Abstract
Purpose of Review:
This article introduces the roles of perinatal DNA methylation in human health and disease, highlights the challenges of tissue and cellular heterogeneity to studying DNA methylation, summarizes approaches to overcome these challenges, and offers recommendations in conducting research in environmental epigenetics.
Recent Findings:
Epigenetic modifications are essential for human development and are labile to environmental influences, especially during gestation. Epigenetic dysregulation is also a hallmark of multiple diseases. Environmental epigenetic studies routinely measure DNA methylation in readily available tissues. However, tissues and cell types exhibit specific epigenetic patterning and heterogeneity between samples complicates epigenetic studies. Failure to account for cell type heterogeneity limits identification of biological mechanisms and biases study results.
Summary:
Tissue-level epigenetic measures represent a convolution of epigenetic signals from individual cell types. Tissue-specific epigenetics is an evolving field and the use of disease-affected target, surrogate, or multiple tissues has inherent trade-offs and affects inference. Likewise, experimental and bioinformatic approaches to accommodate cell type heterogeneity have varying assumptions and inherent trade-offs that affect inference. The relationships between exposure, disease, tissue-level DNA methylation, cell type-specific DNA methylation, and cell type heterogeneity must be carefully considered in study design and analysis. Causal diagrams can inform study design and analytic strategies. Properly addressing cell type heterogeneity limits sources of potential bias, avoids misinterpretation of study results, and allows investigators to distinguish shifts in cell type proportions from direct changes to cellular epigenetic programming, both of which provide insights into environmental disease etiology and aid development of novel methods for prevention and treatment.
Keywords: epigenetics, environment, DNA Methylation, cellular heterogeneity, epidemiology, tissue
Introduction to Epigenetics
Epigenetics refers to the programming of cellular state, memory, or fate not attributable to changes in DNA sequence [1, 2]. Epigenetic modifications include DNA methylation, histone modifications, and non-coding RNAs [3]. DNA methylation describes the methylation of the fifth carbon of the nucleotide cytosine. Advancements in DNA methylation measurement tools, particularly microarrays, have made DNA methylation a popular and relatively inexpensive measure in large studies.
DNA methylation undergoes drastic reprogramming during mammalian development [4, 5] and is a key regulator of cellular differentiation [6]. Epigenetic dysregulation is a hallmark of multiple diseases, including cancer [7], neurodegeneration [8], and cardiovascular disease [9]. Critically, epigenetic modifications are labile to environmental influences, especially during gestation [10]. Understanding how environmental exposures impact epigenetic regulation during development likely will impact our understanding of disease etiology and identify novel methods for prevention and treatment [11]. This article introduces the roles of perinatal DNA methylation in human health and disease, highlights the challenges of tissue and cellular heterogeneity to studying DNA methylation, summarizes approaches to overcome these challenges, and offers recommendations in conducting research in environmental epigenetics.
Epigenetic Alterations During Development
Epigenetics are essential for several aspects of human development. First, DNA methylation undergoes dynamic changes during embryonic development. In the preimplantation embryo, the paternal genome experiences rapid, widespread DNA demethylation. Meanwhile, the maternal genome is passively demethylated to a lesser extent through replication without DNA methylation maintenance [12]. Second, during the reprogramming process, DNA methylation is maintained at specific locations in both the paternal and maternal genomes, termed genomic imprinting [13]. Third, X-chromosome inactivation is a dosage compensation mechanism that randomly transcriptionally silences one of two X chromosomes in females. [14]. Fourth, as tissues differentiate during embryogenesis, they acquire more specialized epigenetic marks. Broad regions of the epigenome are often regulated in concert and we observe larger-scale tissue specific differentially methylated regions [15]. Epigenetic marks help to lock tissues in their differentiation state and maintain tissue identity. Appropriate epigenetic regulation is essential for healthy development and these processes can be dysregulated by environmental exposures.
Tissue Specificity in Environmental Epigenetics
Different tissues have biologically determined epigenetic differences. When designing an epigenetic study, a crucial step is to determine the tissue of interest. A given tissue may be the “target” tissue for a disease process or exposure effects, while another tissue may be a measurable “surrogate” tissue for monitoring biomarkers associated with disease or exposure. Compelling arguments can be made for selecting target tissues (such as brain or lung) based on their direct links to disease. However, the postmortem timing of acquisition for many target tissues, scarcity of exposure and confounder data in most tissue banks, and often modest available sample sizes present challenges for target tissue research [16]. Epigenetic analyses on easily collected surrogate tissues (such as blood or saliva) may be less connected to the disease. Despite this, surrogate tissue research can provide valuable contributions related to etiologic timing through longitudinal sampling, identification of associations with environmental exposures, defining early biomarkers, developing translational utility, and even providing mechanistic insights in some cases [16]. The field of environmental epigenetics is strengthened by evidence from both surrogate and target tissue studies.
Traditionally, studies have investigated a single tissue of interest (either surrogate or target) at a time. Critical investigation of multiple tissues simultaneously is essential to identify similarities in epigenetic profiles across normal, diseased, and exposed tissues. Evidence on DNA methylation correlations across normal and diseased tissues vary. For example, correlation in DNA methylation signatures between surrogate and brain tissue was limited to few informative sites that varied by brain region [17]. Schizophrenia-associated DNA methylation signatures identified in blood and brain only overlapped at 7.9% of positions [18]. To specifically interrogate the utility of surrogate tissues in environmental epigenetics, the Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TARGET) II consortium is studying epigenetic effects of exposure across tissues in perinatal mouse models [19]. Human population studies have also identified environment-associated epigenetic alterations that are detectable across tissues. For example, DNA methylation signatures associated with smoking exposure were first identified in cord blood [20]. A portion of the smoking signatures were then identified in adult blood [21], and a smaller portion were identified in adult lung tissue [22]. Testing epigenetic signatures across tissue types is a powerful approach to disentangle exposure- or disease-related systemic and tissue-specific alterations. A high degree of overlap in epigenetic signatures across target and surrogate tissues due to an exposure would provide support for the hypothesis that surrogate tissue types can provide relevant information about epigenetic alterations in target tissues. Epigenetic signatures specific to an exposure in a target tissue may represent unique effects that are informative of tissue-specific regulation. Understanding the tissue specificity of environmental and disease epigenetics is an important and ongoing field.
Cell Type Specificity in Environmental Epigenetics
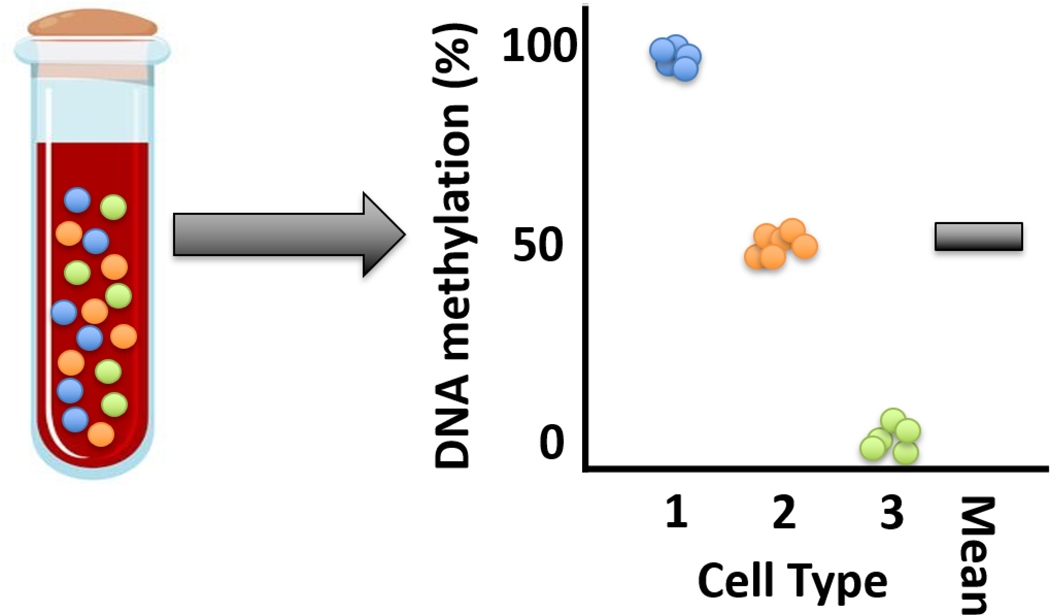
Epigenetic control similarly governs cellular differentiation. Pluripotent stem cells differentiate to myriad cell types by selective regulation of differentiation pathways [5, 6]. Consequently, DNA methylation profiles differ systematically by cell type [23, 24]. Complex cell type mixtures make up tissues. For example, whole blood contains many cell types including T-cells, B-cells, granulocytes, monocytes, and natural killer cells. Thus, tissue-level measures of DNA methylation, such as whole blood DNA methylation, reflect averages across all cells present (Figure 1). Tissue level measurements are therefore “convoluted” by proportions of individual cell types in a tissue [25, 26].
Figure 1.
Conceptual model for understanding tissue measures of DNA methylation as a mixture of signals from cell types. A complex tissue such as whole blood is composed of many individual cell types. Individual circles represent cells, colored by cell type identity. In this example, three cell types compose the tissue. The investigator performs a tissue-level assessment of DNA methylation that averages across cell types. The black bar represents the aggregate observed tissue-level mean DNA methylation signal. The cell type DNA methylation profiles are not observed. The investigator may incorrectly conclude that each genomic locus in each cell type in the sample is uniformly methylated at 50% if they do not consider the cell type heterogeneity of the sample.
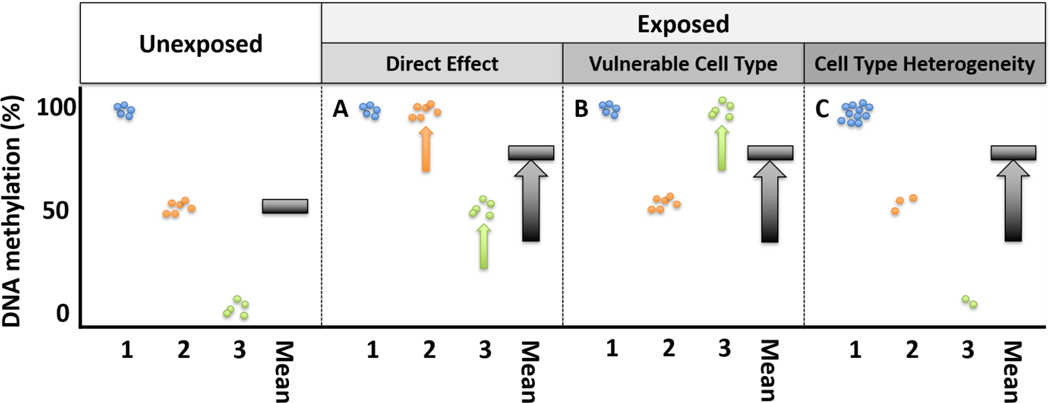
Given tissue measures reflect DNA methylation averages across a mixture of cells, differences in DNA methylation by exposure or disease could have multiple underlying biological mechanisms. There are at least three biological scenarios that lead to the same tissue-level DNA methylation signal (Figure 2). First, the exposure could have a uniform effect on DNA methylation across all cell types, leading to a substantial change in average DNA methylation (Figure 2A). For example, aging has a uniform direct effect on DNA methylation across cell types, termed the epigenetic “clock.” DNA methylation at these positions strongly correlate with age across tissues and cell types [27, 28].
Figure 2.
Conceptual model for differences in tissue DNA methylation by exposure status, considering tissues as mixtures of cell types. Individual circles represent cells, colored by three cell type identities. The black bar represents the observed tissue-level mean DNA methylation signal. A-C represent distinct biological scenarios that could lead to the same exposure-related DNA methylation signal. A. The exposure uniformly increases DNA methylation in each cell type population, which increases the observed DNA methylation signal. B. The exposure directly increases DNA methylation in one vulnerable cell type. C. The exposure does not have a direct effect on DNA methylation and the observed increase in DNA methylation signal is completely mediated by differences in cell type proportion between the exposed and unexposed samples.
Alternatively, a difference in average tissue DNA methylation may be observed when vulnerable cell types exhibit a large shift in DNA methylation (Figure 2B). As an example, a small study of smoking and non-smoking healthy volunteers tested for differences in DNA methylation among sorted blood immune cell subpopulations. In smokers, two DNA methylation sites within the Growth Factor Independent 1 Transcriptional Repressor (GFI1) or F2R Like Thrombin or Trypsin Receptor 3 (F2RL3) genes were hypomethylated in granulocytes but not in peripheral blood mononuclear cells. Further, two sites within the Coproporphyrinogen Oxidase (CPOX) or G Protein-Coupled Receptor 15 (GPR15) genes were hypomethylated in peripheral blood mononuclear cells, including some T cell subtypes, but not in granulocytes [29]. Another small study of smokers and nonsmokers observed cell type-specific associations between smoking and DNA methylation in CD14+ monocytes, CD15+ granulocytes, CD19+ B cells, and CD2+ pan T cells [*30]. These results show cell types may have variable and specific DNA methylation susceptibility to environmental exposures.
In a third plausible scenario, the exposure has no direct effect on DNA methylation in any cell type. The apparent shift in average DNA methylation is attributable to a difference in cell type proportions (Box 1) between exposed and unexposed individuals (Figure 2C). For example, in whole blood, cigarette smoking is associated with DNA methylation at a locus within GPR15. When considering blood immune cells separately, no direct effect of cigarette smoking was observed on GPR15 methylation in GPR15+CD3+ T cells. Instead, smoking led to an increase in the relative proportion of GPR15+CD3+ T cells in whole blood [31]. Exposures can influence DNA methylation measures by causing a shift in cell type proportions, which can have important consequences in the tissue.
Box 1. Definitions of key terms.
Cell type heterogeneity: differences in the proportion of cell types across samples
Epigenotype: the measured or unmeasured epigenetic configuration of a genomic position or region
Tissue-level epigenotype: an epigenotype measurement made at the tissue level, i.e. averaging the epigenotype across all cell types and cells present in a sample
Cell type-specific epigenotype: an epigenotype measurement made on the cell type-specific level by first isolating cell type populations or subpopulations
Tissue-level differences in DNA methylation can be biologically attributed to direct DNA methylation effects across all cells, direct DNA methylation effects in vulnerable cell types, or shifts in cell type heterogeneity. Each scenario represents a unique consequence of an exposure and warrants further investigation. Because bulk tissue-level measures of DNA methylation fail to resolve such biologically distinct mechanisms, observational studies are potentially fraught with incorrect conclusions and misinterpretation [32]. Applying methods to account for cell type heterogeneity is critical to identify underlying biological mechanisms and facilitate proper interpretation.
Methods for Estimating or Accounting for Cell Type Heterogeneity
Accounting for cell type heterogeneity in DNA methylation data allow investigators to distinguish shifts in cellular heterogeneity from direct effects of an exposure on DNA methylation, both of which offer potential insights into disease etiology [25]. There are five main approaches to account for cellular heterogeneity: cellular separation, unbiased single-cell profiling, cell counting, and cellular deconvolution in silico by reference-based or reference-free methods (Table 1). Studies may elect to use one or more of these methods, based on their study design, timing, tissue, and sample or measure availability. The advantages and trade-offs of each method are described below.
Table 1.
Approaches to address cell type heterogeneity in DNA methylation studies
| Approach | Cellular Measure | DNA Methylation Measure | Advantages | Disadvantages | Examples |
|---|---|---|---|---|---|
| Cell type sorting | Direct | Cell type | Measures of DNA methylation within cell types Allows development of cell type-specific reference profiles |
Requires careful sample processing Laborious sorting Cell type specificity limited to prior knowledge |
Adherence Density Antibody-binding: • Flow Cytometry • Magnetic Bead Reviewed in [80] |
| Single-cell technologies | Direct | Single-cell | Measures of DNA methylation at single-cell resolution No a priori knowledge of cell types required |
Requires careful sample processing Expensive Limited throughput Requires single-cell bioinformatics expertise |
Approaches: many, reviewed in [34] Analysis software: BEAT [81], methylpy [82], LIGER [83], Melissa [84] |
| Cell counting | Direct | Tissue | Well-studied and classic experimental techniques | Requires careful sample processing Laborious sorting Subject to error Limited information |
Microscope counting in fresh or histological samples Complete blood counts Other cell count procedures |
| Reference-based deconvolution | Indirect | Tissue | Cell type proportions estimates Few and transparent model assumptions with biological interpretability |
Requires cell type-specific reference profiles Demographically diverse profiles are limited A priori knowledge of cell types required |
Houseman’s constrained projection [85], CIBERSORT [86], IDOL [87], EpiDISH [88] |
| Reference-free deconvolution | Indirect | Tissue | No a priori knowledge of cell types required No cell type-specific reference profiles required |
Cell type proportion estimates generally unavailable Requires delicate model assumptions that can lack biological interpretability |
SVA [48], ISVA [89], RUV [90], FaST-LMM-EWASher [91], RefFreeEWAS 1.0/2.0 [42, 86], ReFACTor [93], BayesCCE [94] |
Direct physical cellular separation is a method to account for cell type heterogeneity in a mixed tissue that requires purifying cells or cell type subpopulations before measuring DNA methylation. Cell sorting technologies such as fluorescence-activated cell sorting or magnetic-activated cell sorting allow the user to isolate cellular subpopulations based on various stains, morphological characteristics, or expression of known cell type markers [33]. A priori knowledge of the distinguishing characteristics of cellular subpopulations present in the tissue is required, however, and represents a key limitation of this approach. Cells must also be processed and separated fresh at the time of sample collection or stored in a way to allow cell membranes to survive freeze-thaw. This can be achieved by using a cryopreservation blood tube or dissociation of solid tissues to a viable single-cell suspension, which is then cryopreserved prior to cell population separation and DNA methylation measurement. Investigators should be cautious as cell types may differentially survive processing. Following cell type separation, DNA methylation is measured in sorted cell types.
Single-cell epigenetics is an emerging technology that accommodates cell type heterogeneity. Single-cell approaches bypass the need for a priori cell type marker identification and generate single-cell epigenetic measures in an unbiased manner. These data can be aggregated at the cell type level using unbiased clustering to quantify epigenetic heterogeneity within and across cell types. Single-cell DNA methylation approaches are being rapidly developed and there is not a current consensus method. Current disadvantages include limited coverage and robustness, labor requirements, and cost [34]. Single-cell technologies may even allow for mechanistic investigation of exposures and DNA methylation within individual cells or cell types and subtypes of tissues, organs, and organisms [35]. Like direct cellular separation, initial sample processing steps apply.
Direct cell counting methods, such as complete blood counting or histopathological cell counting, are used to quantify the relative abundance of cell types in a sample. DNA methylation measures are then made at the tissue level and investigators can adjust for the cell type counts in downstream analyses. This approach requires fresh samples or samples prepared for counting, such as fixed tissues. Direct cell counting allows investigators to test for exposure differences in cell type proportions. Unlike the previous two methods, however, cell counting offers no information about the direct effects of an exposure on DNA methylation. Only cell type proportion estimates of a sample are available and can be used for adjustment or interpretation of a tissue DNA methylation measure.
Indirect cellular deconvolution is a class of methods to account for cell type heterogeneity via in silico estimation of cell type proportions. Deconvolution refers to the bioinformatic process of accounting for differences in intrasample cell type heterogeneity in tissues [36, 37]. Given that the previous three methods require specific laboratory preparation and processing at the time of sample collection, bioinformatic deconvolution is more commonly implemented in observational studies. To leverage DNA methylation data generated from heterogeneous tissues, two classes of deconvolution methods have been developed—reference-based and reference-free.
Reference-based methods are supervised and rely on independently collected cell type-specific DNA methylation profiles to estimate cell type proportions in a tissue sample. Advantages of the reference-based methods include: quantification of cell type proportions, biologically interpretable model components, and few model assumptions [25, 37, 38]. Reference panels are currently available for cord blood [**39–43], umbilical cord tissue [43], adult blood [24, 44], frontal cortex (neuron vs. non-neuron) [45], and broadly epithelial versus fibroblast cell types [46] (Table 2). Disadvantages include a lack of demographically diverse reference samples, a limited number of reference panels, an assumption about constituent cell types, and challenges in identifying methylation sites and regions that discriminate cell types [25, 37, 47]. Similar to direct cell counts, cell type proportions estimated from reference-based deconvolution can be used in regression models when analyzing tissue DNA methylation measures. We recommend that investigators implement reference-based methods when cell type references are available for a tissue of interest.
Table 2.
Summary of cell type-specific reference panels available for reference-based indirect deconvolution
| Tissue | Sample Population | Number of Cell Types | Cell Types Measured | Sorting Technology | DNA Methylation Platform | Reference |
|---|---|---|---|---|---|---|
| Adult Blood | 6 healthy male donors, aged 25–60 | 7 | Neutrophils, Eosinophils Monocytes, B cells, NK cells, CD8+ T cells, CD4+ T cells | Magnetic-activated cell sorting | Illumina 450k microarray | Reinius et al., 2012 [24] |
| 37 healthy donors, aged 19–59 | 6 | Neutrophils, Monocytes, B cells, NK cells, CD8+ T cells, CD4+ T cells | Magnetic-activated cell sorting | Illumina EPIC 850k microarray | Salas et al., 2018 [44] | |
| Umbilical Gord Blood | 11 healthy full-term singleton deliveries | 6 | Granulocytes, Monocytes, B cells, NK cells, CD8+ T cells, CD4+ T cells | Fluorescence-activated cell sorting | Illumina 450k microarray | Gervin et al., 2016 [40] |
| 17 healthy full-term singleton vaginal deliveries | 7 | Granulocytes, Monocytes, B cells, NK cells, CD8+ T cells, CD4+ T cells, Nucleated Red Blood cells | Magnetic-activated cell sorting | Illumina 450k microarray | Bakulski et al., 2016 [41] | |
| 7 elective non-laboring Caesarean-section | 7 | Granulocytes, Monocytes, B cells, NK cells, CD8+ T cells, CD4+ T cells, Nucleated Red Blood cells | Fluorescence-activated cell sorting | Illumina 450k microarray | de Goede et al., 2016 [42] | |
| 14 healthy singleton full-term deliveries from women aged 28–38 | 6 | Granulocytes, Monocytes, B cells, NK cells, CD8+ T cells, CD4+ T cells | Magnetic-activated cell sorting | Illumina EPIC 850k microarray | Lin et al., 2018 [43] | |
| Mixed from above, 34 cell type fractions removed or reclassified from Bakulski, Gervin | 7 | Granulocytes, Monocytes, B cells, NK cells, CD8+ T cells, CD4+ T cells, Nucleated Red Blood cells | Mixed | Mixed | Gervin et al., 2019 [**39] combines Gervin, Bakulski, de Goede, and Lin | |
| Umbilical Gord Tissue | 14 healthy singleton full-term deliveries from women aged 28–38 | 3 | Stromal, Endothelial, and Epithelial | Magnetic-activated cell sorting | Illumina EPIC 850k microarray | Lin et al., 2018 [43] |
| Frontal Cortex | 29 post-mortem major depressive disorder cases and 29 matched controls | 2 | Neuron Non-neuron | Fluorescence-activated cell sorting | Illumina 450k microarray | Guintivano et al., 2013 [45] |
| Broadly Epithelial | ENCODE cell lines and Reinius et al., 2012 [24] dataset | 3 | 11 Epithelial and 7 Fibroblast cell lines, adult blood immune cell types | Secondary Analysis | Illumina 450k microarray | Zheng et al., 2018 [46] |
Reference-free methods are unsupervised methods to account for variation in DNA methylation data, including cell type heterogeneity. This category encompasses many algorithms that account for sources of variation that are unmeasured and unmodeled due biological sources of variation, such as cell type heterogeneity, or nonbiological sources of variation, such as random noise or batch effects in an association study. Reference-free methods, like “surrogate variable analysis”, were originally developed for RNA expression deconvolution [48], and are now applied in epigenome-wide association studies [37]. Advantages of unsupervised methods include no required a priori knowledge of tissue cell types, flexible modelling strategies, and no required cell type references, allowing them to be used in any tissue. Disadvantages include the general inability to estimate intrasample cell type proportions and the large number of delicate model assumptions, including the assumption that the largest driver of variation is due to cell type proportion differences [37, **49, 50]. Following reference-free processing, depending on the specific method, investigators either implement exposure testing on the resulting adjusted DNA methylation matrix or they account for the reference-free “cell types” in regression models when analyzing tissue DNA methylation measures. Reference-free methods are only recommended for tissues lacking adequate references.
Utility of Cellular Heterogeneity in Research Questions and Epidemiologic Modeling
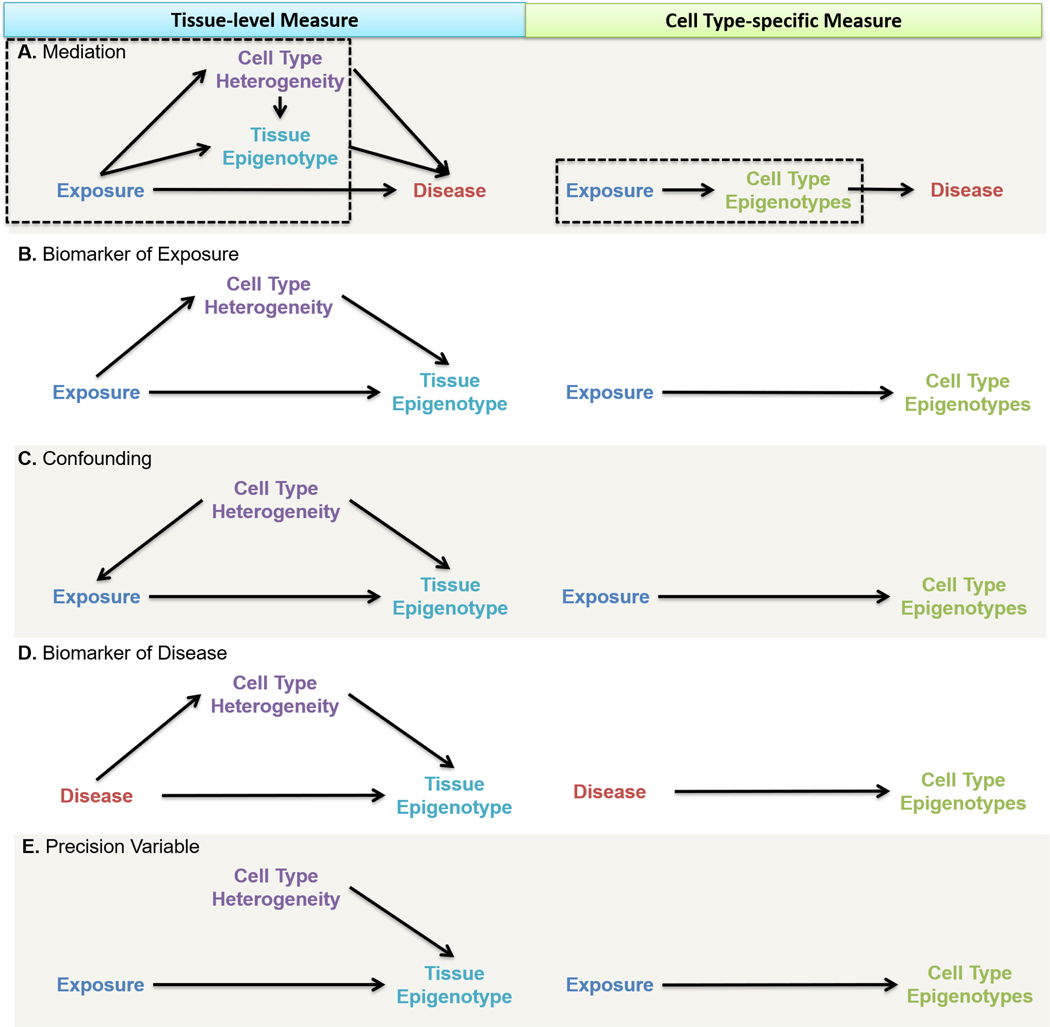
Once cell type heterogeneity has been accounted for, one can ask critical questions about DNA methylation, exposures, and disease. The appropriate approach depends on the study sampling framework, timing of measures, and hypothesized relationships between exposures, cellular heterogeneity, DNA methylation, and disease. Causal diagrams [51] are frequently employed in epidemiologic studies to evaluate and communicate the relationships between key variables and identify appropriate approaches to address bias [52]. Below, we use causal diagrams to describe five study hypotheses involving an exposure, a disease, tissue-level DNA methylation, and cell type-specific DNA methylation epigenotypes (Box 1). These five hypotheses are well-studied in epidemiologic frameworks: mediation, confounding, biomarker of disease, biomarker of exposure, and precision variables. By directly measuring epigenetics within sorted cell types (cell type-specific epigenotypes), researchers simplify casual diagrams and associated statistical models. Simpler causal diagrams require fewer assumptions, use simpler analytic methods, minimize sources of bias, and improve interpretability of study results [51, 52]. In each setting, we demonstrate that studying cell type-specific epigenotypes simplifies the causal diagram and reduces sources of potential bias.
DNA methylation dysregulation is a candidate to mediate early-life environmental exposures and later life health outcomes [53, 54]. Mediation refers to the indirect effect an exposure has on an outcome by acting through an intervening variable [55]. Though perhaps the most biologically compelling, mediation studies were the among rarest study designs according to recent DNA methylation mediation reviews [*56, 57]. Testing mediation requires assumptions under different analytic frameworks, but all models rely on faithfully capturing exposure-mediator and mediator-outcome relationships [58] (Figure 3A—Tissue Epigenotype). These relationships must be identifiable, unconfounded, and free of bias to establish evidence of a causal relationship [59]. Mediation testing in epigenetic perturbations is difficult due to a lack of cell type-specific studies in tissues relevant to the disease process [60] and a lack of observational studies that span the perinatal period until the end of life [61]. One example of a recent mediation-based study design was a case-control study of rheumatoid arthritis and genetic risk that controlled for cell type heterogeneity and removed DNA methylation signatures due to arthritis onset. Ten differentially methylated regions were identified in a mediation analysis [**62]. Recently, a cell type deconvolution algorithm for DNA methylation demonstrated a quantitation of the mediation of phenotypic associations with DNA methylation by cellular heterogeneity in 23 DNA methylation microarray datasets across 13 studies [**49]. Cell type-specific assessment of DNA methylation avoids mediation by cell type heterogeneity altogether (Figure 3A—Cell Type Epigenotypes). Well-designed DNA methylation mediation studies that account for cell type heterogeneity may identify the mechanisms by which environmental exposures affect DNA methylation directly and disease etiology.
Figure 3.
Based on hypothesized relationships between exposure, disease, and DNA methylation, multiple study design scenarios are possible. Measures of DNA methylation can be implemented in directed acyclic graphs for identifying model assumptions and analytic strategies for causal inference. The left column represents a DNA methylation epigenotype aggregated over multiple cell types (e.g., tissue) and the right column represents cell type-specific epigenotypes measures. When measured on the cell type-specific level, the causal link between composition heterogeneity and epigenotype is broken and omitted from the cell type-specific diagram. A. Mediation: The exposure affects disease indirectly through the tissue epigenotype (direct DNA methylation effects across all cells (Figure 2A) or in vulnerable cell types (Figure 2B)) or through cell type heterogeneity (Figure 2C). The exposure may also affect the disease directly. We focus on modeling the tissue epigenotype as the outcome, a subset of the overall causal diagram (Boxed). B. Biomarker of Exposure: The exposure affects the DNA methylation epigenotype directly and indirectly through cell type composition heterogeneity. C. Confounding: Cell type composition affects the level of exposure and directly affects the epigenotype through heterogeneity. D. Biomarker of Disease: The disease state affects the DNA methylation epigenotype directly and indirectly through cell type composition heterogeneity. E. Precision Variable: Cell type composition heterogeneity is independent of the exposure but is a strong predictor of DNA methylation epigenotype.
Now, we focus on modeling the tissue epigenotype as the outcome to understand the role cellular heterogeneity plays in the relationship between exposure and tissue epigenotype in the mediation framework. Typically, investigators do not adjust for a mediator as it represents a contributor to the total effect of the exposure on the outcome. In epigenetic studies, assessing mediation by cell type heterogeneity is essential to distinguish direct intranuclear changes (Figures 2A or 2B) from shifts in cell type heterogeneity (Figure 2C) [38], each of which offers insights into disease etiology [25]. Note that single-cell or cell type-specific assessment is required to distinguish a global direct effect (Figure 2A) from a vulnerable cell type scenario (Figure 2B). When the goal is to identify direct DNA methylation changes, cell type deconvolution and adjustment can block the non-causal pathway that is mediated by cell type heterogeneity. For example, a recent epigenome-wide meta-analysis identified associations robust to cell type adjustment between exposure to maternal smoking in pregnancy and over 6,000 newborn blood DNA methylation sites (Figure 3A—Tissue Epigenotype, boxed) [20]. Again, Cell type-specific assessment of DNA methylation circumvents cell type heterogeneity (Figure 3A—Cell Type Epigenotypes, boxed). Researchers must take care in the design, analysis, and interpretation of epigenetics studies where the exposure is thought to affect cell type heterogeneity, prompting the consideration of a mediation framework.
In a second scenario, epigenetic measures serve as exposure biomarkers. Because DNA methylation is labile to environmental exposures and generally stable once established [63], DNA methylation can serve as a proxy measure of past exposures [64, 65]. Several studies have linked maternal smoking during pregnancy to changes in newborn or later childhood blood DNA methylation, though the potential health consequences of these changes beyond a biomarker is unclear [20, 66, 67]. The framework is identical in structure to the mediation scenario, except that the tissue epigenotype does not affect disease (Figure 3B—Tissue Epigenotype). For similar reasons, cell type-specific epigenotypes should be prioritized over tissue epigenotypes whenever possible (Figure 3B—Cell Type Epigenotypes). DNA methylation could reduce information bias in epidemiological studies by extending the reach of exposure assessment backward in time and more accurately quantifying an individual’s exposure.
In a third scenario, cell type heterogeneity could be a confounder in the relationship between exposure and tissue-level DNA methylation (Figure 3C—Tissue Epigenotype). Confounding refers to a non-causal association between an exposure and outcome due to a shared common cause (the “confounder”) [68]. Most current epigenome-wide association studies implement cell type proportions as adjustment covariates in regression models, with the stated goal to account for potential confounding due to cell type heterogeneity. Cellular heterogeneity may affect the metabolism, storage, or cellular response to an environmental toxicant, impacting biomarker measured toxicant levels. For example, in five cell lines across 20 heavy metal toxicants, an Nrf2-dependent oxidative stress response varied by cell type [69]. Further, we know that cell type heterogeneity predicts tissue-level DNA methylation [70]. The common cause of cell type heterogeneity could therefore distort the measure of association between exposure and tissue-level DNA methylation. As before, evaluating DNA methylation at the cell type level simplifies the causal diagram as well as eliminates the potential for confounding by cell type heterogeneity (Figure 3C—Cell Type Epigenotypes).
Fourth, epigenetic measures can be biomarkers of disease. The application of DNA methylation as a disease biomarker can reduce outcome misclassification, serve as a surrogate endpoint, and monitor disease progression, prognosis, and treatment response [71, 72]. DNA methylation is routinely employed as a disease biomarker in cancers and is being investigated for use in psychiatric conditions and chronic diseases such as cardiovascular disease [73–75]. DNA methylation of placentally derived DNA from maternal plasma has been used as a noninvasive biomarker of aneuploidy [76]. A recent meta-analysis revealed an association between neonatal blood DNA methylation and birthweight, although it is unclear if DNA methylation was a mediating cause of birthweight changes or simply a birthweight biomarker [*77]. DNA methylation is an auspicious vehicle for the promise of precision medicine and might soon be established as the ‘universal’ disease biomarker, given the vast array of disease-specific DNA methylation profiles that are being uncovered [78]. Identification of DNA methylation perturbations related to negative health outcomes will likely identify at-risk individuals and lead to novel preventive and therapeutic strategies [57]. Notice that assessment of DNA methylation as a biomarker of disease is identical in diagram structure to the mediation scenario presented above (Figure 3D—Tissue Epigenotype). Therefore, the same recommendation for assessing cell type-specific epigenotypes can be applied to the use of DNA methylation as a biomarker of disease (Figure 3D—Cell Type Epigenotypes). The use of DNA methylation as a disease biomarker still requires methods to account for cell type heterogeneity.
Finally, cell type heterogeneity may be an important precision variable in epigenetic studies (Figure 3E—Tissue Epigenotype). A precision variable is a predictor of the outcome that is unrelated to the exposure. A precision variable increases statistical efficiency when adjusted for in a model [79]. Cell type proportions are strong predictors of DNA methylation, often accounting for the first principal component of variability in DNA methylation data. Tissue DNA methylation studies may account for cell type proportions in regression models to improve precision in estimating DNA methylation associations with other variables. When cell type-specific DNA methylation is measured, cell type heterogeneity may no longer be relevant to estimating the direct effect of the exposure on cellular DNA methylation (Figure 3E—Cell Type Epigenotypes). However, independence between cell type heterogeneity and cellular DNA methylation may be an unrealistic assumption due to cell-cell interactions in tissues [32]. Even when cell type heterogeneity is unrelated to the exposure, cell type heterogeneity may be an important precision variable.
Summary and Recommendations for Ongoing and Future Studies
DNA methylation is a key regulator of cell differentiation; thus tissues and cell types systematically differ in their DNA methylation profiles. When selecting a tissue type for study, investigators must be thoughtful about the possible utility and scope of inferences in that tissue. Surrogate and target tissues have complementary advantages and disadvantages. Disease target tissues, such as brain, may not be limited or not available for a given study design, though surrogate tissues may still provide insight into disease etiology or onset [16]. When possible, a multi-tissue approach in sampling, biobanking, and measurements will allow for the most robust biological interrogations. Single tissue studies should make comparisons to publicly available data in multiple tissues to extend the reach of insights.
Epigenetic measurements of bulk tissues represent a mixture of individual cell types. This convolution complicates observational and experimental studies, making it impossible to disentangle distinct biological mechanisms that can lead to the same tissue epigenotype measure, particularly when cell type heterogeneity differs between samples. Cell type heterogeneity must be considered and accounted for in any epigenetic study. Direct measures of DNA methylation should be prioritized over indirect methods to faithfully capture the DNA methylation state of each sample without reliance on imperfect indirect methods that “smooth over” inter-sample differences such as in silico deconvolution. Of direct approaches, single-cell measures of DNA methylation show the greatest promise because they unbiasedly account for cell type heterogeneity with the greatest resolution and can later be aggregated at the cell type or sub-cell type level if desired. These methods, however, are not yet widely available for observational human studies. At this time, among indirect deconvolution approaches, referenced-based methods should be prioritized over reference-free methods. Reference-based deconvolution requires fewer assumptions and affords greater transparency and biological interpretability. However, reference-free deconvolution is invaluable when reference data are unavailable or biosampling logistics prevent cell sorting or single-cell approaches. Further studies of deconvolution algorithm performance and collection of high-quality and diverse DNA methylation reference profiles are required to advance indirect deconvolution approaches. Researchers should be transparent in reporting the assumptions and selection criteria for any cell type heterogeneity approach.
At various genomic locations, tissues, and developmental times, DNA methylation is a promising biomarker of exposure and disease, as well as a potential mediator between environmental exposure and health outcomes. The relationships between exposure, disease, tissue-level DNA methylation, cell type-specific DNA methylation, and cell type heterogeneity must be carefully considered in any study design. Subject matter expertise, transparency of model assumptions, and appropriate methods to accommodate and evaluate potential study hypotheses will be required to improve causal inference and interpretability in environmental epigenetics. It is important that investigators clearly state hypotheses and analytic assumptions to generate valid, replicable, and interpretable study results.
Acknowledgments
Funding: K.A.C. was supported by the National Institutes of Health (T32 HG00040) and Michigan State University’s Environmental Influences on Child Health Outcomes program (UG3 OD023285, UH3 OD023285). J.A.C. was supported by the Ravitz Family Foundation, the Forbes Institute for Cancer Discovery at the University of Michigan Rogel Cancer Center, and the National Institutes of Health (grant numbers R01 ES028802, U01 ES026697, P30 ES017885, and P30 CA046592). S.K.P. declares no funding sources. K.M.B. was supported by research grants from the National Institute of Environmental Health Sciences (R01 ES025531; R01 ES025574), National Institute of Aging (R01 AG055406), National Institute on Minority Health and Health Disparities (R01 MD013299), and ALS Association (20-IIA-532).
Footnotes
Ethics declarations
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Conflicts of interest/Competing interests: KC, JC, SKP, and KB declare no conflicts of interest/competing interests in the production of this work.
Conflict of Interest
KB. KC, JC declare no conflicts of interest in the production of this work.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References cited
- 1.Greally JM (2018) A user’s guide to the ambiguous word “epigenetics.” Nat Rev Mol Cell Biol 19:207–208. 10.1038/nrm.2017.135 [DOI] [PubMed] [Google Scholar]
- 2.Deichmann U (2016) Epigenetics: The origins and evolution of a fashionable topic. Developmental Biology 416:249–254. 10.1016/j.ydbio.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 3.Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: A Landscape Takes Shape. Cell 128:635–638. 10.1016/j.cell.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 4.Reik W, Dean W, Walter J (2001) Epigenetic Reprogramming in Mammalian Development. Science 293:1089–1093. 10.1126/science.1063443 [DOI] [PubMed] [Google Scholar]
- 5.Smith ZD, Meissner A (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14:204–220. 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- 6.Khavari DA, Sen GL, Rinn JL (2010) DNA methylation and epigenetic control of cellular differentiation. Cell Cycle 9:3880–3883. 10.4161/cc.9.19.13385 [DOI] [PubMed] [Google Scholar]
- 7.Virani S, Colacino JA, Kim JH, Rozek LS (2012) Cancer Epigenetics: A Brief Review. ILAR J 53:359–369. 10.1093/ilar.53.3-4.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berson A, Nativio R, Berger SL, Bonini NM (2018) Epigenetic Regulation in Neurodegenerative Diseases. Trends in Neurosciences 41:587–598. 10.1016/j.tins.2018.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ordovás JM, Smith CE (2010) Epigenetics and cardiovascular disease. Nat Rev Cardiol 7:510–519. 10.1038/nrcardio.2010.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil R, Fraga MF (2012) Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 13:97–109. 10.1038/nrg3142 [DOI] [PubMed] [Google Scholar]
- 11.Gluckman PD, Hanson MA, Buklijas T, et al. (2009) Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 5:401–408. 10.1038/nrendo.2009.102 [DOI] [PubMed] [Google Scholar]
- 12.Smith ZD, Chan MM, Humm KC, et al. (2014) DNA methylation dynamics of the human preimplantation embryo. Nature 511:611–615. 10.1038/nature13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messerschmidt DM, Knowles BB, Solter D (2014) DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 28:812–828. 10.1101/gad.234294.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaligné R, Heard E (2014) X-chromosome inactivation in development and cancer. FEBS Letters 588:2514–2522. 10.1016/j.febslet.2014.06.023 [DOI] [PubMed] [Google Scholar]
- 15.Doi A, Park I-H, Wen B, et al. (2009) Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41:1350–1353. 10.1038/ng.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakulski KM, Halladay A, Hu VW, et al. (2016) Epigenetic Research in Neuropsychiatric Disorders: the “Tissue Issue.” Curr Behav Neurosci Rep 3:264–274. 10.1007/s40473-016-0083-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannon E, Lunnon K, Schalkwyk L, Mill J (2015) Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics 10:1024–1032. 10.1080/15592294.2015.1100786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walton E, Hass J, Liu J, et al. (2016) Correspondence of DNA Methylation Between Blood and Brain Tissue and Its Application to Schizophrenia Research. Schizophr Bull 42:406–414. 10.1093/schbul/sbv074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang T, Pehrsson EC, Purushotham D, et al. (2018) The NIEHS TaRGET II Consortium and environmental epigenomics. Nat Biotechnol 36:225–227. 10.1038/nbt.4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joubert BR, Felix JF, Yousefi P, et al. (2016) DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. The American Journal of Human Genetics 98:680–696. 10.1016/j.ajhg.2016.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikdar S, Joehanes R, Joubert BR, et al. (2019) Comparison of smoking-related DNA methylation between newborns from prenatal exposure and adults from personal smoking. Epigenomics 11:1487–1500. 10.2217/epi-2019-0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakulski KM, Dou J, Lin N, et al. (2019) DNA methylation signature of smoking in lung cancer is enriched for exposure signatures in newborn and adult blood. Sci Rep 9:. 10.1038/s41598-019-40963-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meissner A, Mikkelsen TS, Gu H, et al. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770. 10.1038/nature07107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinius LE, Acevedo N, Joerink M, et al. (2012) Differential DNA Methylation in Purified Human Blood Cells: Implications for Cell Lineage and Studies on Disease Susceptibility. PLOS ONE 7:e41361 10.1371/journal.pone.0041361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holbrook JD, Huang R-C, Barton SJ, et al. (2017) Is cellular heterogeneity merely a confounder to be removed from epigenome-wide association studies? Epigenomics 9:1143–1150. 10.2217/epi-2017-0032 [DOI] [PubMed] [Google Scholar]
- 26.Jaffe AE, Irizarry RA (2014) Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biology 15:R31 10.1186/gb-2014-15-2-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvath S (2013) DNA methylation age of human tissues and cell types. Genome Biology 14:3156 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jylhävä J, Pedersen NL, Hägg S (2017) Biological Age Predictors. EBioMedicine 21:29–36. 10.1016/j.ebiom.2017.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer M, Fink B, Thürmann L, et al. (2016) Tobacco smoking differently influences cell types of the innate and adaptive immune system—indications from CpG site methylation. Clin Epigenetics 8:. 10.1186/s13148-016-0249-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su D, Wang X, Campbell MR, et al. (2016) Distinct Epigenetic Effects of Tobacco Smoking in Whole Blood and among Leukocyte Subtypes. PLOS ONE 11:e0166486 10.1371/journal.pone.0166486*This study revealed cell type-specific associations between smoking and DNA methylation in multiple leukocyte subpopulations. Further, DNA methylation fine-mapping and discordant gene expression changes provide evidence that disease etiology should be evaluated in a lineage-specific matter.
- 31.Bauer M, Linsel G, Fink B, et al. (2015) A varying T cell subtype explains apparent tobacco smoking induced single CpG hypomethylation in whole blood. Clin Epigenet 7:81 10.1186/s13148-015-0113-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lappalainen T, Greally JM (2017) Associating cellular epigenetic models with human phenotypes. Nat Rev Genet 18:441–451. 10.1038/nrg.2017.32 [DOI] [PubMed] [Google Scholar]
- 33.Herzenberg LA, Parks D, Sahaf B, et al. (2002) The History and Future of the Fluorescence Activated Cell Sorter and Flow Cytometry: A View from Stanford. Clinical Chemistry 48:1819–1827 [PubMed] [Google Scholar]
- 34.Karemaker ID, Vermeulen M (2018) Single-Cell DNA Methylation Profiling: Technologies and Biological Applications. Trends in Biotechnology 36:952–965. 10.1016/j.tibtech.2018.04.002 [DOI] [PubMed] [Google Scholar]
- 35.Tanay A, Regev A (2017) Scaling single-cell genomics from phenomenology to mechanism. Nature 541:331–338. 10.1038/nature21350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen-Orr SS, Gaujoux R (2013) Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Current Opinion in Immunology 25:571–578. 10.1016/j.coi.2013.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teschendorff AE, Zheng SC (2017) Cell-type deconvolution in epigenome-wide association studies: a review and recommendations. Epigenomics 9:757–768. 10.2217/epi-2016-0153 [DOI] [PubMed] [Google Scholar]
- 38.Houseman EA, Kelsey KT, Wiencke JK, Marsit CJ (2015) Cell-composition effects in the analysis of DNA methylation array data: a mathematical perspective. BMC Bioinformatics 16:95 10.1186/s12859-015-0527-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gervin K, Salas LA, Bakulski KM, et al. (2019) Systematic evaluation and validation of reference and library selection methods for deconvolution of cord blood DNA methylation data. Clin Epigenetics 11:. 10.1186/s13148-019-0717-y**This study shows that combining cell type-specific DNA methylation references across multiple studies can improve deconvolution and provides guidelines for conducting reference-based deconvolution in umbilical cord blood that may be extended to other tissues
- 40.Gervin K, Page CM, Aass HCD, et al. (2016) Cell type specific DNA methylation in cord blood: A 450K-reference data set and cell count-based validation of estimated cell type composition. Epigenetics 11:690–698. 10.1080/15592294.2016.1214782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakulski KM, Feinberg JI, Andrews SV, et al. (2016) DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics 11:354–362. 10.1080/15592294.2016.1161875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Goede OM, Lavoie PM, Robinson WP (2016) Characterizing the hypomethylated DNA methylation profile of nucleated red blood cells from cord blood. Epigenomics 8:1481–1494. 10.2217/epi-2016-0069 [DOI] [PubMed] [Google Scholar]
- 43.Lin X, Tan JYL, Teh AL, et al. (2018) Cell type-specific DNA methylation in neonatal cord tissue and cord blood: a 850K-reference panel and comparison of cell types. Epigenetics 13:941–958. 10.1080/15592294.2018.1522929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salas LA, Koestler DC, Butler RA, et al. (2018) An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol 19:1–14. 10.1186/s13059-018-1448-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guintivano J, Aryee MJ, Kaminsky ZA (2013) A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 8:290–302. 10.4161/epi.23924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng SC, Webster AP, Dong D, et al. (2018) A novel cell-type deconvolution algorithm reveals substantial contamination by immune cells in saliva, buccal and cervix. Epigenomics 10:925–940. 10.2217/epi-2018-0037 [DOI] [PubMed] [Google Scholar]
- 47.Liang L, Cookson WOC (2014) Grasping nettles: cellular heterogeneity and other confounders in epigenome-wide association studies. Hum Mol Genet 23:R83–R88. 10.1093/hmg/ddu284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leek JT, Storey JD (2007) Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis. PLOS Genetics 3:e161 10.1371/journal.pgen.0030161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Houseman EA, Kile ML, Christiani DC, et al. (2016) Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics 17:259 10.1186/s12859-016-1140-4**This study introduces an indirect, reference-free deconvolution method with interpretable biological outputs, including cell type proportions, that also explicitly quantitates mediation by cell composition in phenotypic associations with DNA methylation.
- 50.Zheng SC, Beck S, Jaffe AE, et al. (2017) Correcting for cell-type heterogeneity in epigenome-wide association studies: revisiting previous analyses. Nat Methods 14:216–217. 10.1038/nmeth.4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greenland S, Pearl J, Robins J (1999) Causal Diagrams for Epidemiologic Research. Epidemiology 10:37–48 [PubMed] [Google Scholar]
- 52.Shrier I, Platt RW (2008) Reducing bias through directed acyclic graphs. BMC Medical Research Methodology 8:70 10.1186/1471-2288-8-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bianco-Miotto T, Craig JM, Gasser YP, et al. (2017) Epigenetics and DOHaD: from basics to birth and beyond. Journal of Developmental Origins of Health and Disease 8:513–519. 10.1017/S2040174417000733 [DOI] [PubMed] [Google Scholar]
- 54.Godfrey KM, Lillycrop KA, Burdge GC, et al. (2007) Epigenetic Mechanisms and the Mismatch Concept of the Developmental Origins of Health and Disease. Pediatric Research 61:5–10. 10.1203/pdr.0b013e318045bedb [DOI] [PubMed] [Google Scholar]
- 55.Baron RM, Kenny DA (1986) The moderator–mediator variable distinction in social psychological research: Conceptual, strategic, and statistical considerations. Journal of Personality and Social Psychology 51:1173–1182. 10.1037/0022-3514.51.6.1173 [DOI] [PubMed] [Google Scholar]
- 56.Barker ED, Walton E, Cecil CAM (2018) Annual Research Review: DNA methylation as a mediator in the association between risk exposure and child and adolescent psychopathology. Journal of Child Psychology and Psychiatry 59:303–322. 10.1111/jcpp.12782*This article reviews the evidence available to evaluate a DNA methylation conceptual mediation framework for early-life exposures and developmental psychopathology. The article underscores the paucity of longitudinal study designs adequate to assess mediation by DNA methylation.
- 57.Lin VW, Baccarelli AA, Burris HH (2016) Epigenetics—a potential mediator between air pollution and preterm birth. Environ Epigenet 2:. 10.1093/eep/dvv008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imai K, Keele L, Tingley D (2010) A general approach to causal mediation analysis. Psychological Methods 15:309–334. 10.1037/a0020761 [DOI] [PubMed] [Google Scholar]
- 59.VanderWeele TJ (2009) Mediation and mechanism. Eur J Epidemiol 24:217–224. 10.1007/s10654-009-9331-1 [DOI] [PubMed] [Google Scholar]
- 60.Bansal A, Simmons RA (2018) Epigenetics and developmental origins of diabetes: correlation or causation? American Journal of Physiology-Endocrinology and Metabolism 315:E15–E28. 10.1152/ajpendo.00424.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saffery R (2014) Epigenetic Change as the Major Mediator of Fetal Programming in Humans: Are We There Yet? ANM 64:203–207. 10.1159/000365020 [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Aryee MJ, Padyukov L, et al. (2013) Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol 31:142–147. 10.1038/nbt.2487**This study applied a mediation causal inference approach to test whether DNA methylation mediates the genetic risk of rheumatoid arthritis. Causal diagrams were employed to inform study design and analysis. Further, the investigators accounted for cell type heterogeneity by employing a reference-base deconvolution method, explained their choice of target tissue, and ruled out epigenetic changes thought to be a consequence of disease status.
- 63.Meehan RR, Thomson JP, Lentini A, et al. (2018) DNA methylation as a genomic marker of exposure to chemical and environmental agents. Current Opinion in Chemical Biology 45:48–56. 10.1016/j.cbpa.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 64.Shenker N, Ueland P, Polidoro S, et al. (2013) DNA Methylation as a Long-term Biomarker of Exposure to Tobacco Smoke. Epidemiology 24:712–716. 10.1097/EDE.0b013e31829d5cb3 [DOI] [PubMed] [Google Scholar]
- 65.Guerrero-Preston R, Goldman LR, Brebi-Mieville P, et al. (2010) Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics 5:539–546. 10.4161/epi.5.6.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ladd-Acosta C, Shu C, Lee BK, et al. (2016) Presence of an epigenetic signature of prenatal cigarette smoke exposure in childhood. Environmental Research 144:139–148. 10.1016/j.envres.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Breton CV, Siegmund KD, Joubert BR, et al. (2014) Prenatal Tobacco Smoke Exposure Is Associated with Childhood DNA CpG Methylation. PLOS ONE 9:e99716 10.1371/journal.pone.0099716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hernán MA, Hernández-Díaz S, Robins JM (2004) A Structural Approach to Selection Bias. Epidemiology 15:615 10.1097/01.ede.0000135174.63482.43 [DOI] [PubMed] [Google Scholar]
- 69.Simmons SO, Fan C-Y, Yeoman K, et al. (2011) NRF2 Oxidative Stress Induced by Heavy Metals is Cell Type Dependent. Curr Chem Genomics 5:1–12. 10.2174/1875397301105010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michels KB, Binder AM, Dedeurwaerder S, et al. (2013) Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods 10:949–955. 10.1038/nmeth.2632 [DOI] [PubMed] [Google Scholar]
- 71.Schulte PA, Perera FP (2012) Molecular Epidemiology: Principles and Practices. Academic Press [Google Scholar]
- 72.Mayeux R (2004) Biomarkers: Potential Uses and Limitations. NeuroRx 1:182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Udali S, Guarini P, Moruzzi S, et al. (2013) Cardiovascular epigenetics: From DNA methylation to microRNAs. Molecular Aspects of Medicine 34:883–901. 10.1016/j.mam.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 74.Goud Alladi C, Etain B, Bellivier F, Marie-Claire C (2018) DNA Methylation as a Biomarker of Treatment Response Variability in Serious Mental Illnesses: A Systematic Review Focused on Bipolar Disorder, Schizophrenia, and Major Depressive Disorder. International Journal of Molecular Sciences 19:3026 10.3390/ijms19103026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mikeska T, Craig JM (2014) DNA Methylation Biomarkers: Cancer and Beyond. Genes 5:821–864. 10.3390/genes5030821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chu T, Burke B, Bunce K, et al. (2009) A microarray-based approach for the identification of epigenetic biomarkers for the noninvasive diagnosis of fetal disease. Prenatal Diagnosis 29:1020–1030. 10.1002/pd.2335 [DOI] [PubMed] [Google Scholar]
- 77.Küpers LK, Monnereau C, Sharp GC, et al. (2019) Meta-analysis of epigenome-wide association studies in neonates reveals widespread differential DNA methylation associated with birthweight. Nat Commun 10:1–11. 10.1038/s41467-019-09671-3*This meta-analysis shows an association between birthweight and neonatal blood DNA methylation sites. The investigators employed a basic conceptual model to inform careful inference of study results, recognizing the limitations and assumptions of their approach.
- 78.Levenson VV (2010) DNA methylation as a universal biomarker. Expert Review of Molecular Diagnostics 10:481–488. 10.1586/erm.10.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schisterman EF, Cole SR, Platt RW (2009) Overadjustment Bias and Unnecessary Adjustment in Epidemiologic Studies. EpideHmiology 20:488–495. 10.1097/EDE.0b013e3181a819a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tomlinson MJ, Tomlinson S, Yang XB, Kirkham J (2012) Cell separation: Terminology and practical considerations: Journal of Tissue Engineering. 10.1177/2041731412472690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akman K, Haaf T, Gravina S, et al. (2014) Genome-wide quantitative analysis of DNA methylation from bisulfite sequencing data. Bioinformatics 30:1933–1934. 10.1093/bioinformatics/btu142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schultz MD, He Y, Whitaker JW, et al. (2015) Human Body Epigenome Maps Reveal Noncanonical DNA Methylation Variation. Nature 523:212–216. 10.1038/nature14465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Welch JD, Kozareva V, Ferreira A, et al. (2019) Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 177:1873–1887.e17. 10.1016/j.cell.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kapourani C-A, Sanguinetti G (2019) Melissa: Bayesian clustering and imputation of single-cell methylomes. Genome Biology 20:61 10.1186/s13059-019-1665-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Houseman EA, Accomando WP, Koestler DC, et al. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Newman AM, Liu CL, Green MR, et al. (2015) Robust enumeration of cell subsets from tissue expression profiles. Nature Methods 12:453–457. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koestler DC, Jones MJ, Usset J, et al. (2016) Improving cell mixture deconvolution by identifying optimal DNA methylation libraries (IDOL). BMC Bioinformatics 17:120 10.1186/s12859-016-0943-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teschendorff AE, Breeze CE, Zheng SC, Beck S (2017) A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinformatics 18:105 10.1186/s12859-017-1511-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teschendorff AE, Zhuang J, Widschwendter M (2011) Independent surrogate variable analysis to deconvolve confounding factors in large-scale microarray profiling studies. Bioinformatics 27:1496–1505. 10.1093/bioinformatics/btr171 [DOI] [PubMed] [Google Scholar]
- 90.Gagnon-Bartsch JA, Speed TP (2012) Using control genes to correct for unwanted variation in microarray data. Biostatistics 13:539–552. 10.1093/biostatistics/kxr034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zou J, Lippert C, Heckerman D, et al. (2014) Epigenome-wide association studies without the need for cell-type composition. Nat Methods 11:309–311. 10.1038/nmeth.2815 [DOI] [PubMed] [Google Scholar]
- 92.Houseman EA, Molitor J, Marsit CJ (2014) Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics 30:1431–1439. 10.1093/bioinformatics/btu029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rahmani E, Zaitlen N, Baran Y, et al. (2016) Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nature Methods 13:443–445. 10.1038/nmeth.3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rahmani E, Schweiger R, Shenhav L, et al. (2018) BayesCCE: a Bayesian framework for estimating cell-type composition from DNA methylation without the need for methylation reference. Genome Biology 19:141 10.1186/s13059-018-1513-2 [DOI] [PMC free article] [PubMed] [Google Scholar]