Abstract
Background
Caucasian clover (Trifolium ambiguum M. Bieb.) is a strongly rhizomatous, low-crowned perennial leguminous and ground-covering grass. The species may be used as an ornamental plant and is resistant to cold, arid temperatures and grazing due to a well-developed underground rhizome system and a strong clonal reproduction capacity. However, the posttranscriptional mechanism of the development of the rhizome system in caucasian clover has not been comprehensively studied. Additionally, a reference genome for this species has not yet been published, which limits further exploration of many important biological processes in this plant.
Result
We adopted PacBio sequencing and Illumina sequencing to identify differentially expressed genes (DEGs) in five tissues, including taproot (T1), horizontal rhizome (T2), swelling of taproot (T3), rhizome bud (T4) and rhizome bud tip (T5) tissues, in the caucasian clover rhizome. In total, we obtained 19.82 GB clean data and 80,654 nonredundant transcripts were analysed. Additionally, we identified 78,209 open reading frames (ORFs), 65,227 coding sequences (CDSs), 58,276 simple sequence repeats (SSRs), 6821 alternative splicing (AS) events, 2429 long noncoding RNAs (lncRNAs) and 4501 putative transcription factors (TFs) from 64 different families. Compared with other tissues, T5 exhibited more DEGs, and co-upregulated genes in T5 are mainly annotated as involved in phenylpropanoid biosynthesis. We also identified betaine aldehyde dehydrogenase (BADH) as a highly expressed gene-specific to T5. A weighted gene co-expression network analysis (WGCNA) of transcription factors and physiological indicators were combined to reveal 11 hub genes (MEgreen-GA3), three of which belong to the HB-KNOX family, that are up-regulated in T3. We analysed 276 DEGs involved in hormone signalling and transduction, and the largest number of genes are associated with the auxin (IAA) signalling pathway, with significant up-regulation in T2 and T5.
Conclusions
This study contributes to our understanding of gene expression across five different tissues and provides preliminary insight into rhizome growth and development in caucasian clover.
Keywords: Caucasian clover (Trifolium ambiguum M. Bieb.), Full-length transcriptome, RNA-seq, Rhizome, Plant hormone, TFs
Background
Caucasian clover (Trifolium ambiguum M. Bieb.), also known as Kura clover, is a low-crowned, strongly rhizomatous perennial legume [1]; the species originates from the region encompassing Caucasian Russia, eastern Turkey and northern Iran [2]. Caucasian clover can protect lawns as a flowering species and an ornamental plant [3]. Compared with white clover, caucasian clover has lower fiber concentrations, greater protein concentrations and forage digestibility [4] outperforming in high-aluminum soils [5] and providing high-quality fodder during the year when white clover growth is poor [6]. Caucasian clover has deep, semi-woody, usually branched main roots, and many branched roots grow new plantlets, either at the ends or nodes [7–9]. The species can tolerate continuous grazing by cattle (Bos taurus) [7], extreme winter temperatures [8], seasonal moisture deficit and many serious diseases that affect other types of clover [9, 10]. These features are attributed to its prominent primary roots, low-spread crowns and well-developed rhizome systems [11].
In plants without reference genomes, high-throughput RNA sequencing (RNA-seq) technology has become a fast and effective way to master the molecular mechanisms of plants for research purposes [12–14]. This technology has been applied to the study of various rhizomatous species such as sorghum (Sorghum halepense and Sorghum propinquum) [15, 16], bamboo (Phyllostachys praecox) [17], Oryza longistaminata [18–20], Equisetum hyemale [21], Panax ginseng [22], Phragmites australis [23], tropical lotus (Nelumbo nucifera) [24], CangZhu (Atractylodes lacea) [25], Ligusticum chuanxiong [26], ginger (Zingiber officinale) [27], and Miscanthus lutarioriparius [12]. Studies have shown that plant rhizomes are rich in growth regulators, which are related to the energy, metabolism, and hormones pathways of rhizome development. For example, 48 important transcription factors (TFs) belonging to the bHLH, YABBY, NAM, TCP, TALE, and AP2 families are expressed specifically or in abundance in the shoot tip and elongation regions of Oryza longistaminata [18–20].
In recent years, an increasing number of full-length transcriptomes have been generated by Pacific Biosciences (PacBio) sequencing. PacBio sequencing is a single-molecule sequencing technology with a longer read length than second-generation sequencing and an average read length of up to 15 KB, and is also called Single-molecule, real-time (SMRT). This technology not require assembly and can completely retain the entire sequence from the 3′ to 5′ ends of an RNA, but with a higher error rate; moreover, the second-generation stepwise approach can correct mistakes [28–31]. PacBio has been utilized to detect more than 42,280 different splicing isoforms and a large number of alternative splicing (AS) events were found to be associated with the rhizome system and assist genome annotation in orchardgrass [32, 33]. The results indicate that posttranscriptional regulation plays an important role in the rhizome system. Moreover, a combination of Illumina and PacBio sequencing applied to various root tissues, particularly the periderm, has provided a more complete view of the Danshen (Salvia miltiorrhiza) transcriptome [34]. PacBio sequencing and RNA-seq analysis together have also been used to identify differentially expressed transcripts along a developmental gradient from the shoot apex to the fifth internode of Populus P. deltoides×P. euramericana cv.‘Nanlin895’, showing 15,838 differentially expressed transcripts, 1216 of which are TFs [35].
Compared with traditional herbage legumes, such as white clover and red clover, the rhizome is one of the most distinctive characteristics of caucasian clover [9, 36]. The rhizome system has important functions in energy storage, transport and vegetative reproduction [37–40]. We combined Illumina and PacBio sequencing to generate a complete full-length transcriptome of caucasian clover by analysing gene expression in five different tissues and identifying genes related to rhizome development in caucasian clover rhizomes. The obtained results are an excellent reference for further functional characterization to elucidate their roles in rhizome differentiation, growth and development.
Results
Analysis of PacBio sequencing datasets
Transcriptome sequencing of the caucasian clover rhizome was completed, and 19.82 GB of clean data were obtained using one cell. We identified 658,323 reads of inserts (ROIs) with a mean length of 2286 bp, a quality of 0.94, 12 passes from 720,832 polymerase reads with full passes> = 0 and a predicted consensus accuracy> 0.8 (Table 1). In total, the ROIs included 62.87% (449,460) full-length (FL) reads and 29.4% (193,513) non full-length reads of the entire transcriptome sequence from the 5′ to the 3′ end and polyA tail. Additionally, the number of full-length, non-chimeric (FLNC) reads was 441,885, with an average FLNC reads length of 1969 bp (Table 1). The main number distributions of cDNA and ROIs are shown in Additional file 1: Figure S1a and S1b.
Table 1.
Statistics of the PacBio sequencing data, ROI databases and ICE clustering
| cDNA library | 1-6 KB |
|---|---|
| PacBio sequencing | |
| polymerase reads | 720,832 |
| Reads of insert | 658,323 |
| Read bases of insert | 1,505,553,284 |
| Mean read length of insert | 2286 |
| Mean read quality of insert | 0.94 |
| Mean number of passes | 12 |
| Date size (GB) | 19.82 |
| ROI databases | |
| Number of five prime reads | 506,205 |
| Number of three prime reads | 521,059 |
| Number of poly-A reads | 514,174 |
| Number of filtered short reads | 15,350 |
| Number of non-full-length reads | 193,513 |
| Number of full-length reads | 449,460 |
| Number of full-length non-chimeric reads | 441,885 |
| Average full-length non-chimeric read length | 1969 |
| ICE clustering | |
| Number of consensus isoforms | 227,516 |
| Average consensus isoforms read length | 2086 |
| Number of polished high-quality isoforms | 148,836 |
| Number of polished low-quality isoforms | 78,366 |
| Percent of polished high-quality isoforms (%) | 65,42 |
| Non-redundant transcripts | 80,654 |
KB (kilobase): A commonly used length unit for DNA, indicating one thousand base pairs. GB: A unit used to measure the amount of data, where 1 GB = 1,000,000 bp.
As PacBio sequencing results have a high error rate, FLNC reads were clustered using the iterative clustering for error correction (ICE) algorithm and corrected with the Illumina Hiseq2500 platform to correct errors. We generated 227,516 consensus isoforms with an average consensus isoform length of 2086 bp, including 148,836 high-quality isoforms (Table 1). We successfully obtained 80,654 non-redundant transcripts using CD-HIT [41] for caucasian clover rhizomes analysis.
Prediction of ORFs, SSRs and lncRNAs and identification of AS events
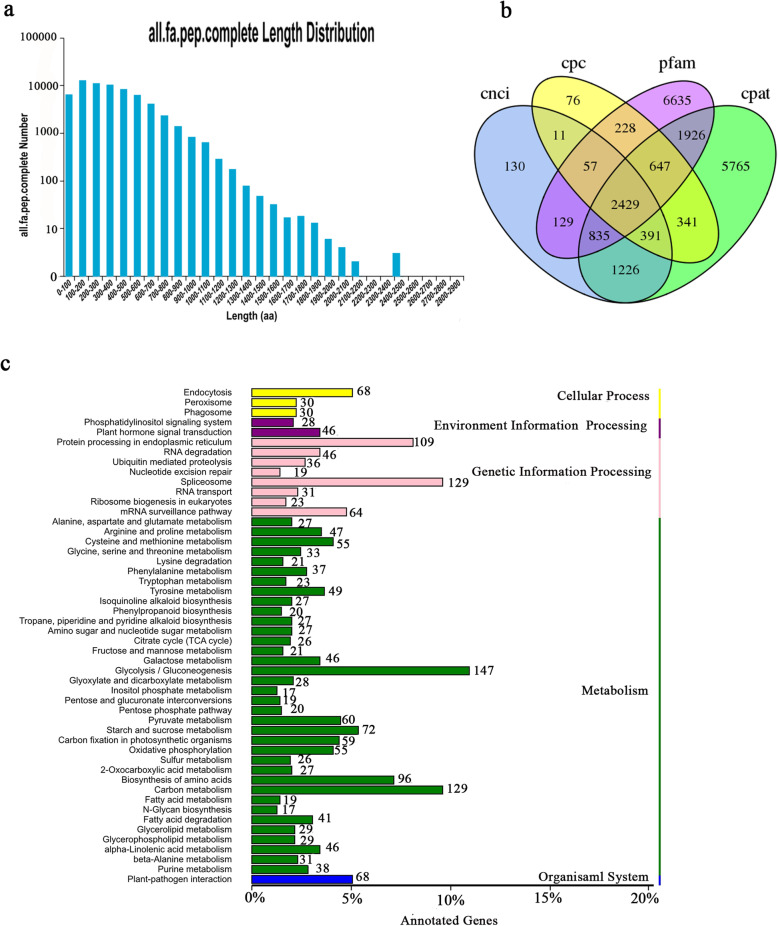
To identify putative protein-coding sequences, we predicted 78,209 open reading frames (ORFs) using TransDecoder. In total, 65,227 coding sequences (CDSs) were identified with start and stop codons, and the distributions of the numbers and lengths of complete CDSs are shown in Fig. 1a. Among them, 12,630 transcripts were distributed in the 100–200-bp range.
Fig. 1.
Prediction of CDS, lncRNAs and AS. a The distribution of CDS lengths with a complete open reading frame. b Venn diagram of the number lncRNAs predicted. c KEGG pathways of genes related to AS
A total of 79,424 sequences (167,351,883 bp) were examined, including 58,276 simple sequence repeats (SSRs) and 36,110 SSR-containing sequences (Additional file 2: Table S1). The number of sequences containing more than one SSR was 13,856, and the number of SSRs present in compound form was 10,041. In addition, most sequences are mononucleotides (33,533), dinucleotides (8610) and trinucleotides (14,026).
In this study, 2429 long noncoding RNAs (lncRNAs) were predicted by a coding potential calculator (CPC), the coding-non-coding index (CNCI), pfam protein structure domain analysis and the coding potential assessment tool (CPAT) (Fig. 1b), revealing candidate lncRNAs for future research.
A total of 6821 AS events were detected. Because no reference genome is available for caucasian clover, we could not identify the types of AS. Nonetheless, as AS is an important mechanism for regulating gene expression and producing proteome diversity, we show the results of these AS events in the KEGG enrichment (Fig. 1c), and the genes were found to be highly enriched in the following categories: “glycolysis/Gluconeogenesis” (147), “spliceosome” (129), “carbon metabolism” (129), “protein processing in endoplasmic reticulum” (109) and “biosynthesis of amino acids” (96).
Transcript annotation
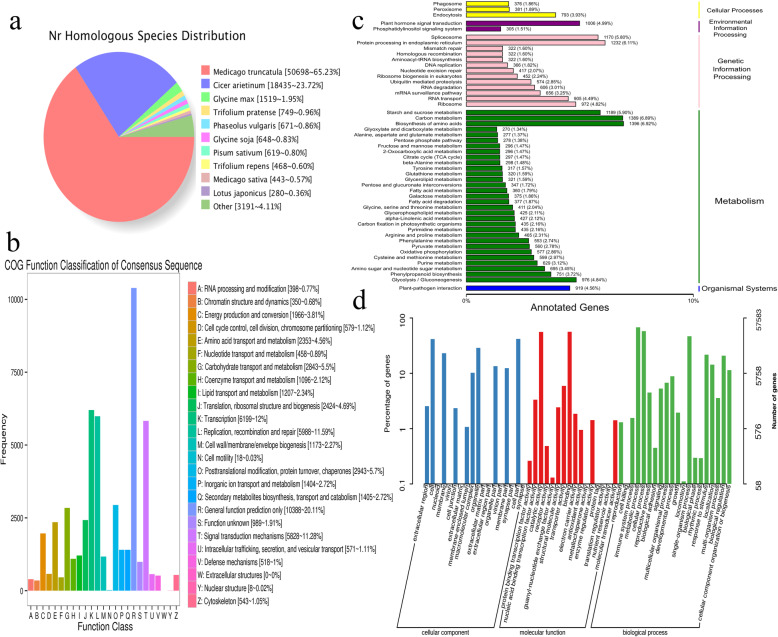
We annotated 77,927 (96.61%) transcripts in at least one of seven databases, including NCBI non-redundant protein (NR), Swiss-Prot (a manually annotated and reviewed protein sequence database), Gene Ontology (GO), Clusters of Orthologous Groups (COG), EuKaryotic Orthologous Groups (KOG), Protein family (Pfam) and Kyoto Encyclopedia of Genes and Genomes (KEGG). The number of detailed annotations for five of the databases (GO, COG, NR, KEGG and Swiss-Prot) is shown in a Venn diagram (Additional file 3: Figure S2).
Through homologous species analysis comparing transcriptome sequences in the NR database, 77,721 transcripts were annotated. Approximately 65.23% (50,689) of the sequences were aligned to Medicago truncatula sequences, followed by Cicer arietinum (23.72%, 18,435) (Fig. 2a).
Fig. 2.
Transcripts annotated in four databases. a NR homologous species distribution diagram of transcripts. b COG function classification of transcripts. c KEGG pathway classification of transcripts. d Distribution of GO terms for all annotated transcripts
All the assembled transcripts were subjected to searches against the COG database to evaluate the effectiveness and completeness of the transcriptome annotation, and the results were divided into 26 main categories (Fig. 2b). The clusters “general function predicted only” (10,388), “transcription” (6199), and “replication, recombination and repair” (5988), represented three of the largest groups, followed by “signal transduction mechanisms” (5828) and “posttranslational modification, protein turnover, and chaperone” (2943).
A total of 33,383 (42.84%) transcripts were matched in the KEGG database and further classified into 128 KEGG pathways (Fig. 2c); “biosynthesis of amino acids” (1396), “carbon metabolism” (1389), “protein processing in endoplasmic reticulum” (1232), “starch and sucrose metabolism” (1189) and “spliceosome” (1170) were the most represented pathways.
Based on the GO analysis, 57,583 transcripts were enriched in the three ontologies (Fig. 2d) “biological process”, “molecular function” and “cellular component”. Transcripts involved in biological processes mainly included “metabolic process” (39,010), “cellular process” (33,383), and “single-organism process” (26,933). In molecular function, transcripts were mainly enriched in “binding” (32,350), “catalytic activity” (32,206), and “transporter activity” (3404). Regarding the “cellular component” category, the major classes of transcripts were related to “cell part” (24,076) “cell” (23,984) and “organelle” (16,648).
Specifically expressed genes and statistics for DEGs
We investigated transcript expression levels in the five tissues, including taproot (T1), horizontal rhizome (T2), swelling of taproot (T3), rhizome bud (T4) and rhizome bud tip (T5) tissues, and T1 had the highest number of expressed genes (76,124), followed by T4 (75.978), T2 (75,885), T3 (74,396) and T2 (74,327) (Additional file 4: Figure S3a and Additional file 4: Figure S3b.). The number of genes co-expressed in each tissue was 68,241. Clean reads obtained by Illumina sequencing were compared with non-redundant transcripts to obtain position information on the transcripts and quantitative expression levels (Additional file 5: Table S2). Fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) values were used to represent the expression levels of genes. To determine specifically expressed genes in tissues and provide insight into the specialized developmental process, these genes (at least two repeats and FPKM> 0.1) with the top 5 average expression levels were selected for investigation (Table 2). Among them, FPKM values were higher in T2 and T5: F01_cb16574_c994/f1p0/1592 (mitogen-activated protein kinase, MAPK3) and F01_cb71 58_ c 94/f1p0/1000 (betaine aldehyde dehydrogenases, BADH).
Table 2.
Specifically expressed genes of the top five FPKMs for each tissue
| Gene | FPKM | Description | |
|---|---|---|---|
| T1 | F01_cb14545_c33/f1p0/902 | 1.04 | Diphosphoinositol-polyphosphate |
| F01_cb17480_c9/f1p0/696 | 0.90 | ADP-ribosylation factor 1 | |
| F01_cb15376_c1/f1p0/1605 | 0.66 | Protein of unknown function | |
| F01_cb14185_c1/f2p0/1231 | 0.50 | Putative DNA-binding protein | |
| F01_cb8972_c16/f1p0/1728 | 0.42 | Cytochrome P450 | |
| T2 | F01_cb16574_c994/f1p0/1592 | 18.39 | Mitogen-activated protein kinase |
| F01_cb17761_c3939/f1p0/3262 | 1.57 | SNF2 family N-terminal domain; | |
| F01_cb10989_c11/f1p0/1938 | 1.21 | Cytochrome P450 | |
| F01_cb16704_c9/f2p2/873 | 0.60 | Plant invertase/pectin methylesterase inhibitor | |
| F01_cb8297_c8/f1p0/2003 | 0.56 | EH-domain-containing protein; | |
| T3 | F01_cb8987_c27/f1p0/456 | 4.07 | ADP-ribosylation factor 1-like |
| F01_cb8782_c40/f44p1/1360 | 3.55 | peroxidase | |
| F01_cb7280_c24/f1p1/2016 | 2.57 | NAC domain-containing protein | |
| F01_cb7489_c3/f1p0/2140 | 2.10 | PHD-finger | |
| F01_cb8820_c25/f1p0/1072 | 2.08 | Ribosomal protein | |
| T4 | F01_cb17761_c89877/f1p0/2957 | 11.28 | Auxin response factor |
| F01_cb17761_c104162/f2p1/2284 | 11.02 | BURP domain | |
| F01_cb16338_c23/f1p0/313 | 5.44 | Calmodulin-like protein | |
| F01_cb17761_c21336/f1p0/3638 | 3.97 | Leucine Rich Repeat | |
| F01_cb1066_c94/f1p0/2823 | 3.35 | Probable galactinol—sucrose galactosyltransferase 2-like | |
| T5 | F01_cb7158_c94/f1p0/1000 | 146.47 | Betaine aldehyde dehydrogenase 1 |
| F01_cb5102_c38/f1p0/684 | 38.70 | Probable protein phosphatase | |
| F01_cb16574_c20256/f2p0/1154 | 36.04 | 60S ribosomal protein L2 | |
| F01_cb9053_c18/f1p0/304 | 30.32 | – | |
| F01_cb11585_c6/f2p0/314 | 20.12 | – |
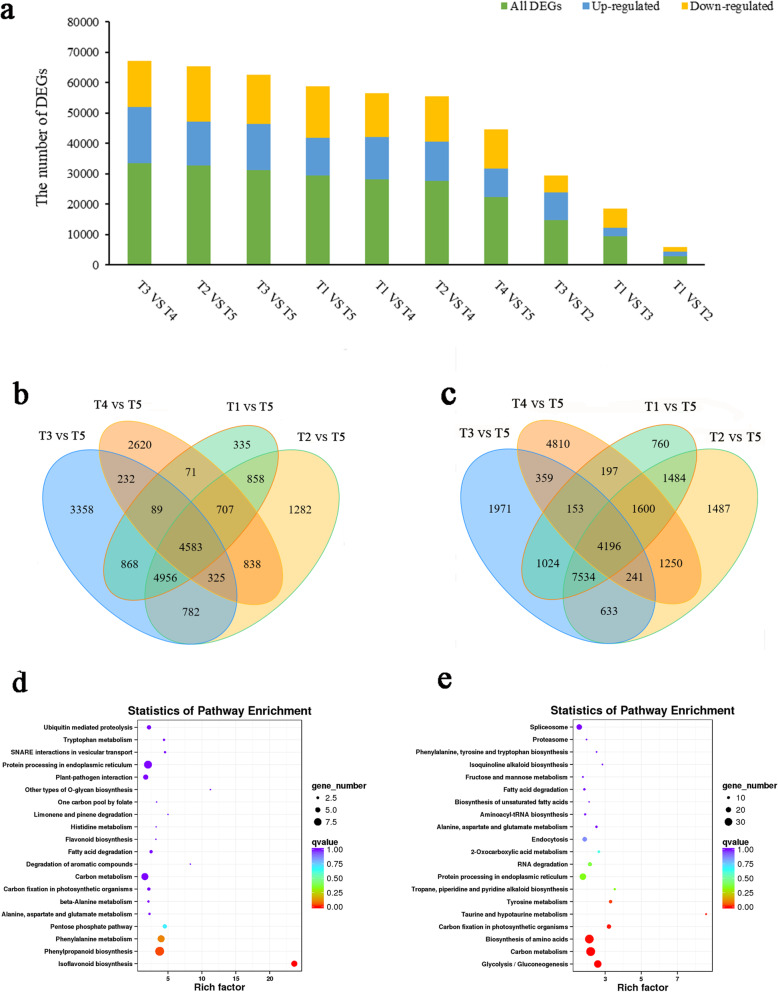
To identify gene expression differences in the development of the caucasian clover rhizome, we focused on identifying differentially expressed genes (DEGs) obtained by Illumina sequencing. As shown in the diagram of the DEGs distribution in different rhizome tissues (Fig. 3a), T3 and T4 had the largest number (33,612) of DEGs, with 18,372 up-regulated and 15,240 down-regulated genes. Moreover, T5 and other tissues (T1, T2, T3 and T4) had more DEGs; 4585 co-upregulated genes and 4196 co-downregulated genes were found in T5 compared with different tissues (Fig. 3b and c). With regard to the up-regulated genes in T5, only 65 were enriched in the KEGG analysis: “phenylpropanoid biosynthesis” (9), “protein processing in endoplasmic reticulum” (8), “phenylalanine metabolism” (7), “carbon metabolism” (7) and “isoflavonoid biosynthesis” (6) (Fig. 3d). For the down-regulated genes in T5, 231 genes were annotated, mainly involved “carbon metabolism” (35), “biosynthesis of amino acids” (34), “glycolysis/gluconeogenesis” (29), “protein processing in endoplasmic reticulum” (25) and “spliceosome” (21) (Fig. 3e).
Fig. 3.
Differentially expressed genes statistics in the rhizome of caucasian clover. a The number of DEGs in different tissues. b Venn diagram showing upregulated genes in tissues compared to T5. c Venn diagram showing down-regulated genes in tissues compared T5. d Co-upregulated genes in KEGG enrichment for T5. e Co-downregulated genes in KEGG enrichment for T5
TFs prediction and WGCNA
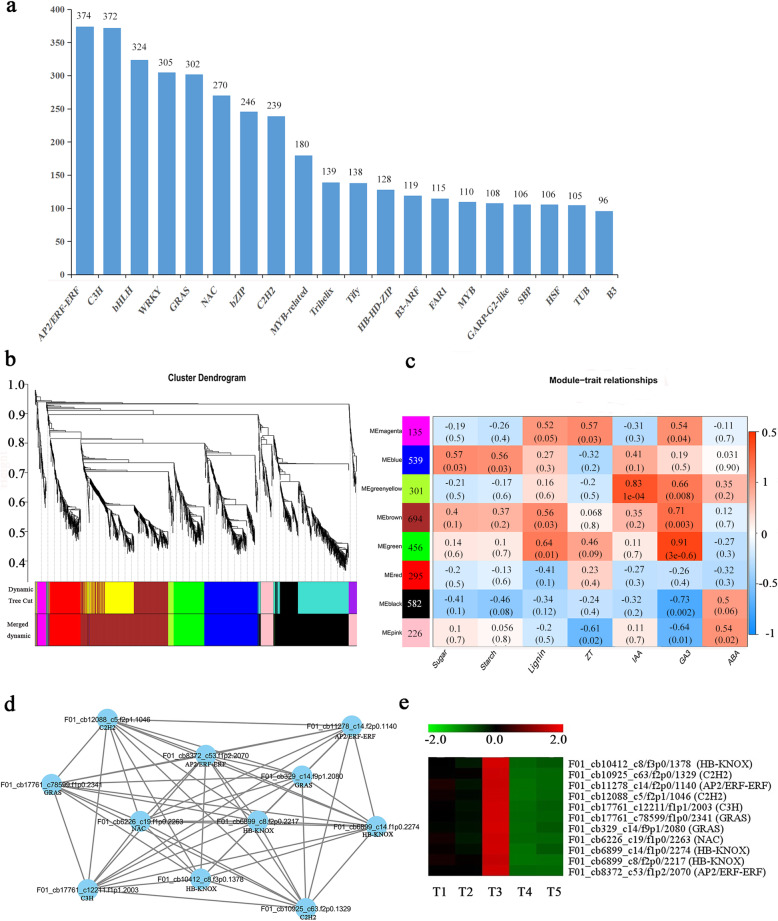
TFs play critical roles in plant growth and development. We examined 4501 putative TFs from 64 different families (Additional file 6: Table S3), and the top 20 TF families are shown in Fig. 4a. Most transcripts belonged to the AP2/ERF-ERF (374), C3H (372), bHLH (324), WRKY (305), GRAS (302), NAC (270), BZIP (246), C2H2 (239), and MYB-related (180) families. These TFs are widely involved in plant growth and responses to stress and are related to rhizome development.
Fig. 4.
The results of TF WGCNA. a Number of top 20 TFs. b Hierarchical cluster tree showing co-expression modules identified by WGCNA. c Module-sample association relationships. d Correlation networks of hub genes in the green module. e Heatmap of hub genes in the green module
We used weighted gene co-expression network analysis (WGCNA) to further explore the relationship between TFs (with filtering of TFs with an FPKM value < 1 and K-ME< 0.7) (Additional file 6: Table S3) and physiological characteristics in the rhizome of caucasian clover (Additional file 7: Table S4). Highly correlated TFs clusters are defined as modules, and TFs within the same cluster are highly correlated. WGCNA identified eight distinct modules (labelled with different colours in Fig. 4b). The correlation coefficients between the characteristic genes of each module of 10 different modules and each different sample (trait) are presented in Fig. 4c. Notably, the auxin (IAA)-trait and GA3-trait were significantly correlated with the MEgreenyellow modules and MEgreen modules (r2 > 0.8, p < 10− 4). Most genes of the green module were up-regulated for six traits, except for abscisic acid (ABA) and most of these TFs were mainly up-regulated in T3 (Additional file 8: Figure S5). We identified 11 hub genes based on the criteria of eigengene-based connectivity (kME) value≥0.99 and an edge weight value ≥0.5 in the green module based on the regulatory network (Fig. 4d). These hub genes mainly belong to the HB-KNOK, AP2/ERF-ERF, GRAS, C2H2, C3H and NAC families (MEgreen-GA3); moreover, these TF families were up-regulated in T1, T2 and T3, particularly in T3 (Fig. 4e) and may be related to the formation of nodules in the rhizome.
Identification of hormone signalling-related genes in rhizome development
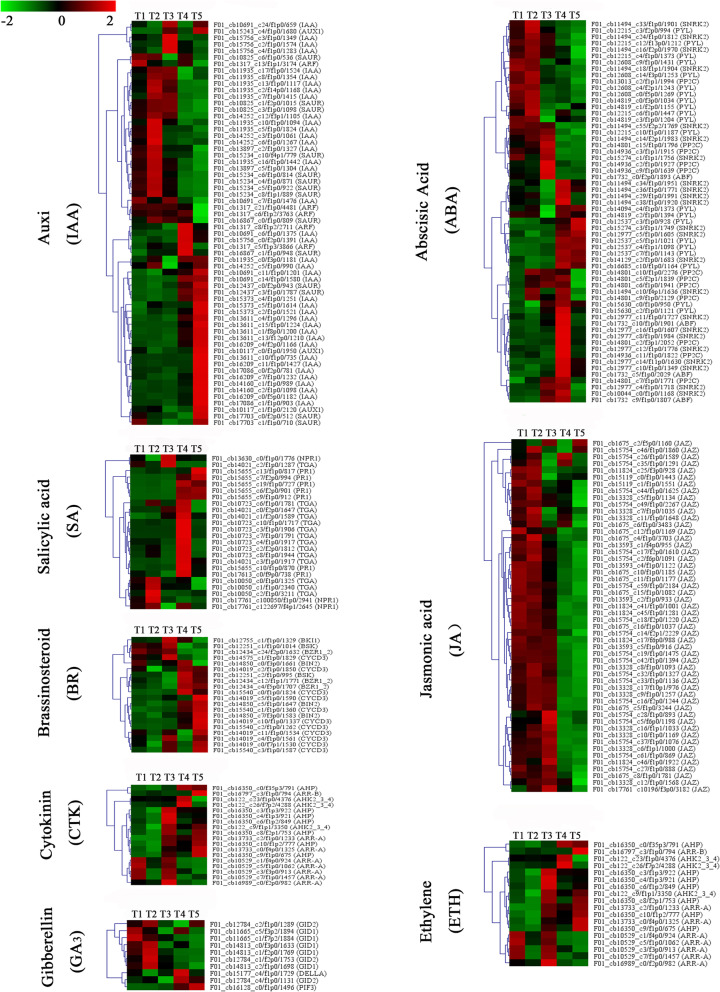
Plant hormones play an important role in all aspects of development. Accordingly, we mapped DEGs to hormone signalling and transduction pathways for caucasian clover and analysed their expression in different tissues. In total, 276 DEGs are related to the synthesis and metabolism of eight hormones, including IAA, ABA, ethylene (ETH), cytokinin (CTK), gibberellic acid (GA), brassinosteroid (BR), jasmonic acid (JA) and salicylic acid (SA) (Fig. 5). The maximum number of genes (62) was found for IAA synthesis and metabolism, followed by ABA at 60, JA at 52, SA at 24, ETH at 28, BR at 20, CTK at 18 and GA at 10. All these significant genes related to hormones synthesis and metabolism exhibited different expression patterns in the different tissues. In addition, More genes were clearly up-regulated in T2 and T5 than other tissues. Regarding SA signalling, almost all genes belonged to the TGA family, with up-regulation only in T4. Most genes associated with BR signalling showed higher levels in T4 and T5. For CTK transduction, all crucial genes associated with the CTK signalling pathway were identified as DEGs. Only three genes showed no up-regulation trend in T3, which may be the tissues in which cells divide in large numbers. Only ten DEGs were associated with GA signalling; five genes were classified as GID1 and significantly up-regulated in T2. Genes related to ABA signalling and transduction displayed no significant change. For JA signalling, 52 genes were annotated as JAZ, with up-regulation in T1, T2 and T3. In contrast, most ETH signalling genes exhibited higher expression in T3, and all were down-regulated in T2.
Fig. 5.
Heatmap of hormone signaling-related genes in the five tissues
Verification of gene expression by qRT-PCR
To confirm the accuracy of the genes obtained by RNA-seq, twelve genes, including six plant hormone signal transduction genes, three TFs and three genes belonging to other classes, were randomly selected for quantitative real-time RT-PCR (qRT-PCR) analysis (three replicates of each gene). Good reproducibility between the qRT-PCR and RNA-seq results was indicated by Pearson’s correlation analysis, thus verifying the accuracy and reliability of the RNA-seq data (Additional file 9: Table S5).
Discussion
The rhizome of caucasian clover is unique among legume species, endowing this plant with particular clonal reproduction characteristics and resistance to stress. In this study, we obtained high-quality transcript sequences for the caucasian clover rhizome by PacBio and Illumina sequencing, and the results will contribute to our understanding of rhizome growth and development and provide a molecular foundation for further study.
AS is a vital mechanism regulating gene expression and producing protein diversity [42]. The numbers of AS events identified for the first time in the caucasian clover rhizome was found to be lower than that in Medicago sativa (7568) [43] but higher than that in Trifolium pratense (5492) [13]. Our study on the special characteristics of the caucasian clover rhizome was hindered by the lack of a reference genome, and the types of AS events are impossible to determine.
Many amino acids (phenylalanine, tyrosine and tryptophan) are not only important components of proteins but are also precursors of many secondary metabolites. These secondary metabolites are crucial for plant growth [44]. Similar to the rhizome of other plants (Oryza longistaminata and Miscanthus lutarioriparius) [12, 19], some basal metabolism plays an important role in the rhizome of caucasian clover, for example, carbon metabolism (1389) and starch and sucrose metabolism (1189)(Fig. 2c).
By analysing the specifically expressed genes in each tissue, we found that MAPK3 (F01_cb16574_c994/f1p0 /1592) is mainly specifically expressed in T2 (Table 2). The MAPK family has been studied in tobacco, and it may be involved in growth, development, and responses to plant hormone and environmental signals [45]. T5 is very different from root tissues (T1, T2 and T3) (Fig. 3a), with more DEGs, and is the site of the formation of new aboveground parts. A specific gene with the highest FPKM in T5 was observed for BADH (F01_cb7158_c94/f1p0/1000) (Table 2). BADH genes are involved in glycine betaine synthesis and act as plant osmotic regulators, with important roles in abiotic stress [46]. Experiments have shown that BADH can increase the tolerance of sweet potato and carrot against abiotic stresses such as salt stress, oxidative stress and low-temperature stress, thus maintaining cell membrane integrity [47, 48]. BADH was specifically expressed at a high level in T5, possibly because T5 is relatively more fragile than other tissues, and BADH may protect T5 to promot the growth of new plants. Thus, we speculate that defence and stress response play a vital role in the development of caucasian clover, which may be the reason for its ability to grow in extreme winter temperatures, or such responses may be a necessary condition for a large rhizome system.
TFs can offer insight into the gene-regulating networks controlling developmental programmes and are recognized as major contributors to a better understanding of root tissue differentiation and root development in response to internal growth regulators as well as environmental signals [49, 50]. The genes involved in hormone metabolism, cellulose synthesis energy, metabolism substance synthesis and transportation stress as well as expansion-related protein genes and TFs such as bHLH, TCP, WRKY, bZIP, MYB and NAC have been reported to in the formation of lotus rhizomes [24]. In addition, the AP2/ERF family accounts for the greatest number in the caucasian clover rhizome (Fig. 4a), and ethylene response factors, such as BBM/PLT4 and PLT1–3, have been described as master regulators of root meristem initiation and maintenance in Arabidopsis thaliana [51, 52]. In Raphanus sativus and Medicago sativa, the abiotic stress response mechanism regulated by AP2/ERF has been carefully studied [53, 54]. The Arabidopsis NAC family member NAC1 transduces IAA signals downstream of TIR1 to promote lateral root development [55]. In the WRKY family, WRKY75 was reported to be involved in regulating the nutrient starvation response and rhizome growth [56]. Notably we performed WGCNA clustering for TFs and found that 3 of 11 hub genes belong to the HB-KNOX family in MEgreen-GA3 (Fig. 4d). GA can suppress the effect of elevated KNOX protein expression, and modifying KNOX may alter the plant structure through local changes in GA levels [57]. In Arabidopsis, the GA20ox1 mRNA level is reduced in leaves overexpressing the KNOX proteins STM or BREVIPEDICELLUS [56]. Moreover, in model plants, such as Arabidopsis, maize and tobacco, KNOX gene expression is confined to the shoot meristem and stem [58]. However, in the underground rhizome of caucasian clover, KNOX genes were identified as hub genes, especially in the main tissues of the swollen taproot (T3). Whether these finding are consistent with KNOX regulation of Arabidopsis meristems, stems and buds is worthy of further investigation [59]. Xi Cheng found that in pear plants co-expressing KNOX and PbKNOX1, these factors are involved in cell wall thickening and lignin biosynthesis, with inhibition of key structural genes involved in lignin synthesis [60].
Growing evidence indicates that hormones affect tillering growth and the formation of storage organs [12, 61]. Yi Kun’s research illustrated that 600 mg·L- 1 GA3 can promote the growth and development of caucasian clover rhizome and increase the contents of endogenous IAA, ZT and GA3 [62]. T1 and T2 showed high GID1 expression and GA may be involved in photoperiod induction and regulation of the formation of storage organs and rhizome elongation [63]; therefore, TI and T2 may be key organs for nutrient storage. In addition, JAZ protein accumulated in roots (T1, T2 and T3). When pathogens invade and abiotic stress occurs, JAZ-MYC is believed to form an immune network, followed by JAZ protein degradation and MYC release [64]. T3 is closely linked to ETH and CTK-mediated pathways and may be responsible for root swelling. IAA has been demonstrated to activate root formation, while CTKs mediate root identity, early primordial disintegration and early loss of bud development initiation [65]. CTKs are conducive to rhizome enlargement but not to rhizome induction.
Genes related to the IAA anabolic pathway were downregulated in T3 and T4. IAA may not participate in root enlargement or induce bud production but may be closely related to lateral root development. T1, T2 and T3 might mainly function as storage organs, providing energy for plant growth. In addition, T4 may be relatively fragile and require the SA pathway to mediate immunity to prevent pathogen infection and to grow new plants. BR signalling is mainly involved in plant growth and plant morphology development, and related genes were upregulated in T5 [66]. T4 and T5 are mainly associated with resistance to stress and secondary metabolic pathways. Of course, hormones are not the only factors that regulate the development of apical meristems and lateral organs; they often cooperate with TFs to balance the maintenance of meristems and organogenesis.
Conclusion
In summary, we provided a full-length transcriptome of the caucasian clover rhizome based on PacBio sequencing and Illumina sequencing, revealing gene expression patterns and annotation for different tissues. We highlighted the role of hormones and TFs in the rhizome of caucasian clover, investigated the expression of hormone pathway-related genes in different tissue of caucasian clover and identified 11 hub genes in TF and GA-related modules by WGCNA. In this study, a set of genes related to rhizome development was identified, laying the foundation for further functional genomics research on rhizome development.
Methods
Plant materials and RNA preparation
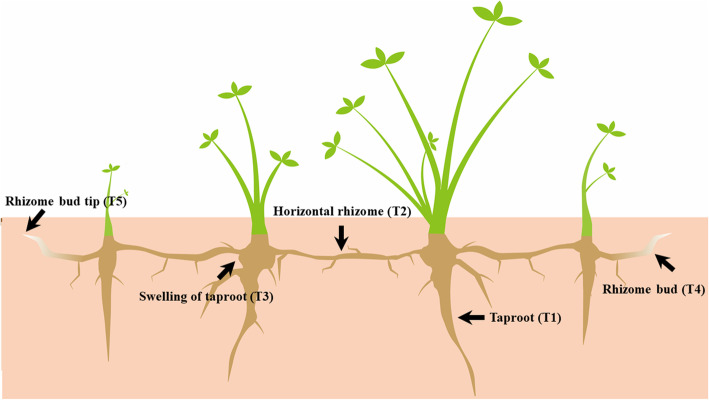
The T1, T2, T3, T4 and T5 tissues of 3-year-old caucasian clover (Fig. 6) were collected from a test field at Northeast Agricultural University (E 126°14′; N 45°05′ in August 2018). We placed each sample collected into a 1.5-ml centrifuge tube with three replicates from five individual plants for each tissue to ensure RNA quantity. Each sample showed a good correlation (R2 > 0.8; Additional file 10: Figure S5). The original sources of the plant materials were introduced from Inner Mongolia Grass Variety Engineering Technology Research Center of Inner Mongolia Agricultural University. The Inner Mongolia Grass Variety Engineering Technology Research Center of Inner Mongolia Agricultural University undertook formal identification of the samples, provided details of the specimens deposited and allowed the collection. The IPNI Life Sciences Identifier (LSID) for caucasian clover is urn:lsid:ipni.org:names:522843–1. Plants were removed from the soil bed, and the roots were washed gently with running water, frozen in liquid nitrogen and immediately stored at − 80 °C. Total RNA was extracted using Trizol. RNA degradation and contamination were monitored by 1.2% agarose gel electrophoresis. The quantity and integrity of the extracted total RNA were determined using a NanoDrop and an Agilent 2100 bioanalyzer (Santa Clara, CA) [13]. For each RNA sample, 1 μg was pooled and sequenced by PacBio single-molecule long-read sequencing (PacBio Sequel, Menlo Park, USA) and Illumina sequencing (Illumina NovaSeq6000, California, U.S.A) in parallel.
Fig. 6.
Schematic graph of tissues collected for PacBio sequencing and Illumina sequencing
PacBio cDNA library preparation and sequencing
Full-length cDNA was synthesized using the SMARTerTM PCR cDNA Synthesis Kit and then subjected to full-length cDNA PCR amplification and repair of cDNA ends. The concentration and quality of the cDNA library were determined using the Qubit 2.0 Fluorometer and an Agilent 2100 bioanalyzer [67]. The 1–6-KB library was sequenced via PacBio Sequel.
Illumina cDNA library construction and sequencing
First, 15 samples of eukaryotic mRNA were enriched with magnetic beads with oligo (dT) and randomly broken into small fragments in fragmentation buffer. First-strand cDNA was synthesized using six-base random hexamers with a small fragment of mRNA as a template. The cDNA was purified by AMPure XP beads after synthesis of second cDNA strand (adding buffer, dNTPs, RNase H and DNA polymerase I). Then, double-stranded cDNA was end repaired, adding a tail and the sequencing linker, and the fragment size was determined by AMPure XP beads [41]. The final cDNA library was assessed by PCR, and the quality of the cDNA library was determined using an Agilent 2100 bioanalyzer. The libraries were sequenced from both 5′ and 3′ ends using Illumina NovaSeq.
PacBio sequencing data analysis
ROI sequencing was extracted using the Iso-seq pipeline with minFullPass = 0 and minPredictedAccuracy = 0.80. In ROIs, reads containing poly(A) tail signals and 5′- and 3′-cDNA primers are regarded as full-length non-chimera (FLNC) transcripts [68]. The consensus sequencing isoform was obtained by using the ICE algorithm. Finally, high-quality FL transcripts were classified with an accuracy greater than 99% using Quiver. CD-HIT (identity > 0.99) was used to remove redundant sequences in high-quality FL transcripts, and ultimately obtain non-redundant transcript sequences [69].
Illumina sequencing data analysis
High-quality clean data (removing containing adapters, poly-N and low-quality reads from the raw data) were used for all downstream analyses [70]. These clean reads were then mapped to the PacBio reference genome sequence and normalized by converting the fragment counts to FPKM values. A differential expression analysis was performed using DESeq (v 1.10.1); a fold change ≥4 and an FDR < 0.01 based on DESeq were considered indicative of differential expression [71].
Detection of SSR, ORFs, AS and lncRNA
SSRs in the transcriptome were identified using MISA (http://pgrc.ipk-gatersleben.de/misa/), and only transcripts ≥500 bp were detected.
TransDecoder software (https://github.com/TransDecoder/TransDecoder/releases) was employed to identify reliable potential CDSs from the transcript sequences based on the ORF length, log-likelihood score, and amino acid sequence comparison in the Pfam database.
We used Iso-SeqTM data directly to run all-vs-all BLAST with high-identity settings [72]. BLAST alignments that met all criteria were considered products of candidate AS events [73], with two HSPs (high segmentation pairs) > 1000 bp in the alignment. Two HSPs have the same forward/reverse orientation, and one sequence should be contiguous in the same alignment or have a small overlap of less than 5 bp. The other sequence should be different to show “AS gap”, and the contiguous sequence should align completely with the different sequences. The AS gap should be greater than 100 bp and at least 100 bp from the 3′/5′ end.
CPC [74], CNCI, CPAT [75] and Pfam [76] were used to distinguish nonprotein-coding RNAs from putative protein-coding RNAs. Transcripts with lengths greater than 200 nt and more than two exons were selected as lncRNA candidate genes.
Functional annotation
Annotation information on the obtained nonredundant transcript sequences was based on BLAST in the following databases: NR, Pfam [76], KOG [77], COG, swiss-Prot [78], KEGG [79] and GO [80].
Real-time RT- PCR
QRT-PCR was conducted in a 10-μl volume containing 0.5 μl of diluted cDNA, 0.2 μl of forward primer, 0.2 μl of reverse primer, and 1 × SYBR Premix Ex Taq II (TaKaRa) with the following conditions: 95 °C for 180 s, followed by 40 cycles of 95 °C for 15 s, 59 °C for 15 s and 72 °C for 15 s. The 2-△△Ct method was used to calculate relative expression levels [81]. All reactions were performed with three replicates. All primers used are shown in Additional file 11: Table S6.
Supplementary information
Additional file 1: Figure S1. The output of ROI. a ROI read length distribution for 1-6 KB size bin. b ROI classification for 1-6 KB.
Additional file 2: Table S1. The prediction of SSRs.
Additional file 3: Figure S2. Venn diagram of the number NR, Swiss-prot, COG, GO and KEGG.
Additional file 4: Figure S3. The number of transcripts. a The number statistics of transcripts in five tissues. b Venn diagram of expressed transcripts among different tissues.
Additional file 5: Table S2. Statistics of comparison results between the Illumina sequencing data and the non-redundant transcripts
Additional file 6: Table S3. The prediction of caucasian clover rhizome transcription factor (TF) family.
Additional file 7: Table S4. The results of different physiological indicators. The bar chart of sugar, starch, lignin, ZT, GA3, IAA and ABA content.
Additional file 8: Figure S4. The heatmap of TFs in green module.
Additional file 9: Table S5. The results of qRT-PCR.
Additional file 10: Figure S5. Heatmap of the Pearson correlation coefficient of each sample.
Additional file 11: Table S6. Primers used for qRT-PCR.
Acknowledgements
We thank the National Natural Science Foundation of China (31802120) for supporting this work and Biomarker Corporation (Beijing, China) for the PacBio and Illumina sequencing and for the raw data analysis.
Abbreviations
- OFRs
Open reading frames
- CDSs
Coding sequences
- SSRs
Simple sequence repeats
- AS
Alternative splicing
- LncRNA
Long non-coding RNA
- TF
Transcription factors
- KB
Kilobase
- ROI
Reads of insert
- ICE: ICE
Iterative clustering for error correction;
- FL
Full-length
- FLNC
Full-length non-chimeric
- CNCI
Coding-Non-Coding Index
- CPC
Coding potential
- CPAT
Coding potential assessment tool calculator
- FLNC
Full-length non chimera
- COG
Clusters of Orthologous Groups
- NR
NCBI non-redundant protein
- GO
Gene Ontology
- KOG
EuKaryotic Orthologous Groups
- Pfam
Protein family
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DEG
Differentially expressed genes
- FPKM
Fragments per kilobase of transcript sequence per million base pairs sequenced
- WGCNA
Weighted gene co-expression network analysis
- IAA
Auxin
- ABA
Abscisic acid
- ETH
Ethylene
- CTK
Cytokinin
- GA
Gibberellic acid
- BR
Brassinosteroid
- JA
Jasmonic acid
- SA
Salicylic acid
- BADHs
Betaine aldehyde dehydrogenases
- MAPK
Mitogen-activated protein kinase
- qRT-PCR
Quantitative real-time RT-PCR
Authors’ contributions
XY designed the experiment and revised the manuscript. KY performed data processing and drafted the manuscript. YZ, YH and XC prepared the materials and performed the experiments. TH and JL assisted in manuscript preparation. GC conceived the study. All authors read and approved the final version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31802120), Research and Demonstration of Large-scale Artificial Grassland Combined Plant and Circular Mode (2017YFD0502106) and Academic Backbone Fund Project of Northeast Agricultural University. The funders did not design the experiment or draft and revise the manuscript.
Availability of data and materials
Raw reads of one combined PacBio library and one Illumina RNAseq library generated in this study are available from BioProject at NCBI (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA586585 and PRJNA588309, respectively.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiujie Yin and Kun Yi contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02637-4.
References
- 1.Black AD, Lucas RJ. Caucasian clover was more productive than white clover in grass mixtures under drought conditions. NZGA. 2000;62:183–188. [Google Scholar]
- 2.Davis PH. Flora of Turkey and the East Aegean Islands. Q Rev Biol. 1965;35(1):103. [Google Scholar]
- 3.Lane I, Watkins E, Spivak M. Turfgrass species affect the establishment and bloom of Kura clover (Trifolium ambiguum) in lawns. Hortscience. 2019;54(5):824. [Google Scholar]
- 4.Andrzejewska J, Contreras-Govea FE, Pastuszka A, Albrechtc KA. Performance of Kura clover compared to that of perennial forage legumes traditionally cultivated in Central Europe. Acta Agr Scand. 2016;66(6):516–522. [Google Scholar]
- 5.Caradus JR, Crush JR, Ouyang L, FRASER W. Evaluation of aluminium-tolerant white clover (Trifolium repens) selections on East Otago upland soils. NZGA. 2001;44:141–150. [Google Scholar]
- 6.Watson RN, Neville FJ, Bell NL, Harris SL. Caucasian clover as a pasture legume for drylanddairying in the coastal Bay of Plenty. NZCA. 1996;58:83–88. [Google Scholar]
- 7.Brummer EC, Moore KJ. Persistence of perennial cool-season grass and legume cultivars under continuous grazing by beef cattle. Agron J. 2000;92(3):466–471. [Google Scholar]
- 8.Sheaffer CC, Marten GC. Kura clover forage yield, forage quality, and stand dynamics. Can J Plant Sci. 1991;71(4):1169–1172. [Google Scholar]
- 9.Taylar NL, Smith RR. Kura clover (Trifolium ambiguum M. B.) breeding, culture, and utilization. Adv Agron. 1997;63(08):153–178. [Google Scholar]
- 10.Andrzejewska J, Contreras-Govea FE, Pastuszka A, Albrecht KA. Performance of Kura clover compared to that of perennial forage legumes traditionally cultivated in Central Europe. Acta Agr Scand B-S P. 2016;66(6):516–522. [Google Scholar]
- 11.Speer GS, Allinson DW. Kura clover (Trifolium ambiguum): legume for forage and soil conservation. Econ Bot. 1985;39(2):165–176. [Google Scholar]
- 12.Hu RB, Yu CJ, Wang XY, Jia CL, Pei SQ, He K, He G, Kong YZ, Zhou GK. De novo transcriptome analysis of Miscanthus lutarioriparius identifies candidate genes in rhizome development. Front Plant Sci. 2017;8:482. doi: 10.3389/fpls.2017.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao YH, Yuan JB, Li SF, Jia SQ, Han LB, Xu LX. Analysis of transcripts and splice isoforms in red clover (Trifolium pratense L.) by single-molecule long-read sequencing. BMC Plant Biol. 2018;18:300–312. doi: 10.1186/s12870-018-1534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang LK, Yan HD, Zhao XX, Zhang XQ, Wang J, Frazier T, Yin G, Huang X, Yan DF, Zang WJ, et al. Identifying differentially expressed genes under heat stress and developing molecular markers in orchardgrass (Dactylis glomerata L.) through transcriptome analysis. Mol Ecol Resour. 2015;151(5):497–509. doi: 10.1111/1755-0998.12418. [DOI] [PubMed] [Google Scholar]
- 15.Jang CS, Kamps TL, Tang H, Bowers JE, Lemke C, Paterson AH. Evolutionary fate of rhizome-specific genes in a non-rhizomatous Sorghum genotype. Heredity. 2009;102(3):266–273. doi: 10.1038/hdy.2008.119. [DOI] [PubMed] [Google Scholar]
- 16.Zhang T, Zhao XQ, Wang WS, Huang LY, Liu XY, Zong Y, Zhu LH, Yang DC, Fu BY, Li ZK. Deep transcriptome sequencing of rhizome and aerial-shoot in Sorghum propinquum. Plant Mol Biol. 2014;84(3):315–327. doi: 10.1007/s11103-013-0135-z. [DOI] [PubMed] [Google Scholar]
- 17.Wang KH, Peng HZ, Lin EP, Jin QY, Hua X, Sheng Y, Bian HW, Ning H, Pan JW, Wang JH. Identification of genes related to the development of bamboo rhizome bud. J Exp Bot. 2010;61(2):551–561. doi: 10.1093/jxb/erp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu FY, Tao DY, Sacks E, Fu BY, Xu P, Li J, Yang Y, Mcnally K, Khush GS, Paterson AH. Convergent evolution of perenniality in rice and sorghum. P Natl Acad Sci USA. 2003;100(7):4050–4054. doi: 10.1073/pnas.0630531100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu F. Identification of rhizome-specific genes by genome-wide differential expression analysis in Oryza longistaminata. BMC Plant Biol. 2011;11(1):18–32. doi: 10.1186/1471-2229-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He RF, Salvato F, Park JJ, Kim MJ, Nelson W, Balbuena TS, Willer M, Crow JA, May GD, Soderlund CA, et al. A systems-wide comparison of red rice (Oryza longistaminata) tissues identifies rhizome specific genes and proteins that are targets for cultivated rice improvement. BMC Plant Biol. 2014;14(1):46–67. doi: 10.1186/1471-2229-14-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balbuena TS, He RF, Salvato F, Gang DR, Thelen JJ. Large-scale proteome comparative analysis of developing rhizomes of the ancient vascular plant Equisetum hyemale. Front Plant Sci. 2012;3:131. doi: 10.3389/fpls.2012.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang B\W, Hahm YT. Transcriptome analysis using de novo RNA-seq to compare ginseng roots cultivated in different environments. Plant Growth Regul. 2018;84(1):149–157. [Google Scholar]
- 23.Ruifeng H, Min-Jeong K, William N, Balbuena TS, Ryan K, Robin K, Crow JA, May GD, Thelen JJ, Soderlund CA. Next-generation sequencing-based transcriptomic and proteomic analysis of the common reed, Phragmites australis (Poaceae), reveals genes involved in invasiveness and rhizome specificity. Am J Bot. 2012;99(2):232–247. doi: 10.3732/ajb.1100429. [DOI] [PubMed] [Google Scholar]
- 24.Yang M, Zhu L, Pan C, Xu L, Liu Y, Ke W, Yang P. Transcriptomic analysis of the regulation of rhizome formation in temperate and tropical Lotus (Nelumbo nucifera) Sci Rep. 2015;5:13059. doi: 10.1038/srep13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang QQ, Huang X, Deng J, Liu HG, Liu YW, Yu K, Huang BS. Differential gene expression between leaf and rhizome in Atractylodes lancea: a comparative transcriptome analysis. Front Plant Sci. 2016;7:348. doi: 10.3389/fpls.2016.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song T, Liu ZB, Li JJ, Zhu QK, Tan R, Chen JS, Zhou JY, Liao H. Comparative transcriptome of rhizome and leaf in Ligusticum Chuanxiong. Plant Syst Evol. 2015;301(8):2073–2085. [Google Scholar]
- 27.Koo HJ, McDowell ET, Ma XQ, Greer KA, Kapteyn J, Xie ZZ, Descour A, Kim H, Yu Y, Kudrna D, et al. Ginger and turmeric expressed sequence tags identify signature genes for rhizome identity and development and the biosynthesis of curcuminoids, gingerols and terpenoids. BMC Plant Biol. 2013;13(1):27–44. doi: 10.1186/1471-2229-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pop M, Salzberg SL. Bioinformatics challenges of new sequencing technology. Trends in Genet. 2008;24(3):142–149. doi: 10.1016/j.tig.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason CE. Faster sequencers, larger datasets, new challenges. Genome Biol. 2012;13(3):314. doi: 10.1186/gb-2012-13-3-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhoads A, Au KF. PacBio sequencing and its applications. Genom Proteom Bioinf. 2015;13(5):278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li PH, Ponnala L, Gandotra N, Wang L, Si YQ, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. The developmental dynamics of the maize leaf transcriptome. Nat Genet. 2010;42(12):1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- 32.Wang T, Wang H, Cai D, Gao Y, Zhang H, Wang Y, Lin C, Ma L, Gu L. Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis) Plant J. 2017;91(4):684–699. doi: 10.1111/tpj.13597. [DOI] [PubMed] [Google Scholar]
- 33.Huang LK, Feng GY, Yan HD, Zhang ZG, Bushman BS, Wang JP, Bombarely A, Li MZ, Yang ZF, Nie G, et al. Genome assembly provides insights into the genome evolution and flowering regulation of orchardgrass. Plant Biotechnol J. 2020;18(2):373–383. doi: 10.1111/pbi.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu ZC, Peters RJ, Weirather J, Luo HM, Liao BS, Zhang X, Zhu YJ, Ji AJ, Zhang B, Hu SN, et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 2015;82(6):951–961. doi: 10.1111/tpj.12865. [DOI] [PubMed] [Google Scholar]
- 35.Chao Q, Gao ZF, Zhang D, Zhao BG, Dong FQ, Fu CX, Liu LJ, Wang BC. The developmental dynamics of the Populus stem transcriptome. Plant Biotechnol J. 2019;17(1):206–219. doi: 10.1111/pbi.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin WH. W HR, Stephen S. physiological responses of five species of Trifolium to drought stress. Chin J Appl Environ Biol. 2011;17(4):580–584. [Google Scholar]
- 37.Schmid B, Bazzaz FA. Clonal integration and population structure in perennials: effects of severing rhizome connections. Ecology. 1987;68(6):2016–2022. doi: 10.2307/1939892. [DOI] [PubMed] [Google Scholar]
- 38.Schmid B, Bazzaz FA. Growth responses of rhizomatous plants to fertilizer application and interference. Oikos. 1992;65(1):13–24. [Google Scholar]
- 39.Humphrey LD, Pyke DA. Clonal foraging in perennial wheatgrasses: a strategy for exploiting patchy soil nutrients. J Eco. 1997;85(5):601–610. [Google Scholar]
- 40.Huber-Sannwald E, Pyke DA, Caldwell MM, Durham S. Effects of nutrient ptches and root systems on the clonal plasticity of a rhizomatous grass. Ecology. 1998;79(7):2267–2280. [Google Scholar]
- 41.Chen H, Feng WW, Chen K, Qiu XC, XU H, Mao GC, Zhao T, Ding YY, XY WU. Transcriptomic analysis reveals potential mechanisms of toxicity in a combined exposure to dibutyl phthalate and diisobutyl phthalate in zebrafish (Danio rerio) ovary. Aquat Toxicol. 2019;216:105290. doi: 10.1016/j.aquatox.2019.105290. [DOI] [PubMed] [Google Scholar]
- 42.Chaudhary S, Khokhar W, Jabre I, Reddy ASN, Byrne LJ, Wilson CM, Syed NH. Alternative splicing and protein diversity: plants versus animals. Front Plant Sci. 2019;10(10):14. doi: 10.3389/fpls.2019.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chao YH, Yuan JB, Guo T, Xu LX, Mu ZY, Han LB. Analysis of transcripts and splice isoforms in Medicago sativa L. by single-molecule long-read sequencing. Plant Mol Biol. 2019;99(3):219–235. doi: 10.1007/s11103-018-0813-y. [DOI] [PubMed] [Google Scholar]
- 44.Tzin V, Galili G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant. 2010;3(6):956–972. doi: 10.1093/mp/ssq048. [DOI] [PubMed] [Google Scholar]
- 45.Zhang S. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001;6:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]
- 46.Fitzgerald TL, Waters DLE, Henry RJ. Betaine aldehyde dehydrogenase in plants. Plant Biol. 2009;11(2):119–130. doi: 10.1111/j.1438-8677.2008.00161.x. [DOI] [PubMed] [Google Scholar]
- 47.Kumar S, Dhingra A, Daniell H. Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance. Plant Physiol. 2004;136(1):2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan WJ, Zhang M, Zhang HX, Zhang P. Improved tolerance to vrious abiotic stresses in tansgenic sweet potato (Ipomoea batatas) expressing spinach betaine aldehyde dehydrogenase. PLoS One. 2012;7(5):14. doi: 10.1371/journal.pone.0037344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin JP, Zhang H, Kong L, Gao G, Luo JC. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42(D1):D1182–D1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montiel G, Gantet P, Jay-Allemand C, Breton C. Transcription factor networks. Pathways to the knowledge of root development. Plant Physiol. 2004;136(3):3478–3485. doi: 10.1104/pp.104.051029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Licausi F, Ohme-Takagi M, Perata P. APETALA/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 2013;199(3):639–649. doi: 10.1111/nph.12291. [DOI] [PubMed] [Google Scholar]
- 52.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Bba-Gene Regul Mech. 2012;1819(2):86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Karanja BK, Xu L, Wang Y, Tang M, M'Mbone Muleke E, Dong J, Liu L. Genome-wide characterization of the AP2/ERF gene family in radish (Raphanus sativus L.): Unveiling evolution and patterns in response to abiotic stresses. Gene. 2019;718:144048. doi: 10.1016/j.gene.2019.144048. [DOI] [PubMed] [Google Scholar]
- 54.Jin XY, Yin XF, Ndayambaza B, Zhang ZS, Min XY, Lin XS, Wang YR, Liu WX. Genome-wide identification and expression profiling of the ERF gene family in Medicago sativa L. under various abiotic stresses. DNA Cell Biol. 2019;13(10):1056-68. [DOI] [PubMed]
- 55.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14(23):3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007;143(4):1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh DP, Filardo FF, Storey R, Jermakow AM, Yamaguchi S, Swain SM. Overexpression of a gibberellin inactivation gene alters seed development, KNOX gene expression, and plant development in Arabidopsis. Physiol Plant. 2010;138(1):74–90. doi: 10.1111/j.1399-3054.2009.01289.x. [DOI] [PubMed] [Google Scholar]
- 58.Hay A, Tsiantis M. KNOX genes: versatile regulators of plant development and diversity. Development. 2010;137(19):3153–3165. doi: 10.1242/dev.030049. [DOI] [PubMed] [Google Scholar]
- 59.Xu L, Shen WH. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol. 2008;18(24):1966–1971. doi: 10.1016/j.cub.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 60.Cheng X, Li ML, Abdullah M, Li GH, Zhang JY, Manzoor MA, Wang H, Jin Q, Jiang TS, Cai YP, et al. In silico genome-wide analysis of the pear (Pyrus bretschneideri) KNOX family and the functional characterization of PbKNOX1, an arabidopsis BREVIPEDICELLUS orthologue gene, involved in cell wall and lignin biosynthesis. Front Genet. 2019;10(17):632. doi: 10.3389/fgene.2019.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17(8):678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 62.Yi K, Zhao YH, Hu Y, Liu JX, He TT, Li X, Song P, Cui GW, Yin XJ. Effect of GA3 and 6 -BA on rhizome segment growth and endogenous hormone content of caucasian clover. Acta Pratacul Sin. 2020;29(2):22–30. [Google Scholar]
- 63.Sun T. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and Development. Plant Physiol. 2010;154(2):567–570. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garrido-Bigotes A, Valenzuela-Riffo F, Figueroa CR. Evolutionary analysis of JAZ proteins in plants: an approach in search of the ancestral sequence. Int J Mol Sci. 2019;20(20):5060. doi: 10.3390/ijms20205060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pernisova M, Grochova M, Konecny T, Plackova L, Harustiakova D, Kakimoto T, Heisler MG, Novak O, Hejatko J. Cytokinin signalling regulates organ identity via the AHK4 receptor in Arabidopsis. Development. 2018;145(14):11. doi: 10.1242/dev.163907. [DOI] [PubMed] [Google Scholar]
- 66.Nie SM, Huang SH, Wang SF, Mao YJ, Liu JW, Ma RL, Wang XF. Enhanced brassinosteroid signaling intensity via SlBRI1 overexpression negatively regulates drought resistance in a manner opposite of that via exogenous BR application in tomato. Plant Physiol Biochem. 2019;138:36–47. doi: 10.1016/j.plaphy.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 67.Gordon SP, Tseng E, Salamov A, Zhang J, Meng X, Zhao Z, Kang D, Underwood J, Grigoriev IV, Figueroa M. Widespread polycistronic transcripts in Fungi revealed by single-molecule mRNA sequencing. PLoS One. 2015;10(7):e0132628. doi: 10.1371/journal.pone.0132628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang YQ, Pu YN, Yin X, Du JC, Zhou ZL, Yang DN, Sun XD, Sun Y, Yang YP. A splice variant of BrrWSD1 in turnip (Brassica rapa var. rapa) and its possible role in wax ester synthesis under drought stress. J Agric Food Chem. 2019;67(40):11077–11088. doi: 10.1021/acs.jafc.9b04069. [DOI] [PubMed] [Google Scholar]
- 69.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 70.Zhang N, Zhang HJ, Zhao B, Sun QQ, Cao YY, Li R, Wu XX, Weesda S, Li L, Ren SX, et al. The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J Pineal Res. 2014;56:39–50. doi: 10.1111/jpi.12095. [DOI] [PubMed] [Google Scholar]
- 71.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):106–118. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu XX, Mei WB, Soltis PS, Soltis DE, Barbazuk WB. Detecting alternatively spliced transcript isoforms from single-molecule long-read sequences without a reference genome. Mol Ecol Resour. 2017;17(6):1243–1256. doi: 10.1111/1755-0998.12670. [DOI] [PubMed] [Google Scholar]
- 74.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L, Park HJ, Dasari S, Wang SQ, Kocher JP, Li W. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41(6):7. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lamb J, Jarmolinska AI, Michel M, Menendez-Hurtado D, Sulkowska JI, Elofsson A. PconsFam: an interactive database of structure predictions of pfam families. J Mol Biol. 2019;431(13):2442–2448. doi: 10.1016/j.jmb.2019.01.047. [DOI] [PubMed] [Google Scholar]
- 77.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5(2):28. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Renaux A, UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46(5):2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2-△△Ct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The output of ROI. a ROI read length distribution for 1-6 KB size bin. b ROI classification for 1-6 KB.
Additional file 2: Table S1. The prediction of SSRs.
Additional file 3: Figure S2. Venn diagram of the number NR, Swiss-prot, COG, GO and KEGG.
Additional file 4: Figure S3. The number of transcripts. a The number statistics of transcripts in five tissues. b Venn diagram of expressed transcripts among different tissues.
Additional file 5: Table S2. Statistics of comparison results between the Illumina sequencing data and the non-redundant transcripts
Additional file 6: Table S3. The prediction of caucasian clover rhizome transcription factor (TF) family.
Additional file 7: Table S4. The results of different physiological indicators. The bar chart of sugar, starch, lignin, ZT, GA3, IAA and ABA content.
Additional file 8: Figure S4. The heatmap of TFs in green module.
Additional file 9: Table S5. The results of qRT-PCR.
Additional file 10: Figure S5. Heatmap of the Pearson correlation coefficient of each sample.
Additional file 11: Table S6. Primers used for qRT-PCR.
Data Availability Statement
Raw reads of one combined PacBio library and one Illumina RNAseq library generated in this study are available from BioProject at NCBI (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA586585 and PRJNA588309, respectively.