Summary
Standardized gene naming is crucial for effective communication about genes, and as genomics becomes increasingly important in healthcare, the need for a consistent language for human genes becomes ever more vital. Here we present the current HUGO Gene Nomenclature Committee (HGNC) guidelines for naming not only protein-coding but also RNA genes and pseudogenes, and outline the changes in approach and ethos that have resulted from the discoveries of the last few decades.
Introduction
The first guidelines for human gene nomenclature were published in 19791, when the Human Gene Nomenclature Committee was originally established and charged with the authority to approve and implement standardized human gene symbols and names. In 1989 the Nomenclature Committee was placed under the auspices of the newly founded Human Genome Organisation (HUGO), becoming the HUGO Gene Nomenclature Committee (HGNC). Subsequent revisions to the nomenclature guidelines were published in 19872,19953, 19974, and 20025. In the intervening years the HGNC has published online updates to the guidelines to reflect the significant changes and increase in knowledge and data during this exciting period in human genomics. Over 40,000 human loci have been named by the HGNC to date; around half are protein-coding genes, and most resources now agree that there are around 19,000-20,000 protein-coding genes in the human genome, considerably lower than some earlier estimates. As well as naming protein-coding genes, significant progress has been made in different classes of RNA genes and pseudogenes. All approved human gene symbols can be found in the online HGNC database (https://www.genenames.org/)6.
The philosophy of the HGNC used to be that “gene nomenclature should evolve with new technology” and that symbol changes, if supported by most researchers working on a gene, were considered if they reflected new functional information. Since the advent of clinical genomics such changes have much wider impacts, and it is impossible to reach all clinicians, patients, charities and other parties interested in genes. Therefore, the stability of gene symbols, particularly those associated with disease, is now a key priority for the HGNC. Nevertheless, novel information can still be encapsulated in the gene name without changing the gene symbol.
As human gene symbols are also routinely transferred to homologous vertebrate genes, including in our sister project the Vertebrate Gene Nomenclature Committee (VGNC), we now avoid references to human-specific traits in nomenclature wherever possible.
We strongly advise researchers to contact us whenever they are considering naming a novel gene, or renaming an existing gene or group of genes, of all locus types, not only for protein-coding genes. It is not always possible to approve the symbol requested, but we strive to work with researchers to find an acceptable alternative. Requesting an approved symbol ensures that your published symbol is present in our and other biomedical databases. We further encourage journal editors and reviewers to check that approved nomenclature is being used and require that authors contact the HGNC prior to publication for any novel symbols. Submitters should bear in mind that the HGNC is committed to minimal future changes to gene symbols and that we do not take publication precedence into account when approving nomenclature.
Readers should note that the following are guidelines and recommendations (Box 1), not strict rules. We are aware of numerous exceptional legacy symbols and names that remain approved. The HGNC considers the naming of each and every gene on a case-by-case basis, and deviations from these guidelines may be made given sufficient evidence that the nomenclature will ultimately aid communication and data retrieval.
Box 1:
A summary of the guidelines is:
Each gene is assigned a unique symbol, HGNC ID and descriptive name.
Symbols contain only uppercase Latin letters and Arabic numerals.
Symbols should not be the same as commonly used abbreviations
Nomenclature should not contain reference to any species or “G” for gene.
Nomenclature should not be offensive or pejorative.
Gene naming
For many years the HGNC has maintained the definition of a gene as “a DNA segment that contributes to phenotype/function. In the absence of demonstrated function a gene may be characterized by sequence, transcription or homology”. As there is still no universally agreed alternative we continue to use this definition.
Ideally gene symbols are short, memorable and pronounceable, and most gene names are long form descriptions of the symbol. Names should be brief, specific and convey something about the character or function of the gene product(s), but not attempt to describe everything known. Each gene is assigned only one symbol; the HGNC does not routinely name isoforms (i.e. alternate transcripts or splice variants). This means no separate symbols for protein-coding or non-coding RNA isoforms of a protein-coding locus or alternative transcripts from a non-coding RNA locus (Box 2).
Box 2:
HGNC does not provide official nomenclature for the following:
sequence variant nomenclature, which is the responsibility of the Human Genome Variation Society (HGVS)7. They provide recommendations for defining variations found in DNA, RNA and protein sequences, and endorse the use of HGNC gene symbols within their notation.
products of gene translocations or fusions: we are not aware of official naming guidelines for these. SYMBOL1-SYMBOL2 is widely used, but we use this format for readthrough transcripts (see section 2.4) and hence would specifically not recommend this for translocations or fusions. We recommend the format SYMBOL1/SYMBOL2, which has been used in some publications, e.g. BCR/ABL1.
protein nomenclature: we have no authority over naming proteins, but coordinate closely with specialist groups who name specific subsets of proteins, such as the Enzyme Commission. The recently devised International Protein Nomenclature Guidelines (https://www.ncbi.nlm.nih.gov/genome/doc/internatprot_nomenguide/) were written with the involvement of the HGNC, and in agreement with these guidelines we recommend that “protein and gene symbols should use the same abbreviation”. We further advise that proteins are referenced using non-italicised gene symbols, to distinguish them from genes.
nomenclature for regulatory genomic elements such as promoters, enhancers and transcription factor binding sites. We also do not provide nomenclature for transposable element insertions in the human genome. Protein-coding and long non-coding RNA genes that fit the criteria outlined in Mayer et al.8 may be named as ERV-derived genes, but ERV insertions will not be named.
nomenclature for human loci associated with clinical phenotypes and complex traits. While HGNC historically named these, this activity has been taken over by OMIM9. All HGNC entries with the locus type “phenotype only” now have the status “entry withdrawn”. Note that some uncharacterized genes shown to be causative for a specific phenotype adopted the phenotype symbol and name. Where these phenotypic symbols have become entrenched in the literature, we aim to update the corresponding gene names to reflect an aspect of the normal function of the gene and its products, e.g. TSC1, “tuberous sclerosis 1” is now TSC1, “TSC complex subunit 1”.
Where authors wish to use their own isoform notation, we advise stating clearly that this notation denotes an isoform of a particular gene and quoting the HGNC symbol for that gene.
In exceptional circumstances, and following community demand, separate symbols have been approved for gene segments in complex loci, i.e. the UGT1 locus, the clustered protocadherins at 5q31 and the immunoglobulin and T cell receptor families. Putative bicistronic loci may be assigned separate symbols to represent the distinct gene products. For example, PYURF, “PIGY upstream reading frame” is encoded by the same transcript as PIGY, “phosphatidylinositol glycan anchor biosynthesis class Y”.
Table 1 summarizes key factors considered when assigning gene nomenclature. Additionally, Supplementary table 1 lists characters recommended for specific usage in symbols, Supplementary table 2 highlights specific conventions used in gene names, and Supplementary tables 3 and 4 provide Greek-to-Latin alphabet conversions and single letter amino acid symbols, respectively.
Table 1:
| Symbols | Names |
|---|---|
| Must be unique within a given genome | Should be brief and specific |
| Must not be offensive or pejorative (ideally in any language) | |
| Must not use superscripts or subscripts or punctuation* | Should minimize punctuation; commas, hyphens and parentheses are included for clarity** |
| Must only contain uppercase*** Latin letters and Arabic numerals | Must be written in American English |
| Must start with a letter | Must start with a lowercase letter (unless starting with an eponymous term or capitalised abbreviation) |
| Should not include “G” for gene, “H” for human, Roman numerals or Greek letters | Should not include the words “gene” or “human” |
| Should not spell proper names or common words or match commonly used abbreviations | Should start with the same letter as the symbol (to facilitate alphabetical listing and grouping) |
| Should avoid duplicating symbols in other species (unless orthologous) | Should not reference: any species, taxa, tissue specificity, molecular weight, chromosomal location, human-specific features and phenotypes, familial terms |
| Some letters or combinations of letters are used in a symbol to give a specific meaning, and their use for other meanings should be avoided where possible (see supplementary table 1). | Descriptive modifiers usually follow the main part of the name, to enable the use of a common root symbol for a gene group, e.g. ACADM “acyl-CoA dehydrogenase medium chain” and ACADS “acyl-CoA dehydrogenase short chain”. |
see Supplementary Table 2 for punctuation exceptions in symbols
exceptions on punctuation are made for enzyme names
sole exception of C#orfs
Gene naming by biotype
Protein-coding genes
We aim to name protein-coding genes based on a key normal function of the gene product. Many protein-coding genes of known function are named in collaboration with internationally recognized bodies composed of experts in a specific field. Where possible, related genes are named using a common root symbol to enable grouping, typically based on sequence homology, shared function or membership of protein complexes.
Gene group members should be designated by Arabic numerals placed immediately after the root symbol, e.g. KLF1, KLF2, KLF3. More rarely single-letter suffixes may be used, e.g. LDHA, LDHB, LDHC. Some large gene families may include a variety of number/letter combinations to indicate subgroupings, e.g. CYP1A1, CYP21A2, CYP51A1 (cytochrome P450 superfamily members).
For genes involved in specific immune processes, or encoding an enzyme, receptor or ion channel, we consult with specialist nomenclature groups (see Supplementary Note). For other major gene groups we consult a panel of advisors when naming new members and discussing proposed nomenclature updates. A list of our specialist advisors is provided on our website6 and we welcome suggestions of new experts for specific gene groups.
In the absence of functional data, protein-coding genes may be named in the following ways: (1) Based on recognized structural domains and motifs encoded by the gene (e.g. ABHD1 “abhydrolase domain containing 1”, HEATR1 “HEAT repeat containing 1”). As these features can provide insight into the character of the gene product, this type of symbol is commonly retained even after the normal function of the gene product has been elucidated, though further information may be added to the gene name; (2) Based on homologous genes within the human genome. Where naming is based on characterized homologs, genes of unknown function are given the next symbol within a designated series but with a different gene name format, e.g. CASTOR3, “CASTOR family member 3” rather than “cytosolic arginine sensor for mTORC1 subunit 3”. The placeholder root symbol FAM (“family with sequence similarity”) is used when there is no information available for any of the homologous genes. Each homologous family has a unique FAM number, e.g. FAM3, and each family member is distinguished by a letter or letter and number, e.g. FAM3A, FAM3C2P. Note that this root can be applied to both protein-coding and non-coding gene families; (3) Based on homologous genes from another species. Where there is a 1-to-1 ortholog, the same/equivalent symbol will be approved, e.g. human CDC45 “cell division cycle 45” based on S. cerevisiae CDC45. A unique number or letter suffix is added if there is more than one human homolog, e.g. UNC45A and UNC45B are co-orthologs of C. elegans unc-45. Gene names are updated to be appropriate for vertebrates, e.g. “unc-45 myosin chaperone” instead of “UNCoordinated 45”; (4) Based only on the presence of an open reading frame. Genes of unknown function that fit none of the above criteria are designated by the chromosome of origin, the letters “orf” for open reading frame (in lower case to prevent confusion between “O” and the numeral “0”, which may be part of the chromosome number) and a number in a series, e.g. C3orf18, “chromosome 3 open reading frame 18”. In cases where the coding potential of the locus is in doubt we include the word “putative” in the name, e.g. “chromosome 18 putative open reading frame 15”.
Historically, genes of unknown function identified by the Human cDNA project at the Kazusa DNA Research Institute10 have been named using the KIAA# identifiers assigned by this project.
Pseudogenes
We define a pseudogene as a sequence that is incapable of producing a functional protein product but has a high level of homology to a functional gene. In general, we only name pseudogenes that retain homology to a significant proportion of the functional ancestral gene.
The majority of pseudogenes are processed and named based on a specific parent gene, e.g. DPP3P1, “DPP3 pseudogene 1”. Such pseudogene numbering is usually species-specific and hence orthology cannot be inferred from identical pseudogene symbols in different species.
Pseudogenes that retain most of the coding sequence compared to other family members (and are usually unprocessed) are named as a new family member with a “P” suffix, e.g. CBWD4P, “COBW domain containing 4, pseudogene”. This naming format is also used for genes that are pseudogenized relative to their functional ortholog in another species, e.g. ADAM24P, “ADAM metallopeptidase domain 24, pseudogene” is the pseudogenized ortholog of mouse Adam24. Note, rarely such pseudogenes do not include the “P” if the symbol is well established, e.g. UOX, “urate oxidase (pseudogene)”.
A small number of genes are currently pseudogenized in the reference genome, but known to have coding alleles segregating in the population. Such loci are given the locus type “protein-coding” and indicated by including “(gene/pseudogene)” at the end of the gene name, e.g. CASP12, “caspase 12 (gene/pseudogene)”.
Non-coding RNA genes
We name non-coding RNA (ncRNA) genes according to their RNA type, please see our recent review11. For small RNAs where an expert resource exists, we follow their naming schema, e.g. miRBase12 for microRNAs and the genomic tRNA database (GtRNAdb)13 for tRNAs. Other classes of ncRNA such as small nuclear RNAs are named in collaboration with specialist advisors.
For long non-coding RNAs (lncRNAs), wherever possible we name these based on a key function or characteristic of the encoded RNA. Where functional information is not available, a systematic nomenclature is applied, see Figure 1.
Figure 1:
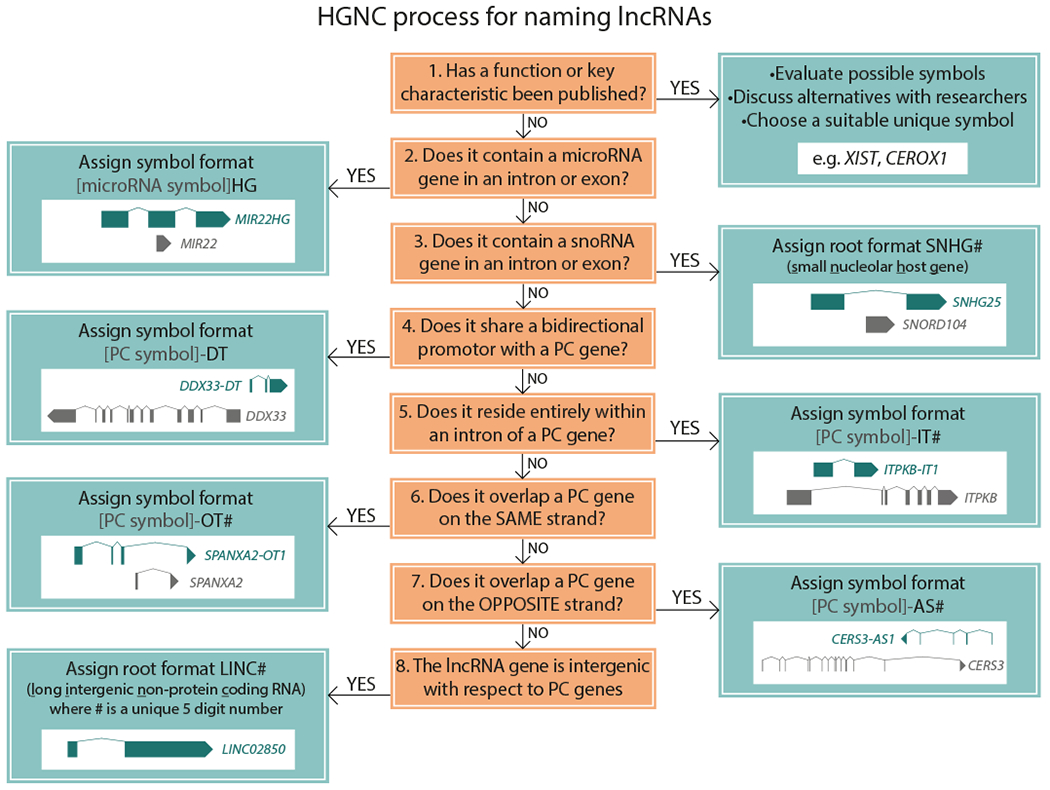
The HGNC has a systematic process for naming long non-coding (lnc)RNA genes. In the absence of suitable published information, lncRNA genes are named based on genomic context. Abbreviations are as follows: HG: host gene, PC: protein-coding, DT: divergent transcript (used for lncRNA genes that share a promoter with a PC gene), IT: intronic transcript, OT: overlapping transcript, AS: antisense RNA, LINC: long intergenic non-protein coding RNA.
Readthrough transcripts
Readthrough transcripts are normally produced from adjacent loci and include coding and/or non-coding parts of two (or more) genes. The HGNC only names readthrough transcripts that are consistently annotated by both the RefSeq annotators at NCBI14 and the GENCODE annotators at Ensembl15. These transcripts have the locus type “readthrough transcript” and are symbolized using the two (or more) symbols from the parent genes, separated by a hyphen, e.g. INS-IGF2, and the name “[symbol] readthrough”, e.g. “INS-IGF2 readthrough”. The name may also include additional information about the potential coding status of the transcript, such as “(NMD candidate)”.
Gene segments
For specific complex loci the HGNC assigns symbols to individual gene segments, solely based on community request. Examples of this are the immunoglobulins and T-cell receptors, the UGT1 locus and clustered protocadherins.
Genomic regions
The HGNC previously named genomic regions referenced in the literature, such as XIC, “X chromosome inactivation center”, and gene clusters were assigned symbols suffixed with the “@” character, e.g. HOXA@, “homeobox A cluster”. We no longer routinely provide symbols for genomic regions but some, such as those for fragile sites, have been retained where they have been used in publications and this information would otherwise be lost.
Genes only found within subsets of the population
Historically, the HGNC has only approved symbols for genes that are on the human reference genome. Rare exceptions have been made when requested by particular communities, e.g. structural variants within the HLA and KIR gene families, both of which have dedicated nomenclature committees. Future naming of structural variants will be restricted to those on alternate loci that have been incorporated into the human reference genome by the Genome Reference Consortium (GRC, https://www.ncbi.nlm.nih.gov/grc). The underscore character is reserved for genes annotated on alternate reference loci, e.g. GTF2H2C_2 is a second copy of GTF2H2C on a 5q13.2 alternate reference locus; APOBEC3A_B is a deletion hybrid on a 22q13 alternate reference locus that includes exons from both the APOBEC3A and APOBEC3B parent genes.
Status
All HGNC gene records have a status: the vast majority are “approved”, but when new evidence shows that a previously named gene is no longer considered to be real the entry changes to the status “entry withdrawn”. Wherever possible we avoid reusing symbols from “entry withdrawn” records, as this can cause considerable confusion.
Naming across vertebrates
We recommend that orthologous genes across vertebrate (and where appropriate, non-vertebrate) species should have the same gene symbol.
The Vertebrate Gene Nomenclature Committee
The Vertebrate Gene Nomenclature Committee (VGNC, https://vertebrate.genenames.org/) is an extension of the HGNC responsible for assigning standardized names to genes in vertebrate species that currently lack a nomenclature committee. The VGNC coordinates with the five established existing vertebrate nomenclature committees, MGNC (mouse)16, RGNC (rat, https://rgd.mcw.edu/nomen/nomen.shtml), CGNC (chicken)17, XNC (Xenopus frog)18 and ZNC (zebrafish)19, to ensure vertebrate genes are named in line with their human homologs.
Orthologs of human C#orf# genes are assigned the human symbol with the other species chromosome number as a prefix and an H denoting human. Therefore, as the ortholog of human C1orf100 is on cow chromosome 16, the cow symbol is C16H1orf100 with the corresponding gene name “chromosome 16 C1orf100 homolog”.
Gene families with a complex evolutionary history should ideally be named with the help of an expert in the field, as has already been implemented for the olfactory receptor20 and cytochrome P450 gene families.
Species designation
To distinguish the species of origin for homologous genes with the same gene symbol, we recommend citing the NCBI taxonomy ID21, as well as either the current name or the
GenBank common name, e.g. Taxonomy ID: 9598 and either Pan troglodytes or chimpanzee.
Nomenclature updates
While we are committed to minimizing symbol changes some updates will still be appropriate. All requests for change are considered on a case-by-case basis and often involve community consultation. We anticipate most future changes will fall into one of the following categories.
Symbol updates for placeholders
FAMs, C#orfs and KIAAs are regarded as placeholder symbols and updated with structure and/or function-based designations whenever possible. However, where specific placeholder symbols have become entrenched in the literature, we may make exceptions and retain the placeholder, while updating the gene name, e.g. FAM20B has been retained with the updated gene name FAM20B glycosaminoglycan xylosylkinase.
Replacing underused and problematic nomenclature
We may consider updating symbols that have been rarely/never published, are not suitable for transfer to other vertebrates, and/or have been widely used but could cause significant problems. Examples are shown in Box 3.
Box 3:
Scenarios that may merit a symbol change include:
adoption of a more appropriate/popular alias, e.g. RNASEN was updated to DROSHA (drosha ribonuclease III) due to overwhelming community usage.
domain or motif-based nomenclature, e.g. TMEM206 (transmembrane protein 206) is now PACC1 (proton activated chloride channel 1).
phenotype/disease-based nomenclature, e.g. CASC4 (cancer susceptibility candidate 4) was renamed GOLM2 (golgi membrane protein 2), consistent with its paralog GOLM1.
location-based nomenclature, e.g. TWISTNB (TWIST neighbour) is now POLR1F (RNA polymerase I subunit F).
pejorative symbols, e.g. DOPEY1 was renamed to DOP1A (DOP1 leucine zipper like protein A).
misleading/incorrect nomenclature, e.g. OTX3 was initially named erroneously as an OTX family member and has been renamed as DMBX1 (diencephalon/mesencephalon homeobox 1).
symbols that affect data handling and retrieval, e.g. all symbols that auto-converted to dates in Microsoft Excel have been changed (SEPT1 is now SEPTIN1; MARCH1 is now MARCHF1 etc); tRNA synthetase symbols that were also common words have been changed (WARS is now WARS1, CARS is now CARS1, etc.).
Gene symbol usage
The HGNC endorses the use of italics to denote genes, alleles and RNAs to distinguish them from proteins.
We advise that authors quote the approved gene symbol at least once in the abstract of any publication. Every gene with an approved symbol also has a unique HGNC ID in the format HGNC:number (e.g. gene symbol BRAF, HGNC ID HGNC:1097). While we aim to minimize symbol changes some updates are inevitable and sometimes an approved symbol can be used to denote a different gene in the literature; therefore we advise quoting the HGNC ID for each gene to avoid ambiguity. HGNC IDs are associated with the gene sequence and do not change unless the gene structure undergoes extreme alteration (i.e. merged with another locus or split into multiple loci). This ensures effective and reliable tracking of data regardless of any nomenclature changes.
Supplementary Material
Acknowledgements
Many thanks to all current and former members of the HGNC team, in particular the late Professor Sue Povey who was HGNC’s PI from 1996-2007, our specialist advisors and advisory board members past and present. The HGNC relies heavily on the expertise and feedback of researchers and are grateful for all input we receive. The HGNC is currently funded by the National Human Genome Research Institute (NHGRI) grant U24HG003345 (to EAB) and Wellcome Trust grant 208349/Z/17/Z (to EAB).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Shows TB et al. International system for human gene nomenclature (ISGN, 1979). Cytogenet Cell Genet. 25, 96–116 (1979). [DOI] [PubMed] [Google Scholar]
- 2.Shows TB et al. Guidelines for human gene nomenclature. An international system for human gene nomenclature (ISGN, 1987). Cytogenet Cell Genet. 46, 11–28 (1987). [DOI] [PubMed] [Google Scholar]
- 3.McAlpine P Genetic nomenclature guide. Human. Trends Genet. Mar, 39–42 (1995). [PubMed] [Google Scholar]
- 4.White JA et al. Guidelines for human gene nomenclature. Genomics 45, 468–471 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Wain HM, Bruford EA, Lovering RC, Lush MJ, Wright MW, Povey S Guidelines for Human Gene Nomenclature. Genomics 79, 464–470 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Braschi B et al. Genenames.org: the HGNC and VGNC resources in 2019. Nucleic Acids Res. 47, D786–D792 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Dunnen JT Describing Sequence Variants Using HGVS Nomenclature. Methods Mol Biol. 1492, 243–251 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Mayer J, Blomberg J, Seal RL A revised nomenclature for transcribed human endogenous retroviral loci. Mob DNA 2, 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amberger JS, Bocchini CA, Scott AF, Hamosh A OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 47, D1038–D1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagase T, Koga H, Ohara O Kazusa mammalian cDNA resources: towards functional characterization of KIAA gene products. Brief Funct Genomic Proteomic. 5, 4–7 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Seal RS et al. A guide to naming human non-coding RNA genes. EMBO J. 39(6), e103777 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozomara A, Birgaoanu M, Griffiths-Jones S miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan PP, Lowe TM GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Leary NA et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankish A et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maltais LJ, Blake JA, Eppig JT, Davisson MT Rules and guidelines for mouse gene nomenclature: a condensed version. Genomics 45, 471–476 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Burt DW et al. The Chicken Gene Nomenclature Committee Report. BMC Genomics 10, S5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James-Zorn C et al. Xenbase: Core features, data acquisition, and data processing. Genesis 53, 486–497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruzicka L et al. The Zebrafish Information Network: new support for non-coding genes, richer Gene Ontology annotations and the Alliance of Genome Resources. Nucleic Acids Res. 47, D867–D873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olender T et al. A unified nomenclature for vertebrate olfactory receptors. BMC Evol. Biol. 20, 42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Federhen S The NCBI Taxonomy Database. Nucleic Acids Res. 40, D136–D143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


