Abstract
Pulque is a culturally important 4,000-year-old traditional Mexican fermented drink. Pulque is produced by adding fresh aguamiel (agave sap) to mature pulque, resulting in a mixture of microbial communities and chemical compositions. We performed shotgun metagenomic sequencing of five stages of pulque fermentation to characterize organismal and functional diversity. We identified 6 genera (Acinetobacter, Lactobacillus, Lactococcus, Leuconostoc, Saccharomyces and Zymomonas) and 10 species (Acinetobacter boissieri, Acinetobacter nectaris, Lactobacillus sanfranciscensis, Lactococcus lactis, Lactococcus piscium, Lactococcus plantarum, Leuconostoc citreum, Leuconostoc gelidum, Zymomonas mobilis and Saccharomyces cerevisiae) that were present ≥ 1% in at least one stage of pulque fermentation. The abundance of genera and species changed during fermentation and was associated with a decrease in sucrose and increases in ethanol and lactic acid, suggesting that resource competition shapes organismal diversity. We also predicted functional profiles, based on organismal gene content, for each fermentation stage and identified an abundance of genes associated with the biosynthesis of folate, an essential B-vitamin. Additionally, we investigated the evolutionary relationships of S. cerevisiae and Z. mobilis, two of the major microbial species found in pulque. For S. cerevisiae, we used a metagenomics assembly approach to identify S. cerevisiae scaffolds from pulque, and performed phylogenetic analysis of these sequences along with a collection of 158 S. cerevisiae strains. This analysis suggests that S. cerevisiae from pulque is most closely related to Asian strains isolated from sake and bioethanol. Lastly, we isolated and sequenced the whole-genomes of three strains of Z. mobilis from pulque and compared their relationship to seven previously sequenced isolates. Our results suggest pulque strains may represent a distinct lineage of Z. mobilis.
Subject terms: Metagenomics, Microbiology, Food microbiology
Introduction
Humans have utilized bacteria, yeasts, and molds for millennia in the production of traditionally fermented foods and beverages1,2. Microbial fermentation was a key innovation as it played a crucial role in improving food preservation, nutritional content, consistency, and flavor1,3. Fermented foods are typically produced by a community of microbes that fluctuate in abundance and diversity during the fermentation process4–9. The traditional artisanal practice of backslopping, the addition of a small quantity of a mature fermented food to the beginning of a new fermentation, has led to the reproducible formation of these beneficial microbial communities, and their associated metabolic transformations of food and beverages1,2,10.
Pulque is a traditional Mexican alcoholic beverage produced by fermenting aguamiel (agave sap)11,12. Archaeological chemistry evidence suggests that the origins of pulque date to at least 1,500 years ago13, and historical documents suggest that pulque was being produced as far as 4,000 years ago11,12. Pulque has served as a sacred beverage during religious ritual, is heralded as a nutritionally-rich supplement, and is a source of cultural pride and identity11,12. Modern production of pulque has remained nearly identical to production during pre-Hispanic times11. Briefly, aguamiel is extracted from mature agave plants, this fresh collected aguamiel is mixed with mature pulque, and the mixture is then fermented in vats from three hours to as long as 12 days11.
Pulque fermentation is carried out by a suite of microorganisms that produce three metabolites distinctive to pulque11,14. Lactic acid bacteria and acetic acid bacteria produce the characteristic acidity, which ranges from a pH of 3.5–4.211. The ethanol content of pulque, which ranges between 4 and 7%, is produced through the metabolism of sugars by yeasts (Saccharomyces sp., Kluyveromyces sp. etc.) and the bacteria Zymomonas mobilis11,15. Lastly, species from the lactic acid bacteria genus Leuconostoc produce extracellular polysaccharides (EPS) resulting in pulque’s distinctive viscosity16. Lactic acid bacteria and S. cerevisiae have been isolated from aguamiel17–19, which likely represent the major source of microorganisms for pulque fermentation. The source of Z. mobilis in pulque is less clear, as the specie has not been directly isolated from or detected in aguamiel. However, aguamiel’s high sugar content suggests that it is a suitable natural habitat20,21. Alternatively, the identification of Z. mobilis from honey bees22 and the practice of filtering aguamiel to remove insects and other debris suggests a possible insect transmission11.
Studies focused on the microbial diversity of pulque have used traditional culture-based approaches14,15,23, and more recently, high-throughput 16S/ITS rDNA metagenomics sequencing24,25. However, whole-genome shotgun metagenomic sequencing has not been applied to study the microbial and metabolic diversity of pulque fermentation. The advantages of shotgun metagenomic sequencing over amplicon sequencing include (i) the absence of amplification bias, (ii) the joint and comprehensive examination of the entire microbial community (bacterial and fungal), and (iii) the ability to draw functional inferences from gene abundance data26,27. Here, we conducted shotgun metagenomic sequencing across five distinct stages of pulque fermentation to investigate microbial and functional diversity.
Results
Metagenomics DNA extraction and sequencing of pulque fermentation
We extracted metagenomic DNA of four technical replicates from fresh aguamiel (AM), a mixture of fresh AM with mature pulque (0 h and start of fermentation, T0), 3 h into fermentation (T3), 6 h into fermentation (T6), and 12 h into fermentation (mature pulque, PQ). Our DNA extraction protocol yielded high molecular weight DNA suitable for shotgun metagenomics sequencing (Supplementary Figure S1). As a quality control step to demonstrate the integrity of metagenomic DNA extraction, we successfully amplified the 16S rDNA (V3–V4) locus in each of the technical replicates (Supplementary Figure S2). We generated a total of 18,750,182, 7,798,353, 17,691,690, 20,946,800, and 25,133,620 read pairs across the technical replicates for the AM, T0, T3, T6, and PQ stages, respectively. Species abundance between technical replicates was highly correlated (all comparisons r > 0.99). Thus, we combined technical replicate data for each fermentation stage for subsequent analysis.
Taxonomic profiling of microbial community during pulque fermentation
Taxonomic profile and relative abundance of the microbial community was assessed during stages of pre-fermentation (AM) and fermentation (T0, T3, T6, and PQ). We used MetaPhlAn28 and Kaiju29 for taxonomic classification. Both methods were highly agreeable at the genus- and species-levels (Supplementary Tables S1–4). For instance, all of the major 14 genera identified with MetaPhlan were also identified with Kaiju, and Pearson correlations (r) of percent genus abundance across fermentation stages between the two methods averaged 0.70. Considering the strong agreement between methods, we primarily report results from Kaiju because it uses a larger database for taxonomic classification and thus, has increased resolution at the species-level29.
Viruses and archaea made up less than 0.4% and 0.2% of assigned reads across all stages of pulque fermentation. Bacteria made up 99%, 85%, 75%, 92%, and 82% and fungi made up 0.6%, 14%, 25%, 8%, and 18% of assigned reads across the AM, T0, T3, T6, and PQ stages, respectively. Six, 8, and 31 genera and 10, 15, and 56 species were present ≥ 1%, ≥ 0.5%, and ≥ 0.1% in at least one fermentation stage, respectively (Supplementary Tables 3 and 5). Six of the 56 species present ≥ 0.1% in at least one fermentation stage were fungal (Kluyveromyces marxianus, Saccharomyces arboricola, S. cerevisiae, S. cerevisiae x Saccharomyces kudriavzevii, Saccharomyces eubayanus and S. kudriavzevii).
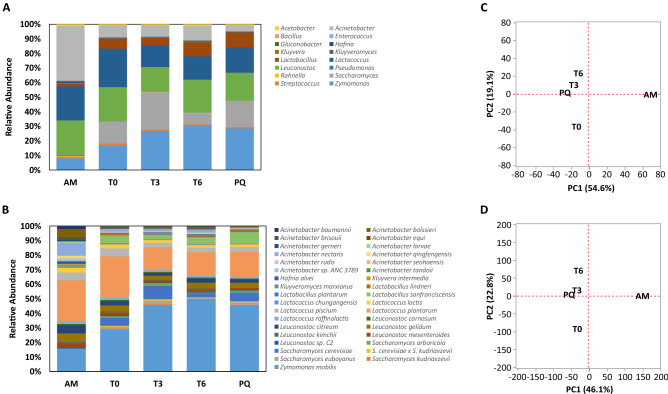
Overall, AM showed the highest diversity (Simpson’s diversity (D) and Shannon’s diversity (H)) (DAM = 9.03, HAM = 2.86), followed by T0 (DT0 = 5.88, HT0 = 2.43), while the T3, T6, and PQ stages showed lower levels of diversity (DT3 = 4.19, HT3 = 2.21, DT6 = 3.71, HT6 = 2.15 and DPQ = 4.10, HPQ = 2.13). At the genus-level, AM was dominated by Acinetobacter (21.95%), Leuconostoc (13.92%), Lactococcus (13.72%), Zymomonas (4.77%) and Lactobacillus (0.97%), and the most abundant species were Lactococcus plantarum (8.50%), Z. mobilis (4.78%), Acinetobacter nectaris (2.68%), Leuconostoc gelidum (1.77%), Leuconostoc citreum (1.68%), Leuconostoc piscium (1.65%), Acinetobacter boissieri (1.59%) and Lactococcus lactis (0.97%) (Fig. 1, Supplementary Tables S2 and S4). The most abundant genera during the T0 stage was Lactococcus (19.52%), Leuconostoc (17.34%), Zymomonas (12.57%), Saccharomyces (10.79%), Acinetobacter (5.73%) and Lactobacillus (4.94%) and the most abundant species were Z. mobilis (12.57%), L. plantarum (12.11%), L. piscium (2.44%), Saccharomyces cerevisiae (2.42%), Lactobacillus sanfranciscensis (2.22%), L. gelidum (1.77%), L. citreum (1.68%), and L. lactis (0.97%) (Fig. 1, Supplementary Tables S2 and S4). The T3 stage was dominated by the genera Zymomonas (19.91%), Saccharomyces (19.08%), Leuconostoc (12.38%), Lactococcus (11.25%), Acinetobacter (5.45%) and Lactobacillus (3.61%) and the species Z. mobilis (19.91.2%), L. plantarum (7.08%), S. cerevisiae (4.05%), Lactobacillus sanfranciscensis (1.41%), L. piscium (1.31%), L. gelidum (1.29%), and Saccharomyces eubayanus (0.99%) (Fig. 1, Supplementary Tables S2 and S4). The most abundant genera during the T6 stage were Zymomonas (22.27%), Leuconostoc (16.50%), Lactococcus (11.73%), Acinetobacter (7.17%), Lactobacillus (7.08%), and Saccharomyces (5.65%) and the most abundant species were Z. mobilis (22.27%), L. plantarum (7.33%), L. sanfranciscensis (2.20%), L. gelidum (1.72%), L. piscium (1.39%), S. cerevisiae (1.25%), and L. citreum (1.20%) (Fig. 1, Supplementary Tables S2 and S4). Finally, the most abundant genera in the PQ stage were Zymomonas (21.48%), Leuconostoc (14.30%), Saccharomyces (13.51%), Lactococcus (13.03%), Lactobacillus (7.53%), and Acinetobacter (2.85%) and the species Z. mobilis (21.48%), Lactococcus plantarum (8.11%), L. sanfranciscensis (3.71%), S. cerevisiae (2.86%), L. piscium (1.57%), L. gelidum (1.47%) and L. citreum (1.04%) (Fig. 1, Supplementary Tables S2 and S4). Principal Component Analysis of relative organismal abundance at the genus and species level showed nearly identical patterns and clear separation between AM from all fermentation stages as well as separation between T0 from T3, T6 and PQ (Fig. 1C, D, Supplementary Figure S3).
Figure 1.
Organismal diversity during pulque fermentation. (A) Genus-level and (B) Species-level relative abundance during pulque fermentation of organisms present ≥ 0.2% in at least one fermentation stage as estimated by Kaiju29. Columns represent relative abundance of organisms (Y-axis) per fermentation stage (X-axis). AM = Aguamiel, T0 = pulque and aguamiel mixture, T3 = 3-h fermentation, T6 = 6-h fermentation, and PQ = ~12-h fermentation (mature pulque). Values are reported as percentages. Principal component analysis (PCA) of relative organismal abundance for genera (C) and species (D). In both comparisons, principal components 1 and 2 explain > 68% of variance.
Next, we evaluated the temporal patterns of the major genera and species (present ≥ 1% in at least one fermentation stage) and their associations with the main fermentative products of pulque11 (i.e. acetic acid, lactic acid, ethanol, and polysaccharides from available sugars in AM). Pearson correlation values for all combinations of metabolites and the 35 species present ≥ 0.1% in at least one fermentation stage are reported in Supplementary Table S5. Acinetobacter, Lactococcus and Leuconostoc were highly abundant in the AM stage (> 13%). Both Lactococcus and Leuconostoc fluctuated slightly during fermentation but were present at approximately the same abundance in PQ (Fig. 2). Conversely, Acinetobacter decreased sharply after AM, eventually reaching 2.85% in PQ (Fig. 2A). Acinetobacter abundance was positively associated with sucrose (r = 0.91, p value = 0.031) and negatively associated with fructose (r = −0.89, p value = 0.046) and ethanol (r = −0.93, p value = 0.022) (Fig. 2). Ten species of Acinetobacter displayed identical significant negative associations with ethanol and fructose, and significant positive associations with sucrose Supplementary Table S5. Zymomonas, Saccharomyces, and Lactobacillus were all present < 4% in AM, but by the end of fermentation (PQ) reached 21.48%, 13.51% and 7.53%, respectively (Fig. 2). Notably, the increase in relative abundance of Zymomonas was positively correlated with the increased production of ethanol (r = 0.95, p value = 0.014), fructose (r = 0.96, p value = 0.010) and lactate (r = 0.95, p value = 0.0143), and negatively correlated with sucrose (r = −0.99, p value = 0.0016) (Fig. 2). Zymomonas and Z. mobilis were the only genus or species significantly positively correlated with fructose (Supplementary Table S5). The increase in relative abundance in Lactobacillus was positively correlated with ethanol (r = 0.95, p value = 0.0137) and lactate (r = 0.95, p value = 0.0128). Saccharomyces was at low relative abundance during AM (0.033%) and fluctuated during fermentation until reaching 13.51% in PQ. Interestingly, Saccharomyces abundance was not significantly correlated with sugar or ethanol abundance, reinforcing the role of Zymomonas in ethanol production during pulque fermentation. At the species-level, the yeast Kluyveromyces marxianus was also significantly positively associated with ethanol abundance (r = 0.88, p value = 0.049) (Supplementary Table S5).
Figure 2.
Temporal patterns of chemical and organismal abundance during pulque fermentation. (A) Concentration of the main sugars and (B) ethanol, lactic and acetic acid (B) during pulque fermentation (red square—sucrose, blue triangle—glucose, green down triangle—fructose, red filled down triangle—ethanol, green filled diamond—acetic acid, blue filled circle—lactic acid). (C) Relative abundance of the dominating genera (Y-axis) during pulque fermentation (X-axis). AM = Aguamiel, T0 = pulque and aguamiel mixture, T3 = 3-h fermentation, T6 = 6-h fermentation, and PQ = ~ 12 h fermentation (mature pulque). (D) Heat map of Pearson correlations (r) between species abundance for organisms present ≥ 0.2% in at least one fermentation stage and chemical concentrations across pulque fermentation.
Functional profiling during pulque fermentation
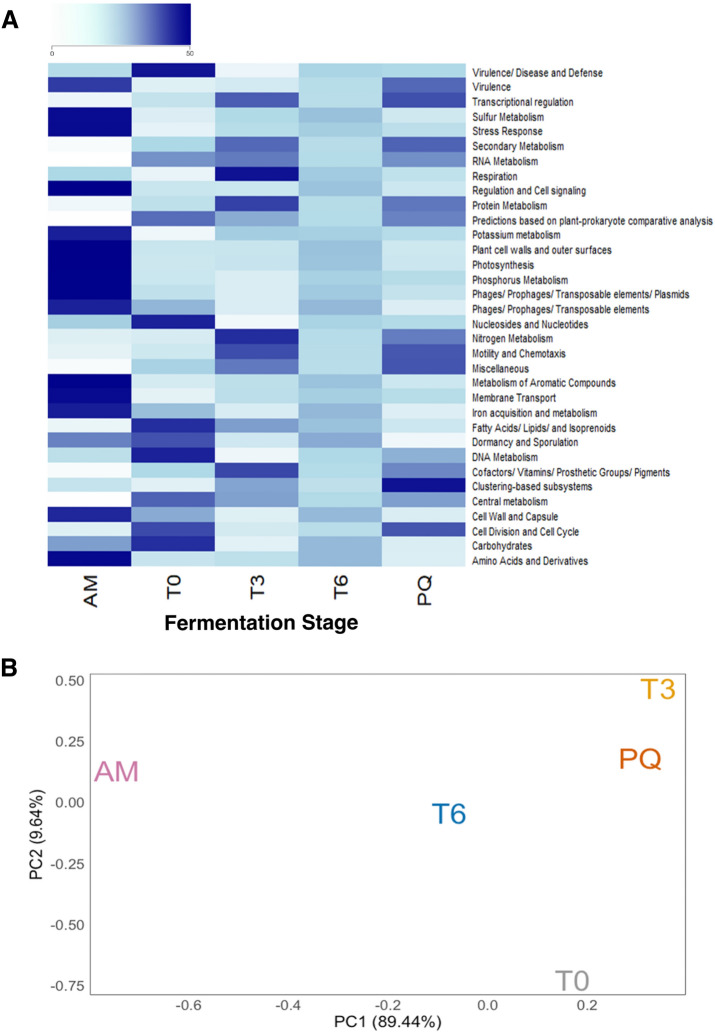
We used the Super-Focus pipeline to evaluate the functional profile of each stage30. We identified 34, 183, 1,153, and 17,544 functional clusters for level 1, level 2, level 3, and the seed level, respectively (Fig. 3A, Supplementary Figure S4A). Consistent with the taxon abundance profiles (Fig. 1), PCA of functional cluster relative abundance showed clear separation between AM to all fermentation stages (Fig. 3B, Supplementary Fig. 4B).
Figure 3.
Metagenomic functional profiles change during pulque fermentation. (A) Heatmap of relative abundance for level-1 functional groups as defined by SUPER-FOCUS30 (X-axis) during pulque fermentation (Y-axis), where dark blue indicates high abundance and light blue indicates low abundance. (B) Principal Component Analysis (PCA) of functional profiles. Principal components 1 and 2 explain > 99% of variance.
Next, we compared the functional profiles of AM versus PQ, and each sequential stage of fermentation (AM vs. T0, T0 vs. T3, T3 vs. T6, and T6 vs. PQ). Presence/absence data for each Super-Focus subsystem across 26 of the 29 bacterial genera present in the Super-Focus database are reported in Supplementary Data S1. For level-2, there were 85 functional groups that displayed significant differences in abundance between AM versus PQ, representing pre-fermentation and final fermentation (Supplementary Table S6). Sixty-three of the significant functional groups were more abundant in AM and 22 functional groups were more abundant in PQ. Interestingly, many of the functional groups enriched in AM were associated with bacterial defense and stress response (e.g. “Bacteriocins, Ribosomally Synthesized Antibacterial Peptides”, “General Stress Response and Stationary Phase Response”, “Osmotic Stress”, “Pathogenicity Islands”, “Phages, Prophages”, “Programmed Cell Death and Toxin-antitoxin Systems”, “Transposable Elements”, “Bacteriophage integration/excision/lysogeny” etc.). Functional groups enriched in PQ included “Carotenoid Biosynthesis”, “Folate and Pterines”, and “Proteolytic Pathway” (Supplementary Table S6).
AM versus T0 compares fresh aguamiel to the backslopping stage, where fresh aguamiel is added to a container with fermented pulque (in the vessel where fermentation takes place). The T0 stage introduces water, sugars (mainly sucrose) (Fig. 2B) and microorganisms present in the fresh aguamiel. Between AM and T0 we identified 71 functional groups in level-2 that were significantly different in abundance. Fifty-four functional groups had a high abundance in AM and 17 had a high abundance in T0 (Supplementary Table S7). Several of the functional groups that were more abundant in AM were associated with primary metabolism, bacterial defense and stress response (e.g. “A bicyclomycin resistance protein, a helicase, and a pseudouridine synthase “, “Fatty Acid Metabolic Cluster”, “General Stress Response and Stationary Phase Response”, “Pathogenicity Islands”, “Programmed Cell Death and Toxin-antitoxin Systems”, “Shiga Toxin Cluster”, “Transposable Elements”). Functional groups with elevated abundance in T0 included “Carotenoid Biosynthesis”, “Proteolytic Pathway”, “Biosynthesis of Phenylpropanoids” and “Lactate Racemization” (Supplementary Table S7).
The T0 versus T3 comparison revealed 27 functional groups with differential abundance, with 14 and 13 at higher abundance in T0 and T3, respectively (Supplementary Table S8S8). Some of the functional groups enhanced in T0 were “Gram-positive Cell Wall Components”, “Acid Stress” and “Bacterial Checkpoint Control”. In T3, we observed elevated abundance of the functional groups “Carotenoid Biosynthesis”, “Inorganic Sulfur assimilation”, and “Proteolytic Pathway”. The T3 versus T6 comparison revealed only 12 functional groups with differential abundance, with 8 and 4 groups displaying elevated abundance in T3 and T6, respectively. Functional groups with higher abundance in T3 included “Polysaccharides” and “Proteolytic Pathway” while functional groups higher in abundance in T6 included “Invasion and Intracellular Resistance” and “Lactate Racemization” (Supplementary Table S9). Finally, 28 functional groups showed differential abundance in T6 versus PQ. We found 24 functional groups that displayed elevated abundance in T6 and 4 functional groups that displayed elevated abundance in PQ. Functional groups elevated in T6 included “Programmed Cell Death and Toxin-antitoxin Systems”, “Metabolism of Central Aromatic Intermediates”, “Lactate racemization”, “Triacylglycerols”. Functional groups elevated in PQ included “Proteolytic pathway”, “Toxins and Superantigens”, “Nucleotide Sugars” and “Plant Alkaloids” (Supplementary Table S10).
Evolutionary relationship of metagenomics assembly of S. cerevisiae from pulque
Because S. cerevisiae is a prominent species found in many fermented foods and beverages, and S. cerevisiae evolutionary history is well-studied31,32, we aimed to assess how S. cerevisiae from pulque is related to other strains isolated from fermentations and other sources. We performed a metagenomics assembly of Illumina reads from PQ using metaSpades33 in order to identify and extract scaffolds assigned to S. cerevisiae. Using Autometa34 and Metabat35, we identified three bins containing S. cerevisiae scaffolds. We recovered 368, 190, and 184 scaffolds totaling 5.07 Mb (Pulque_Metabat03), 5.19 Mb (Pulque_Metabat08), and 3.50 Mb (Pulque_Autometa) in which we predicted 2,753, 2,805, and 1,922 protein-coding genes.
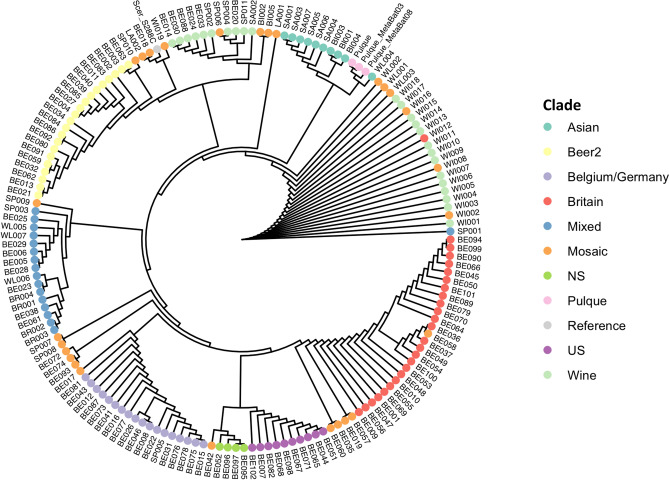
We analyzed the S. cerevisiae pulque metagenomic assembly data with 157 S. cerevisiae strains from Gallone et al.31, and the SC288 reference genome36. We used OrthoFinder37 to identify 7,232 ortholog groups. This set of translated coding sequences were used to construct a phylogenetic tree using the Species Tree from All Genes algorithm (STAG)38. Importantly, this analysis was highly agreeable with results from Gallone et al.31, and placed the majority of isolates into their major lineages (Fig. 4). The S. cerevisiae metagenomic assembly was positioned within the Asian clade with samples isolated from sake and bio-ethanol (Fig. 4).
Figure 4.
Evolutionary relationship of Saccharomyces cerevisiae metagenomics assembly from pulque and isolates from fermented beverages. Phylogenetic tree of 157 isolates from Gallone et al.31, and the S288C reference genome with translated CDS sequences predicted from scaffolds assigned as S. cerevisiae using Autometa (Pulque) and MetaBat2 (Pulque_MetaBat03 and Pulque_MetaBat08). Clades are labeled as in Gallone et al.31. The S. cerevisiae metagenomics assembly from pulque is nested within the “Asian” clade.
Evolutionary relationship of Z. mobilis strains isolated from pulque
We isolated three strains of Z. mobilis from pulque from Huitzilac, a town in Morelos State, Mexico (altitude of 2,550 m in a cold weather mountainous region) and sequenced their genomes in order to determine their evolutionary relationships to other sequenced strains. The batch of pulque from which these strains were isolated from was used in the T0 backslopping stage of our experiment, when fresh aguamiel was mixed with mature pulque. Cumulative genome assembly size ranged from 2.07 to 2.49 Mb, with GC content ranging from 32 to 36%. Using the PhAME pipeline39, we identified 74,825 polymorphic sites between the genomes of the three Z. mobilis strains from pulque and an additional 7 previously sequenced Z. mobilis genomes40–45. Phylogenetic analysis revealed that the pulque isolates we sequenced were monophyletic with a previously sequenced strain ATCC 10988, which was also isolated from pulque (Fig. 5). This relationship was supported with 100% bootstrap support. The pulque clade showed a close proximity with strain ATCC 31821 from Brazil that was originally isolated from sugarcane45. Strains NCIMB 11163 from England and strain NRRL B-1960 from Scotland isolated from beer and cider, respectively, were clustered together. Strains NRRL-B-14023 and NRRL-B-12526, which are clones, composed another group. Strain ATCC 29191 isolated from palm sap in Zaire displayed the most divergent patterns of polymorphism. Phylogenetic network analysis reinforced these relationships (Supplementary Figure S5), as well as the results of a previous phylogenetic analysis of a subset of these strains46.
Figure 5.

Evolutionary relationship of Zymomonas mobilis isolates. Maximum likelihood phylogenetic tree of Z. mobilis isolates with complete genomes or whole-genome sequence data reconstructed from 74,825 genome-wide SNPs using PhaME pipeline39. Bootstrap values are reported for each node. Strains T32, D5, AM2 and ATCC10988 were isolated from pulque and are monophyletic.
Presence of Z. mobilis strains isolated from pulque across fermentation
We assessed the relatively proportion of the sequenced Z. mobilis isolates across fermentation. We carried out two independent methods to identify and quantify the relative abundance of the AM2 and D5/T32 Z. mobilis genotypes from our metagenomics data. D5 the T32 isolates are very closely related and may represent a clonal lineage or the same isolate (Fig. 5). Thus, we grouped these isolates together. First, we identified 380 single nucleotide polymorphisms (SNPs) that differentiate the AM2 and D5/T32 genotypes. We then calculated allele frequency for each locus across fermentation stages. The averaged allele frequencies suggests that the relative frequency of AM2 genotype to the D5/T32 genotype ranges from 67 to 73% (Supplementary Figure S6A). Additionally, we identified lineage specific genes between the AM2 and D5/T32 genotypes and calculated their average coverage across fermentation stages. The trend of this analysis agrees with the SNP analysis, and suggests that the relative frequency of the AM2 genotype ranges from 80%-88%, while the D5/T32 genotype ranges from 12 to 20% (Supplementary Figure S6B). Taken together, our results suggests that the Z. mobilis genotypes were relatively stable across fermentation and that the AM2 genotype was more prevalent in our fermentation than the D5/T32 genotype.
Discussion
We used shotgun metagenomics sequencing to analyze the microbial and functional diversity during pulque fermentation, and metagenomic assembly and whole-genome sequencing to investigate the relationship of S. cerevisiae and Z. mobilis strains from pulque, respectively. These analyses yielded several key findings, which collectively shed light on microbial and functional diversity during pulque fermentation. First, we provide the first direct evidence of Z. mobilis in aguamiel (Figs. 1, 5). Although aguamiel has been a suspected reservoir of Z. mobilis, isolation and detection have remained elusive17,21,24,25,47. For instance, Z. mobilis was not detected in aguamiel from Agave atrovirens and Agave salmiana across four seasons using 16S rDNA sequencing and denaturing gradient gel electrophoresis24, or from aguamiel from three different locations in Hidalgo State using high-throughput 16S rDNA amplicon sequencing25. However, Z. mobilis was identified from aguamiel using selective media, though without validation of species identity with 16S rDNA sequencing12. These results suggest that the presence of Z. mobilis in aguamiel may be variable, and perhaps dependent on a number of factors including agave species, geography, environmental conditions etc.
We identified a cohesive set of organisms that were consistently present across most or all stages of pulque fermentation. These organisms included several species of lactic acid bacteria, acetic acid bacteria, Z. mobilis, and S. cerevisiae (Figs. 1, 2C, D). Interestingly, the acetic acid bacteria genus Acinetobacter made up the highest percentage of organisms in aguamiel (when sucrose concentration is highest), followed by Leuconostoc and Lactococcus (Figs. 1, 2). The abundance of Acinetobacter dropped sharply from 22 to ~ 2.8% during fermentation, while the lactic acid bacteria genera Leuconostoc and Lactococcus remained relatively stable across fermentation (Figs. 1, 2). Importantly, these genera have been found in high abundance in aguamiel and pulque23–25,48. Temporal shifts in microbial composition during fermentation are consistent with most traditionally fermented foods and beverages4,10. In pulque, shifts in microbial abundance likely reflect changes in resource competition (e.g. sucrose consumption, acid and alcohol production and tolerance etc.) (Fig. 2A, B). For example, strains of S. cerevisiae isolated from pulque show higher levels of ethanol tolerance than strains isolated from aguamiel where ethanol content is lower18.
In pulque, the native microbial community is likely related to the traditional non-aseptic conditions during the collection, transportation, and fermentation of aguamiel11,49. For instance, the identification of Z. mobilis from honeybees22 and the practice of filtering aguamiel to remove insects and other debris11 suggests a possible insect transmission of microbial community members. Consistent with these fermentation practices, our analysis showed that the aguamiel stage differed most compared with the other fermentation stages, and was the most diverse (Fig. 1C, D). It is clear that the microbial community of aguamiel is variable24, and these differences could translate into regional, seasonal, and environmental differences in the microbial communities of pulque.
There is a long history reporting the nutritional benefits associated with low-level consumption of pulque. For instance, pulque is a major source of vitamin C, and was used to treat scurvy in Mexican penitentiary inmates in the late 1800s11. Supporting this observation, more recently, a study of dietary patterns in rural central Mexico revealed that pulque is the most important source of ascorbic acid50. Another early study of pulque from 1946 in the indigenous Otomí population in Hidalgo state demonstrated that pulque was the second most important food in the diet after tortilla, because it provided substantial amounts of calories, total protein, thiamin, riboflavin, niacin, vitamin C, calcium, and iron11. More recent analysis supports the importance of pulque as a dietary source of iron and folates50,51. Folic acid deficiency during pregnancy can lead to neural tube defects52, and, importantly, pulque intake is a strong indicator of folate status in rural Central Mexican populations50. Our functional enrichment analysis suggests that microbial genes involved in folate biosynthesis are present in all major bacterial genera and significantly more abundant in pulque than in aguamiel (Supplementary Table S6, Supplementary Data S1). However, it is important to note that while low or moderate pulque intake during pregnancy and lactation may have some positive health benefits, heavy pulque intake during lactation is significantly associated with adverse postnatal growth53.
Though pulque fermentation was dominated by bacteria, we identified 6 fungal species that were present ≥ 1% in at least one stage of fermentation (Fig. 1, Supplementary Table S4). In agreement with previous work23, we observed a drastic increase in yeast abundance when mature pulque was mixed with fresh aguamiel (T0 stage) (Figs. 1, 2). S. cerevisiae was the most abundant species at each stage, but S. eubayanus, S. arboricola, S. kudriavzevii, and S. cerevisiae x S. kudriavzevii hybrids were all detected ≥ 0.1% during all stages of fermentation with the exception of AM. Interestingly, S. eubayanus and S. kudriavzevii are both cryotolerant species54,55. The thermotolerant yeast K. marxianus was also present at ≥ 0.1% during all stages of fermentation. K. marxianus has been previously identified from aguamiel and pulque18,25,56,57, as well as from agave used to ferment other traditionally distilled beverages15, and can ferment a more diverse and complex set of substrates than Saccharomyces species58–61. The observation that Saccahromyces species were not significantly correlated with ethanol production, but K. marxianus was, may suggest unequal roles for yeasts in alcohol production during pulque fermentation.
Lastly, because Z. mobilis is the dominant species in pulque and contributes to the alcohol content of pulque11,14,23, we isolated three Z. mobilis strains from pulque to understand their evolutionary relationship with previously sequenced isolates. Using maximum likelihood phylogenetic analysis and phylogenetic network analysis of ~ 74825 SNPs scattered across the genome, our results show a distinct “pulque” clade made up of the three strains sequenced here, and a previously sequenced strain isolated from pulque (ATCC 10988) (Fig. 5, Supplementary Figure S5). The presence of monophyletic groups from particular fermented food sources is indicative of microbial domestication31,62,63. However, we acknowledge that our results could be the outcome of sampling bias. Assessing whether Z. mobilis strains isolated from pulque represent a domesticated lineage will require more extensive sampling of pulque and environmental strains across Mexico and globally, as well as phenotypic work to asses characteristics unique to pulque derived isolates.
Materials and methods
Laboratory pulque fermentation
Pulque (PQ) (approximately 12 h of overnight fermentation) and fresh extracted aguamiel (AM) samples were collected from the town of Huitzilac, Morelos State, Mexico (altitude of 2,550 m in a cold weather mountainous region, 19°01′42″N 99°16′02″W). Samples were placed in sterile plastic bags and transported immediately to the laboratory. A laboratory fermentation was carried out in a 5 L sterile plastic container and was initiated by mixing collected fermented pulque and fresh aguamiel (3:2 v/v) (as recommended by the local pulque supplier). Aliquots for metagenomic DNA extraction were sampled immediately after mixing PQ and fresh AM (0 h, T0), at 3 h (T3), at 6 h (T6), and from AM and fermented pulque (PQ). The concentration of sucrose, glucose, fructose, ethanol, lactic-, and acetic acids were determined in all samples using a Waters HPLC system, equipped with an Aminex column for fermentation analysis as previously reported23.
Illumina shotgun sequencing of pulque fermentation
Four 10 mL aliquot from each fermentation stage (AM, T0, T3, T6 and PQ) was centrifuged at 10,000×g for 40 min at 4° C to sediment the total cells present in each sample. The pellet was washed with 10 mL of 1× PBS buffer 3 times. Next, cells were successively lysed by resuspending the pellet in 9.5 mL TE buffer and 0.5 mL SDS 10% and adding (i) 5 mg crystalline lysozyme from chicken egg white (Sigma) for 40 min at 42° C, and (ii) by adding 3 µL of 20 mg/mL of proteinase K (Sigma) for 10 min at 60° C. Lysates were treated with 1.8 mL NaCl 5 M and 1.5 mL of CTAB/NaCl and incubated for 10 min at 65° C. DNA was extracted by adding one volume of chloroform/isoamyl alcohol (24:1). DNA recovered from the aqueous phase was purified via silica gel spin filtration (Collection Tube 2 of the MoBio kit). After this step, we followed the instructions in the MoBio Kit. (MoBio Cat. no. 12224-250). Finally, DNA resuspended with TRIS buffer pH 8.0.
Samples were sequenced on a MiSeq Illumina Sequencer (Instituto Nacional de Medicina Genómica, Mexico City) producing paired-end 145 bp reads. Four technical replicates were sequenced at each stage (AM, T0, T3, T6, and PQ). Raw sequencing data was improved as follows: first identical read pairs, which likely represent PCR duplicates, were collapsed using Tally64. Next, residual adapter sequences were trimmed from reads using Trim Galore v0.4.3 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with the “--stringency = 1” parameter. Trim galore was also used to trim reads at bases with quality scores < Q30. Read pairs were discarded when one read was < 50 bp. Quality improved read sets were inspected via FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Raw metagenomics is available in the NCBI Sequence Read Archive under BioProject accession PRJNA603591.
Bioinformatic analysis of microbial composition and functional profiles
Microbial composition and abundance for all stages and all replicates was predicted using MetaPhlan 2.028 and Kaiju v1.7.329. MetaPhlan relies on unique clade specific markers from a database of over 3,000 genomes to make taxonomic predictions at various levels. We used the BowTie2 --bt2_ps “very-sensitive” preset parameters, --tax_lev ‘a’ for prediction of all taxonomic levels, “--min_cu_len 2000” for minimum total nucleotide length for the markers in the clade, and “--stat_q 0,1” for quantile value. Kaiju was also used for taxonomic classification of shotgun metagenomics reads. Kaiju searches reads against the NCBI NR database and, because of the database size, has better resolution at the species level. Kaiju was run using the “greedy” algorithm against the NCBI BLAST nr + euk database which consists of over 100 million NCBI NR protein sequences from bacteria, archaea, viruses, fungi and other microbial eukaryotes.
Metagenomic data was also used to predict the functional profile of each stage and each replicate, using SUPER-FOCUS30. This tool reports functional annotation for 4 subsystem levels (levels 1–3 and seed function) using CD-HIT65. Briefly, SUPER-FOCUS identifies the taxonomic profile of the data and creates a database with the subsystems for predicted organism. Metagenomic data was aligned against the database using RAPSearch266. Sequences with e-values ≤ 1e−5, a minimum identity of 60%, and an alignment length ≥ 15 amino acids were retained. Output with the subsystem levels ranges from general function (level 1) to a specific function (seed level). We identified level 2 functional groups that were significantly different between AM versus PQ, AM versus T0, T0 versus T3, T3 versus T6, and T6 versus PQ. We considered functional groups significantly different in abundance when (a) there was ≥ 1.5-fold difference in relative abundance between groups, and (b) Pearson's chi-squared test with absolute counts for each group yielded p values ≤ 0.000273 (Bonferroni corrected p value cutoff; p value = 0.05/183 level 2 categories). Pearson's chi-squared test was performed in JMP Pro 14.0.0. To identify the SUPERFOCUS subsystem presence and absence patterns across the major genera identified with Kaiju, we extracted organism to pathway information from the “organisms2subsystem.txt” file and linked pathway numbers to subsystems using the “database_PKs.txt” file, both of which were obtained from the “db” directory in the SUPERFOCUS package.
Statistical analysis and data visualization
Statistical analyses were performed in R.3.2.267 and JMP Pro 14.0.0. Principal component analysis (PCA) was performed using the prcomp function to compare species relative abundance and functional group relative abundance across all stages. Data visualization was performed using ggplot268, Heatplus69, ggfortify70 and RColorBrewer71 packages.
Evolutionary analysis of assembled S. cerevisiae metagenomics scaffolds from pulque
We used the cleaned and trimmed metagenomics read sets from the PQ fermentation stage to perform a metagenomics assembly using MetaSPAdes33, with a k-mer range of 33, 43, 53, 63 and 73. Contigs or scaffolds ≥ 2,000 bp were retained for subsequent analysis. Autometa34 and MetaBAT235 binning algorithms were used to independently recover contigs and scaffolds identified as S. cerevisiae.
We analyzed S. cerevisiae contigs/scaffolds identified from the metagenomic data with 157 S. cerevisiae strains from Gallone et al.31 and the reference 288C genome. To eliminate bias in gene prediction stemming from different approaches, we performed gene prediction in all genome assemblies with Augustus72 using the S. cerevisiae training set. Translated coding sequence (CDS) files were used to identify orthologous groups with Orthofinder37 and to perform phylogenetic analysis. Orthofinder was run using diamond v0.8.2273 and distant matrices were inferred by dendroblast74. The species tree was inferred from unrooted orthogroup gene trees and a consensus tree was estimated using the STAG algorithm38. Tree visualization was performed using the ggtree75 R package.
Whole-genome analysis of Z. mobilis isolates from pulque
We isolated three strains of Z. mobilis from the laboratory pulque fermentation described above. We modified a previously described method for Zymomonas enrichment and isolation76. Briefly, a 5 mL aliquot of pulque was taken after 3 h of fermentation, transferred to a 50 mL Falcon tube containing 30 mL of enrichment broth76 and incubated at 30° C for 5 h. Serial dilutions were performed and plated in agar Zm containing 3 g/L malt extract, 3 g/L yeast extract, 20 g/L glucose, 5 g/L peptone (all reagents from DIFCO) and 1 µg/mL of cycloheximide (Fluka). Plates were incubated overnight (ON) in an anaerobic jar at 30 °C. Colonies were transferred to a fresh agar Zm plate and incubated in an anaerobic jar at 30 °C ON. Colonies were verified visually by microscopy for purity and gram stain. Selected colonies were cultured in 3 mL of Zm broth (pH 6.8) at 30 °C, ON and screened for ethanol smell and gas production. Chromosomal DNA of selected colonies was extracted with the UltraClean Microbial DNA extraction kit (MoBio). 16S rDNA was PCR amplified and sequenced as described previously23 and resultant sequences were identified in the NCBI non-redundant database.
Library preparation and whole-genome paired-end 152 bp Illumina sequencing was conducted at Macrogen (Rockville, Maryland). Improvement of raw sequencing data was performed as follows: First identical read pairs, which likely represent PCR duplicates, were collapsed using Tally64. Next, residual adapter sequences were trimmed from reads using TrimGalore v0.4.3 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with the “--stringency = 1” parameter. TrimGalore was also used to trim reads at bases with quality scores < Q30. Read pairs were discarded when one read was < 50 bp. Finally, error correction was performed with SPAdes v3.1077. We used the Unicycler pipeline78, which uses SPAdes v3.10, to assemble the Z. mobilis genomes. We used the “--careful” “-k 21,35,47,57,67,73,81,85,91,95” parameters in SPAdes for genome assembly. Raw whole-genome Illumina sequence data is available in the NCBI Sequence Read Archive under BioProject accession PRJNA603591.
Evolutionary relationships of Z. mobilis isolates
We used the PhaME package39 to identify single nucleotide polymorphisms between seven publicly available Z. mobilis genomes and the three Z. mobilis genome we isolated from pulque. Strains extracted from the NCBI database included (strain (GenBank accession number)): ATCC31821 (ASM710v1), ATCC10988 (ASM17525v2), ATCC29191 (ASM27775v1), NRRL B-14023 (ASM57616v1), NRRL B-12526 (ASM57612v1), NRRL B-1960 (ASM215884v1), NCIMB 11163 (ASM2424v1)40–45. We used the ATCC31821 genome as the reference during PHaME analysis. The alignment consisted of 74,825 polymorphic sites. This data was used to construct a maximum likelihood phylogenetic tree using RaxML79. We implemented the generalized time reversible model (GTR) with 100 bootstrap replicates. Additionally, we used SplitTree4 to build a phylogenetic network in order to assess recombination between isolates80. We used the NeighborNet method with 100 bootstrap replicates.
Measuring the relative abundance of sequenced Z. mobilis genotypes during fermentation
We predicted the relative proportion of the sequenced Z. mobilis isolates across each stage of fermentation. D5 the T32 isolates are very closely related and may represent a clonal lineage or the same isolate (Fig. 5). Thus, we grouped these isolates together. We performed two independent analyses to identify and quantify the relative abundance of the AM2 and D5/T32 genotypes. First, reads for each isolate were mapped against the reference Z. mobilis ATCC10988 genome44 using bwa mem v0.7.1581 with default settings. We chose this reference genome because it was isolate from pulque and closely related to our sequenced strains (Fig. 5). Next, sorted bam files were generated with samtools v1.4.182 and read group information was added to each sorted bam file using the bamaddrg program (https://github.com/ekg/bamaddrg). We then used freebayes v1.3.1 to identify variants using the parameters “--ploidy 1” and “-C 40”83. Next, we used vcftools v0.1.1484 to filter our variant file with the following parameters “-remove-indels”, “--remove-filtered-all”, “--min-meanDP 20”, “--minQ 20”, “--recode” and “--recode-INFO-all”. Finally, we used GATK to convert the VCF file to table format85. Using these criteria, we identified 380 SNP that differentiated the AM2 and D5/T32 genotypes. Next, using the procedure above, we mapped metagenomics reads for each fermentation stage against the Z. mobilis ATCC10988 genome and used the samtools “mpileup” function to quantify allele frequency at each of the 380 polymorphic sites. We averaged the 380 allele frequencies for the AM2 and D5/T32 genotypes to quantify the relative proportion of each genotype.
Additionally, we used the LS-BSR pipeline86 with default settings to identify lineage-specific genes between the genome assemblies of AM2, D5, and ATCC10988. We used the ATCC10988 genome because it was the reference genome used for metagenomics mapping and genes uniquely shared by AM2 and ATCC10988 or D5 and ATCC10988 could have falsely inflated relative abundance estimates. We used the “compare_BSR.py” script included in LS-BSR software to detect genes that were unique to ATCC10988, AM2 and D5. We identified 24 genes unique to AM2 and 7 genes unique to D5. We collected all consensus genes between the three isolates and merged them into a single fasta file which was used as a mapping reference file for the metagenomics read sets. Using the approach above, we mapped read sets from each fermentation stage against the gene consensus fasta file, and then used the samtools “depth” function to calculate read depth across the 31 genes unique to either AM2 or D5. We averaged read depth values for each gene across fermentation stages and divided the AM2 average read depth by the sum of the averaged value of AM2 and D5 average read depths for each stage. This value gives the relative abundance estimate of the AM2 genotype.
Supplementary information
Acknowledgements
This work was supported by Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT), UNAM projects IN207914 and IN207917.
Author contributions
K.C.V., M.G.-G., J.T., A.E., and J.G.G. designed the experiments and analysis, and wrote and reviewed the manuscript.
Data availability
Raw Illumina shotgun metagenomics data from aguamiel and pulque fermentation and Raw whole-genome Illumina sequence data from three Z. mobilis strains isolated from pulque are available through the NCBI Sequence Read Archive under BioProject accession PRJNA603591.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Adelfo Escalante, Email: adelfo@ibt.unam.mx.
John G. Gibbons, Email: jggibbons@umass.edu
Supplementary information
is available for this paper at 10.1038/s41598-020-71864-4.
References
- 1.Gibbons JG, Rinker DC. The genomics of microbial domestication in the fermented food environment. Curr. Opin. Genet. Dev. 2015;35:1–8. doi: 10.1016/j.gde.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steensels J, Gallone B, Voordeckers K, Verstrepen KJ. Domestication of industrial microbes. Curr. Biol. 2019;29:R381–R393. doi: 10.1016/j.cub.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 3.Douglas GL, Klaenhammer TR. Genomic evolution of domesticated microorganisms. Annu. Rev. Food Sci. Technol. 2010;1:397–414. doi: 10.1146/annurev.food.102308.124134. [DOI] [PubMed] [Google Scholar]
- 4.Bokulich NA, Ohta M, Lee M, Mills DA. Indigenous Bacteria and Fungi Drive Traditional Kimoto Sake Fermentations. Appl. Environ. Microb. 2014;80:5522–5529. doi: 10.1128/Aem.00663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kergourlay G, Taminiau B, Daube G, Verges MCC. Metagenomic insights into the dynamics of microbial communities in food. Int. J. Food. Microbiol. 2015;213:31–39. doi: 10.1016/j.ijfoodmicro.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Jung JY, et al. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 2011;77:2264–2274. doi: 10.1128/AEM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Sullivan DJ, et al. Temporal and spatial differences in microbial composition during the manufacture of a continental-type cheese. Appl. Environ. Microbiol. 2015;81:2525–2533. doi: 10.1128/AEM.04054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamang JP, et al. Fermented foods in a global age: East meets West. Comprehen. Rev. Food Sci. Food Saf. 2020 doi: 10.1111/1541-4337.12520. [DOI] [PubMed] [Google Scholar]
- 9.Tamang JP, Watanabe K, Holzapfel WH. Review: Diversity of Microorganisms in Global Fermented Foods and Beverages. Front. Microbiol. 2016 doi: 10.3389/fmicb.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe BE, Dutton RJ. Fermented foods as experimentally tractable microbial ecosystems. Cell. 2015;161:49–55. doi: 10.1016/j.cell.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 11.Escalante A, et al. Pulque, a Traditional Mexican alcoholic fermented beverage: Historical, microbiological, and technical aspects. Front. Microbiol. 2016;7:1026. doi: 10.3389/fmicb.2016.01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valadez-Blanco R, Bravo-Villa G, Santos-Sanchez NF, Velasco-Almendarez SI, Montville TJ. The artisanal production of pulque, a traditional beverage of the Mexican Highlands. Probiotics Antimicro. 2012;4:140–144. doi: 10.1007/s12602-012-9096-9. [DOI] [PubMed] [Google Scholar]
- 13.Correa-Ascencio M, Robertson IG, Cabrera-Cortes O, Cabrera-Castro R, Evershed RP. Pulque production from fermented agave sap as a dietary supplement in Prehispanic Mesoamerica. Proc. Natl. Acad. Sci. USA. 2014;111:14223–14228. doi: 10.1073/pnas.1408339111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escalante A, et al. Characterization of bacterial diversity in Pulque, a traditional Mexican alcoholic fermented beverage, as determined by 16S rDNA analysis. Fems Microbiol. Lett. 2004;235:273–279. doi: 10.1016/j.femsle.2004.04.045. [DOI] [PubMed] [Google Scholar]
- 15.Lappe-Oliveras P, et al. Yeasts associated with the production of Mexican alcoholic nondistilled and distilled Agave beverages. Fems Yeast Res. 2008;8:1037–1052. doi: 10.1111/j.1567-1364.2008.00430.x. [DOI] [PubMed] [Google Scholar]
- 16.Torres-Rodriguez I, et al. Screening and characterization of extracellular polysaccharides produced by Leuconostoc kimchii isolated from traditional fermented pulque beverage. Springerplus. 2014 doi: 10.1186/2193-1801-3-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escalante-Minakata P, Blaschek HP, de la Rosa APB, Santos L, De Leon-Rodriguez A. Identification of yeast and bacteria involved in the mezcal fermentation of Agave salmiana. Lett. Appl. Microbiol. 2008;46:626–630. doi: 10.1111/j.1472-765X.2008.02359.x. [DOI] [PubMed] [Google Scholar]
- 18.Estrada-Godina AR, et al. Isolation and identification of killer yeasts from agave sap (aguamiel) and pulque. World J. Microb. Biot. 2001;17:557–560. doi: 10.1023/A:1012210106203. [DOI] [Google Scholar]
- 19.Narvaez-Zapata JA, Rojas-Herrera RA, Rodriguez-Luna IC, Larralde-Corona CP. Culture-independent analysis of lactic acid bacteria diversity associated with mezcal fermentation. Curr. Microbiol. 2010;61:444–450. doi: 10.1007/s00284-010-9636-z. [DOI] [PubMed] [Google Scholar]
- 20.Swings J, De Ley J. The biology of Zymomonas. Bacteriol Rev. 1977;41:1–46. doi: 10.1128/MMBR.41.1.1-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir PM. The ecology of Zymomonas: A review. Folia Microbiol. (Praha) 2016;61:385–392. doi: 10.1007/s12223-016-0447-x. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz-Argueso T, Rodriguez-Navarro A. Microbiology of ripening honey. Appl. Microbiol. 1975;30:893–896. doi: 10.1128/AEM.30.6.893-896.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escalante A, et al. Analysis of bacterial community during the fermentation of pulque, a traditional Mexican alcoholic beverage, using a polyphasic approach. Int. J. Food Microbiol. 2008;124:126–134. doi: 10.1016/j.ijfoodmicro.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Morales SLV, et al. Metagenomic microbial diversity in Aguamiel from two agave species during 4-year seasons. Food Biotechnol. 2019;33:1–16. doi: 10.1080/08905436.2018.1547200. [DOI] [Google Scholar]
- 25.Rocha-Arriaga, C. et al. Deep microbial community profiling along the fermentation process of pulque, a major biocultural resource of Mexico. bioRxiv10.1101/718999 (2019). [DOI] [PubMed]
- 26.De Filippis F, Parente E, Ercolini D. Metagenomics insights into food fermentations. Microb. Biotechnol. 2017;10:91–102. doi: 10.1111/1751-7915.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh AM, Crispie F, Claesson MJ, Cotter PD. Translating Omics to Food Microbiology. Annu. Rev. Food. Sci. Technol. 2017;8:113–134. doi: 10.1146/annurev-food-030216-025729. [DOI] [PubMed] [Google Scholar]
- 28.Truong DT, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 29.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016;7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva GGZ, Green KT, Dutilh BE, Edwards RA. SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data. Bioinformatics. 2016;32:354–361. doi: 10.1093/bioinformatics/btv584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallone B, et al. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell. 2016;166:1397. doi: 10.1016/j.cell.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peter J, et al. Genome evolution across 1011 Saccharomyces cerevisiae isolates. Nature. 2018;556:339. doi: 10.1038/s41586-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller IJ, et al. Autometa: automated extraction of microbial genomes from individual shotgun metagenomes. Nucleic Acids Res. 2019;47:e57. doi: 10.1093/nar/gkz148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang DD, et al. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. Peerj. 2019;7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goffeau A, et al. Life with 6000 genes. Science. 1996;274(546):563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 37.Emms DM, Kelly S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emms, D. M. & Kelly, S. STAG: Species tree inference from all genes. bioRxiv. 10.1101/267914 (2018).
- 39.Ahmed, S. A., Lo, C. C., Davenport, K. W. & Chain, P. S. G. From raw reads to trees: Whole genome SNP phylogenetics across the tree of life. bioRxiv. 10.1101/032250 (2015).
- 40.Chacon-Vargas, K. et al. Genome sequence of Zymomonas mobilis subsp. mobilis NRRL B-1960. Genome Announc.. 10.1128/genomeA.00562-17 (2017). [DOI] [PMC free article] [PubMed]
- 41.Desiniotis A, et al. Complete genome sequence of the ethanol-producing Zymomonas mobilis subsp mobilis Centrotype ATCC 29191. J. Bacteriol. 2012;194:5966–5967. doi: 10.1128/Jb.01398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kouvelis VN, et al. Complete genome sequence of the ethanol producer Zymomonas mobilis NCIMB 11163. J. Bacteriol. 2009;191:7140–7141. doi: 10.1128/Jb.01084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kouvelis, V. N. et al. Finished genome of Zymomonas mobilis subsp. mobilis Strain CP4, an applied ethanol producer. Genome Announc.. 10.1128/genomeA.00845-13 (2014). [DOI] [PMC free article] [PubMed]
- 44.Pappas, K. M. et al. Genome sequence of the ethanol-producing Zymomonas mobilis subsp. mobilis lectotype strain ATCC 10988. J. Bacteriol.193, 5051–5052. 10.1128/JB.05395-11 (2011). [DOI] [PMC free article] [PubMed]
- 45.Seo JS, et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 2005;23:63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C, et al. Genome comparison of different Zymomonas mobilis strains provides insights on conservation of the evolution. PLoS ONE. 2018;13:e0195994. doi: 10.1371/journal.pone.0195994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enriquez-Salazar MI, et al. Microbial diversity and biochemical profile of aguamiel collected from Agave salmiana and A-atrovirens during different seasons of year. Food Sci. Biotechnol. 2017;26:1003–1011. doi: 10.1007/s10068-017-0141-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diana CR, Humberto HS, Jorge YF. Probiotic properties of leuconostoc mesenteroides isolated from Aguamiel of Agave salmiana. Probiotics Antimicro. 2015;7:107–117. doi: 10.1007/s12602-015-9187-5. [DOI] [PubMed] [Google Scholar]
- 49.Villarreal-Morales SL, Montanez-Saenz JC, Aguilar-Gonzalez CN, Rodriguez-Herrera R. Metagenomics of traditional beverages. Handb. Food Bioeng. 2018;14:301–326. doi: 10.1016/B978-0-12-811443-8.00011-6. [DOI] [Google Scholar]
- 50.Backstrand JR, Allen LH, Black AK, de Mata M, Pelto GH. Diet and iron status of nonpregnant women in rural Central Mexico. Am. J. Clin. Nutr. 2002;76:156–164. doi: 10.1093/ajcn/76.1.156. [DOI] [PubMed] [Google Scholar]
- 51.Backstrand JR, Allen LH, Martinez E, Pelto GH. Maternal consumption of pulque, a traditional central Mexican alcoholic beverage: Relationships to infant growth and development. Public Health Nutr. 2001;4:883–891. doi: 10.1079/phn2001130. [DOI] [PubMed] [Google Scholar]
- 52.Berry, R. J. et al. Prevention of neural-tube defects with folic acid in China. China–U.S. collaborative project for neural tube defect prevention. N. Engl. J. Med.341, 1485–1490. 10.1056/NEJM199911113412001 (1999). [DOI] [PubMed]
- 53.Backstrand JR, Goodman AH, Allen LH, Pelto GH. Pulque intake during pregnancy and lactation in rural Mexico: Alcohol and child growth from 1 to 57 months. Eur. J. Clin. Nutr. 2004;58:1626–1634. doi: 10.1038/sj.ejcn.1602019. [DOI] [PubMed] [Google Scholar]
- 54.Boynton PJ, Greig D. The ecology and evolution of non-domesticated Saccharomyces species. Yeast. 2014;31:449–462. doi: 10.1002/yea.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hittinger CT. Saccharomyces diversity and evolution: A budding model genus. Trends Genet. 2013;29:309–317. doi: 10.1016/j.tig.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Cruz-Guerrero, A. E., Olvera, J. L., García-Garibay, M. & Gómez-Ruiz, L. Inulinase-hyperproducing strains of Kluyveromyces sp. isolated from aguamiel (Agave sap) and pulque. World j. Microbiol. Biotechnol.22 (2006).***
- 57.Villarreal-Morales, S. L. et al. Metagenomic microbial diversity in Aguamiel from two agave species during 4-year seasons. Food Biotechnol.33 (2018).***
- 58.Fonseca GG, Heinzle E, Wittmann C, Gombert AK. The yeast Kluyveromyces marxianus and its biotechnological potential. Appl. Microbiol. Biotechnol. 2008;79:339–354. doi: 10.1007/s00253-008-1458-6. [DOI] [PubMed] [Google Scholar]
- 59.Lertwattanasakul N, et al. Genetic basis of the highly efficient yeast Kluyveromyces marxianus: complete genome sequence and transcriptome analyses. Biotechnol. Biofuels. 2015;8:47. doi: 10.1186/s13068-015-0227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margaritis A, Bajpai P. Direct fermentation of d-xylose to ethanol by Kluyveromyces marxianus strains. Appl. Environ. Microbiol. 1982;44:1039–1041. doi: 10.1128/AEM.44.5.1039-1041.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nonklang S, et al. High-temperature ethanol fermentation and transformation with linear DNA in the thermotolerant yeast Kluyveromyces marxianus DMKU3-1042. Appl. Environ. Microbiol. 2008;74:7514–7521. doi: 10.1128/AEM.01854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dumas E, et al. Independent domestication events in the blue-cheese fungus Penicillium roqueforti. Mol Ecol. 2018;29:2639–2660. doi: 10.1111/mec.15359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gibbons, J. G. et al. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr. Bio.l22, 1403–1409. 10.1016/j.cub.2012.05.033 (2012). [DOI] [PMC free article] [PubMed]
- 64.Davis MP, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods. 2013;63:41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li WZ, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 66.Zhao YA, Tang HX, Ye YZ. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2012;28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R: A Language and Environment for Statistical Computing (2018).
- 68.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Berlin: Springer; 2009. [Google Scholar]
- 69.Ploner, A. Heatplus: Heatmaps with Row and/or Column Covariates and Colored Clusters (2019). https://github.com/alexploner/Heatplus.
- 70.Horikoshi , M. & Tang, Y. C. ggfortify: Data Visualization Tools for Statistical Analysis Results (2016). https://github.com/sinhrks/ggfortify.
- 71.Neuwirth, E. RColorBrewer: ColorBrewer Palettes (2014). https://github.com/cran/RColorBrewer
- 72.Stanke, M. & Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics19 Suppl 2, ii215–ii225. 10.1093/bioinformatics/btg1080 (2003). [DOI] [PubMed]
- 73.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 74.Kelly S, Maini PK. DendroBLAST: approximate phylogenetic trees in the absence of multiple sequence alignments. PLoS ONE. 2013;8:e58537. doi: 10.1371/journal.pone.0058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu G, Lam TT, Zhu H, Guan Y. Two methods for mapping and visualizing associated data on phylogeny using Ggtree. Mol. Biol. Evol. 2018;35:3041–3043. doi: 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Obire O. Activity of Zymomonas species in palm-sap obtained from three areas in Edo State, Nigeria. J. Appl. Sci. Environ. Manag. 2005;9:25–30. [Google Scholar]
- 77.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol.13, 10.1371/journal.pcbi.1005595 (2017). [DOI] [PMC free article] [PubMed]
- 79.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 81.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marth GT, et al. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- 84.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sahl JW, Caporaso JG, Rasko DA, Keim P. The large-scale blast score ratio (LS-BSR) pipeline: A method to rapidly compare genetic content between bacterial genomes. Peerj. 2014 doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw Illumina shotgun metagenomics data from aguamiel and pulque fermentation and Raw whole-genome Illumina sequence data from three Z. mobilis strains isolated from pulque are available through the NCBI Sequence Read Archive under BioProject accession PRJNA603591.