Abstract
Background
Streptomycete bacteria are prolific producers of specialized metabolites, many of which have clinically relevant bioactivity. A striking feature of their genomes is the expansion of gene families that encode the same enzymatic function. Genes that undergo expansion events, either by horizontal gene transfer or duplication, can have a range of fates: genes can be lost, or they can undergo neo-functionalization or sub-functionalization. To test whether expanded gene families in Streptomyces exhibit differential expression, an RNA-Seq approach was used to examine cultures of wild-type Streptomyces coelicolor grown with either glucose or tween as the sole carbon source.
Results
RNA-Seq analysis showed that two-thirds of genes within expanded gene families show transcriptional differences when strains were grown on tween compared to glucose. In addition, expression of specialized metabolite gene clusters (actinorhodin, isorenieratane, coelichelin and a cryptic NRPS) was also influenced by carbon source.
Conclusions
Expression of genes encoding the same enzymatic function had transcriptional differences when grown on different carbon sources. This transcriptional divergence enables partitioning to function under different physiological conditions. These approaches can inform metabolic engineering of industrial Streptomyces strains and may help develop cultivation conditions to activate the so-called silent biosynthetic gene clusters.
Keywords: Streptomyces, RNA-Seq, central carbon metabolism, metabolic plasticity, gene redundancy, silent biosynthetic clusters, metabolic engineering
Introduction
Streptomycete bacteria are major source of clinically useful bioactive natural products including antibiotics, immunosuppressive and anti-cancer agents. A remarkable feature of their genomes is that there are often several genes that appear to encode the same biochemical function [1, 2]. This is often referred to as ‘genetic redundancy’, where two or more genes are performing the same biochemical function [3]. While genetic redundancy does occur in nature, many so called ‘redundant’ genes have evolved divergent functions and provide the functional diversity and evolutionary robustness that can be observed in genomes from a wide range of organisms [4]. There are two main mechanisms that contribute to the expansion of gene families within genomes – the duplication of genes or horizontal gene-transfer events [4–6]. Such gene-family expansions are well known in streptomycete regulatory genes [7–10] and in specialized metabolite biosynthesis [11, 12]. Yet, there has been limited attention focussed on the genes of central metabolism despite a number of gene-expansion events being identified from biochemical and phylogenomic studies. There is an increasing appreciation that gene-expansion events in central metabolism may facilitate the evolution of specialized metabolites [2, 13–17]. Whilst expansion of gene families is widespread, expression differences, gene dosage, cofactor variation, allostery or substrate affinities are seldom taken into account during the construction of metabolic models for Streptomyces . As a result, homologous functions often combined into a single flux pathway that may not reflect the physiological nature of each gene product. This is especially stark when attempting to understand the supply of precursor molecules from central metabolism to the production of specialised metabolites.
It was hypothesized that gene families that have undergone gene-expansion events would exhibit different expression profiles if they have diverged functionally. To understand the role of these gene expansions in Streptomyces and their impact on central metabolism and specialised metabolism, an RNA-Seq approach was taken using S. coelicolor grown either on glucose or tween as sole carbon source. Cultures were compared at a single point during growth (mid-log phase) to understand how transcription of central metabolic genes varies when the cultures are growing primarily via glycolysis (glucose as the sole carbon source) or gluconeogenically (with the mono-oleate tween). It was found that expanded gene families exhibit different responses to growth on different carbon sources. These data will enable prioritization of targets for metabolic engineering and will be informative for the construction of metabolic models.
Results and Discussion
Expanded gene families in carbon metabolism exhibit different transcriptional profiles
To investigate the differences between glycolytic and gluconeogenic growth conditions, S. coelicolor M145 [an A3(2) strain] was grown respectively on glucose and Tween40 as sole carbon source. Growth of liquid cultures was monitored by cell dry weight and samples were removed for total RNA extraction at mid-exponential phase, which for glucose was 19 h and for tween was 36 h (Fig. S1, available in the online version of this article). None of the pigmented antibiotics (actinorhodin or undecylprodigiosin) could be detected at time points RNA was harvested. The specific growth rate of S. coelicolor M145 was 0.18 h−1 when grown on glucose and 0.1 h−1 for growth on tween (see Fig. S1).
RNA samples from these cultures showed global effects on transcription, when analysed by RNA-Seq, when the strain was grown on different carbon sources. Growth on tween resulted in 644 genes being differentially expressed when compared to growth on glucose. Of the 644 differentially expressed genes, 37 % were predicted to encode hypothetical proteins, 9 % encoded regulatory genes, 12 % encoded central carbon metabolic enzymes, 6 % were transporters, 4 % were associated with specialized metabolism, 2 % encoded genes associated with stress metabolism and 1 % of genes were associated with DNA replication, nitrogen metabolism and genes associated with metal metabolism (see Tables S1–6). Growth on Tween40 resulted in an upregulation of genes associated with gluconeogenesis and fatty acid degradation when compared to cultures grown on glucose (Fig. 1c, Tables S1–S6).
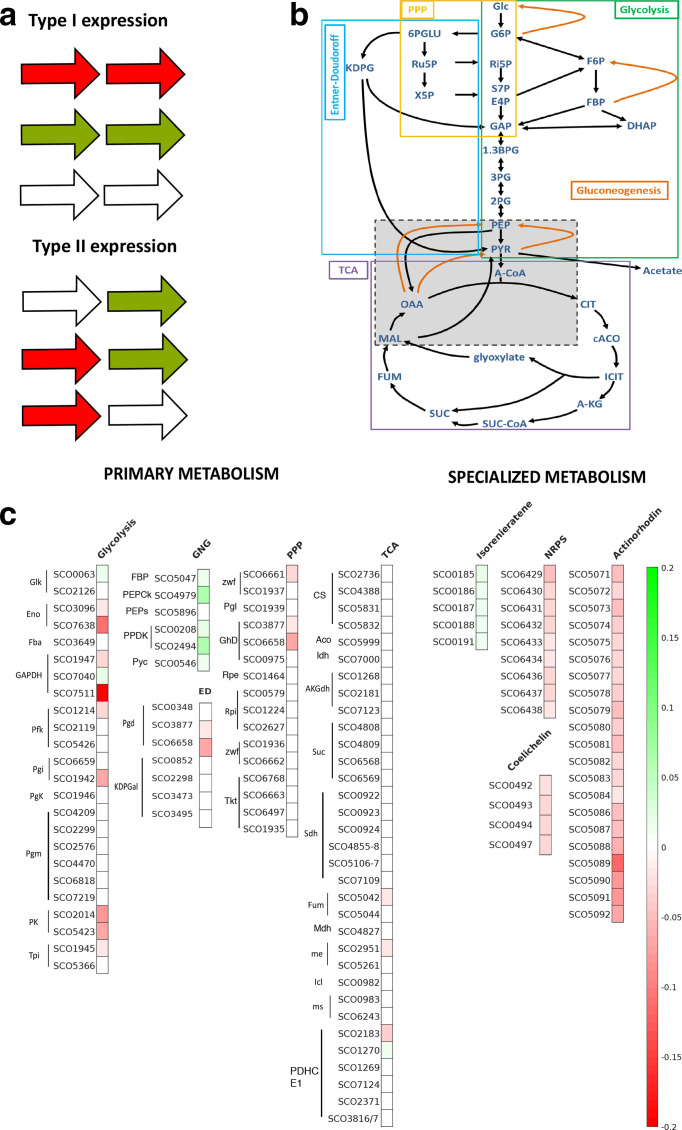
Fig. 1.
Carbon source-dependent expression of genes in S. coelicolor. (a) Schematic overview of expression patterns observed for expanded gene families type I expression, where genes behaved in the same manner under the different growth conditions and type II, where members of an expanded gene family exhibited differential gene expression across multiple gene-family members (green=upregulated; red=downregulated and white=no expression change). (b) Schematic overview with main metabolites of the central carbon metabolism grouped according to pathways: glycolysis (green), pentose phosphate pathway (PPP, yellow), Entner–Doudoroff pathway (not complete in S. coelicolor; blue), tricarboxylic acid cycle (TCA, purple), gluconeogenesis (orange arrows). (c) Visualization of gene expression using Heatmap by pathway (gene expression was normalized to maximum and minimum values of the entire dataset, and heat map colouring was based on maximum and minimum values of genes represented, where green is upregulated and red is downregulated). Legend for abbreviations: Catalytic functions: Glk=glucose kinase, Zwf=Glucose-6-phosphate 1-dehydrogenase, Pgl=6-phosphogluconolactonase, GhD=6 -phosphogluconate dehydrogenase, Rpi=Ribose -5-phosphate isomerase, Rpe=Ribulose 5-Phosphate 3-Epimerase, Tal=transaldolase, Tkt=transketolase, Pgd=6-phosphogluconate dehydratase, KDPGal=KDPG aldolase, Eno=enolase, Fba=fructose-1,6-bisphosphate aldolase, Gap=glyceraldehyde-3-phosphate dehydrogenase, Pfk=phosphofructokinase, Pgi=Phosphoglucose isomerase, Pgk=Phosphoglycerate kinase, Pgm=Phosphoglycerate mutase, Pyk=pyruvate kinase, Tpi=triosephosphate isomerase, FBPase=FBP bisphosphatase, PEPCk=PEP carboxykinase, PPS=PEP synthase, PPDK=pyruvate phosphate dikinase, Pyc=Pyruvate carboxylase, CS=citrate synthase, Aco=aconitase, Idh=isocitrate dehydrogenase, AKGdh=Alpha-ketoglutarate dehydrogenase, Suc=succinyl CoA synthetase, Sdh=succinate dehydrogenase, Fum=Fumarase, Mdh=malate dehydrogenase, me=malic enzyme, Icl=isocitrate lyase, Ms=malate synthase, PDHC=pyruvate dehydrogenase complex. Metabolites: Glc=Glucose, G6P=Glucose-6-Phosphate, 6-PGLU=6-phosphogluconate, Ru5P=Ribulose-5-phosphate, X5P=Xylose-5-Phosphate, KDPG=2-keto-3-deoxy-6-phosphogluconate, F6P=Fructose-6-Phosphate, FBP=Fructose 1.6-bisphosphate, DHAP=dihydroxyacetone phosphate, Ri5P=Ribose-5-Phosphate, S7P=Seduheptulose-7-Phosphate, E4P=Erythrose-4-Phosphate, GAP=glyceraldehyde-3-phosphate, 1.3BGP=1.3-bisphosphoglycerate, 3PG=3-phosphoglycerate, 2PG=2-phosphoglycerate, PEP-=Phosphoenolpyruvate, PYR=Pyruvate, ACoA=Acetyl CoA, Cit=Citrate, cAco=cisAconitate, ICit=Isocitrate, A-KG-= α-Ketoglutarate, SucCoA=Succinyl CoA, Suc=Succinate, Fum=Fumarate, Mal=Malate, OAA=Oxaloacetate.
Gene-ontology (GO) enrichment analysis (using PANTER http://www.pantherdb.org; geneontology.org [18]), was used to investigate the enrichment of specific gene ontologies between the two growth conditions. Analyses were performed for genes upregulated on Tween and one for genes downregulated on Tween. The glycolytic genes are downregulated when S. coelicolor was grown on tween as would be expected, with GO enrichment analysis highlighting tpiA (SCO1945), pyk1 (SCO2014) and pyk2 (SCO5423), one of the three GAPDH (SCO7511) and a single copy of the pyruvate dehydrogenase E1 complex (SCO2183), it was noted that all genes belonged in the oxidoreductase gene-ontology grouping.
GO enrichment analysis for genes upregulated during growth on Tween were found to be those associated with fatty acid catabolism, gluconeogenesis (pyc; SCO0546) and another of the three GAPDH homologues (SCO7040).
To examine transcription of expanded gene families, two categories of expression profiles were considered to reflect changes across the gene family that are predicted to encode the same enzymatic function: type 1, where all genes within an expanded gene family behaved similarly under the growth conditions, and type II where members of an expanded gene family exhibit differential gene expression, i.e. one or more member of the family increased, while expression of other members of the gene family, increased to a different degree, decreased or remained the same (Fig. 1a and Table S1). We identified 34 enzymatic reactions in central metabolism whose expression differed when cultures were grown on tween or glucose. Of these, 21 had more than one enzyme predicted to encode the same enzymatic function, i.e. were expanded gene families (Fig. 1, Table S1). When the expression profiles of these genes were examined further, 9/21 enzymatic reactions had families that showed type I expression profiles and 12/21 expanded gene families exhibited type II patterns of expression. This indicates that despite their identity in genome annotation, members of some expanded gene families are not redundant in function but have distinctive physiological roles [2, 13, 19]. It is unlikely that functional differences can be attributed at the transcriptional level. However, the data enable prioritization of targets for metabolic engineering, such as modulating the expression poorly expressed members of gene families.
The glycolysis pathway, unsurprisingly, showed reduced expression when the cultures were grown on Tween40 as sole carbon source. Only two glycolytic enzyme families exhibited type II expression, with an increase in expression for the minor glucose kinase (SCO0063; fivefold increase in expression), whereas expression of the primary glk (SCO2126) remained unchanged under both conditions. GAPDH also showed type II expression profiles, with expression of SCO7040 increased on tween, whereas the other two copies of the genes encoding GAPDH had reduced expression (SCO1947 and SCO7511). A previous proteomic-based study also identified an increase in abundance of the SCO7040 protein when grown on a non-glucose carbon source (fructose; [19]). The two PK genes, as we have previously demonstrated, exhibited type I expression [2], with both copies being downregulated when cultures are grown on tween.
Overall, genes involved in gluconeogenesis were upregulated when cultures were grown on tween compared to glucose – as would be expected. The only expanded gene family encoding gluconeogenic function is that encoding the two PPDKs, which both displayed substantially increased expression (type I expression), with ppdk1 (SCO0208) upregulated 11-fold up and ppdk2 (SCO2494) upregulated 30-fold (ppdk2). This suggests that, under these conditions, S. coelicolor may be using unconventional gluconeogenic routes for anaplerotic reactions rather than via the glyoxylate shunt, as expression of ICL and both MS enzymes remain unchanged during growth on either carbon source.
Expression of the pentose phosphate pathway showed little differential expression under the two conditions studied. Type II expression profiles were observed for zwf, with expression of SCO6661 reduced when cells were grown on tween, whereas expression of SCO1937 was unchanged between the two conditions. This was consistent with the work of Gubbens et al. [19], where SCO6661 was downregulated when cultures were grown on fructose rather than glucose. There was no evidence of changes in transcriptional activity for the putative Entner–Doudoroff (ED) pathway genes for KDPG aldolase (SCO2298, SCO3473 and SCO3495). However, reduced expression of two of the three phosphogluconate dehydratase homologues (SCO3877 and SCO6658) was observed when cultures were grown on tween (Fig. 1c). Whilst it was reported previously that there is no active ED pathway in S. coelicolor [20] these data suggest that putative phosphogluconate dehydratase homologues do respond to changes in carbon source.
In general, expression of genes encoding enzymes of the tricarboxylic acid (TCA) cycle remained stable under both growth conditions as would be expected, given this core part of metabolism is required for biosynthesis under all physiological conditions. The exception was expanded gene families, which exhibited differential expression (type II): expression of the fumarase (SCO5042) was reduced twofold on tween whilst SCO5044 expression remained unchanged; one copy of the malic enzyme (SCO2951) showed a twofold decrease in expression, compared to SCO5261, a second malic enzyme, which remained unchanged under both conditions; the extensive expansion in S. coelicolor of genes encoding the PHDCE1 subunit exhibited a range of expression changes – expression profiles for SCO1269, SCO7124, SCO2371 and SCO3816/3817 were the same for both carbon sources, whereas SCO2183 had a 3.5-fold decrease and SCO1270 a sixfold increase on tween.
As expected fatty acid utilization genes were expressed more when S. coelicolor was grown on tween, especially cholesterol esterase (SCO5420), which had an increase of almost 37-fold. The enzyme catalyses the hydrolysis of the head group of tween from the fatty acid palmitate, which are then utilized as carbon source [21–23]. The other genes from this pathway showed between 3- to 12-fold increase in expression (Table S2).
To verify the RNA-Seq data, we performed qPCR on the RNA samples used for the RNA-Seq experiment using primer pairs for five different genes – pyk1 (SCO2014) and pyk2 (SCO5423) from glycolysis, ppdk1 and ppdk2 from gluconeogenesis with primary sigma factor hrdB (SCO5820) as control. These two pairs of genes were chosen as representative of expanded families, identified in a previous study [2]. RNA-Seq data showed that expression was strongly up or downregulated under the chosen conditions, whilst hrdB (SCO5820, as expected) showed no difference under the two conditions tested. The fold-change difference in the qPCR data was similar to that observed in RNA-Seq experiments, corroborating the wider results (Figs S2 and S3).
Carbon source influences specialized metabolite gene expression
Genes involved in isorenieratene biosynthesis (SCO0185-0191), showed an increase in transcription in cultures grown on tween, with a three to eightfold increase in expression across the entire operon. Isorenieratene is associated with blue-light exposure, with the pathway present in green photosynthetic bacteria and a few actinobacteria [24]. Isorenieratene is a carotenoid with antioxidative properties; it is synthesized via the mevalonate-independent pathway (MEP/DOXP pathway) from basic precursors of GAP and pyruvate, derived from glycolysis to form Isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAP). These metabolites then enter the carotenoid biosynthetic pathway via phytoene, lycopene and beta-carotene. The MEP/DOXP pathway also had increased expression of some genes (SCO6768 – 2.2-fold, SCO5250 – 4.4-fold) when grown on glucose (Table S2). Given the only difference between the cultures was the carbon source, we hypothesize that cultures experienced oxidative stress. This is in part supported by the GO enrichment analysis where 51 genes with GO terms matching ‘oxidoreductases’ exhibiting increased expression on Tween as the sole carbon source, with a sevenfold increase in expression of a superoxide mutase (SCO0999) and an eightfold increase of expression of a putative bacterioferritin co-migratory protein (SCO7353; Tables S2 and S4). Caution should be exercised however as many genes in primary metabolism also share GOs with ‘oxidoreductases’.
A non-ribosomal peptide synthetase (NRPS) pathway (SCO6429-6438), the siderophore coelichelin biosynthetic gene cluster (SCO0491-0498) and the actinorhodin cluster (SCO5071-5092) all had decreased expression when grown on tween compared to glucose by two to fivefold, two to threefold and three to 11-fold, respectively (Fig. 1, Table S2). This may reflect a tighter control of entry into specialized metabolism when cultures are grown on tween rather than glucose, although further experiments are needed to confirm this.
Studies such as this can inform metabolic modelling approaches for strain improvement enabling expression data and metabolic flux analysis to be taken into account with respect to isoenzymes within expanded gene families rather than combining the activities of all copies of a gene family into a single flux [15]. The identification of metabolic engineering targets can often be complicated through the presence of multiple genes that putatively encode the same enzymatic function in streptomycetes. Studies such as this enables the identification of transcriptionally active genes under relevant conditions to be identified and prioritized for manipulation. This is exemplified by GAPDH in S. coelicolor where three genes are annotated as having this function, with one (SCO7040) increasing expression on tween when the other two copies show decreased expression under the same conditions. This knowledge will help inform choices and enable prioritization of metabolic engineering targets for industrial streptomycetes and the supporting dataset will also inform researchers on the role of hypothetical proteins and regulators, during growth on two different carbon sources that drive metabolism along two different pathways.
Methods
Bacterial strains and growth conditions
S. coelicolor A3(2) M145 [25] was used throughout the study. Spores were germinated in 50 ml of 2× YT medium in flasks containing a metal spring and were incubated for up to 8 h at 30°C and 250 r.p.m. until emerging germ tubes were visible under a microscope [25]. The cultures were harvested by centrifugation, washed twice with 0.25 M TES buffer (pH 7.2) and pellets were resuspended in media. Growth curves were performed at 400 ml scale with minimal medium [26] in 2 l flasks containing a metal spring at 30 °C, shaken at 250 r.p.m. Carbon source added was adjusted to ensure that each culture contained 166.5 mM equivalent of carbon.
RNA extraction and sample preparation
S. coelicolor biomass (15 ml sample) from liquid cultures was harvested by centrifugation (5 min, 4 °C, 4000 g). Cell pellets were resuspended in an equal volume of RNAprotect (Qiagen) for 5 min at room temperature. Following centrifugation (5 min, 4 °C, 6000 g) biomass was then resuspended in 1 ml 1× TE buffer containing 15 mg ml−1 lysozyme. Tubes were vortexed for 10 s and incubated at room temperature for 60 min whilst shaking. Then, 1 ml RLT buffer (Qiagen RNA Isolation Kit)+10 µl β-mercaptoethanol was added and vortexed and a phenol chloroform extraction followed by an ethanol precipitation was carried out. The sample was then purified using a commercial RNA isolation Kit (Qiagen). The isolated RNA was treated with RNAse free DNase (Ambion, Life Technologies) as specified by the manufacturer.
Quantification of RNA was carried out using Qubit (Life Technologies). The quality and integrity of the RNA was assessed using a Bioanalyzer (Agilent). Furthermore, the RNA samples were also used as templates for a generic PCR in order to check for DNA contamination using primers for hrdB (SCO5820).
To enrich the samples for mRNA, rRNA depletion was performed (rRNA depletion Kit Ribo Zero Magnetic Kit for Gram-positive bacteria; Epicentre [Illumina]) according to the manufacturer’s instructions. The rRNA-depleted samples were then precipitated with ethanol and resuspended according to the manufacturer’s instructions. The quality and integrity of the samples were then analysed on the BioAnalizer (Agilent) and the concentration was determined using Qubit (Life Technologies).
Library preparation
cDNA synthesis and library preparation was carried out using the Ion Total RNA-Seq Kit v2 Revision E from Ion Torrent, Life Technologies. The manufacturer’s protocol for less than 100 ng rRNA depleted samples was followed. The yield and size distribution of the amplified cDNA was assessed using BioAnalizer (Agilent). The three samples harvested from glucose cultures and three samples from tween grown cultures were barcoded and pooled to a concentration of 20 pM as specified by the manufacturer. The Ion OneTouch 2 system using the Ion PGM Template OT200 Kit (Life Technologies) was used for template preparation which includes the steps of emulsification, amplification and enrichment of the library. The libraries were checked using the quality control assay for the Qubit (Life Technologies).
RNA sequencing
Sequencing of the samples was carried out using a Ion Torrent Personal Genome Machine System (PGM; Life Technologies) on a 316v2 chip following the procedures in the manual, Ion PGM Sequencing 200 Kit v2 User Guide Revision 3.0. All sequence data were deposited on the Sequence Read Archive (SRA) under the BioProject: PRJNA566372 (https://www.ncbi.nlm.nih.gov/sra/PRJNA566372).
Data analysis
Sequencing data were downloaded from the Ion Torrent server version Torrent Suite 4.0.2 in Fastq format. Reads were trimmed and low-quality data removed. Reads were mapped to the reference genome of S. coelicolor [1]; GenBank: NC_003888.3. The data were analysed using CLC Genomics Workbench (Version 7.5, Qiagen). The software showed on average 99.8 and 99.7% alignment of the reads to the reference sequence. The CLC differential gene-expression tool was used to determine differential gene expression. To examine differential gene-expression analysis within the dataset, the cut-off was set at a P-value of 0.05. Genewise dispersions are estimated by conditional maximum likelihood using the total count for the gene of interest followed by empirical Bayes to obtain a consensus value [27, 28]. The differential expression is then assessed using Fisher’s exact test adjusted to over-dispersed data [29]. The raw data output from CLC Genomics Suite can be found in Table S8, and a list of all differentially expressed genes is shown in Table S5. For the heatmap representation in Fig. 1c, all differentially expressed genes were normalized to the maximum (+1) and minimum (−1) of all significantly different expressed genes and this can be found in Table S6. The code to create Fig. 1c can be found in Code S1 (https://doi.org/10.6084/m9.figshare.10008914.v3). The colour code was expressed relative to the highest and lowest expression change in the data shown ranging from green to red, respectively.
Gene-ontology enrichment analysis was performed using Panther classification from gene ontology at NCBI (https://www.ncbi.nlm.nih.gov/pubmed/23868073) and summarized in Table S7.
qPCR
qPCR was performed on the same samples as the RNA-Seq in order to provide independent confirmation of the data obtained. Each primer pair was tested using genomic DNA as the template. cDNA was synthesized from the RNA samples using qPCRBIO cDNA synthesis Kit (PCR Biosystems) following the manufacturer’s instructions. Quantification of the cDNA was carried out using the QuantiFluor ssDNA system (Promega) with the Qubit Fluorometer (Life Technologies).
All cDNA samples were diluted to a concentration of 10 ng µl−1 and each reaction contained 10 ng of cDNA. Samples were mixed with qPCRBio MasterMix and the respective primers (Table S9; Kit 2× qPCRBIO SyGreen Mix Lo-ROX from PCRBIOSYSTEMS). Using a Corbett Research 6000 (Qiagen) machine, PCR reactions were subjected to a three-stage thermocycling reaction (one 3 min step at 95 °C, followed by 40 cycles of 5 s at 95 °C and one 25 s step at 60 °C. Each reaction was carried out in duplicate and a no template control was included for each set of primers. To allow quantification, standard curves for each gene were prepared (in triplicate) using purified PCR product from a genomic DNA PCR. This template was diluted to create seven different standards ranging from 101 to 107 molecules/per reaction (Fig. S3) and was used to calculate the concentrations of the unknown samples obtained in the RNA sequencing.
Funding information
This work was funded through PhD studentships from the Scottish Universities Life Sciences Alliance (SULSA) to J.K.S. and R.R.
Acknowledgements
The authors would like to thank Jan Schniete for writing the MATLAB code for the heatmap representation in Fig. 1(c). P.A.H. would like to acknowledge the support of NERC (grant: NE/M001415/1) and BBSRC/NPRONET (grant: NPRONET POC045). P.A.H. and P.R.H. acknowledge SULSA funding for PhD studentships to J.K.S. and R.R.
Author contributions
Conceptualization: J.K.S., P.A.H., N.P.T., L.K. Data curation: J.K.S. & R.R. Formal analysis: J.K.S. & R.R. Funding acquisition: P.A.H., P.R.H., I.S.H. Methodology: J.K.S., R.R., P.A.H., P.R.H., N.P.T., L.K. Project administration: P.A.H. & P.R.H. Supervision: P.A.H., I.S.H. & P.R.H. Writing – original draft: J.K.S and P.A.H. Writing – review and editing: J.K.S., R.R., L.K., N.P.T., I.S.H., P.R.H., P.A.H.
Conflicts of interest
The author declare no competing financial or non-financial
interests.
Footnotes
Nine supplementary tables are available with the online version of this article.
References
- 1.Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 2.Schniete JK, Cruz-Morales P, Selem-Mojica N, Fernández-Martínez LT, Hunter IS, et al. Expanding Primary Metabolism Helps Generate the Metabolic Robustness To Facilitate Antibiotic Biosynthesis in Streptomyces . mBio. 2018;9:e02283–17. doi: 10.1128/mBio.02283-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 4.Wagner A. Gene duplications, robustness and evolutionary innovations. Bioessays. 2008b;30:367–373. doi: 10.1002/bies.20728. [DOI] [PubMed] [Google Scholar]
- 5.Wagner A. Robustness and evolvability: a paradox resolved. Proc Biol Sci. 2008a;275:91–100. doi: 10.1098/rspb.2007.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Treangen TJ, Rocha EPC. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7:e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush MJ. The actinobacterial WhiB-like (Wbl) family of transcription factors. Mol Microbiol. 2018;110:663–676. doi: 10.1111/mmi.14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chater KF, Chandra G. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol Rev. 2006;30:651–672. doi: 10.1111/j.1574-6976.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- 9.Clark LC, Hoskisson PA. Duplication and evolution of devA-like genes in Streptomyces has resulted in distinct developmental roles. PLoS One. 2011;6:e25049. doi: 10.1371/journal.pone.0025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girard G, Traag BA, Sangal V, Mascini N, Hoskisson PA, et al. A novel taxonomic marker that discriminates between morphologically complex actinomycetes. Open Biol. 2013;3:130073. doi: 10.1098/rsob.130073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenke-Kodama H, Sandmann A, Müller R, Dittmann E. Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol. 2005;22:2027–2039. doi: 10.1093/molbev/msi193. [DOI] [PubMed] [Google Scholar]
- 12.Ridley CP, Lee HY, Khosla C. Evolution of polyketide synthases in bacteria. Proc Natl Acad Sci U S A. 2008;105:4595–4600. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borodina I, Siebring J, Zhang J, Smith CP, van Keulen G, et al. Antibiotic overproduction in Streptomyces coelicolor A3 2 mediated by phosphofructokinase deletion. J Biol Chem. 2008;283:25186–25199. doi: 10.1074/jbc.M803105200. [DOI] [PubMed] [Google Scholar]
- 14.Cruz-Morales P, Kopp JF, Martínez-Guerrero C, Yáñez-Guerra LA, Selem-Mojica N, et al. Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model streptomycetes. Genome Biol Evol. 2016;8:1906–1916. doi: 10.1093/gbe/evw125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernández-Martínez LT, Hoskisson PA. Expanding, integrating, sensing and responding: the role of primary metabolism in specialised metabolite production. Curr Opin Microbiol. 2019;51:16–21. doi: 10.1016/j.mib.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Noda-García L, Barona-Gómez F. Enzyme evolution beyond gene duplication: a model for incorporating horizontal gene transfer. Mob Genet Elements. 2013;3:e26439. doi: 10.4161/mge.26439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noda-García L, Camacho-Zarco AR, Medina-Ruíz S, Gaytán P, Carrillo-Tripp M, et al. Evolution of substrate specificity in a recipient's enzyme following horizontal gene transfer. Mol Biol Evol. 2013;30:2024–2034. doi: 10.1093/molbev/mst115. [DOI] [PubMed] [Google Scholar]
- 18.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-Scale gene function analysis with the Panther classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gubbens J, Janus M, Florea BI, Overkleeft HS, van Wezel GP. Identification of glucose kinase-dependent and -independent pathways for carbon control of primary metabolism, development and antibiotic production in Streptomyces coelicolor by quantitative proteomics. Mol Microbiol. 2012;86:1490–1507. doi: 10.1111/mmi.12072. [DOI] [PubMed] [Google Scholar]
- 20.Gunnarsson N, Mortensen UH, Sosio M, Nielsen J. Identification of the Entner-Doudoroff pathway in an antibiotic-producing actinomycete species. Mol Microbiol. 2004;52:895–902. doi: 10.1111/j.1365-2958.2004.04028.x. [DOI] [PubMed] [Google Scholar]
- 21.Plou FJ, Ferrer M, Nuero OM, Calvo MV, Alcalde M, et al. Analysis of Tween 80 as an esterase/ lipase substrate for lipolytic activity assay. Biotechnology Techniques. 1998;12:183–186. doi: 10.1023/A:1008809105270. [DOI] [Google Scholar]
- 22.Pratt J, Cooley JD, Purdy CW, Straus DC. Lipase activity from strains of Pasteurella multocida. Curr Microbiol. 2000;40:306–309. doi: 10.1007/s002849910061. [DOI] [PubMed] [Google Scholar]
- 23.Sakai Y, Hayatsu M, Hayano K. Use of Tween 20 as a substrate for assay of lipase activity in soils. Soil Science and Plant Nutrition. 2002;48:729–734. doi: 10.1080/00380768.2002.10409263. [DOI] [Google Scholar]
- 24.Krügel H, Krubasik P, Weber K, Saluz HP, Sandmann G. Functional analysis of genes from Streptomyces griseus involved in the synthesis of isorenieratene, a carotenoid with aromatic end groups, revealed a novel type of carotenoid desaturase. Biochim Biophys Acta. 1999;1439:57–64. doi: 10.1016/S1388-1981(99)00075-X. [DOI] [PubMed] [Google Scholar]
- 25.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Foundation; 2000. [Google Scholar]
- 26.Hobbs G, Frazer C, Gardner DJ, Cullum J, Oliver S. Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol. 1989;31:272–277. doi: 10.1007/BF00258408. [DOI] [Google Scholar]
- 27.Smyth GK, Verbyla AP. A conditional likelihood approach to residual maximum likelihood estimation in generalized linear models on JSTOR. J R Stat Soc. 1996;58:565–572. doi: 10.1111/j.2517-6161.1996.tb02101.x. [DOI] [Google Scholar]
- 28.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 29.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]