Abstract
Isolation of exosomes from biological samples provides a minimally-invasive alternative for basic understanding, diagnosis, and prognosis of metastatic cancers. The biology and clinical values of exosomes are under intensive investigation, yet most studies are limited by technical challenges in recovering these exosomes with heterogeneous sizes and cargos from biological samples. We report a novel method based on “particle ferrohydrodynamics” and its associated microfluidic device, termed as the FerroChip, which can separate exosome-like nanoparticles from microliters of cell culture media and human serum in a label-free, continuous-flow and size-dependent manner, and achieves a high recovery rate (94.3%) and a high purity (87.9%). Separated exosome-like nanoparticles had diameters, morphology and protein expressions that were consistent with other reports. This method, upon further molecular characterization, could potentially facilitate basic understanding of exosomes and its clinical application in blood liquid biopsy.
Graphical Abstract

Particle ferrohydrodynamics and its device (FerroChip) enables label-free and size-dependent separation of exosome-like nanoparticles with high recovery rate and purity.
Introduction
Progress on understanding the communication between tumor cells and their microenvironments are crucial for the development of new diagnostic methods and therapeutic strategies.1–3 Such communication, traditionally known to occur through direct cell-cell contacts and soluble factor secreted by tumor cells, is now revealed to have an additional mechanism involving extracellular vesicles (EVs).4, 5 EVs consist of a heterogeneous population of lipid-encapsulated vesicles that transmit functional proteins and nucleic acids, with multiple subtypes including exosomes that are extracellular membrane vesicles of endosomal origin, having a physical diameter typically within the range of 30 – 150 nm.4–7 Other subtypes of EVs include microvesicles that are shed directly by budding from the cellular plasma membrane, with a physical diameter ranges from 100 – 1000 nm.4–7 Growing evidence shows that EVs participate in the metastatic spreading of cancers, in which tumor-derived EVs aid the establishment of pre-metastatic niche and facilitates tumor progression.8–13 As a result, circulating tumor exosomes containing tumor-specific molecular messages could hold promising clinical utilities as next-generation diagnostic and prognostic biomarkers in liquid biopsy.4, 8, 9
Despite the rapid progress in the understanding of the biology, function, and clinical utilities of extracellular vesicles, the heterogeneous sizes and cargos of these nanoscale vesicles and the technical limitations in separating pure exosomal subpopulation have hindered the characterization of their molecular signatures and its clinical utility. Various strategies have been developed to isolate EV subpopulations, especially exosomes. These strategies include benchtop methods that rely on either differential ultra-centrifugation or polymer-based precipitation. Even though these methods can handle milliliters to liters of samples, the recovery rate, purity and integrity of separated exosomes, as well as their labor-intensiveness and long processing time remain to be improved.6 Microfluidic methods emerged as a promising alternative to the benchtop methods in separating exosomes in microliters to milliliters of samples, with high purity or recovery rate while keeping the cost affordable and the processing time short. Technologies of microfluidic exosome separation can be divided into two categories, with the label-based ones relying on surface markers of exosomes for selective separation of specific exosomal subpopulations with high purity.14–20 On the other hand, label-free separation explores the size difference between exosomes and other EVs, including deterministic lateral displacement,21 pinched-flow fractionation,22 asymmetric-flow field-flow fractionation,23 filtration,24–28 nanowire trapping,29, 30 viscoelastic separation,31 and acoustic separation.32 Label-free separation of exosomes was not biased by the use of specific biomarkers and therefore could lead to exosome separation with high recovery rate. In this paper we report a new label-free ferrohydrodynamic method for high recovery and purity separation of exosome-like nanoparticles, which are defined as having diameters of 30 – 150 nm, and morphology and protein expressions consistent with exosomes. This method relies on the physical principle of “particle ferrohydrodynamics” and can separate exosome-like nanoparticles from biological samples with a 94.3% recovery rate and 87.9% purity.
Ferrohydrodynamics describes the mechanics and motion of a magnetizable liquid (e.g., ferrofluids) influenced by strong forces of magnetic polarization.33 In the context of this paper, we explore “particle ferrohydrodynamics” that refers to the mechanics and motion of an immersed diamagnetic object (either a solid particle or an extracellular vesicle) with close to zero magnetic susceptibility in a magnetizable liquid under an externally applied magnetic field. Particle ferrohydrodynamics is a physical process that drives the immersed object’s movement as a result of the interaction between the magnetic field and the magnetic liquid surrounding the object. The magnetic liquid itself, often a ferrofluid, consists of a colloidally stable suspension of magnetic nanoparticles. Under a non-uniform magnetic field, these magnetic nanoparticles exhibit a gradient of particle concentration while maintaining its stability against irreversible agglomeration thanks to the surfactants on their surfaces. In the particle ferrohydrodynamic process, the magnetic nanoparticles within a ferrofluid continuously collide with the immersed diamagnetic object and generate pressure across the object’s surface. This leads to an imbalance of pressure across the object’s surface, and a net movement of the object in the opposite direction of nanoparticle concentration, as well as the magnetic field gradient. This phenomenon of an immersed diamagnetic object moving in the opposite direction of the magnetic field gradient is referred to as “diamagnetophoresis” or “negative magnetophoresis” in the literature.34 Particle ferrohydrodynamics was applied in microfluidic systems for micron-sized particle and cellular manipulation, such as isolating extremely rare circulating tumor cells from cancer patients’ blood.35, 36 However, ferrohydrodynamic particle manipulation was thought to be limited to micron-sized objects, because the carrier magnetic liquid consist of nanoparticles with a diameter of ~10 nm. It was thought that the size of the immersed object needed to be significantly larger than the magnetic nanoparticles themselves. Indeed, the smallest objects that were experimentally manipulated in ferrofluids to date was limited to 1 μm, such as polymer particles of 1 μm in diameter,37 and Escherichia coli cells with the short axis of 0.5 – 1 μm and the long axis of 2 – 4 μm.38 In this paper, we successfully demonstrated a label-free ferrohydrodynamic method that can separate exosome-like particles with diameters of 30 – 150 nm from biological samples. To the best of our knowledge, this is the first time that nanosized particles were separated via “particle ferrohydrodynamics” in microfluidic devices.
The rest of the paper is structured as following. We first calculated the theoretical limit of the smallest particles that could be manipulated via particle ferrohydrodynamics, and determined the feasibility of applying particle ferrohydrodynamics in microfluidic setting for exosomal separation. We then developed a prototype device termed as the FerroChip, performed systematic optimization of key factors influencing the performance of the FerroChip, and determined parameters for high purity and recovery rate exosomal separation. Finally, we challenged the FerroChip with both cell culture media and human serum for its ability to separate exosome-like nanoparticles.
Results and discussion
Ferrohydrodynamic nanoparticle manipulation
We estimate the smallest nanoparticle that can be ferrohydrodynamically manipulated by comparing quasi-static effects originated from thermal diffusion and ferrohydrodynamic motion, assuming that (1) ferrofluids are a continuous magnetic medium without significant phase separation under an externally applied magnetic field, and (2) the concentration of ferrofluids is small enough so that interparticle interaction can be neglected (Figure 1a). Ferrofluids at room temperature are colloidal suspensions of magnetic nanoparticles that are stable against externally applied magnetic fields because the magnetic nanoparticles are on the order of 10 nm in diameter. Ferrofluids used in this study had a volume fraction of magnetic materials of 0.3%, therefore they could be considered non-interacting. The theoretical estimate considers a one-dimensional case with a ferrofluid suspension of height L, and immersed particles uniformly dispersed along L. The characteristic time of an immersed diamagnetic object migrating across L is determined by the shorter of either diffusive time τD or ferrohydrodynamic migration time τM. The diffusion time of the immersed object in ferrofluids is τD= L2 / 2D, where is the diffusion coefficient, d is diamagnetic object’s diameter, η is ferrofluid viscosity, T is temperature, and kB is Boltzmann constant. Ferrohydrodynamic migration time of the same diamagnetic object is determined by the Stokes velocity through solving 3πηdvM + FM = 0, where FM = −μ0V(M · ∇)H is ferrohydrodynamic force acting on the particle, μ0 is permeability of free space, V is object’s volume, M is non-linear magnetization of the dilute ferrofluid that is typically modeled through a Langevin function, and H is magnetic field strength at the center of the particle. This yields a characteristic time associated with ferrohydrodynamic motion τM = l × L/ D, where has the dimension of length and is indicative of the spatial scale of ferrohydrodynamic migration. The total characteristic time τ of the diamagnetic object migrating across length L is thus . The faster process, whether it is diffusive or ferrohydrodynamic, decides the actual time τ. When τD = τM, one obtains the critical diameter of the diamagnetic object that experiences both diffusion and ferrohydrodynamic migration equally, which is .
Figure 1.
Overview of the label-free ferrohydrodynamic nanoparticle separation principle and the FerroChip device design. a (top panel) Schematic of an extracellular vesicle (EV, 30 – 1000 nm in diameter) experiencing both diffusion and “particle ferrohydrodynamics” in a colloidally-stable magnetic nanoparticle (~10 nm particle diameter) suspension (i.e., ferrofluids). The magnetization of the unlabeled EV MEV is near zero and much less than its surrounding ferrofluids Mferrofluid. The ferrohydrodynamic force on the EV is generated from magnetic nanoparticle-induced pressure imbalance on the vesicle’s surface, which is proportional to the EV’s volume. The color bar indicates the relative amplitude of the magnetic field strength. Red arrows show the direction of vesicle movement, small black arrows on the EV’s surface show the direction of magnetic nanoparticle-induced surface pressure. (bottom panel) The relationship between characteristic migration time of a diamagnetic nanoparticle in ferrofluids and the diameter of the nanoparticle. The migration of nanoparticles is affected by both diffusion and “particle ferrohydrodynamics”, and determined by the faster process out of the two. For nanoparticles with a diameter of less than ~30 nm, the diffusive process dominates the migration; for nanoparticles with a diameter of larger than ~30 nm, the ferrohydrodynamic process dominates the migration. b Work principle of a label-free continuous-flow EV focusing/separation device, termed as FerroChip. In the focusing mode of device operation, samples of EVs were premixed with a dilute ferrofluid and entered a straight microchannel at a relatively slow flow rate and with a uniform distribution across the channel width. A symmetric magnetic field with its minimum in the middle of the microchannel was used to direct EVs towards the center of the microchannel, effectively focusing them into a narrow stream. In the separation mode, EVs entered the channel predominately through the regions close to the channel wall due to a sheath flow, with a relatively fast flow rate. The same magnetic field was used to direct unlabeled EVs from the sidewall region towards the channel center for continuous collection. EVs of various sizes migrated towards the center of the microchannel, with varying speeds that depended on their sizes. Large EVs migrated to the channel center at a faster speed than smaller exosomes. Yellow arrows with gradients indicate the distribution of magnetic fields in the microchannel. c Top-view schematic drawing of the FerroChip’s microchannel. Initial samples of nanoparticles and/or EVs were injected into the channel from inlet 1. The samples, after first going through a debris filter that removed large debris, entered a straight channel (labeled as focusing/separation region) which focused or separated nanoparticles and/or EVs based on their sizes. Processed samples were collected from middle and side outlets for characterizations. When the FerroChip operates in focusing mode, inlet 2 (sheath flow) is not used. When the FerroChip operates in separation mode, inlet 2 provides a ferrofluid sheath flow so that nanoparticles and/or EVs entered from the top and bottom walls of the channel. The width, height and length of the microchannel are 1200 μm, 150 μm and 53 mm, respectively. d A photo of a microchannel (left, blue ink indicating the channel geometry) and assembled FerroChip with four permanent magnets in quadrupole configuration inside a holder (right). The microfluidic device and permanent magnets were placed within an aluminum manifold during its operation. Scale bars: 1 cm.
Figure 1a shows that this critical diameter at room temperature (23°C) is ~30 nm, calculated from relevant parameters in a typical microfluidic setting, including a migration distance L = 500 μm, a ferrofluid magnetization M = ~1100 A/m, corresponding to the 0.3% (v/v) maghemite particle based ferrofluid used in this study, and a gradient of magnetic field strength ∇H = ~8×108 A/m2, corresponding to a gradient of flux density ∇B = 1000 T/m. However, Figure 1a shows that for a diamagnetic nanoparticle of 30 nm diameter to migrate across 500 μm distance, the estimated characteristic time would be unrealistically long (on the order of 104 seconds) in a microfluidic device. On the other hand, when the nanoparticle diameter increases to ~100 nm, ferrohydrodynamic motion becomes a much faster process than diffusion, leading to a significant reduction of the characteristic time to the order of ~101 – 102 seconds, making it feasible to manipulate them in microfluidic devices. These theoretical estimates of nanoparticle sizes and characteristic times in particle ferrohydrodynamics, taken together with the constraints from the operation of microfluidic devices, demonstrates that it is possible to ferrohydrodynamically separate diamagnetic nanoparticles that have diameters in the range of 100 – 1000 nm, and also offers guidance on designing optimal microfluidic devices for this purpose.
Overview of FerroChip design and operation
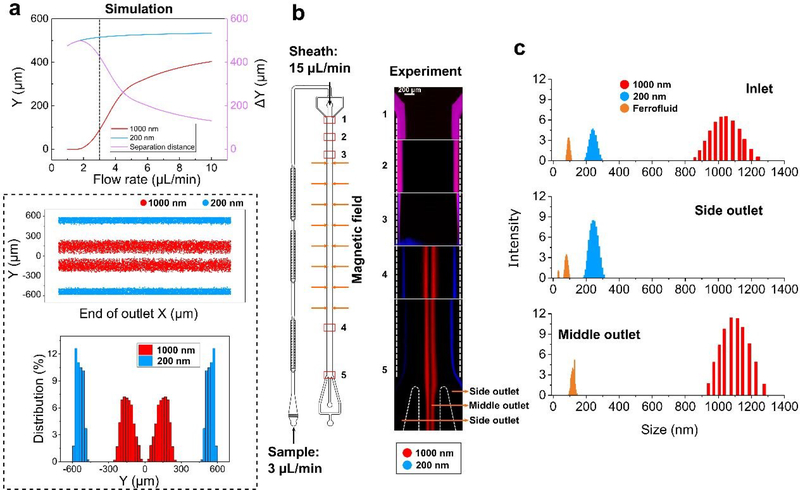
We designed a microfluidic device, termed as FerroChip, which incorporated the particle ferrohydrodynamic working principle to effectively either focus or separate diamagnetic nanoparticles based on their size alone. Figure 1b illustrates the device design of a prototype FerroChip. In the focusing mode of device operation (Figure 1b, top panel), samples of either diamagnetic nanoparticles or extracellular vesicles (EVs) are premixed with a dilute ferrofluid and enter a straight microchannel with a uniform distribution across the channel width. The magnetic fields from a quadrupole array of permanent magnets, the flow velocity of the nanoparticles or EVs, together with the concentration of the ferrofluid, are chosen so that the diamagnetic nanoparticles or EVs will ferrohydrodynamically migrate towards the center of the microchannel regardless of their sizes, effectively focusing all of them into a narrow stream. In the separation mode of operation (Figure 1b, bottom panel), premixed samples of diamagnetic nanoparticles or EVs with ferrofluids enter the straight microchannel through predominately the regions close to the channel wall, due to the effect of a ferrofluid sheath flow. The magnetic fields, and the flow velocity of the nanoparticles or EVs, together with the concentration of the ferrofluid, are chosen so that the ferrohydrodynamic migration of nanoparticles or EVs towards the center of the microchannel depends on their sizes. Larger diamagnetic nanoparticles or EVs migrate with a faster speed while smaller ones with a slower speed, resulting in a spatial separation of the differently sized objects at the outlets of the channel. Figures 1c and 1d show the prototype microchannel that is capable of both nanoparticles and EVs focusing and separation and an assembled FerroChip in use.
The theoretical estimate of size-dependent characteristic time presented in the previous section has provided a framework for understanding the dominant effects in particle ferrohydrodynamics. Ferrohydrodynamic motion of particles becomes the dominant effect at room temperature as the diameter of particles exceeds ~100 nm. With the FerroChip, we intended to apply the particle ferrohydrodynamic principle to focus or separate diamagnetic nanoparticles or EVs in ferrofluids. For that purpose, we characterized the performance of the FerroChip using three metrics, including the nanoparticle/EV-processing throughput, nanoparticle/EV recovery rate, nanoparticle/EV purity after processing, which were consistently used and reported in evaluating exosome isolation techniques.6, 39 For the FerroChip, the parameters affecting these three metrics include device geometry, magnetic field and its gradient, sample flow rates, and ferrofluid properties. These parameters were coupled and needed to be optimized systematically. Hence, we developed a physical model that takes into consideration the effects of particle ferrohydrodynamics in microfluidic settings, which allowed us to optimize relevant device geometries and operating parameters. This physical model predicted three-dimensional (3D) trajectories of diamagnetic nanoparticles or EVs under laminar flow conditions inside a microchannel. Ferrohydrodynamic force and hydrodynamic drag force were considered in simulating the particle trajectories. In our simulation, we chose to neglect the diffusive effect because the diffusive effect starts to become weaker than the ferrohydrodynamic effect when the diamagnetic particle diameter exceeded ~30 nm (Figure 1a). For instance, the diffusive time is ~36 times longer than the ferrohydrodynamic migration time for a diamagnetic particle of 100 nm in diameter. Because the diameter range of the diamagnetic particles in this study is 30 – 1000 nm, we can neglect the diffusive effect in our model while still simulate the particle migration accurately. This model provided analytical and quick design optimization to determine the above-mentioned variables and parameters depending on the design constraints.
Validation of ferrohydrodynamic motion through nanoparticles focusing
We first validated the ferrohydrodynamic manipulation in the FerroChip through focusing nanoparticles of diameters ranging from 100 nm to 1000 nm, with a goal of understanding the size-dependence of ferrohydrodynamic particle migration at relevant sample flow rates (1 – 10 μL/minute). The range of sample flow rates represents typically reported data from existing microfluidic EVs separation technologies.6, 39 The range of particle diameters (100 – 1000 nm) coincides with major populations of EVs that include microvesicles (100 – 1000 nm) and exosomes (30 – 150 nm).5–7
Validation focused on the effects of device geometry, magnetic field, and its gradient, sample flow rates, as well as ferrofluid concentration on the effect of diamagnetic nanoparticle focusing. Firstly, we determined the dimension of focusing region of the microchannel by balancing a need of processing ~100 μL of EVs sample within one hour, and a need to maintain laminar flow in the device. Final microchannel dimensions (55×1.2×0.15 mm, L×W×H) were optimized so that Reynold’s number was on the order of 0.03 when the sample flow rate was 100 μL h−1, ensuring laminar flow condition. The prototype microchannel is shown in Figure 1c. Secondly, the amplitude of ferrohydrodynamic force on diamagnetic nanoparticles is proportional to the amplitude of the magnetic field gradient. In order to maximize the field gradient, we adopted a quadrupole magnet configuration in the FerroChip design that could be optimized to generate the needed magnetic flux density and its gradient. Using four permanent magnets (38.1 mm by 6.35 mm by 6.35 mm, N52 neodymium magnet) in a quadrupole configuration shown in Figure 2a, a magnetic flux density of up to 0.5 T in the x-y plane (z = 0), and a magnetic flux density gradient of 1272 T m−1 in the y-z plane (x = 0) were obtained. Under the magnetic field of the quadrupole magnets, simulation of trajectories of 520 nm polystyrene particles in ferrofluids were conducted to study the effectiveness of FerroChip in focusing these particles. A sample inlet flow rate of 3 μL min−1 and a ferrofluid concentration of 0.3% (v/v) were used in the simulation. Results in Figure 2a show that 520 nm particles could be focused in all three dimensions in the prototype device. Experimental results in Figure 2b confirmed that ferrohydrodynamic effects under this quadrupole magnet configuration were significant enough so that 520 nm particles were successfully focused into a narrow stream in the FerroChip.
Figure 2.
System optimizations of the FerroChip for continuous-flow nanoparticle focusing. a Optimization of high magnetic flux densities and high magnetic flux density gradients in the FerroChip for nanoparticle focusing. Using four permanent magnets (38.1 mm by 6.35 mm by 6.35 mm, N52 neodymium magnet) in a quadrupole configuration shown here, a magnetic flux density of up to 0.5 T in the x-y plane (z = 0), and a magnetic flux density gradient of 1272 T m−1 in the y-z plane (x = 0) were obtained. Under the magnetic field of the quadrupole magnets, simulation of submicron particle trajectories for 520 nm polystyrene beads in ferrofluids in x-y plane (left), x-z plane (middle), and y-z plane (right), were conducted to study the effectiveness of FerroChip in focusing 520 nm diamagnetic beads. A sample inlet flow rate of 3 μL min−1 and a ferrofluid concentration of 0.3% (v/v) were used in the simulation. Results show that 520 nm beads could be focused in all three dimensions in the prototype device. Triangles in the trajectory plots indicate the starting points of particles. b (Left panel) Experimental bright-field images show that 520 nm diamagnetic polystyrene beads inside a microchannel can be focused into a narrow stream in the presence of the ferrofluid and quadrupole magnets, using optimized parameters from a. (Right panel) Experimental fluorescence images confirm the focusing effects on 520 nm polystyrene. A sample inlet flow rate of 3 μL min−1 and a ferrofluid concentration of 0.3% (v/v) were used in b. Scale bar: 200 μm.
The remaining study explored the effect of sample flow rates and ferrofluid concentration on the focusing effect. We defined an output – the width of focused diamagnetic particle streams (see Figure 2a for coordinates) as a way to quantify the focusing effect. This output was optimized using parameters including sample flow rates (1–30 μL min−1) and ferrofluid concentrations (0–0.4% v/v). The goal was to maximize the focusing effects, which translated to minimizing the stream width. Figure 3a shows that simulation and experimental data agreed reasonably well, and both of which indicated that the stream widths had a monotonic relationship with the flow rates. Faster flow rates reduced the residual time of diamagnetic nanoparticles in the microchannel, thereby decreasing the ferrohydrodynamic migration of the particles, which in turn increased the particle stream width. As the size of diamagnetic nanoparticle increased, the width of stream decreased, due to the fact that ferrohydrodynamic force is proportional to the volume of diamagnetic particles. We further optimized the ferrofluid concentration and found that a higher concentration could lead to a higher magnitude of the diamagnetic force on nanoparticles therefore a larger focusing effect (Figure 3b).
Figure 3.
Optimizations of nanoparticle focusing in the FerroChip, with parameters including particle diameter, flow rate, and ferrofluid concentration. a Optimizations of nanoparticles focusing with parameters including nanoparticle diameters and flow rates through both simulations and experiments. Simulation results agreed with experimental results reasonably well. Both confirmed that the ferrohydrodynamic focusing effect on nanoparticles depended on the diameters of particles in ferrofluids. As the particle size increased, the focusing effect became more pronounced. Additionally, as the flow rates in the microchannel increased, the focusing effect decreased. Particle stream width of the y-axis label indicates the width of the focused particle stream. Orange color shaded area indicates the width of the collection outlet in the FerroChip. A panel of fluorescent images captured during the focusing of 100 nm, 380 nm, 520 nm, and 1000 nm polystyrene beads at a flow rate of 5 μL min−1 illustrates that focusing effect depended on particle sizes. b Optimizations of nanoparticles focusing with the parameter of ferrofluid concentration through both simulations and experiments. Simulation results agreed with experimental results reasonably well. Both confirm that the ferrohydrodynamic focusing effect on submicron beads depended on the ferrofluid concentration. As the ferrofluid concentration increased, the focusing effect became more pronounced. A panel of fluorescent images captured during the focusing of 520 nm polystyrene beads at 0.05%, 0.1%, 0.2% and 0.3% (v/v) ferrofluid concentration illustrates that focusing effect depended on ferrofluid concentrations. Flow rate was 5 μL min−1. Scale bar is 200 μm in all images.
Optimization of FerroChip for exosome-like nanoparticles separation
We then optimized FerroChip design and its operating parameters for label-free separation of exosomes in biological fluids, with a goal of isolating exosomes based on their size differences from large extracellular vesicles. The range of diameters of exosomes (30 – 150 nm) was reported to be smaller than that of apoptotic bodies (>1000 nm) and microvesicles (100 – 1000 nm).5–7 We aimed to exploit this size difference between exosomes and other extracellular vesicles for a high recovery rate and purity separation of exosomes with a clinically relevant throughput. In quantitative terms, the performance goals for the FerroChip included: (1) an exosome recovery rate of > 90%; (2) a sample processing throughput of ~1 μL min−1, and (3) a purity of exosome of > 90% after separation. These metrics were chosen as performance targets after a survey of existing microfluidic exosome separation methods (see Table 2).6, 39
Table 2.
A survey of existing microfluidic exosome separation methods
| Type | Methods | Throughput | Recovery rate of exosomes | Purity of recovered exosomes | Biological samples used in the study | Reference |
|---|---|---|---|---|---|---|
| Label-free (size-based) | Nano-DLD | 0.012 μL /h | N/A | N/A | Human urine | 48 |
| Acoustics | 240 μL/h | 82% | 98.4% | Whole blood, cell culture medium | 47 | |
| Pinched-flow fractionation | 1200 μL/h | N/A | N/A | Cell culture medium | 45 | |
| AF4 | 267–1333 μL /h | N/A | N/A | Cell culture medium | 49 | |
| Nanowire trapping | 600 μL /h | 10% | N/A | Mixture of BSA, liposome and beads | 46 | |
| Viscoelastic | 200 μL/h | >80% | > 90% | Cell culture medium, pure serum | 31 | |
| ExoTIC | 5000 μL /h | >90% | N/A | Plasma, urine, lavage, cell culture medium | 24 | |
| Double filtration | 2400 μL/h | 92% | N/A | Cell culture medium, urine | 25 | |
| Exodisc-B/P | 180–900 μL/h | 76–88% | N/A | Whole blood, plasma, and cell culture medium | 26 | |
| FerroChip | 60–180 μL/h | 94.3% | 87.9% | Cell culture medium and serum | This work | |
| Label-based | iMER | 240 μL / h | 93% | N/A | Serum | 44 |
| CD63-modified herringbone groves | ∼ 800 μL / h | 42–94% | N/A | Serum and cell culture medium | 42 | |
| CD41-modified mica surface | 72 μL / h | N/A | N/A | Human plasma, | 41 | |
| ExoChip | 240 μL / h | N/A | N/A | Serum | 17 | |
| nPLEX | 500 μL / h | N/A | N/A | Ascites fluid | 43 | |
| Nano-IMEX | 3 μL / h | N/A | N/A | Diluted plasma | 19 |
Through the optimization of FerroChip design and its operating parameters in the previous section, we determined the following parameters that were appropriate for exosome separation. (1) FerroChip device geometry and dimensions: the top view of the device is depicted in Figure 1c, with optimized channel dimensions (55 × 1.2 × 0.15 mm, L × W × H) for laminar flow condition and clinically relevant throughput. (2) Magnetic field parameters: a magnetic flux density of up to 0.5 T, and a magnetic flux density gradient of up to 1272 T m−1 were achieved via assembling four neodymium permanent magnets in quadrupole configuration. (3) Ferrofluid concentration: a 0.3% volume fraction of maghemite nanoparticle based ferrofluid was used. The corresponding viscosity of this ferrofluids was 1.68 mPa∙s at 23°C. (4) Sample flow rates: exosomes were represented by 200 nm particle and other larger extracellular vesicles were represented by 1000 nm particles. The mean diameter of exosomes was chosen to be slightly larger than 150 nm to ensure all particles smaller than 200 nm to be separated from larger extracellular vesicles. With these two diameters, we determined through simulations that a sample flow rate of 3 μL minute−1, and a sheath flow rate of 15 μL minute−1, would yield significant separation of the two species in the FerroChip (Figure 4a). For the simulations, we calculated two outputs – a deflection in the y-direction for EVs (see Figure 2a for coordinates), denoted as Y, and a separation distance between exosome (200 nm) and large EVs (1000 nm), denoted as ΔY. Both outputs were optimized using a sample flow rate of (0–10 μL min−1, i.e., 0–600 μL h−1). The goal was to maximize the exosome recovery rate and minimize large EV contamination, which translated to maximizing ΔY and the flow rate simultaneously. Figure 4a shows that separation distance ΔY was close to the maximum when using a ferrofluid with 0.3% magnetic volume fraction, and a flow rate of 3 μL min−1 (i.e., 180 μL h−1). Figure 4a also shows a distribution of simulated EV locations at the outlets. The distribution confirms that 200 nm exosomes and 1000 nm large EVs can be ferrohydrodynamically separated in the FerroChip, and 100% of exosomes can be recovered with none of the large EV contamination.
Figure 4.
Optimizations of the FerroChip for the separation of exosome-like nanoparticles (30 – 150 nm in diameter) from large extracellular vesicles (200 – 1000 nm in diameter). a Simulation results of the separation of 200 nm and 1000 nm polystyrene beads, at different flow rates (1 – 10 μL min−1) in a 0.3% (v/v) ferrofluid. We considered 1 – 3 μL min−1 to be optimal flow rates because they correspond to more than 400 μm separation distance in simulation. A 3 μL min−1 optimal flow rate was chosen to separate 200 nm and 1000 nm polystyrene beads from each other. The ratio of particle flow and sheath flow was 1:5 (sample – 3 μL min−1; sheath – 15 μL min−1). Other simulation parameters included a ferrofluid concentration of 0.3%, and the same quadrupole magnet configuration in previous figures. Using these parameters, we simulated the distribution of 200 nm and 1000 nm beads at the outlets of the FerroChip. Particle trajectories from the simulation results predicted that the two differently sized particles could be completely separated. b Experimental images of the separation of a mixture of 200 nm (blue) and 1000 nm (red) polystyrene particles in the FerroChip. Fluorescent images taken at different locations along the microchannel showed that 200 nm particles (blue) were completely separated from 1000 nm particles (red). Both particles were collected for subsequent size characterization. Broad blue fluorescent signals across the channel at the location #3 were from the shade of the permanent magnets, not fluorescent particles. c Particle size distributions measured by Dynamic Light Scattering (DLS) indicate a well-separated 1000 nm and 200 nm particles after FerroChip processing.
Using optimized device geometry and operating parameters, we studied FerroChip’s performance in separating 200 nm diamagnetic nanoparticles from a mixture of 200 nm and 1000 nm particles. A typical separation process can be visualized in Figure 4b, in which premixed blue fluorescent 200 nm diamagnetic particles and red fluorescent 1000 nm diamagnetic particles with 1 to 1 ratio were processed in a FerroChip device at a sample flow rate of 3 μL min−1 and a sheath flow rate of 15 μL min−1. Fluorescent images from the separation process show that before ferrohydrodynamic separation, both 200 nm and 1000 nm particles were mixed and remained close to the sidewall of the microchannel (observation windows 1, 2 and 3 in Figure 4b). After ferrohydrodynamic separation, blue 200 nm particles migrated slightly away from the sidewall due to a weak ferrohydrodynamic effect on them and exited the channel through the side outlets. On the other hand, red 1000 nm particles experienced a significantly larger ferrohydrodynamic effect and migrated to the center of the channel and exited through the middle outlet (observation windows 4 and 5 in Figure 4b). This experimental result matched the simulation result in Figure 4a very well. From the fluorescent image analysis, we confirmed that 200 nm particles could be recovered at 100% recovery rate from the side outlets, and the purity of 200 nm particles from the side outlets were 100% as none of the larger 1000 nm particles exited through side outlets. We further confirmed this result by collecting samples from FerroChip outlets and characterized their particle size distribution using dynamic light scattering (DLS). Figure 4c shows that particle sample from the FerroChip inlet was a mixture of 200 nm and 1000 nm particles; sample collected from the side outlet were exclusively 200 nm particles; sample collected from the middle outlet were exclusively 1000 nm particles. We note that maghemite particles in ferrofluids appeared in inlets and outlets too. The mean diameters of maghemite particles (~100 nm) from these spectra were larger than individual maghemite particles (~10 nm), likely due to particle agglomeration induced by the dilution process that disrupted the surfactant concentration. Additional simulation and experimental data of 200 nm and 520 nm diamagnetic particles in the supplementary information (Figure S1) shows these two could be separated in the FerroChip as well. We also studied the theoretical size resolution of the FerroChip in separating nanoparticles and found the solution to be ~100 nm (supplementary information Figure S2).
In summary, we showed that the performance of FerroChip devices in separating 200 nm and 1000 nm particles met or exceeded the goals, including: (1) a complete 200 nm particle recovery rate of 100%; (2) a sample processing throughput of 3 μL min−1 (180 μL h−1), and (3) a purity of 200 nm particles of 100% after separation. This performance enabled us to continue the device characterization using biological samples. We note that the particles used in this experiment had a very narrow size distribution, which led to better-than-expected recovery rate and purity performance. When dealing with exosomes and other EVs in biological samples, their size polydispersity will lead to a decrease in the exosomal recovery rate and purity.
Validation of FerroChip for exosome-like nanoparticles separation using biological samples
We characterized the FerroChip’s performance in separating exosomes from biological samples. The device geometry and operating parameters remained to be the same as in the previous section, except we further decreased the sample flow rate to 1 μL min−1 (60 μL hour−1) to maximize the separation between exosomes and large EVs (see supplementary information Figure S1). This was determined after considering the polydispersity of sizes of exosomes and large EVs.
We first challenged the FerroChip with extracellular media collected from cultured breast cancer cell line MDA-MB-231 and characterized the exosomal recovery rate and purity of the FerroChip. The EVs from this media were first enriched and separated into two groups based on their sizes using ultracentrifugation and a commercial kit (ExoQuick-TC, see Materials and Methods). The first group contained large EVs (diameters: 200 nm to 1000 nm) that were stained with PKH 26 red fluorescence, and the second group contained small exosomes (diameters: 30 nm to 150 nm) that were stained with PKH 67 green fluorescence. Size profiles of the exosomes and large EVs were measured by atomic force microscopy and provided in the supplementary information (Figure S3). These two groups of EVs were then mixed together at a ratio of 1 to 1 (20 μL large EVs and 20 μL of exosomes resuspended in 1 mL of ferrofluids) and processed using the FerroChip at a sample flow rate of 1 μL min−1 and a sheath flow rate of 5 μL min−1 (Figure 5a). After device processing, samples collected from middle and side outlets were analyzed for exosomal recovery and purity using two methods including super-resolution imaging and atomic force microscopy. In these analyses, we defined exosome-like nanoparticles to have diameters of 30 – 150 nm, and morphology and protein expressions consistent with other reports.5 Firstly, from a super-resolution imaging analysis of 730 particles, we found that the recovery rate of exosome-like particles, defined as the number of PKH 67 green fluorescent particles found in the side outlets over the number of PKH 67 green fluorescent particles found in all outlets of the FerroChip, was 92.6%, and the purity of recovered exosome-like particles, defined as the number of PKH 67 green fluorescent particles found in the side outlets over the sum of PKH 67 green fluorescent particles and PKH 26 red fluorescent particles found in the side outlets of the FerroChip, was 91.1% (Table 1). Secondly, the separation of the exosomes and large EVs was analyzed by atomic force microscopy (AFM, representative images in supplementary information Figure S4). From an atomic force microscopic image analysis of 1581 particles, we found that the recovery rate of exosome-like particles with diameter less than 150 nm was 96%, and the purity was 84.7% (Table 1). The average recovery rate between these two characterization methods was 94.3% and the average purity was 87.9%. In addition, we used transmission electron microscopy (TEM) to characterize the diameter distribution of the separated exosome-like particles from the FerroChip. Figure 5e shows that the diameter distribution of these separated exosome-like nanoparticles was 45 ± 26 nm, with a minimal diameter of 17 nm and a maximal diameter of 254 nm. This diameter range was consistent with established exosome diameters (30 – 150 nm).5 Conventional transmission electron microscopy with negative staining of the exosome output from the FerroChip also revealed that most separated exosomes had an artifactual cup-shaped morphology caused by shrinking (Figure 5d), consistent with their established morphology in literature.5 Magnetic nanoparticles were removed from the exosome solution prior to the TEM. This process, described in Materials and Methods, did not appear to affect the morphology of the exosomes. We used Western blot analysis to examine the expression of exosomal protein markers in the samples collected from the side outlets and the middle outlet. We analyzed the expression of EV membrane tetraspanin CD63 and heat shock 70 kDa protein, HSP70. The sample collected from the side outlets showed a high expression of CD63 and HSP70 (Figure 5c), confirming that the majority of exosomes were separated from the initial EV mixture into the side outlets. The EVs collected from the middle outlet showed a low level of CD63, indicating that a small quantity of exosomes also exited the device through the middle outlet. In addition to the separation of exosomes from the extracellular media, we also demonstrated an effective focusing effect on EVs in the FerroChip (see supplementary information Figure S5).
Figure 5.
Characterization of the FerroChip in separating exosome-like nanoparticles from biological samples. a We chose to use a 1 μL min−1 sample flow rate in order to separate exosome-like nanoparticles (30 – 150 nm in diameter) from extracellular media and human serum. This sample flow rate maximized the separation between exosome-like nanoparticles and large EVs (200 – 1000 nm in diameter). The ratio of sample flow and sheath flow was 1:5 (sample – 1 μL min−1; sheath –5 μL min−1). Other device and operating parameters remained the same as in previous figures. b Super-resolution microscopic images of exosome-like nanoparticles (PKH 67 green fluorescence) and large EVs (PKH 26 red fluorescence) from the mixture (pre-separation), and the samples after FerroChip processing (large EVs and exosome outlets). A total of 730 particles were analyzed. c Western blot analysis of CD63 and HSP70 protein levels in large EVs and exosome-like nanoparticles that were processed by the FerroChip. d Transmission electron microscopy (TEM) images of exosome-like nanoparticles collected from the FerroChip. e Size distribution of separated exosome-like nanoparticles from TEM images (n=755). f Immunofluorescence images of separated exosome-like nanoparticles from human serum. Three channels were used in immunofluorescent staining, including EpCAM (green), CD24 (red), and CD63 (cyan). g Venn diagram depicts the percentage of antibody presence on the surfaces of separated exosome-like nanoparticles (n=12000) from human serum. EpCAM+ alone: 5%; CD24+ alone: 52%; CD63+ alone: 21%; EpCAM+ and CD24+:4%; EpCAM+ / CD63+: 2%; CD24+ / CD63+: 14%; CD63+ / CD24+ / EpCAM+: 2%.
Table 1.
Summary of recovery rate and purity of exosome-like particles separated from MDA-MB-231 breast cancer cell culture media using the FerroChip. Atomic force microscopy characterization used a maximum ferret diameter of 150 nm to differentiate exosome-like particles and large extracellular vesicles, i.e., diameter of exosome-like particles is less than 150 nm, while diameter of large extracellular vesicles is larger than 150 nm. Super-resolution microscopy characterization used two different fluorescent colors to differentiate exosome-like particles and large extracellular vesicles. Recovery rate is defined as the number of exosome-like particles found in the side outlets over exosome-like particles found in all outlets of the FerroChip. Purity is defined as the number of exosome-like particles found in the side outlets over the sum of exosomes-like particles and large extracellular vesicles found in the side outlets of the FerroChip.
| Recovery rate | Purity | |
|---|---|---|
| AFM (n=1581) | 96.0% | 84.7% |
| Super-resolution microscopy (n=730) | 92.6% | 91.1% |
| Average of the above two methods | 94.3% | 87.9% |
We further challenged the FerroChip with serum from a healthy human’s blood. We aimed to show that the FerroChip could separate exosome-like nanoparticles from the serum. For this purpose, we processed 100 μL of serum using the FerroChip at a sample flow rate of 1 μL min−1 and a sheath flow rate of 5 μL min−1. Exosomes separated from the FerroChip were investigated for their molecular signatures through the profiling of three antibodies. We chose these antibodies based on prior studies, including CD63 that was particularly enriched on the surface of exosomes,5, 30, 40 and two putative cancer makers – epithelial cell adhesion molecule (EpCAM) and CD24.30, 40 Both of these cancer markers were not only detected in tumor-derived exosomes, but also in samples from healthy subjects.30 We found that all three markers, CD63, EpCAM, and CD24 were detected in different combinations on the exosomes’ surface from the human serum (Figure 5f). Figure 5g shows the Venn diagram depicting the percentage of each antibody’s presence – EpCAM+ alone: 5%; CD24+ alone: 52%; CD63+ alone: 21%; EpCAM+ / CD24+: 4%; EpCAM+ / CD63+: 2%; CD24+ / CD63+: 14%; CD63+ / CD24+ / EpCAM+: 2%. We also challenged the FerroChip with extracellular media from 7 cancer cell lines and investigated the antibody presence on the surfaces of recovered exosomes. The presence of these antibodies was heterogeneous across the cell lines (see supplementary information Figure S6). These information together indicate that using a label-based exosome separation strategy will result in partial recovery and highlights a need for label-free strategies include the FerroChip.
Advantages and limitations of the FerroChip in separating exosome-like particles
We compared the FerroChip’s performance of separating exosome-like nanoparticles to existing microfluidic methods. In doing so, we used three metrics including the sample-processing throughput, exosomal particle recovery rate, and purity of recovered exosomes. These three metrics were well established in the current literature of exosome separation.6, 39 Through validations with both spike-in samples and biological samples using these operating parameters, the performance of the FerroChip in separating exosome-like particles was determined to have: (1) a recovery rate of 94.3%; (2) a purity of 87.9% in recovered particles; (3) a sample processing throughput of 60 μL h−1. We compared these results to recently published microfluidic methods (Table 2) and found that the FerroChip had a better combined recovery rate and purity than existing methods.17, 19, 31, 41–49
On the other hand, the FerroChip presented in this study have the following limitations in separating exosome-like nanoparticles from biological fluids. Firstly, the sample processing throughput of the FerroChip was limited to ~60 μL h−1. In the process of separating exosome-like particles from biological fluids, a sheath flow of ferrofluids was introduced along with the sample which resulted in a dilution of separated particles. Because of this limitation, FerroChip may be more suitable for applications such as blood liquid biopsy where the starting sample volume is usually small (~100 μL) and the tumor-derived exosomes are abundant (>109/mL).20, 30, 39, 50 Secondly, the impact of ferrofluids on the physiology of extracellular vesicles needs further studies. While in previous studies we have demonstrated the ferrofluids had minimally detrimental effects on mammalian cells,35, 36, 51 and in this study, the biophysical diameter, morphology and common protein expressions appeared to be consistent with other reports, further molecular characterizations are still needed to assess the quality of the exosome-like particles from the FerroChip in order to adapt this method for biological applications. Lastly, the FerroChip relies on the size-dependent ferrohydrodynamic force to separate larger EVs from smaller exosomes. As a result, it cannot distinguish exosomes from particulate impurities that have similar sizes in a biological fluid, which may lead to decreased purity of separated exosomes.
Conclusion
In this paper we developed a new label-free ferrohydrodynamic method and its prototype devices (FerroChip) for a size-based separation of exosome-like particles from biological samples. FerroChip relies on particle ferrohydrodynamics of nanoscale extracellular vesicles in a biocompatible ferrofluid to separate smaller exosome-like nanoparticles from larger extracellular vesicles (EVs). Particle ferrohydrodynamics developed in the past was limited to micron-sized particle and cellular separations. Here we successfully demonstrated both in theory and with experiments that particle ferrohydrodynamics could be applied to exosome-like nanoparticles separation in microfluidic devices with ~100 nm size resolution. The developed FerroChip was used to separate exosome-like nanoparticles, defined as particles of 30 – 150 nm and having morphology and protein markers that were consistent exosomes, from biological fluids. FerroChip’s separation had a recovery rate of 94.3% and a purity of 87.9% in recovered exosome-like nanoparticles. This is the first time that nanosized particles were separated via particle ferrohydrodynamics in microfluidic devices.
Materials and Methods
Ferrohydrodynamic modeling in microfluidic devices
We adopted a previously developed analytical model in this study to simulate the trajectories of immersed nanoparticles/EVs in ferrofluids in a three-dimensional (3D) manner.35, 51 This model provides a fast prediction of three-dimensional ferrohydrodynamic transport of nanoparticles and EVs inside a microfluidic channel coupled with quadruple configurations of permanent magnets. Trajectories of the nanoparticles/EVs in the FerroChip were obtained by (1) first computing the three-dimensional magnetic force via an experimentally verified and analytically computed distribution of magnetic fields as well as their gradients, together with a nonlinear Langevin magnetization model of the ferrofluid that considers ferrofluids to be a continuous medium, and the magnetic nanoparticles inside the ferrofluid to be non-interacting, (2) secondly computing the ferrohydrodynamic motion of diamagnetic nanoparticles/EVs through the governing equations of nanoparticles/EVs in laminar flow conditions, using analytical expressions of magnetic forces and hydrodynamic viscous drag forces. The script for this model was developed and solved in MATLAB (MathWorks Inc., Natick, MA).
Ferrofluids synthesis, characterization and composition
Water-based ferrofluid with maghemite nanoparticles was synthesized by a chemical co-precipitation method following an established protocol.51 Transmission electron microscopy (TEM; FEI, Eindhoven, the Netherlands) were used to characterize the size and morphology of maghemite nanoparticles. The viscosity of ferrofluids was measured with a compact rheometer (Anton Paar, Ashland, VA) at room temperature. Magnetic properties (volume fraction of magnetic materials and saturation magnetization) of the ferrofluid were characterized using a vibrating sample magnetometer (VSM; MicroSense, Lowell, MA). The diameter distribution of the maghemite nanoparticles in the ferrofluid is 10.91 ± 4.86 nm. The surfactant on the nanoparticles is a graft copolymer (Atlox 4913, Croda, Inc., Edison, NJ). Ferrofluid was adjusted to biocompatible by changing the pH to 7 with sodium hydroxide and balancing the osmotic pressure with Hank’s Balanced Salt Solution (Thermo Fisher Scientific, Waltham, MA). The concentration of ferrofluid was measured to be 0.3% (v/v) and the corresponding viscosity of this ferrofluids was 1.68 mPa∙s at 23°C.
Cell culture and extracellular vesicles preparation
Extracellular vesicles were prepared from cell lines: human breast cancer (MDA-MB-231, ATCC, Manassas, VA) and lung cancer (A549, ATCC, Manassas, VA). Cell lines were cultured following the manufacturer’s recommended protocol. The culture medium was supplemented with 10% exosome-free FBS (Thermo Fisher Scientific, Waltham, MA). The extracellular vesicles were isolated from the cell culture supernatant. The collected supernatant was centrifuged at 500g for 5 minutes to remove cells. To collect extracellular vesicles smaller than 800 nm, the supernatant was processed by membrane filtration (800 nm pores, Pall Corporation), followed by adding 20% ExoQuick-TC reagent (System Biosciences, CA). The mixture was incubated overnight at 4°C, followed by centrifugation at 1,500g for 30 minutes. Extracellular vesicles were resuspended in 200 μL using sterile 1×PBS. To collect large extracellular vesicles (diameters: 200 nm to 1000 nm), the collected culture supernatant was processed by centrifugation at 12,000g for 90 minutes at room temperature. Large extracellular vesicles were collected by resuspending the pellet in 200 μL 1×PBS. Smaller exosomes (diameters: 30 nm to 150 nm) were obtained by mixing the remaining supernatant with 20% ExoQuick-TC reagent, following the manufacturer’s protocol. The exosomes were resuspended in 200 μL 1×PBS. The extracellular vesicles (both large extracellular vesicles and small exosomes) were then fluorescently stained with PKH 67 or PKH 26 (Sigma-Aldrich, St. Louis, MO) following the manufacturer’s protocol.
FerroChip fabrication and assembly
The microfluidic channel of the FerroChip contains a filtration channel and particle manipulation (focusing or separation) channel. Using standard soft lithography techniques, the devices were made of polydimethylsiloxane (PDMS) with a channel height of 150 μm, measured by a profilometer (Veeco Instruments, Chadds Ford, PA). The fabricated microchannel was placed in the quadrupole permanent magnet array and held in a custom-made aluminum manifold. Each magnet was 38.1 mm in length, 6.35 mm in both width and thickness, with a residual magnetic flux density of 1.48 T.
Microfluidic experiment setup and procedure
The FerroChip was first flushed by 70% ethanol for 10 minutes, followed by priming with 1×PBS supplemented with 0.5% (w/v) bovine serum albumin (BSA) and 2 mM EDTA (Thermo Fisher Scientific, Waltham, MA) for 10 minutes with a flow rate of 100 μL/min. Sample ferrofluids that contained polystyrene particles or extracellular vesicles were supplemented with 0.5% (w/v) BSA to prevent particle aggregation. Sample fluids and sheath fluids were injected into the microfluidic inlets using individually controlled syringe pumps (Chemyx, Stafford, TX) at variable flow rates. The FerroChip was placed on the stage of an inverted microscope (Axio Observer, Carl Zeiss, Germany) for observation and recording. Images and videos of particles were obtained from a CCD camera (Carl Zeiss, Germany).
Removal of magnetic nanoparticles from exosomes
After FerroChip processing and before characterizations, the maghemite nanoparticles in ferrofluids could be removed by increasing the pH of the ferrofluid to 7.5 – 8 with sodium bicarbonate. Briefly, the sample was collected in an Eppendorf tube and the tube was placed into a customized magnet array. 0.1 M NaHCO3 was added into the solution until its pH reached to 7.5 – 8. The solution was incubated for 3 hours at room temperature. The maghemite nanoparticles formed clusters and were attracted by the magnet array. Supernatant of the solution was removed, and the resulted solution was centrifuged at 1000g for 5 minutes to further remove nanoparticle clusters.
Extracellular vesicles characterization
Dynamic Light Scattering (DLS)
Polystyrene submicron particles were measured by a Zetasizer Nano ZS Analyzer (Malvern Panalytical Ltd, United Kingdom). 5 μL of the collected sample from FerroChip outlets was diluted with 20 mL filtered DI water and measured at room temperature.
Atomic Force Microscopy (AFM)
Collected extracellular vesicles were imaged by a multimode-8 AFM (Bruker, Billerica, MA) using the tapping mode with the DNP-S10 probe.
Super-resolution microscopy
Collected extracellular vesicles were imaged by a Zeiss ELYRA S1 (SR-SIM) super-resolution microscope (Carl Zeiss, Germany).
Transmission electron microscopy (TEM)
Collected extracellular vesicles were fixed with 2% paraformaldehyde for 10 min at room temperature and then dropped onto electron microscope grids. After incubation for 20 min, the grids were transferred to a 50 μL drop of 2.5% glutaraldehyde in sodium cacodylate buffer for 10 min. The grid was transferred to a 100 μL drop of filtered distilled water and washed 3 times. After drying for 20 min, the extracellular vesicles were stained with 1% uranyl acetate for 1 min. Collected extracellular vesicles were imaged by JEOL JEM1011 (JOEL, Inc., Peabody, MA) transmission electron microscope (TEM) at 80kV.
Western blot
Exosomes collected from the side channel of the FerroChip were lysed using RIPA buffer with protease inhibitor cocktail added. The protein concentration was quantified using a standard Bradford protein assay and 20 μg of protein was loaded for each well. Laemmli buffer (with Beta-mercaptoethanol) was added and the sample was heated at 95°C for 5 minutes and chilled on ice before loading onto gel (Bio-Rad Laboratories, Hercules, CA). Standard SDS-PAGE electrophoresis was performed, and the protein lysates were transferred onto PVDF membrane (Santa Cruz Biotechnology, Inc., Dallas, TX). The membrane was then incubated overnight with exosome primary antibodies against CD63 and HSP70 (Santa Cruz Biotechnology, Inc., Dallas, TX following incubation with Goat-Rabbit-HRP secondary antibody (Santa Cruz Biotechnology, Inc., Dallas, TX). Blot was incubated with Western ECL substrate (Bio-Rad Laboratories, Hercules, CA) to enhance the signal, and the blot was visualized with ChemiDoc MP Imaging System (Bio-Rad Laboratories, Hercules, CA).
Live subject statement
All experiments in this study were performed in compliance with the regulations of the United States Office for Human Research Protections, and the University of Georgia Human Subjects Office. Human whole blood from healthy subjects was purchased from a commercial vendor (Zen-Bio, Research Triangle, NC) without any identifying information. Blood samples are collected from an United States Food and Drug Administration regulated blood bank. All procedures, polices, forms and consents used in the donor screening, collection and manufacturing process are submitted to the FDA for review and approval through the Prior Approval Supplement process described in 21 CFR 601.12(b). The blood sample used in this study, for which the authors or sponsors have no access to identifying information, do not meet the definition of human subjects (45 CFR 46) and are not regulated by the Office of Human Subjects Protections and would not be subject to institutional review board (IRB).
Blood sample preparation
The purchased blood sample was centrifuged at 3,000g for 15 minutes to remove cells. The supernatant was mixed with a ferrofluid (volume fraction 0.6%) with a 1:1 ratio. Pluronic F-68 non-ionic surfactant (Thermo Fisher Scientific, Waltham, MA) was added before processing.
Immunofluorescence staining of exosomes
Isolated exosomes from human blood were plated onto a poly-L-lysine coated glass slide with a customized cell collection chamber for 24 hours at 4°C. The collected exosomes were fixed with 4% (w/v) PFA solution (Santa Cruz Biotechnology, Inc., Dallas, TX), following permeabilization with Triton X-100 buffer (Alfa Aesar, Haverhill, MA) for 10 minutes. The nonspecific binding sites of exosomes were blocked with a blocking reagent (Santa Cruz Biotechnology, Inc., Dallas, TX) before immunostaining with primary antibodies, including anti-EpCAM, anti-CD24, and anti-CD63 (Santa Cruz Biotechnology, Inc., Dallas, TX). The stained exosomes were washed with PBS and covered with mounting medium before imaging.
Supplementary Material
Acknowledgments
We thank Mr. Yongwook Kim and Professor Sergiy Minko for their help in AFM imaging. We thank the Biomedical Microscopy Core facility personnel for their help in super-resolution microscopy. We also thank the Georgia Electron Microscopy personnel for their help in the TEM imaging. This material is based upon work supported by the National Science Foundation under Grant Nos. 1150042, 1659525 and 1648035; by the National Institute of General Medical Sciences of the National Institutes of Health under Award No. R21GM104528; and the National Center for Advancing Translational Sciences of the National Institutes of Health under Award No. UL1TR002378. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of interest
The University of Georgia filed patent protection for FerroChip technology.
This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication.
Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available.
You can find more information about Accepted Manuscripts in the Information for Authors.
Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains.
References
- 1.Massague J and Obenauf AC, Nature, 2016, 529, 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert AW, Pattabiraman DR and Weinberg RA, Cell, 2017, 168, 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roussos ET, Condeelis JS and Patsialou A, Nat Rev Cancer, 2011, 11, 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW and Simpson RJ, Nat Rev Clin Oncol, 2018, 15, 617–638. [DOI] [PubMed] [Google Scholar]
- 5.van Niel G, D’Angelo G and Raposo G, Nat Rev Mol Cell Biol, 2018, 19, 213–228. [DOI] [PubMed] [Google Scholar]
- 6.Contreras-Naranjo JC, Wu HJ and Ugaz VM, Lab Chip, 2017, 17, 3558–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathieu M, Martin-Jaular L, Lavieu G and Thery C, Nat Cell Biol, 2019, 21, 9–17. [DOI] [PubMed] [Google Scholar]
- 8.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang Y, Bissell MJ, Cox TR, Giaccia AJ, Erler JT, Hiratsuka S, Ghajar CM and Lyden D, Nat Rev Cancer, 2017, 17, 302–317. [DOI] [PubMed] [Google Scholar]
- 9.Celia-Terrassa T and Kang Y, Nat Cell Biol, 2018, 20, 868–877. [DOI] [PubMed] [Google Scholar]
- 10.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, T, Theilen TM, Garcia-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J and Lyden D, Nat Cell Biol, 2015, 17, 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguado BA, Bushnell GG, Rao SS, Jeruss JS and Shea LD, Nat Biomed Eng, 2017, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J and Lyden D, Nat Med, 2012, 18, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J and Lyden D, Nature, 2015, 527, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H and Weissleder R, Nat Commun, 2015, 6, 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M and Irimia D, Lab Chip, 2010, 10, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashcroft BA, de Sonneville J, Yuana Y, Osanto S, Bertina R, Kuil ME and Oosterkamp TH, Biomed Microdevices, 2012, 14, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanwar SS, Dunlay CJ, Simeone DM and Nagrath S, Lab on a Chip, 2014, 14, 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Im H, Shao HL, Park YI, Peterson VM, Castro CM, Weissleder R and Lee H, Nat Biotechnol, 2014, 32, 490–U219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, He M and Zeng Y, Lab on a Chip, 2016, 16, 3033–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan Y, Cheng G, Liu X, Hao SJ, Nisic M, Zhu CD, Xia YQ, Li WQ, Wang ZG, Zhang WL, Rice SJ, Sebastian A, Albert I, Belani CP and Zheng SY, Nat Biomed Eng, 2017, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, Austin RH, Stolovitzky G and Astier Y, Nat Nanotechnol, 2016, 11, 936–940. [DOI] [PubMed] [Google Scholar]
- 22.Shin S, Han D, Park MC, Mun J, Choi J, Chun H, Kim S and Hong JW, Sci Rep-Uk, 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H and Lyden D, Nat Protoc, 2019, 14, 1027–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu F, Vermesh O, Mani V, Ge TJ, Madsen SJ, Sabour A, Hsu EC, Gowrishankar G, Kanada M, Jokerst JV, Sierra RG, Chang E, Lau K, Sridhar K, Bermudez A, Pitteri SJ, Stoyanova T, Sinclair R, Nair VS, Gambhir SS and Demirci U, ACS Nano, 2017, 11, 10712–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang LG, Kong MQ, Zhou S, Sheng YF, Wang P, Yu T, Inci F, Kuo WP, Li LJ, Demirci U and Wang S, Sci Rep, 2017, 7, 46224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunkara V, Kim CJ, Park J, Woo HK, Kim D, Ha HK, Kim MH, Son Y, Kim JR and Cho YK, Theranostics, 2019, 9, 1851–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong X, Chi J, Zheng L, Ma B, Li Z, Wang S, Zhao C and Liu H, Lab on a Chip, 2019, 19, 2897–2904. [DOI] [PubMed] [Google Scholar]
- 28.Woo HK, Sunkara V, Park J, Kim TH, Han JR, Kim CJ, Choi HI, Kim YK and Cho YK, ACS Nano, 2017, 11, 1360–1370. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, Godin B, Zhang JX and Liu X, Lab Chip, 2013, 13, 2879–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK and Zeng Y, Nat Biomed Eng, 2019, 3, 438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu C, Guo J, Tian F, Yang N, Yan F, Ding Y, Wei J, Hu G, Nie G and Sun J, ACS nano, 2017, 11, 6968–6976. [DOI] [PubMed] [Google Scholar]
- 32.Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, Li H, Li P, Quinn D, Dao M, Suresh S, Sadovsky Y and Huang TJ, Proc Natl Acad Sci U S A, 2017, 114, 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosensweig RE, Ferrohydrodynamics, Cambridge University Press, Cambridge, 1985. [Google Scholar]
- 34.Zhao W, Cheng R, Miller JR and Mao L, Adv Funct Mater, 2016, 26, 3916–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao WJ, Cheng R, Jenkins BD, Zhu TT, Okonkwo NE, Jones CE, Davis MB, Kavuri SK, Hao ZL, Schroeder C and Mao LD, Lab on a Chip, 2017, 17, 3097–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao W, Liu Y, Jenkins BD, Cheng R, Harris BN, Zhang W, Xie J, Murrow JR, Hodgson J, Egan M, Bankey A, Nikolinakos PG, Ali HY, Meichner K, Newman LA, Davis MB and Mao L, Lab Chip, 2019, 19, 1860–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erb RM, Son HS, Samanta B, Rotello VM and Yellen BB, Nature, 2009, 457, 999–1002. [DOI] [PubMed] [Google Scholar]
- 38.Zhu T, Cheng R, Lee SA, Rajaraman E, Eiteman MA, Querec TD, Unger ER and Mao L, Microfluid Nanofluidics, 2012, 13, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He M and Zeng Y, Jala-J Lab Autom, 2016, 21, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R and Lee H, Nat Biotechnol, 2014, 32, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashcroft B, De Sonneville J, Yuana Y, Osanto S, Bertina R, Kuil M and Oosterkamp T, Biomed Microdevices, 2012, 14, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Skog J, Hsu C-H, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M and Irimia D, Lab on a Chip, 2010, 10, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R and Lee H, Nature biotechnology, 2014, 32, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H and Weissleder R, Nature communications, 2015, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin S, Han D, Park MC, Mun JY, Choi J, Chun H, Kim S and Hong JW, Sci Rep-Uk, 2017, 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Wu H.-j., Fine D, Schmulen J, Hu Y, Godin B, Zhang JXand Liu X, Lab on a Chip, 2013, 13, 2879–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu M, Ouyang Y, Wang Z, Zhang R, Huang P-H, Chen C, Li H, Li P, Quinn D and Dao M, Proceedings of the National Academy of Sciences, 2017, 114, 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, Austin RH, Stolovitzky G and Astier Y, Nature nanotechnology, 2016, 11, 936. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H and Lyden D, Nature protocols, 2019, 14, 1027–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu H, Liao C, Zuo P, Liu Z and Ye BC, Anal Chem, 2018, 90, 13451–13458. [DOI] [PubMed] [Google Scholar]
- 51.Zhao WJ, Cheng R, Lim SH, Miller JR, Zhang WZ, Tang W, Xie J and Mao LD, Lab on a Chip, 2017, 17, 2243–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.