Abstract
Being able to replicate research results is the hallmark of science. Replication of research findings using computational models should, in principle, be possible. In this manuscript, we assess code sharing and model documentation practices of 7500 publications about individual-based and agent-based models. The code availability increased over the years, up to 18% in 2018.
Model documentation does not include all the elements that could improve the transparency of the models, such as mathematical equations, flow charts, and pseudocode. We find that articles with equations and flow charts being cited more among other model papers, probably because the model documentation is more transparent.
The practices of code sharing improve slowly over time, partly due to the emergence of more public repositories and archives, and code availability requirements by journals and sponsors. However, a significant change in norms and habits need to happen before computational modeling becomes a reproducible science.
Keywords: Reuse, Open science, Replicability, Agent-based modeling, Individual-based modeling
Highlights
-
•
Lack code sharing of computational science limits reuse and reproducibility.
-
•
A database of 7500 articles of agent-based and individual-based models was created.
-
•
Only 11% of articles shared code, although there is an upward trend.
-
•
Better documentation practices increase the citations to model papers by other model papers.
-
•
Major changes in practices are needed on code sharing within computational modeling research.
1. Introduction
Isaac Newton famously wrote: “If I have seen further it is by standing on the shoulders of Giants.”, which emphasized an essential component of science, namely the accumulation of knowledge by reusing and improving knowledge built-up by others. Ideally, scholars will document and share the methods used to come to the insights they report in academic publications.
Although sharing of methodological details can be now done at a much lower effort than in Newton's era due to widely available digital technologies, there remains a significant lack of sharing of computational models. There have also been broader concerns raised about the reproducibility of computational sciences (Barnes, 2010; Peng, 2011; Hutton et al., 2016). For starters, authors do not share their source code in the majority of publications (Collberg and Proebsting, 2016; Janssen, 2017). Even if source code is provided, it is not always a given that it can be compiled and executed due to changing dependencies, or that the published results can be exactly reproduced. A computational model's dependencies are the system packages and libraries that a researcher's code rely on, which often continue to evolve in ways such that future versions are backwards incompatible, causing errors in compilation or execution or worse, affecting the outputs of a model (Bogart et al., 2015).
In this article, we analyze a dataset of 7500 articles on individual and agent-based models. An earlier version of this catalog examined 2300 publications and found that just 10% of these publications on agent-based models provided information about the source code used to generate the results (Janssen, 2017). We have now extended this database to agent-based and individual-based models, covering 7500 publications among more than 1500 academic journals. The new version goes beyond the documentation of code sharing practices in publications as we now contact authors to request that they share their code. The database includes metadata on where code is being made available and how the model was documented. Authors are sent an email and can provide feedback and update the database with metadata about where the model code can be currently found, whether it is the original implementation or a replication, even if the model code was not available in the original publication.
In the rest of the paper, we describe the database and the methodology to add select relevant articles and add metadata. We also present findings of an analysis of this dataset. Subsequently, we discuss the next steps of the development and use of the database.
2. Methodology
For more than a decade, we have been involved in creating an archive where scholars can easily deposit computational model code and documentation and make them accessible to others (Rollins et al., 2014). The CoMSES Model Library has more than 780 publicly available models with over 40,000 downloads a year, but we are cognizant that building a tool does not mean that people will use it. With that in mind, we began to investigate the practice of model code-sharing in academic publications. We focus on agent-based and individual-based models, a subset of computational models, because of our expertise in this domain. The goal was to map the code sharing practices and contact authors to increase the availability of code of published models.
Another primary goal of this database is to develop an ecosystem of references to publications and model code that enable scholars to easily find relevant models and model code, and build on existing work instead of reinventing the wheel. We have designed it to be open access so that anyone interested can easily find information about previously published models, and we also allow users to provide feedback and improve the content. We have bootstrapped the contents of this database by manually inspecting the publications on agent based and individual based models and entering categorical metadata on model code availability and documentation for these publications.
There are many places and formats in which model code can be shared – an appendix of a journal article, an author's personal or institutional webpage, a GitHub repository, or a digital repository like the CoMSES Net Model Library. Publications often mention that code is available upon request, which we do not include in our analysis since the code is not being made publicly available. Other researchers have also found that such requests are typically not honored (Stodden et al., 2018). Sharing code on a personal or institutional website or as an appendix of a journal is a good start, but it is not a good long-term solution. We believe that the only acceptable long-term solution is for model code to be documented and archived in a citable, trusted digital repository that adheres to the FORCE11 Software Citation Principles (Smith et al., 2016). Deposited model code should receive a permanent identifier e.g., a Digital Object Identifier (DOI) that should be used as a software citation in publications that reference the given model – this permanent identifier should resolve to a durable web landing page with descriptive metadata and links to files corresponding to the exact version of the computational model that produced the results claimed in the publication. Publishing code in a GitHub repository and referencing the GitHub repository as a citation is not an acceptable solution as it does not make clear which version of the code in the repository was actually used for the publication, and GitHub repositories do not provide any guarantees of permanence and can be deleted by their owner at will. That said, GitHub repositories can be integrated with a trusted digital repository like Zenodo (2020) to create a citable software archive. Trusted digital repositories provide guarantees for digital permanence and have backup and contingency plans for migrating content should the repository cease operations (Lin et al., 2020).
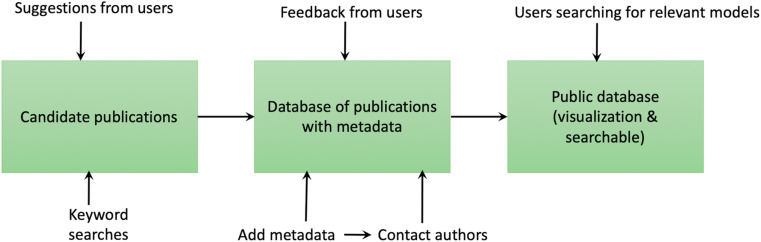
After creating a database of model publications and links to model code, we contacted authors to inform them about best practices on archival of model code (Fig. 1 ). We also asked them to check the metadata of their publication(s) in our database and requested that they provide us with information on the availability of their model code if it was one of the about 85% of publications that were tagged as having not made their code available (i.e., links to the model code were not explicitly referenced within the publication or the links were dead).
Fig. 1.
Workflow of to update the database.
This database is currently live, and new publications continue to be added by using keyword searches and user recommendations (https://catalog.comses.net/). Our team verifies user submissions for relevance to ensure that user submitted publications are appropriate and adds their metadata to this living dataset. Users can also explore the searchable database and visualize basic trends.
3. Creating the database
To seed the initial version of this database with relevant publications and high-quality metadata, including citation references to other publications, we made use of the ISI Web of Science. We used the search term “agent-based model*” and “individual-based model*” several times to build our database, with the last update on August 31, 2019. The terms “agent-based model*” and “individual-based models*” could be used in the title, abstract, or keywords. All publications were evaluated to verify that it was about an agent-based or individual-based model. Reviews, conference abstracts, analytical models, or presented conceptual models were discarded. This resulted in 7500 publications that report on a model and results of model simulations.
For each publication, we checked whether the model code was made available. If so, we tested whether the URL was still available, classified the kind of model code sharing used, and stored the URL. Types of model sharing we distinguish include archives (e.g., the CoMSES Model Library, Zenodo, Open Science Framework), web-based version control repositories (e.g., GitHub, BitBucket, SourceForge), Journal, Personal or Organizational (e.g., Dropbox, Google Drive, a personal or institutional webpage), and framework websites (e.g., the NetLogo Modeling Commons, Cormas).
We rely on the information we found in the publication, and we recognize that the model code may have been published online but not mentioned in the article or can be provided by authors upon request. This is one of the reasons we contact the authors in the latter steps of our workflow.
After we determined whether the model publication provided access to the model code, we recorded which programming platform or language was used, and which external sponsors funded the research. Finally, we used the classification from Müller et al. (2014) to record how the model was described in the main article and appendixes:
-
•
Narrative. How was the model description organized? Did it use a standard protocol called Overview-Design-Details (ODD) (Grimm et al., 2006, 2020), or did it use a non-prescriptive narrative?
-
•
Visualized Relationships. How were the relationships visualized? Did it include flow charts, a Unified Modeling Language (UML) diagram, or provide an explicit depiction of an ontology that describes entities and their structural interrelationships.
-
•
Code and formal description. How were the algorithmic procedures documented? Did the authors provide the source code? Did they describe the model in pseudocode or use mathematical equations to describe (parts) of the model?
One benefit of using the ISI Web of Science is the inclusion of references for each article. This information was included in our database, and as such, we will be able to perform citation and network analysis of the database.
4. Results
We present the basic statistics of the current version of the database containing 7500 publications on individual and agent-based models. Those articles were published in more than 1500 journals, which demonstrate the breadth of the application of this type of modeling, but also the fragmentation of the field.
Fig. 2 shows the number of publications in our database for the date of publication. In Fig. 2, we also show the fraction of publications of which code is available. From the whole database, we have 840 publications for which code is available, 11.2% of all articles. The increasing fraction of papers where code is available and the rapid increase of the absolute number of publications a year demonstrate the exponential growth of the number of models for which code is available.
Fig. 2.
Number of publications over time, as well as the fraction of publications for which code is available.
Table 1 lists the most popular journals in the database (See also Appendix 1 for additional descriptive statistics). Those include a substantial number of ecology journals that report on findings from individual-based models and complexity related journals where the use of individual-based and agent-based modeling is a standard method. There is a substantial variation in the percentage of papers that share their model code. The Journal of Artificial Societies and Social Simulation, an open-access journal that recommends sharing model code, has the highest level with 45%. Meanwhile Physica A does not provide any details on model code and relies on mathematical descriptions of their models being sufficient for replication.
Table 1.
Model code availability of the 15 most popular journals in the database.
| Rank | Name | Number publications | % code available |
|---|---|---|---|
| 1 | Ecological Modeling | 498 | 14.9% |
| 2 | PLOS ONE | 298 | 23.5% |
| 3 | Journal of Artificial Societies and Social Simulation | 250 | 45.6% |
| 4 | Physica A | 183 | 0% |
| 5 | Journal of Theoretical Biology | 175 | 8.0% |
| 6 | Environmental Modeling & Software | 90 | 37.8% |
| 7 | American Naturalist | 77 | 26.0% |
| 8 | Marine Ecology Progress Series | 69 | 2.9% |
| 9 | Advances in Complex Systems | 65 | 15.4% |
| 10 | Proceedings of the Royal Society B | 58 | 13.8% |
| 11 | Canadian Journal of Fisheries an Aquatic Sciences | 57 | 0% |
| 12 | Journal of Economic Dynamics & Control | 53 | 7.5% |
| 13 | Ecological Applications | 53 | 5.7% |
| 14 | Computers Environment and Urban Systems | 52 | 7.7% |
| 15 | Scientific Reports | 49 | 20.4% |
All publications describe in one way or another the model for which they report computational results. Using the classification from Müller et al. (2014), we try to get an overview of how models are described in the publications. Note that we did not evaluate how well the model was described, as such an exercise would be beyond the scope of our efforts. Table 2 shows that mathematical descriptions and flow charts are popular ways to describe a model. Since we distinguish 9.5% of all publications which use the ODD protocol of Grimm et al. (2006) and additional ODD variations, and those who do not use the ODD protocol, we provided in Table 2 information on potential differences. If scholars use the ODD protocol, they are more likely to use flow charts and pseudocode and more likely to share the code. Those correlations do not imply causation of using more ways to describe models and to share model code, as they may relate to the attributes of scholars adopting the ODD protocol. Moreover, we found that many publications which use the ODD protocol in name did not provide a detailed description of the model, which is acknowledged by Grimm et al. (2020) as one of the challenges of the adoption for the ODD protocol.
Table 2.
Methods to describe a model in publication (including appendices). We distinguish between publications using the ODD or not the ODD protocol to describe the model.
| ODD | No ODD | |
|---|---|---|
| Mathematical description | 54.1% | 56.5% |
| Flow charts | 56.8% | 31.7% |
| Pseudo code | 12.2% | 6.8% |
| Source code | 32.8% | 8.9% |
| UML | 4.1% | 1.4% |
Although all publications report simulation results, the majority of the publications do not mention which modeling platform or programming language was used for their study. From 42.9% of the articles which provided information, Table 3 lists the most popular modeling languages. It is no surprise that NetLogo and Repast are popular platforms since they are specifically targeted for individual-based and agent-based modeling. General modeling languages such as R and Matlab are also used frequently due to the wide application in modeling communities. Basic programming languages like C and Java are frequently mentioned, but this is an underrepresentation. For example, a Java model using Repast libraries might be listed as a Repast model based on the references in the article. What might be interesting from Table 3 is the prominence of Python, which is not used in a prominent ABM modeling platform yet, and it's a high percentage of code sharing. Perhaps this reflects the new generation of modelers who adopt new languages and different practices of code sharing. Furthermore, it is notable that there are a substantial number of individual-based and agent-based models implemented in Fortran.
Table 3.
Model code availability for the most common platforms.
| Rank | Simulation Platform | Number of publications | % code available |
|---|---|---|---|
| 1 | NetLogo | 891 | 36.3% |
| 2 | C, C++, C#, Objective C | 432 | 25.2% |
| 3 | Matlab | 297 | 22.2% |
| 4 | Java | 277 | 30.7% |
| 5 | Repast | 243 | 18.1% |
| 6 | R | 242 | 33.5% |
| 7 | AnyLogic | 121 | 8.3% |
| 8 | Python | 110 | 47.3% |
| 9 | Mason | 70 | 21.4% |
| 10 | Fortran | 52 | 32.7% |
| 11 | Swarm | 48 | 10.4% |
| 12 | Cormas | 40 | 25% |
| 13 | Visual Basic | 33 | 24.2% |
| 14 | MatSim | 21 | 0% |
| 15 | Mathematica | 18 | 11.1% |
Whether an article mentions which platform or language is used for the implementation of the simulation model is a predictor of the kind of model description that is given (Table 4 ). Those who do not mention what software is used to implement the model rely on mathematical descriptions. In contrast, more diverse ways of describing the model are used when the implementation software is mentioned. A possible explanation in differences in model descriptions are differences in awareness of the importance of sharing details of the model implementation leading to the published results. Better journal policies could improve the quality of model documentation and code sharing (Stodden et al., 2018).
Table 4.
Methods to describe a model in publication (including appendices). We distinguish between publications mentioning the modeling platform or computer language used to implement the model.
| Platform mentioned | Platform not mentioned | |
|---|---|---|
| Mathematical description | 50.9% | 60.3% |
| Flow charts | 42.7% | 27.7 |
| Pseudo code | 10.1% | 5.2% |
| Source code | 26.1% | 0% |
| UML | 3.0% | 0.7% |
| ODD | 15.8% | 4.8% |
58.9% of the publications mention one or more external sponsors that provided financial support for the research leading to the publication. Increasingly funding agencies require data from the research being made available publicly after the research findings are published. There might be differences in enforcement of compliance, but in general, the level of code availability is very low (Table 5 ). Code is not always seen as research data and might be overlooked in data availability guidelines.
Table 5.
Model code availability for the 15 most common external sponsors.
| Rank | Sponsor | Number of pubs | % code available |
|---|---|---|---|
| 1 | United States National Science Foundation (NSF) | 754 | 14.3% |
| 2 | European Union | 549 | 10.2% |
| 3 | United States National Institutes of Health (NIH) | 326 | 12.3% |
| 4 | National Natural Science Foundation of China | 255 | 5.9% |
| 5 | Natural Sciences and Engineering Research Council of Canada (NSERC) | 158 | 9.5% |
| 6 | German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) | 132 | 13.6% |
| 7 | United Kingdom Engineering and Physical Sciences Research Council (EPSRC) | 123 | 14.6% |
| 8 | United Kingdom Natural Environment Research Council (NERC) | 110 | 9.1% |
| 9 | National Oceanic and Atmospheric Administration (NOAA) | 105 | 1.9% |
| 10 | United States Department of Energy (DOE) | 102 | 6.9% |
| 11 | Australian Research Council (ARC) | 89 | 18.0% |
| 12 | French National Research Agency (ANR) | 87 | 10.3% |
| 13 | German Federal Ministry of Education and Research (BMBF) | 83 | 9.6% |
| 14 | Netherlands Organization for Scientific Research (NWO) | 74 | 10.8% |
| 15 | United States Department of Agriculture (USDA) | 64 | 15.6% |
Where do authors deposit their code as reported in their article? Table 6 demonstrates that there is a wide variety of where the model code is archived. Using public archives is the golden standard, and 236 articles, 3.1% of the database, archives their code in permanent archives. CoMSES is the most commonly used archived, which is not surprising as this archive has targeted this user group. 172 articles have been archived in repositories, especially Github, which provides public access but might not be permanent, and it might not be obvious which version is used for the publication. 261 articles have model code included as an appendix, whether it is a code file or a print of the model code. Frequently those appendixes are publicly available, but not always. 142 articles have their code being stored on their personal websites or even Dropbox links. Those model code depositions are in danger to become missing in the coming years since they are not sustainable solutions. Finally, 30 articles have their model code is available in a library maintained by one of the platforms.
Table 6.
Locations where articles archive their model code. We see 0.5 for a few options since some articles use different type of locations to store their code, and when an article uses both the CoMSES Model Library and the NetLogo Modeling Commons, we give them both 0.5 points.
| Category | Sub category | Frequency |
|---|---|---|
| Archive | CoMSES | 165.5 |
| Datadryad | 26 | |
| Figshare | 20 | |
| Zenodo | 14 | |
| Open Science Framework | 5 | |
| University archives | 3 | |
| Data verse | 2 | |
| Repository | Github | 141 |
| Sourceforge | 14 | |
| Bitbucket | 8 | |
| Gitlab | 5 | |
| Google code | 4 | |
| Journal | Journal | 261 |
| Personal or Organizational | Own website | 126 |
| Dropbox | 5 | |
| Researchgate | 2 | |
| Googledrive | 3 | |
| Amazon Cloudfront | 6 | |
| Platform | Netlogo | 19.5 |
| Cormas | 6 | |
| R Cran | 4 |
Fig. 3 shows the fraction of the publications storing the code in different locations. We see a rapid increase in archives and repositories in recent years. These are much preferred to the alternatives, but we still see a stable share of publications storing their code as appendices of journals and personal websites.
Fig. 3.
Stacked shared of type of location where code is stored.
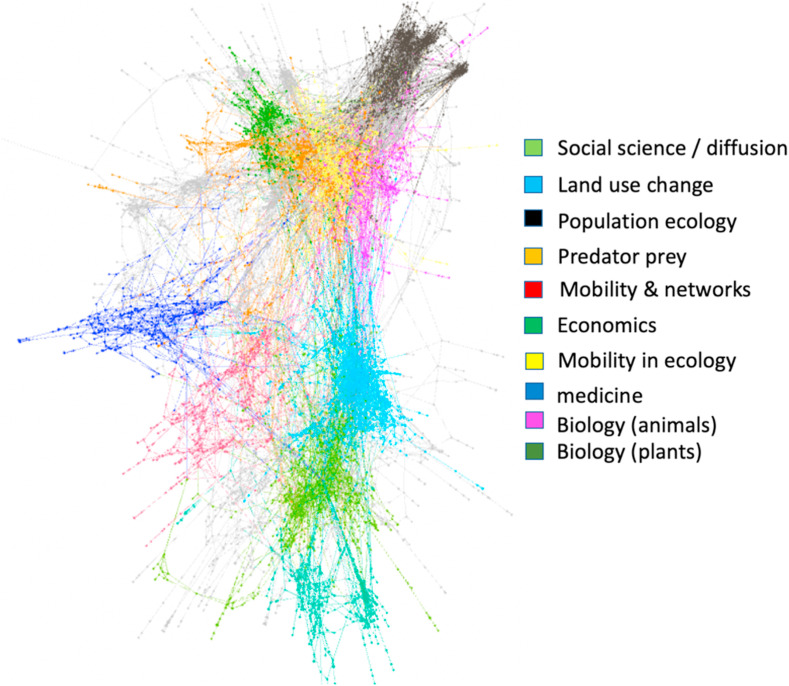
We now look at the 7500 articles from a network perspective. The links refer to references of one publication to another publication. Using the ForceAtlas2 algorithm in the Gephi software, we visualized the network of publications (Jacomy et al., 2014). This visualization of the network is to uses a modularity algorithm to find groups of nodes that are more connected with each other than others (Blondel et al., 2008). Fig. 4 shows the nodes of the 10 biggest clusters. Looking at the topics of the publications in the clusters that are cited most, we identify a topic for each of the clusters. We see that the top of the network is dominated by ecology-oriented models, and economics at the bottom of the network. There is also a large medicine application cluster at the left of the network.
Fig. 4.
Network visualization with nodes colored for the main clusters in the network.
As a final step in the analysis, we ask whether the way scholars document their models and share their code affects whether their model is used. A common question we get on our efforts to document code sharing is whether model papers who share their code get cited more. There are many reasons papers get cited, so it would be remarkable if code sharing will have a significant impact.
With our current database we have we cannot test whether there is an increase of general citations due to code sharing. However, we can evaluate whether there are characteristics for publications in the database which explain why they have more citations by other model papers. We are aware that a citation does not mean a reuse of the model or an endorsement of the quality of the model in the cited publication.
In order to evaluate whether there is any effect of citation by other model papers within the database, we performed a linear regression to explain the number of citations as a function of various factors, and we find that it is the way the model is described, not the code availability, that impacts the number of citations (Table 7 ). This is in line with the Milkowski et al. (2018).
Table 7.
Regression analysis of the number of citations as function of a number of dependencies. Each cell contain the estimate of the coefficient in a linear regression and the standard deviation. *p < 0.05; **p < 0.01; ***p < 0.001.
| Variable | Estimates | Estimates |
|---|---|---|
| constant | 579.331 (18.434)*** | 590.421 (18.553)*** |
| Number of authors | 0.107 (0.022)*** | 0.105 (0.022)*** |
| Year publication | −0.287 (0.009)*** | −0.293 (0.009)*** |
| Code available | 0.154 (0.149) | −0.163 (0.164) |
| Flow charts | 0.356 (0.100)*** | |
| Mathematical equations | 0.404 (0.094)*** | |
| Pseudocode | −0.057 (0.179) | |
| Language/Platform mentioned | 0.399 (0.105)*** | |
| ODD | 0.489 (0.165)** | |
| N | 7500 | 7500 |
| R2 | 0.1178 | 0.1252 |
Since the rapid increase in the number of papers in recent years as well as the fraction of publication for which model code is available, we have to be careful with this analysis. It takes some time for publications to gain citations due to the slow nature of publications. Hence the year of publication is the best predictor for the number of citations. Beyond that, better documentation is a good predictor. In a few years, this analysis should be repeated if a larger number of model publications are available with years of model code availability. Doing such an analysis might focus on a specific field, such as ecological modeling, since there are different citation cultures among different fields of study. In such a future analysis in a specific field, an a control variable might be included for the reputation or impact factor of the journal where the article is published.
5. Discussion
Results published from computational science should, in principle, be reproducible, and the code should be available to be reused and built on. Unfortunately, this is not what is happening. We looked at the practice of documenting agent-based and individual-based models and how code is made available. We see an increase in the fraction of publications for which code is available (about 18% in 2018). This is a promising trend, but since in many cases those models are available on a personal website or as an appendix to the journal article (only available for subscribers), current availability does not mean availability in the long run. Only about 3% of the model publications in recent years archive their model in public archives where the focus is on the preservation of the code in the longer term.
The importance of proper documentation and code availability is illustrated during the COVID-19 pandemic (Squazzoni et al., 2020; Barton et al., 2020). A large number of several projections of the COVID-19 crisis are difficult to put into context since there is no transparency about the underlying assumptions. Since policymakers are primarily guided in their decision making in the COVID-19 crisis by model results, the lack of practicing model transparency is disappointing.
Some scholars may argue that this extra level of archiving and documenting is too much of extra work, and if models would be useful, their code will become available somehow. Many articles indeed mention that code is available upon request to the authors. Since the Fall of 2019, we started sending out emails to corresponding authors to request code used in their publications, and all corresponding authors in our database an email has been send to. In this email, we provide information on our project, best practices on model archiving and list which publications are in our database, and whether we have been able to locate the model code or not (Appendix 2). For less than 1% of the emails, we received a response. Some responses included URLs where code is currently available, and other responses indicated that the author no longer had access to the code.
Increasing use of empirical data of human subjects to make empirical-grounded agent-based models could be a concern for sharing model code on models of human social dynamics. However, sharing processed data about human subjects is a common practice in the social science (Yoon and Kim, 2017). There are ethical and moral objections raised to share data in the social sciences, and alternative policies for data sharing are part of the debate (Mauthner and Parry, 2013; Tenopir et al., 2015). There is not one policy that may work for all types of data or code.
What can we to do to improve to model documentation and code availability so we can build on the work of others? There is no lack of technical solutions, such as public archives. Journals and sponsors also start to improve their requirements, but those requirements could be more precise about code availability. At COMSES Net, we have created an open code badge that various journals have begun to adopt to signal best practices and provide durable links to model code (https://www.comses.net/resources/open-code-badge/).
Although we see some positive trends in the fraction of code availability in publications, the transition towards a desirable level of practice will be a long-term process. It requires a change in norms and habits. In fact, research on adoption of data sharing in the social sciences suggest that personal motivations and perceived normative pressure are more critical in the data sharing behavior than availability of repositories or pressures from funding agencies and journals (Kim and Adler, 2015). The new generation of scholars using computational models must be trained in the tools available to derive best practices. Besides code sharing and documentation, this should include containerization (e.g. Docker), which can preserve software dependencies, to be able to reproduce computational results in the longer term. This could be integrated with education by letting students work in groups and build on existing model code available.
In sum, there is a slow process of improvement in code availability in the field of agent-based and individual-based models. Still, more concerted action is needed to improve the adoption of best practices. This could be done by enforcement of best practices by journals and sponsors, and by making this topic a common topic in modeling graduate courses and beyond.
Data availability
The data from the 7500 articles explored in this article are available at https://osf.io/y2vs3/?view_only=c0ec40bd9de54e1b88e1438656fb84f0.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Rachael Gokool, Yee-Yang Hsieh, Juan Rodriguez, and Christy Contreras for entering metadata, for Christine Nguyen and Dhuvil Patel for software development, and Cindy Huang for contacting corresponding authors. We acknowledge financial support for this work from the National Science Foundation, grant numbers 0909394 and 1210856.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envsoft.2020.104873.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Barnes N. Publish your computer code: it is good enough. Nature. 2010;467:753. doi: 10.1038/467753a. [DOI] [PubMed] [Google Scholar]
- Barton M., Alberti M., Ames D., Atkinson J.A., Bales J., Burke E., Chen M., Diallo S.Y., Earn D.J., Fath B., Feng Z., Gibbons C., Heffernan J., Hammond R., Houser H., Hovmand P.S., Kopainsky B., Mabry P.L., Mair C., Meier P., Niles R., Nosek B., Nathaniel O., Suzanne P., Polhill G., Prosser L., Robinson E., Rosenzweig C., Sankaran S., Stange K., Tucker G. Call for transparency of COVID-19 models. Science. 2020;368:6490. doi: 10.1126/science.abb8637. [DOI] [PubMed] [Google Scholar]
- Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. Theor. Exp. 2008;10:P10008. doi: 10.1088/1743-5468/2008/10/P10008/. [DOI] [Google Scholar]
- Bogart C., Kästner C., Herbsleb J. 2015 30th IEEE/ACM International Conference on Automated Software Engineering Workshop. 2015. When it breaks, it breaks: how ecosystem developers reason about the stability of dependencies; pp. 86–89. [Google Scholar]
- Collberg C., Proebsting R.A. Repeatability in computer systems research. Commun. ACM. 2016;59(3):62–69. [Google Scholar]
- Grimm V., Berger U., Bastiansen F., Eliassen S., Ginot V., Giske J., Goss-Custard J., Grand T., Heinz S.K., Huse G., Huth A., Jepsen J.U., Jørgensen C., Mooij W.M., Müller B., Pe'er G., Piou C., Railsback S.F., Robbins A.M., Robbins M.M., Rossmanith E., Rüger N., Strand E., Souissi S., Stillman R.A., Vabø R., Visser U., DeAngelis D.L. A standard protocol for describing individual-based and agent-based models. Ecological Modeling. 2006;198:115–126. [Google Scholar]
- Grimm V., Railsback S.F., Vincenot C.E., Berger U., Gallagher C., DeAngelis D.L., Edmonds B., Ge J., Giske J., Groeneveld J., Johnston A.S.A., Milles A., Nabe-Nielsen J., Polhill J.G., Radchuk V., Rohwäder M.-S., Stillman R.A., Thiele J.C., Ayllón D. The ODD protocol for describing agent-based and other simulation models: a second update to improve clarity, replication and structural realism. J. Artif. Soc. Soc. Simulat. 2020;23(2):7. doi: 10.18564/jasss.4259. http://jasss.soc.surrey.ac.uk/23/2/7.html [DOI] [Google Scholar]
- Hutton C., Wagener T., Freer J., Han D., Duffy C., Arheimer B. Most computational hydrology is not reproducible, so is it really science? Water Resour. Res. 2016;52:7548–7555. doi: 10.1002/2016WR019285. [DOI] [Google Scholar]
- Jacomy M., Venturini T., Heymann S., Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PloS One. 2014;9(6) doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen M.A. The practice of archiving model code of agent-based models. J. Artif. Soc. Soc. Simulat. 2017;20(1):2. doi: 10.18564/jasss.3317. http://jasss.soc.surrey.ac.uk/20/1/2.html [DOI] [Google Scholar]
- Kim Y., Adler M. Social scientists' data sharing behaviors: investigating the roles of individual motivations, institutional pressures, and data repositories. Int. J. Inf. Manag. 2015;35(4):408–418. [Google Scholar]
- Lin D., Crabtree J., Dillo I., Downs R.R., Edmunds R., Giaretta D., De Giusti M., L'Hours H., Hugo W., Jenkyns R., Khodiyar V., Martone M.E., Mokrane M., Navale V., Petters J., Sierman B., Sokolova D.V., Stockhause M., Westbrook J. The TRUST Principles for digital repositories. Scientific Data. 2020;7:144. doi: 10.1038/s41597-020-0486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkowski M., Hensel W.M., Hohol M. Replicability or reproducibility? On the replication cirses in computational neuroscience and sharing only relevant detail. J. Comput. Neurosci. 2018;45(3):163–172. doi: 10.1007/s10827-018-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Balbi S., Buchmann C.M., de Sousa L., Dressler G., Groeneveld J., Klassert C.J., Bao Le Q., Millington J.D.A., Nolzen H., Parker D.C., Polhill J.G., Schlüter M., Schulze J., Schwarz N., Sun Z., Taillandier P., Weise H. Standardised and transparent model descriptions for agent-based models: current status and prospects. Environ. Model. Software. 2014;55:156–163. [Google Scholar]
- Mauthner N.S., Parry O. Open access digital data sharing: principles, policies and practices. Soc. Epistemol. 2013;27(1):47–67. [Google Scholar]
- Peng R.D. Reproducible research in computational science. Science. 2011;334:1226–1227. doi: 10.1126/science.1213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins N.D., Barton C.M., Bergin S., Janssen M.A., Lee A. A computational model library for publishing model documentation and code. Environ. Model. Software. 2014;61:59–64. [Google Scholar]
- Smith A.M., Katz D.S., Niemeyer K.E., Force11 Software Citation Working Group Software citation principles. PeerJ Computer Science. 2016;2:e86. doi: 10.7717/peerj-cs.86. [DOI] [Google Scholar]
- Squazzoni F., Polhill J.G., Edmonds B., Ahrweiler P., Antosz P., Scholz G., Chappin É., Borit M., Verhagen H., Giardini F., Gilbert N. Computational models that matter during a global pandemic outbreak: a call to actions. J. Artif. Soc. Soc. Simulat. 2020;23(2):10. doi: 10.18564/jasss.4298. http://jasss.soc.surrey.ac.uk/23/2/10.html [DOI] [Google Scholar]
- Stodden V., Seiler J., Ma Z. An empirical analysis of journal policy effectiveness for computational reproducibility. Proceedings of the National Academy of Sciences USA. 2018;115(11):2584–2589. doi: 10.1073/pnas.1708290115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenopir C., Dalton E.D., Allard S., Frame M., Pjesivac I., Birch B., Pollock D., Dorsett K. Changes in data sharing and data reuse practices and perceptions among scientists worldwide. PloS One. 2015;10(8) doi: 10.1371/journal.pone.0134826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon A., Kim Y. Social scientists' data reuse behaviors: exploring the roles of attitudinal beliefs, attitudes, norms, and data repositories. Libr. Inf. Sci. Res. 2017;39(3):224–233. [Google Scholar]
- Zenodo . 2020. Making Your Code Citable.https://guides.github.com/activities/citable-code/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from the 7500 articles explored in this article are available at https://osf.io/y2vs3/?view_only=c0ec40bd9de54e1b88e1438656fb84f0.