter is that cellular identity tends to become less constrained, or even lost. This observation matches a number of recent insights, outside of single-cell transcriptomics. For example, single-cell investigations of global chromatin modification data revealed an increased epigenetic variation with aging (Cheung et al., 2018). Moreover, the loss of cell identity due to chromatin modifiers attending double-strand breaks was recently suggested to be one of the main drivers of aging (Yang et al., 2019). This proposal connects transcriptional heterogeneity to cellular senescence, which is often caused by double-strand breaks and other kinds of DNA damage.
There is a lack of interventional studies supported by single-cell transcriptomics. This topic will be fascinating to investigate, promising insights into the drug response for different cell types for the same individual at identical conditions. Another prospect lies in the investigation of transcriptomic profiles that are preserved when bringing these cells in culture. This ties observational studies to experimental follow-up investigations, given that a conserved “context-free” subset of characteristic gene expression changes may be a primary target for new drugs to modulate aging-associated (and other) phenotypes (Struckmann et al., 2020). And likely, such sets of co-regulated genes, established for specific cell types, will be recognized as motifs in their own right, with their own functional annotation, in analogy to what domain databases have contributed to our understanding of biological sequences.
Ten years back, de Magalhaes et al. (2010) reviewed the state of Next-Generation Sequencing in aging research in this journal. Technological progress has been breathtaking since then, and Next-Generation Sequencing has become possible at single-cell resolution, on a large scale. Aging affects every cell, and to some degree every cell is affected differently by aging. In turn, all cells together make up the aging metazoan organism, and their contribution to the aging phenotype is not uniform. It will be exciting to watch the progress that can be made by the magnifying glass provided by scRNA-seq, to aging research and beyond.
Funding
GF is supported by the BMBF (FKZ 01ZX1903A, 03V0396 and 1DS19049), the European Commission (Aging with elegans, grant agreement 633589), the DFG (FU-583/5-1), Karls Erdbeerhof, Rövershagen, Germany, and the Interdisziplinäre Fakultät (Department AGIS) of Rostock University.
Declaration of Competing Interest
The authors declare that no competing interests exist.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.arr.2020.101156.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Aarts M., Georgilis A., Beniazza M., Beolchi P., Banito A., Carroll T., Kulisic M., Kaemena D.F., Dharmalingam G., Martin N., Reik W., Zuber J., Kaji K., Chandra T., Gil J. Coupling shRNA screens with single-cell RNA-seq identifies a dual role for mTOR in reprogramming-induced senescence. Genes Dev. 2017;31:2085–2098. doi: 10.1101/gad.297796.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelaal T., Michielsen L., Cats D., Hoogduin D., Mei H., Reinders M.J.T., Mahfouz A. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 2019;20:194. doi: 10.1186/s13059-019-1795-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almanzar N., Antony J., Baghel A.S., Bakerman I., Bansal I., Barres B.A., et al. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020;583:590–595. doi: 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelidis I., Simon L.M., Fernandez I.E., Strunz M., Mayr C.H., Greiffo F.R., Tsitsiridis G., Ansari M., Graf E., Strom T.M., Nagendran M., Desai T., Eickelberg O., Mann M., Theis F.J., Schiller H.B. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 2019;10:963. doi: 10.1038/s41467-019-08831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar M.P., Brandt R.M., Putavet D.A., Klein J.D., Derks K.W., Bourgeois B.R., et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169(132–147):e116. doi: 10.1016/j.cell.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar R., Hartmann C.H., Rodriguez K.A., Denny A.D., Busuttil R.A., Dolle M.E., Calder R.B., Chisholm G.B., Pollock B.H., Klein C.A., Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Baker D.J., Childs B.G., Durik M., Wijers M.E., Sieben C.J., Zhong J., et al. Naturally occurring p16-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A., Rashid S.T., Acosta J.C., Li S., Pereira C.F., Geti I., Pinho S., Silva J.C., Azuara V., Walsh M., Vallier L., Gil J. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruzzo G., Hayer K.E., Kim E.J., Di Camillo B., FitzGerald G.A., Grant G.R. Simulation-based comprehensive benchmarking of RNA-seq aligners. Nat. Methods. 2017;14:135–139. doi: 10.1038/nmeth.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatsirisupachai K., Palmer D., Ferreira S., de Magalhaes J.P. A human tissue-specific transcriptomic analysis reveals a complex relationship between aging, cancer, and cellular senescence. Aging Cell. 2019;18 doi: 10.1111/acel.13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Pan X., Zhang Y.-H., Huang T., Cai Y.-D. Analysis of gene expression differences between different pancreatic cells. ACS Omega. 2019;4:6421–6435. [Google Scholar]
- Cheung P., Vallania F., Warsinske H.C., Donato M., Schaffert S., Chang S.E., Dvorak M., Dekker C.L., Davis M.M., Utz P.J., Khatri P., Kuo A.J. Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell. 2018;173(1385–1397):e1314. doi: 10.1016/j.cell.2018.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M.B., Risso D., Wagner A., DeTomaso D., Ngai J., Purdom E., Dudoit S., Yosef N. Performance assessment and selection of normalization procedures for single-cell RNA-Seq. Cell Syst. 2019;8(315–328):e318. doi: 10.1016/j.cels.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.J., Sedivy J.M. Involvement of the INK4a/Arf gene locus in senescence. Aging Cell. 2003;2:145–150. doi: 10.1046/j.1474-9728.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- Coppe J.P., Desprez P.Y., Krtolica A., Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keizer P.L. The fountain of youth by targeting senescent cells? Trends Mol. Med. 2017;23:6–17. doi: 10.1016/j.molmed.2016.11.006. [DOI] [PubMed] [Google Scholar]
- de Magalhaes J.P., Finch C.E., Janssens G. Next-generation sequencing in aging research: emerging applications, problems, pitfalls and possible solutions. Ageing Res. Rev. 2010;9:315–323. doi: 10.1016/j.arr.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelly C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., Peacocke M., Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Adiconis X., Simmons S.K., Kowalczyk M.S., Hession C.C., Marjanovic N.D., Hughes T.K., Wadsworth M.H., Burks T., Nguyen L.T., Kwon J.Y.H., Barak B., Ge W., Kedaigle A.J., Carroll S., Li S., Hacohen N., Rozenblatt-Rosen O., Shalek A.K., Villani A.-C., Regev A., Levin J.Z. Systematic comparative analysis of single cell RNA-sequencing methods. bioRxiv. 2019 [Google Scholar]
- Dong M., Thennavan A., Urrutia E., Li Y., Perou C.M., Zou F., Jiang Y. SCDC: bulk gene expression deconvolution by multiple single-cell RNA sequencing references. Brief Bioinform. 2020 doi: 10.1093/bib/bbz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duo A., Robinson M.D., Soneson C. A systematic performance evaluation of clustering methods for single-cell RNA-seq data. F1000Research. 2018;7:1141. doi: 10.12688/f1000research.15666.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyada E., Bolisetty M., Laise P., Flynn W.F., Courtois E.T., Burkhart R.A., Teinor J.A., Belleau P., Biffi G., Lucito M.S., Sivajothi S., Armstrong T.D., Engle D.D., Yu K.H., Hao Y., Wolfgang C.L., Park Y., Preall J., Jaffee E.M., Califano A., Robson P., Tuveson D.A. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyahu Y., Hekselman I., Eizenberg-Magar I., Berner O., Strominger I., Schiller M., Mittal K., Nemirovsky A., Eremenko E., Vital A., Simonovsky E., Chalifa-Caspi V., Friedman N., Yeger-Lotem E., Monsonego A. Aging promotes reorganization of the CD4 T cell landscape toward extreme regulatory and effector phenotypes. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aaw8330. eaaw8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enge M., Arda H.E., Mignardi M., Beausang J., Bottino R., Kim S.K., Quake S.R. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell. 2017;171(321-330):e314. doi: 10.1016/j.cell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P.G., Munoz-Aguirre M., Reverter F., Sa Godinho C.P., Sousa A., Amadoz A., et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat. Commun. 2018;9:490. doi: 10.1038/s41467-017-02772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C., Bonafe M., Valensin S., Olivieri F., De Luca M., Ottaviani E., De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Franceschi C., Garagnani P., Vitale G., Capri M., Salvioli S. Inflammaging and’ Garb-aging’. Trends Endocrinol. Metab. 2017;28:199–212. doi: 10.1016/j.tem.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Franceschi C., Zaikin A., Gordleeva S., Ivanchenko M., Bonifazi F., Storci G., Bonafe M. Inflammaging 2018: an update and a model. Semin. Immunol. 2018;40:1–5. doi: 10.1016/j.smim.2018.10.008. [DOI] [PubMed] [Google Scholar]
- Fuellen G., Jansen L., Cohen A.A., Luyten W., Gogol M., Simm A., Saul N., Cirulli F., Berry A., Antal P., Kohling R., Wouters B., Moller S. Health and aging: unifying concepts, scores, biomarkers and pathways. Aging Dis. 2019;10:883–900. doi: 10.14336/AD.2018.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuellen G., Liesenfeld O., Kowald A., Barrantes I., Bastian M., Simm A., Jansen L., Tietz-Latza A., Quandt D., Franceschi C., Walter M. The preventive strategy for pandemics in the elderly is to collect in advance samples & data to counteract chronic inflammation (inflammaging) Ageing Res. Rev. 2020 doi: 10.1016/j.arr.2020.101091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawel D.R., Serra-Musach J., Lilja S., Aagesen J., Arenas A., Asking B., Bengner M., Bjorkander J., Biggs S., Ernerudh J., Hjortswang H., Karlsson J.E., Kopsen M., Lee E.J., Lentini A., Li X., Magnusson M., Martinez-Enguita D., Matussek A., Nestor C.E., Schafer S., Seifert O., Sonmez C., Stjernman H., Tjarnberg A., Wu S., Akesson K., Shalek A.K., Stenmarker M., Zhang H., Gustafsson M., Benson M. A validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome Med. 2019;11:47. doi: 10.1186/s13073-019-0657-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain P.-L., Sonrel A., Robinson M.D. pipeComp, a general framework for the evaluation of computational pipelines, reveals performant single-cell RNA-seq preprocessing tools. Genome Biology. 2020;21:227. doi: 10.1186/s13059-020-02136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V., Adams P.D., Alimonti A., Bennett D.C., Bischof O., Bishop C., Campisi J., Collado M., Evangelou K., Ferbeyre G., Gil J., Hara E., Krizhanovsky V., Jurk D., Maier A.B., Narita M., Niedernhofer L., Passos J.F., Robbins P.D., Schmitt C.A., Sedivy J., Vougas K., von Zglinicki T., Zhou D., Serrano M., Demaria M. Cellular senescence: defining a path forward. Cell. 2019;179:813–827. doi: 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- Grover A., Sanjuan-Pla A., Thongjuea S., Carrelha J., Giustacchini A., Gambardella A., Macaulay I., Mancini E., Luis T.C., Mead A., Jacobsen S.E., Nerlov C. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 2016;7:11075. doi: 10.1038/ncomms11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A.J., Gergits F., Segel M., Nemesh J., Marsh S.E., Saunders A., Macosko E., Ginhoux F., Chen J., Franklin R.J.M., Piao X., McCarroll S.A., Stevens B. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50(253–271):e256. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza B., Ng S.R., Prakadan S.M., Delgado F.F., Chin C.R., King E.M., et al. Optofluidic real-time cell sorter for longitudinal CTC studies in mouse models of cancer. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2232–2236. doi: 10.1073/pnas.1814102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Cao W., Huang J., Zou X., Han Z.G. Optimal Gene filtering for Single-Cell data (OGFSC)-a gene filtering algorithm for single-cell RNA-seq data. Bioinformatics. 2019;35:2602–2609. doi: 10.1093/bioinformatics/bty1016. [DOI] [PubMed] [Google Scholar]
- Harley C.B. Telomere loss: mitotic clock or genetic time bomb? Mutat. Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- Harley C.B., Futcher A.B., Greider C.W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Hayflick L., Moorhead P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Hernandez-Segura A., de Jong T.V., Melov S., Guryev V., Campisi J., Demaria M. Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol. 2017;27(2652–2660):e2654. doi: 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Herraez I., Evano B., Stubbs T., Commere P.H., Jan Bonder M., Clark S., Andrews S., Tajbakhsh S., Reik W. Ageing affects DNA methylation drift and transcriptional cell-to-cell variability in mouse muscle stem cells. Nat. Commun. 2019;10:4361. doi: 10.1038/s41467-019-12293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt N.J., Kang S.W.S., Lockwood G.P., Le Couteur D.G., Cogger V.C. Hallmarks of aging in the liver. Comput. Struct. Biotechnol. J. 2019;17:1151–1161. doi: 10.1016/j.csbj.2019.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Işıldak U., Somel M., Thornton J.M., Dönertaş H.M. Gene expression heterogeneity during brain development and aging: temporal changes and functional consequences. bioRxiv. 2019 [Google Scholar]
- Kimmel J.C., Penland L., Rubinstein N.D., Hendrickson D.G., Kelley D.R., Rosenthal A.Z. Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome Res. 2019;29:2088–2103. doi: 10.1101/gr.253880.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Nyul-Toth A., Balasubramanian P., Tarantini S., Ahire C., DelFavero J., Yabluchanskiy A., Csipo T., Farkas E., Wiley G., Garman L., Csiszar A., Ungvari Z. Single-cell RNA sequencing identifies senescent cerebromicrovascular endothelial cells in the aged mouse brain. Geroscience. 2020;42:429–444. doi: 10.1007/s11357-020-00177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Jimenez C.P., Eling N., Chen H.C., Vallejos C.A., Kolodziejczyk A.A., Connor F., Stojic L., Rayner T.F., Stubbington M.J.T., Teichmann S.A., de la Roche M., Marioni J.C., Odom D.T. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science. 2017;355:1433–1436. doi: 10.1126/science.aah4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard A., McCoach C.E., Rotow J.K., Harris L., Haderk F., Kerr L., Yu E.A., Schenk E.L., Tan W., Zee A., Tan M., Gui P., Lea T., Wu W., Urisman A., Jones K., Sit R., Kolli P.K., Seeley E., Gesthalter Y., Le D.D., Yamauchi K.A., Naeger D., Thomas N.J., Gupta A., Gonzalez M., Do H., Tan L., Gomez-Sjoberg R., Gubens M., Jahan T., Kratz J.R., Jablons D., Neff N., Doebele R.C., Weissman J., Blakely C.M., Darmanis S., Bivona T.G. Heterogeneity and targeted therapy-induced adaptations in lung cancer revealed by longitudinal single-cell RNA sequencing. bioRxiv. 2019 [Google Scholar]
- McHugh D., Gil J. Senescence and aging: causes, consequences, and therapeutic avenues. J. Cell Biol. 2018;217:65–77. doi: 10.1083/jcb.201708092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereu E., Lafzi A., Moutinho C., Ziegenhain C., McCarthy D.J., Álvarez-Varela A., et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0469-4. [DOI] [PubMed] [Google Scholar]
- Mohammadi S., Davila-Velderrain J., Kellis M. Reconstruction of cell-type-specific interactomes at single-cell resolution. Cell Syst. 2019;9(559–568):e554. doi: 10.1016/j.cels.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsiani C., Bacalini M.G., Santoro A., Garagnani P., Collura S., D’Errico A., de Eguileor M., Grazi G.L., Cescon M., Franceschi C., Capri M. The peculiar aging of human liver: a geroscience perspective within transplant context. Ageing Res. Rev. 2019;51:24–34. doi: 10.1016/j.arr.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Ozdemir B.C., Pentcheva-Hoang T., Carstens J.L., Zheng X., Wu C.C., Simpson T.R., Laklai H., Sugimoto H., Kahlert C., Novitskiy S.V., De Jesus-Acosta A., Sharma P., Heidari P., Mahmood U., Chin L., Moses H.L., Weaver V.M., Maitra A., Allison J.P., LeBleu V.S., Kalluri R. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N., Almouzni G., Gorski S.A., Aerts S., Amit I., Bertero M.G., Bock C., Bredenoord A.L., Cavalli G., Chiocca S., Clevers H., De Strooper B., Eggert A., Ellenberg J., Fernández X.M., Figlerowicz M., Gasser S.M., Hubner N., Kjems J., Knoblich J.A., Krabbe G., Lichter P., Linnarsson S., Marine J.-C., Marioni J., Marti-Renom M.A., Netea M.G., Nickel D., Nollmann M., Novak H.R., Parkinson H., Piccolo S., Pinheiro I., Pombo A., Popp C., Reik W., Roman-Roman S., Rosenstiel P., Schultze J.L., Stegle O., Tanay A., Testa G., Thanos D., Theis F.J., Torres-Padilla M.-L., Valencia A., Vallot C., van Oudenaarden A., Vidal M., Voet T. LifeTime and improving European healthcare through cell-based interceptive medicine. Nature. 2020 doi: 10.1038/s41586-020-2715-9. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyfman P.A., Walter J.M., Joshi N., Anekalla K.R., McQuattie-Pimentel A.C., Chiu S., Fernandez R., Akbarpour M., Chen C.I., Ren Z., Verma R., Abdala-Valencia H., Nam K., Chi M., Han S., Gonzalez-Gonzalez F.J., Soberanes S., Watanabe S., Williams K.J.N., Flozak A.S., Nicholson T.T., Morgan V.K., Winter D.R., Hinchcliff M., Hrusch C.L., Guzy R.D., Bonham C.A., Sperling A.I., Bag R., Hamanaka R.B., Mutlu G.M., Yeldandi A.V., Marshall S.A., Shilatifard A., Amaral L.A.N., Perlman H., Sznajder J.I., Argento A.C., Gillespie C.T., Dematte J., Jain M., Singer B.D., Ridge K.M., Lam A.P., Bharat A., Bhorade S.M., Gottardi C.J., Budinger G.R.S., Misharin A.V. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim A.D., Oberstein P.E., Thomas D.H., Mirek E.T., Palermo C.F., Sastra S.A., Dekleva E.N., Saunders T., Becerra C.P., Tattersall I.W., Westphalen C.B., Kitajewski J., Fernandez-Barrena M.G., Fernandez-Zapico M.E., Iacobuzio-Donahue C., Olive K.P., Stanger B.Z. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhme D. Evidence for a relationship between longevity of mammalian species and life spans of normal fibroblasts in vitro and erythrocytes in vivo. Proc. Natl. Acad. Sci. U. S. A. 1981;78:5009–5013. doi: 10.1073/pnas.78.8.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T., et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schussler-Fiorenza Rose S.M., Contrepois K., Moneghetti K.J., Zhou W., Mishra T., Mataraso S., et al. A longitudinal big data approach for precision health. Nat. Med. 2019;25:792–804. doi: 10.1038/s41591-019-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé-Boldo L., Raddatz G., Schütz S., Mallm J.-P., Rippe K., Lonsdorf A.S., Rodríguez-Paredes M., Lyko F. Single-cell transcriptomes of the aging human skin reveal loss of fibroblast priming. bioRxiv. 2019 doi: 10.1038/s42003-020-0922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Robinson M.D. Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods. 2018;15:255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- Struckmann S., Ernst M., Fischer S., Mah N., Fuellen G., Moller S. Scoring functions for drug-effect similarity. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Coppe J.P., Lam E.W. Cellular senescence: the sought or the unwanted? Trends Mol. Med. 2018 doi: 10.1016/j.molmed.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Sun S., Zhu J., Ma Y., Zhou X. Accuracy, robustness and scalability of dimensionality reduction methods for single cell RNAseq analysis. Genome Biology. 2019;20:269. doi: 10.1186/s13059-019-1898-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V., Natarajan K.N., Ly L.H., Miragaia R.J., Labalette C., Macaulay I.C., Cvejic A., Teichmann S.A. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods. 2017;14:381–387. doi: 10.1038/nmeth.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V., Vento-Tormo R., Teichmann S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018;13:599–604. doi: 10.1038/nprot.2017.149. [DOI] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A., Lao K., Surani M.A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Tang H., Geng A., Zhang T., Wang C., Jiang Y., Mao Z. Single senescent cell sequencing reveals heterogeneity in senescent cells induced by telomere erosion. Protein Cell. 2019;10:370–375. doi: 10.1007/s13238-018-0591-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P.J., Shah A., Ntranos V., Van Gool F., Atkinson M., Bhushan A. Targeted elimination of senescent Beta cells prevents type 1 diabetes. Cell Metab. 2019;29(1045-1060):e1010. doi: 10.1016/j.cmet.2019.01.021. [DOI] [PubMed] [Google Scholar]
- Tian L., Dong X., Freytag S., Le Cao K.A., Su S., JalalAbadi A., Amann-Zalcenstein D., Weber T.S., Seidi A., Jabbari J.S., Naik S.H., Ritchie M.E. Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat. Methods. 2019;16:479–487. doi: 10.1038/s41592-019-0425-8. [DOI] [PubMed] [Google Scholar]
- Tran H.T.N., Ang K.S., Chevrier M., Zhang X., Lee N.Y.S., Goh M., Chen J. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21:12. doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen J.M. Senolytic therapies for healthy longevity. Science. 2019;364:636–637. doi: 10.1126/science.aaw1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieth B., Parekh S., Ziegenhain C., Enard W., Hellmann I. A systematic evaluation of single cell RNA-seq analysis pipelines. Nat. Commun. 2019;10:4667. doi: 10.1038/s41467-019-12266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldera Lupa D.M., Kalfalah F., Safferling K., Boukamp P., Poschmann G., Volpi E., et al. Characterization of skin aging-associated secreted proteins (SAASP) produced by dermal fibroblasts isolated from intrinsically aged human skin. J. Invest. Dermatol. 2015;135:1954–1968. doi: 10.1038/jid.2015.120. [DOI] [PubMed] [Google Scholar]
- Wiley C.D., Flynn J.M., Morrissey C., Lebofsky R., Shuga J., Dong X., Unger M.A., Vijg J., Melov S., Campisi J. Analysis of individual cells identifies cell-to-cell variability following induction of cellular senescence. Aging Cell. 2017;16:1043–1050. doi: 10.1111/acel.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.C., Yao J., Ho K.S., Lambowitz A.M., Wilke C.O. Limitations of alignment-free tools in total RNA-seq quantification. BMC Genomics. 2018;19:510. doi: 10.1186/s12864-018-4869-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmus R., Uyar B., Osberg B., Franke V., Gosdschan A., Wreczycka K., Ronen J., Akalin A. PiGx: reproducible genomics analysis pipelines with GNU Guix. Gigascience. 2018;7 doi: 10.1093/gigascience/giy123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximerakis M., Lipnick S.L., Innes B.T., Simmons S.K., Adiconis X., Dionne D., Mayweather B.A., Nguyen L., Niziolek Z., Ozek C., Butty V.L., Isserlin R., Buchanan S.M., Levine S.S., Regev A., Bader G.D., Levin J.Z., Rubin L.L. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 2019;22:1696–1708. doi: 10.1038/s41593-019-0491-3. [DOI] [PubMed] [Google Scholar]
- Xu M., Pirtskhalava T., Farr J.N., Weigand B.M., Palmer A.K., Weivoda M.M., Inman C.L., Ogrodnik M.B., Hachfeld C.M., Fraser D.G., Onken J.L., Johnson K.O., Verzosa G.C., Langhi L.G.P., Weigl M., Giorgadze N., LeBrasseur N.K., Miller J.D., Jurk D., Singh R.J., Allison D.B., Ejima K., Hubbard G.B., Ikeno Y., Cubro H., Garovic V.D., Hou X., Weroha S.J., Robbins P.D., Niedernhofer L.J., Khosla S., Tchkonia T., Kirkland J.L. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 2018 doi: 10.1038/s41591-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H., Fraifeld V.E. The role of cellular senescence in aging through the prism of Koch-like criteria. Ageing Res. Rev. 2018;41:18–33. doi: 10.1016/j.arr.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Yang J.-H., Griffin P.T., Vera D.L., Apostolides J.K., Hayano M., Meer M.V., Salfati E.L., Su Q., Munding E.M., Blanchette M., Bhakta M., Dou Z., Xu C., Pippin J.W., Creswell M.L., O’Connell B.L., Green R.E., Garcia B.A., Berger S.L., Oberdoerffer P., Shankland S.J., Gladyshev V.N., Rajman L.A., Pfenning A.R., Sinclair D.A. Erosion of the epigenetic landscape and loss of cellular identity as a cause of aging in mammals. bioRxiv. 2019 [Google Scholar]
- Yao C., Guan X., Carraro G., Parimon T., Liu X., Huang G., Soukiasian H.J., David G., Weigt S.S., Belperio J.A., Chen P., Jiang D., Noble P.W., Stripp B.R. Senescence of alveolar stem cells drives progressive pulmonary fibrosis. bioRxiv. 2019 doi: 10.1164/rccm.202004-1274OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia L., Phipson B., Oshlack A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.J., Pisco A.O., Darmanis S., Zou J. Mouse aging cell atlas analysis reveals global and cell type specific aging signatures. bioRxiv. 2019 doi: 10.7554/eLife.62293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhang S., Yan P., Ren J., Song M., Li J., et al. A single-cell transcriptomic landscape of primate arterial aging. Nat. Commun. 2020;11:2202. doi: 10.1038/s41467-020-15997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang S. Comparison of computational methods for imputing single-cell RNA-Sequencing data. IEEEACM Trans. Comput. Biol. Bioinform. 2020;17:376–389. doi: 10.1109/TCBB.2018.2848633. [DOI] [PubMed] [Google Scholar]
- Zirkel A., Nikolic M., Sofiadis K., Mallm J.P., Brackley C.A., Gothe H., Drechsel O., Becker C., Altmuller J., Josipovic N., Georgomanolis T., Brant L., Franzen J., Koker M., Gusmao E.G., Costa I.G., Ullrich R.T., Wagner W., Roukos V., Nurnberg P., Marenduzzo D., Rippe K., Papantonis A. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell. 2018;70(730–744):e736. doi: 10.1016/j.molcel.2018.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.